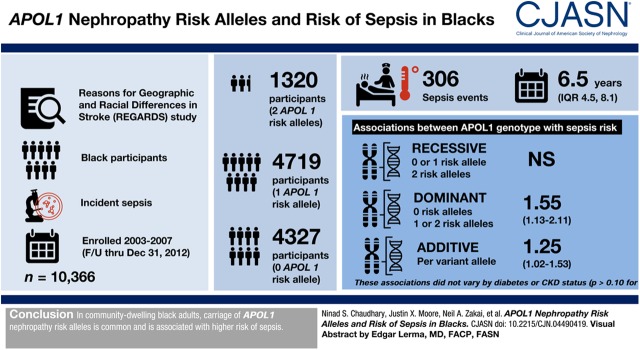
Visual Abstract
Keywords: chronic kidney disease, clinical epidemiology, ethnicity, genetic renal disease, adult, alleles, proportional hazards models, African Americans, apolipoprotein L1, confidence intervals, independent living, follow-up studies, genetic models, odds ratio, risk, chronic renal insufficiency, genotype, sepsis, diabetes mellitus, stroke, humans
Abstract
Background and objectives
apo L1 (APOL1) nephropathy risk alleles are associated with CKD in blacks. Although APOL1 has innate immune functions, little is known about the association of APOL1 genotypes with risk of infectious outcomes, such as sepsis. The objective of this study was to examine the associations of APOL1 nephropathy risk alleles with risk of sepsis in black adults.
Design, setting, participants, & measurements
We assessed the association of APOL1 risk alleles with incident sepsis in 10,366 black participants of the Reasons for Geographic and Racial Differences in Stroke study enrolled between 2003 and 2007 with follow-up through December 31, 2012. In Cox models adjusted for demographics, comorbid conditions, and principal components ancestry, we examined the association of APOL1 risk alleles with incident sepsis using recessive (comparing zero or one versus two risk alleles), dominant (zero versus one or two risk alleles), and additive genetic models. We also examined models stratified by diabetes and CKD status.
Results
A total of 1320 (13%) participants had two APOL1 risk alleles, 4719 (46%) had one risk allele, and 4327 (42%) participants had zero risk alleles. A total of 306 sepsis events occurred over a median 6.5 years (interquartile range, 4.5–8.1). There were no statistically significant associations of APOL1 genotype with sepsis risk under recessive genetic models. APOL1 genotypes were associated with sepsis risk under dominant (hazard ratio, 1.55; 95% confidence interval, 1.13 to 2.11) and additive (hazard ratio per variant allele copy, 1.25; 95% confidence interval, 1.02 to 1.53) genetic models adjusted for covariates and ancestry. These associations did not vary by diabetes or CKD status (Pinteraction>0.10 for both).
Conclusions
In community-dwelling black adults, carriage of APOL1 nephropathy risk alleles are common and associated with higher risk of sepsis.
Introduction
Apolipoprotein L1 (APOL1) is a constituent of HDL complexes that play a vital role in lysing trypanosomes that cause African sleeping sickness (1). Two coding variants in APOL1, known as G1 and G2, are present at high frequency in individuals of recent African descent, likely because they encode amino acid changes in the serum resistance–associated domain of APOL1 allowing it to evade inactivation by circulating trypanosomes (2). Although this imparts resistance against acute trypanosomiasis, carriage of two APOL1 risk alleles is associated with a greater risk for the development and progression of CKD in blacks (2,3).
Emerging evidence hints that APOL1 may play a broader role in immune function beyond protecting against trypanosomiasis. The expression of APOL1 is upregulated by inflammatory cytokines such as TNF-α, suggesting that APOL1 expression is part of a generalized immune response to infection (4,5). Further, APOL1 may help protect against Leishmania infection (6) and HIV-1 replication (7–9).
These data suggest that APOL1 has broad actions promoting innate immunity. Whether these actions differ by APOL1 genotype is unknown. Determining the relationship between APOL1 risk alleles and infection is important for clarifying the extent to which adverse effects of APOL1 risk alleles on kidney health may be counter-balanced by potential survival advantages from lowering infection risk in individuals of recent African descent. Accordingly, we examined the association of APOL1 nephropathy risk alleles with risk of community-acquired sepsis in black participants of the Reasons for Geographic and Racial Differences in Stroke (REGARDS) study (10). We hypothesized that APOL1 risk alleles may be associated with lower risk of sepsis, and that these associations may be modified by diabetes and CKD status.
Materials and Methods
Study Design
We performed an analysis of APOL1 genotypes and risk of sepsis events in the REGARDS cohort. The institutional review boards at all participating institutions approved this study. The REGARDS study obtained informed consent from all participants during the baseline visit, including separate consent for genetics research.
Data Source
The REGARDS study recruited 30,239 individuals between 2003 and 2007 and was designed to determine the reasons for racial and geographic differences in stroke mortality in the contiguous United States (10). In a separate ancillary study, we utilized REGARDS to assess risk factors for sepsis outcomes (11). At baseline, REGARDS included participants at least 45 years of age, with 45% men and 41% blacks. The study team collected detailed information at baseline in the in-home visit including participant demographics, health behaviors, chronic medical conditions, physical status, diet, and medications (10).
Genotyping
A total of 10,872 black study participants were genotyped for APOL1 nephropathy risk alleles (G1 [rs71785313] and G2 [rs73885319]) using TaqMan SNP Genotyping Assays (Applied Biosystems/Thermo Fisher Scientific). Of those, 196 were missing genotype data for rs71785313, 98 were missing data for rs73885319, and 27 were missing data for both APOL1 alleles, leaving 10,605 participants with available genotype data. After excluding 239 participants with missing follow-up data, the final analytic cohort consisted of 10,366 participants. A subset of participants had available genomic array data (Illumina exome chip) to estimate population substructure (n=6714) (12,13). In sensitivity analyses, ten principal components of ancestry were generated by EIGENSTRAT to adjust for ancestry in this subset.
Identification of Sepsis Events
The primary outcome of this study was a hospitalization for a first sepsis event (incident sepsis) through December 31, 2012. Participants were contacted by telephone every 6 months to identify the date, location, and attributed reason for all hospitalizations. From these reports, study personnel obtained hospital records for all hospitalizations attributed to a serious infection using the International Classification of Disease taxonomy of Angus et al. (14). Two trained research personnel performed a structured review of hospital records to confirm the presence of a serious infection as a major reason for hospital presentation and to identify pertinent physiologic measures and laboratory test results. We defined a sepsis event as a hospital admission for a serious infection with the presence of at least two Systemic Inflammatory Response Syndrome criteria, including heart rate >90 beats/min, fever (temperature>38.3°C or <36°C), tachypnea (>20 breaths/min or PCO2<32 mm Hg), and leukocytosis (white blood cells >12,000 or <4000 cells/mm3 or >10% band forms), and at least two sepsis-related organ dysfunctions (15,16). We used vital signs and laboratory test results for the initial 28 hours of hospitalization—because of our focus on community-acquired (as opposed to hospital-acquired) sepsis, we did not include sepsis developing at later time points during hospitalization. Among the entire REGARDS cohort, initial review of 1349 hospital records indicated excellent inter-rater agreement for presence of serious infection (κ=0.92) and the presence of sepsis (κ=0.90) upon hospital presentation.
Participant Characteristics
Participant characteristics included socio-demographics, health behaviors, chronic medical conditions, and specific biomarkers. Socio-demographics included age, race, sex, income, education, and geographic location. Health behaviors included tobacco and alcohol use. Chronic medical conditions included atrial fibrillation, chronic lung disease, coronary artery disease, deep venous thrombosis (DVT), diabetes, dyslipidemia, hypertension, myocardial infarction, obesity, peripheral artery disease, and stroke. Biomarkers measured at baseline included urine albumin-to-creatinine ratio (ACR), serum creatinine, and high-sensitivity C-reactive protein. Serum creatinine was calibrated to an international isotope dilution mass spectroscopic–traceable standard, measured by colorimetric reflectance spectrophotometry. eGFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration equation (17). Albumin and creatinine were measured in a random spot urine specimen by nephelometry (BN ProSpec Nephelometer; Dade Behring, Marburg, Germany) and Modular-P chemistry analyzer (Roche/Hitachi, Indianapolis, IN), respectively. Spot ACR was calculated in milligrams per gram. Prevalent CKD was defined as an eGFR<60 ml/min per 1.73 m2 or an ACR≥30 mg/g.
Statistical Analyses
We compared baseline socio-demographics, health behaviors, chronic medical conditions, and biomarkers by APOL1 risk genotype using chi-squared tests for categoric characteristics and t tests for continuous variables, and Wilcoxon rank-sum test for continuous variables without normal distribution. We considered P values <0.05 as statistically significant.
We fit Cox proportional hazard models with time to first sepsis as the primary end point. Individuals were censored at the time of their event, death, loss to follow-up, or end of follow-up. We adjusted the models sequentially for demographics (age, sex, income, education), covariates that met the P value criterion of ≤0.10 on the bivariate APOL1 analysis (tobacco use, DVT, dyslipidemia, and peripheral artery disease), and known sepsis risk factors (diabetes, coronary heart disease, systolic BP, diastolic BP, eGFR, and ACR). In the subset with available data on ancestry, we further adjusted the models for principal components ancestry in a sensitivity analysis similar to previous work in REGARDS (18). The initial model, assuming a recessive genetic effect, compared individuals with two APOL1 risk alleles (G1/G1, G2/G2, or G1/G2, representing high risk) with those with zero or one risk alleles (G1/G0, G2/G0, or G0/G0, representing low risk). In addition, we examined additive (0<1<2 risk alleles) and dominant (zero versus one or two risk alleles) genetic models. As secondary analyses, we further adjusted for the development of ESKD before the development of the sepsis event, and determined the association of APOL1 risk alleles with sepsis-related mortality, defined as death within 30 days of the sepsis hospitalization. Given prior work showing that the association of APOL1 nephropathy risk alleles with outcomes differs by diabetes status and presence or absence of CKD (18), we further examined models stratified by diabetes and CKD status and tested for differences in the association of APOL1 genotypes with sepsis outcomes by diabetes and CKD using interaction terms. Owing to the potential differences in the effects of G1 versus G2 alleles on innate immunity on the basis of prior literature (19), we conducted sensitivity analyses using G1-specific genetic models and, separately, G2-specific genetic models. Finally, we examined whether the type and number of sepsis-related organ dysfunctions differed by APOL1 genotype. We performed all statistical analyses using SAS version 9.4.
Results
Study Population and Baseline Characteristics
Among 10,366 participants, 1320 (13%) had two APOL1 risk alleles, 4719 (46%) had one risk allele (2906 had the G1/G0 genotype and 1813 the G2/G0 genotype), and 4327 (42%) participants had zero risk alleles in accordance with Hardy–Weinberg expectations for genotype frequency distributions. Table 1 compares demographic and clinical characteristics of participants with two versus zero or one risk alleles. As compared with participants with zero or one risk allele, participants with two risk alleles were younger, were less likely to have a history of DVT, had higher urine ACR, and had lower eGFR at baseline.
Table 1.
Summary statistics of participants’ characteristics by APOL1 risk genotype group, among 10,366 black Reasons for Geographic and Racial Differences in Stroke participants
| Characteristic | APOL1 Risk Genotype | ||
|---|---|---|---|
| 0 Risk Alleles (n=4327) | 1 Risk Allele (n=4719) | 2 Risk Alleles (n=1320) | |
| Agea | 64 (9) | 64 (9) | 63 (9) |
| Sex (%) | |||
| Male | 1658 (38) | 1840 (39) | 504 (38) |
| Female | 2669 (62) | 2879 (61) | 816 (62) |
| Education (%) | |||
| ≤ High school | 815 (19) | 960 (20) | 254 (19) |
| High school graduate | 1215 (28) | 1292 (27) | 368 (28) |
| Some college | 1159 (27) | 1236 (26) | 361 (27) |
| ≥ College graduate | 1132 (26) | 1227 (26) | 335 (25) |
| Income (%) | |||
| ≤$20,000 | 1136 (26) | 1259 (27) | 366 (28) |
| $20,000–$34,000 | 1189 (28) | 1220 (26) | 320 (24) |
| $35,000–$74,000 | 1106 (26) | 1198 (25) | 363 (28) |
| ≥$75,000 | 382 (9) | 448 (10) | 115 (9) |
| Refused | 514 (12) | 594 (13) | 156 (12) |
| Region | |||
| Stroke belt | 1410 (33) | 1606 (34) | 442 (34) |
| Stroke buckle | 786 (18) | 812 (17) | 232 (18) |
| Nonbelt | 2131 (49) | 2301 (49) | 646 (49) |
| Tobacco use (%) | |||
| Never | 1913 (44) | 2126 (45) | 630 (48) |
| Past | 1646 (38) | 1727 (37) | 469 (36) |
| Current | 751 (17) | 846 (18) | 210 (16) |
| Alcohol use (%) | |||
| None | 3001 (71) | 3326 (72) | 948 (73) |
| Moderate | 1099 (26) | 1150 (25) | 315 (24) |
| Heavy | 102 (2) | 122 (3) | 28 (2) |
| Chronic medical conditions | |||
| Atrial fibrillation | 334 (8) | 360 (8) | 104 (8) |
| Chronic lung disease | 344 (8) | 378 (8) | 93 (7) |
| Coronary heart disease | 668 (16) | 714 (16) | 190 (15) |
| Deep vein thrombosis | 221 (5) | 234 (5) | 49 (4) |
| Diabetes | 1294 (30) | 1460 (3) | 404 (31) |
| Dyslipidemia | 2276 (54) | 2567 (55) | 735 (56) |
| Hypertension | 3048 (71) | 3406 (72) | 947 (72) |
| Myocardial infarction | 509 (12) | 534 (12) | 137 (11) |
| Obesity | 2702 (63) | 2969 (63) | 837 (64) |
| Peripheral artery disease | 102 (2) | 118 (3) | 21 (2) |
| Stroke | 355 (8) | 350 (7) | 100 (8) |
| Biomarkersbc | |||
| ACR | 7.6 (4.6–18.3) | 8.0 (4.7–20.1) | 9.2 (4.9–26.0) |
| eGFR | 93 (76–107) | 91 (73–107) | 89 (72–106) |
| eGFR<60 ml/min per 1.73 m2 (%) | 518 (12) | 567 (12) | 179 (14) |
| hsCRP | 2076 (48.7) | 2307 (49.7) | 637 (48.9) |
Data are given as n (%) except where indicated. Significance was determined using chi-squared test, t test, or Wilcoxon tests. ACR, albumin-to-creatinine ratio; hsCRP, high-sensitivity C-reactive protein.
Mean (SD).
Median (interquartile range).
eGFR in ml/min per 1.73 m2; ACR in milligrams per gram; hsCRP in milligrams per deciliter.
Associations of APOL1 Risk Genotype with Incident Sepsis
A total of 306 sepsis events were observed over a median follow-up period of 6.5 years (interquartile range, 4.5–8.1). There were no associations of the APOL1 genotype with sepsis in crude or multivariable-adjusted recessive genetic models (Table 2). However, under a dominant genetic model, as compared with carriage of no APOL1 risk alleles, carriage of one or two risk alleles was associated with higher risk of sepsis in the crude model (hazard ratio [HR], 1.43; 95% confidence interval [95% CI], 1.31 to 1.82), after adjustment for age, sex, income, tobacco use, DVT, dyslipidemia, peripheral artery disease, diabetes, coronary heart disease, systolic BP, diastolic BP, eGFR, and ACR (HR, 1.48; 95% CI, 1.14 to 1.91), and after additional adjustment for principal components ancestry (HR, 1.55; 95% CI, 1.13 to 2.11). Similarly, in additive genetic models, there was a direct association between greater number of APOL1 risk alleles and higher risk of sepsis in crude and multivariable-adjusted models (HR per variant allele copy, 1.25; 95% CI, 1.02 to 1.53 in the fully adjusted model). Results were not materially changed when further adjusting for the development of ESKD before the development of sepsis (Supplemental Table 1). Further, there was no statistically significant association of APOL1 risk alleles with sepsis-related mortality (Supplemental Table 2). The association of APOL1 risk alleles with sepsis in G1- and G2-specific recessive, additive, and dominant genetics models was consistent with models that combined G1 and G2 risk alleles (Supplemental Tables 3–7). There were no statistically significant differences in the type and number of sepsis-related organ dysfunctions by APOL1 genotype (Supplemental Tables 8 and 9).
Table 2.
Hazard ratios and 95% confidence intervals for the association between APOL1 risk genotype and incident sepsis in blacks
| APOL1 Genotype Modela | No. Events (%)b | Crude | Model 1 | Model 2 | Model 3 |
|---|---|---|---|---|---|
| Recessive model | |||||
| 0 or 1 risk allele | 259 (3) | — | — | — | |
| 2 risk alleles | 47 (4) | 1.23 (0.90 to 1.68) | 1.27 (0.93 to 1.73) | 1.21 (0.86 to 1.69) | 1.06 (0.70 to 1.61) |
| Dominant model | |||||
| 0 risk alleles | 101 (2) | — | — | — | |
| 1 or 2 risk alleles | 205 (3) | 1.43 (1.31 to 1.82) | 1.46 (1.15 to 1.85) | 1.48 (1.14 to 1.91) | 1.55 (1.13 to 2.11) |
| Additive model | |||||
| Per variant allele copy | 306 (3) | 1.26 (1.07 to 1.48) | 1.28 (1.09 to 1.50) | 1.27 (1.07 to 1.51) | 1.25 (1.02 to 1.53) |
Model 1: adjusted for sex, age, income, education. Model 2: Model 1+adjusted for tobacco, deep vein thrombosis, dyslipidemia, peripheral artery disease, eGFR, albumin-to-creatinine ratio, systolic BP, diastolic BP, prevalent comorbidities (diabetes, coronary heart disease, stroke, dyslipidemia). Model 3: Model 2+adjusted for principal components ancestry. —, reference.
Estimated from Cox proportional hazard models.
Row percentage, proportion of participants experiencing a sepsis event (total n=306).
Stratified Analyses
The association between APOL1 and sepsis risk did not vary by diabetes (Table 3) or CKD (Table 4), Pinteraction>0.10 for both. As in the primary analyses, the recessive genetic models were NS across all strata. The magnitude and statistical strength of the dominant and additive genetic models with sepsis risk were consistent with the primary models.
Table 3.
Hazard ratios and 95% confidence intervals for the association between APOL1 risk genotype group and incident sepsis among blacks stratified by presence or absence of diabetes
| APOL1 Genotype Modela | No. Events (%)b | Crude | Model 1 | Model 2 | Model 3 |
|---|---|---|---|---|---|
| Diabetes | |||||
| Recessive model | |||||
| 0 or 1 risk allele | 132 (5) | — | — | — | |
| 2 risk alleles | 28 (7) | 1.41 (0.94 to 2.12) | 1.45 (0.96 to 2.18) | 1.49 (0.96 to 2.31) | 1.31 (0.77 to 2.22) |
| Dominant model | |||||
| 0 risk alleles | 48 (4) | — | — | — | |
| 1 or 2 risk alleles | 112 (6) | 1.60 (1.14 to 2.24) | 1.62 (1.15 to 2.27) | 1.67 (1.15 to 2.41) | 1.54 (1.00 to 2.38) |
| Additive model | |||||
| Per variant allele copy | 160 (5) | 1.38 (1.10 to 1.71) | 1.39 (1.11 to 1.74) | 1.42 (1.12 to 1.80) | 1.31 (0.99 to 1.74) |
| No Diabetes | |||||
| Recessive model | |||||
| 0 or 1 risk allele | 127 (2) | — | — | — | |
| 2 risk alleles | 19 (2) | 1.02 (0.63 to 1.66) | 1.06 (0.65 to 1.71) | 0.90 (0.52 to 1.54) | 0.76 (0.38 to 1.52) |
| Dominant model | |||||
| 0 risk alleles | 53 (2) | — | — | — | |
| 1 or 2 risk alleles | 93 (2) | 1.26 (0.90 to 1.76) | 1.28 (0.91 to 1.80) | 1.31 (0.91 to 1.89) | 1.67 (1.06 to 2.64) |
| Additive model | |||||
| Per variant allele copy | 146 (2) | 1.13 (0.89 to 1.43) | 1.15 (0.91 to 1.46) | 1.12 (0.87 to 1.44) | 1.21 (0.90 to 1.64) |
Model 1: adjusted for sex, age, income, education. Model 2: Model 1+adjusted for tobacco, deep vein thrombosis, dyslipidemia, peripheral artery disease, eGFR, albumin-to-creatinine ratio, systolic BP, diastolic BP, prevalent comorbidities (coronary heart disease, stroke, dyslipidemia). Model 3: Model 2+adjusted for principal components ancestry. —, reference.
Estimated from Cox proportional hazard models.
Row percentage, proportion of participants experiencing a sepsis event.
Table 4.
Hazard ratios and 95% confidence intervals for the association between APOL1 risk genotype group and incident sepsis among blacks stratified by presence or absence of CKD
| APOL1 Genotype Modela | No. Events (%)b | Crude | Model 1 | Model 2 | Model 3 |
|---|---|---|---|---|---|
| CKD | |||||
| Recessive model | |||||
| 0 or 1 risk allele | 141 (6) | — | — | — | |
| 2 risk alleles | 24 (6) | 0.96 (0.62 to 1.48) | 1.01 (0.66 to 1.57) | 1.14 (0.73 to 1.79) | 1.18 (0.70 to 1.99) |
| Dominant model | |||||
| 0 risk alleles | 53 (5) | — | — | — | |
| 1 or 2 risk alleles | 112 (7) | 1.38 (0.99 to 1.91) | 1.41 (1.01 to 1.96) | 1.66 (1.16 to 2.37) | 1.73 (1.13 to 2.65) |
| Additive model | |||||
| Per variant allele copy | 165 (6) | 1.14 (0.92 to 1.42) | 1.17 (0.94 to 1.46) | 1.31 (1.04 to 1.65) | 1.34 (1.02 to 1.76) |
| No CKD | |||||
| Recessive model | |||||
| 0 or 1 risk allele | 118 (2) | — | — | — | |
| 2 risk alleles | 23 (3) | 1.41 (0.90 to 2.21) | 1.46 (0.93 to 2.28) | 1.30 (0.77 to 2.18) | 0.92 (0.45 to 1.86) |
| Dominant model | |||||
| 0 risk alleles | 48 (2) | — | — | — | |
| 1 or 2 risk alleles | 93 (2) | 1.41 (0.99 to 2.00) | 1.41 (1.00 to 2.00) | 1.30 (0.89 to 1.90) | 1.45 (0.91 to 2.31) |
| Additive model | |||||
| Per variant allele copy | 141 (2) | 1.30 (1.02 to 1.65) | 1.32 (1.04 to 1.67) | 1.23 (0.94 to 1.60) | 1.19 (0.86 to 1.65) |
Model 1: adjusted for sex, age, income, education. Model 2: Model 1+adjusted for tobacco, deep vein thrombosis, dyslipidemia, peripheral artery disease, eGFR, albumin to creatinine ratio, systolic BP, diastolic BP, prevalent comorbidities (diabetes, coronary heart disease, stroke, dyslipidemia). Model 3: Model 2+adjusted for principal components ancestry. —, reference.
Estimated from Cox proportional hazard models.
Row percentage, proportion of participants experiencing a sepsis event.
Discussion
In this study of community-dwelling black adults, carriage of APOL1 nephropathy risk alleles was associated with higher risk of developing sepsis independently of traditional risk factors and genetic ancestry.
Although APOL1 has several biologic functions related to innate immunity, relatively few studies examined the association of APOL1 genotypes with infections apart from trypanosomiasis (5,7–9,20,21). Our findings suggest that whereas APOL1 risk variants confer protection against parasitic infections (trypanosomiasis, leishmaniasis), carriage of even one APOL1 risk variant heightens the risk for severe infections complicated by organ dysfunction (sepsis). The reasons for these findings are unclear, but we can speculate about several possibilities. APOL1 expression is strongly upregulated in response to proinflammatory cytokines such as IFN-γ and TNF (22). APOL1, along with other family members of apo L, is involved in cellular apoptotic processes (23,24). One hypothesis is that the intense inflammatory response characteristic of sepsis increases APOL1 expression in a variety of tissues, resulting in cytotoxicity and glomerular and tubular damage and, thereby, contributes to systemic organ dysfunction. Another potential mechanism may relate to differential effects of APOL1 risk alleles on neutrophil apoptosis observed in patients with sepsis (25). A recent study assessing the neutrophil profile of patients with sepsis observed that APOL1 RNA and protein expression is downregulated, delaying neutrophil apoptosis in patients with sepsis (26). Similarly, another study showed downregulation of APOL1 expression in patients with sepsis (27). This is somewhat surprising, because sepsis is associated with an increased circulating level of IFN, which by itself would drive increased APOL1 expression. Further mechanistic studies on the role of APOL1 regulation and function in sepsis, addressing the role of the common and nephropathy risk alleles, may clarify these issues.
The association of APOL1 risk alleles with sepsis did not follow the inheritance pattern observed for CKD, in which increased risk fits recessive but not dominant or additive genetic models. In contrast, the association of APOL1 risk variants with increased risk of sepsis was observed only in dominant and additive genetic models, suggesting that the carriage of even one risk allele confers increased risk for sepsis. This is consistent with the finding that carriage of only one APOL1 risk allele is needed to confer protection against trypanosomiasis, indicating that having either one or two APOL1 risk alleles is consequential for infectious outcomes in blacks. If confirmed, this has important public health implications in that G1 and G2 alleles are common, with 45%–50% of blacks carrying one risk allele and another 10%–15% carrying two. Understanding the biologic mechanisms underlying this association may provide novel insights into factors that enhance sepsis risk in community-dwelling blacks.
Our study has multiple strengths, including its large sample size, detailed information on health comorbidities, long-term follow-up of sepsis in community-dwelling adults, and genetic ancestry information in a large subset of participants. Despite these strengths, our findings require careful consideration in the presence of several limitations. This is a hypothesis-generating study and does not establish any causal association in spite of measuring a genotype-outcome association. The mechanisms of disease progression due to presence of APOL1 variants are not established, limiting us from adjusting for all possible confounding effects. We did not have sufficient follow-up measures of kidney function, so we could not determine the degree to which kidney function decline in black participants with two risk alleles may have contributed to the development of sepsis. Nonetheless, the observation that individuals carrying one risk allele had higher risk of sepsis as compared with those with zero risk alleles suggests that reduced kidney function (which in most settings is not associated with carriage of one risk allele) does not explain the association of APOL1 risk alleles with sepsis risk. Next, our study evaluated community-acquired infections developing into sepsis, and so, therefore, does not provide insight into the effect of APOL1 risk alleles on hospital-acquired infections. We also did not have sufficient information on the use of immunosuppressive medications to account for this in multivariable modeling. Finally, it is possible that some sepsis events were missed because hospital admissions for infection were identified by the participants during follow-up.
Carriage of one or two APOL1 risk alleles was associated with heightened risk of sepsis in community-dwelling black adults. These findings indicate a potential role of APOL1 variants in sepsis, and future validation studies followed by mechanistic studies should be planned to better understand the functional role of APOL1 risk genotypes in community-acquired sepsis. Other studies could also address whether those with APOL1 risk alleles have more unfavorable outcomes after sepsis.
Disclosures
Dr. Gutiérrez reports receiving grant support and consulting fees from Keryx Biopharmaceuticals, grant support and consulting fees from Amgen, and grant support from GSK, outside of the submitted work. Dr. Kopp reports receiving nonfinancial research project support from Merck. Dr. Naik reports receiving funding from Elsevier. Dr. Chaudhary, Dr. Irvin, Dr. Lange, Dr. Moore, and Dr. Winkler have nothing to disclose.
Funding
Dr. Gutiérrez was supported by grants from the National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK) (K24DK116180) and NINDS (R01NS080850). Dr. Kopp was supported by the Intramural Research Program, NIDDK, NIH. Dr. Wang was supported by a grant from the National Institute of Nursing Research (R01 NR012726). Dr. Winkler was funded in part with federal funds from the National Cancer Institute (NCI), NIH, under contract HHSN26120080001E, and supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research. Dr. Zakai was supported by the Reasons for Geographic and Racial Differences in Stroke grant from the National Heart, Lung, and Blood Institute (K08 HL096841). This research project is supported by a cooperative agreement cofunded by NINDS and NIA, NIH, US DHHS (U01 NS041588). This work was also supported by grants from the Center for Clinical and Translational Science and the Lister Hill Center for Health Policy of the University of Alabama at Birmingham.
Supplementary Material
Acknowledgments
The authors thank the other investigators, the staff, and the participants of the Reasons for Geographic and Racial Differences in Stroke (REGARDS) study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at http://www.regardsstudy.org.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke (NINDS) or the National Institute on Aging (NIA). The content of this publication does not necessarily reflect the views or policies of the National Institutes of Health (NIH), or the Department of Health and Human Services (DHHS), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related editorial, “APOL1 Nephropathy Risk Variant Associations with Diseases beyond the Kidney: APOL1 and Sepsis,” on pages 1684–1686.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.04490419/-/DCSupplemental.
Supplemental Table 1. Hazard ratios (HRs) and 95% confidence intervals (95% CIs) for the association between APOL1 genotype and incident sepsis in blacks in the REGARDS study adjusting for the development of ESKD before sepsis.
Supplemental Table 2. Odds ratios (ORs) and associated 95% confidence intervals (95% CIs) for the association between APOL1 risk genotype group and incident 30-day sepsis mortality in blacks with sepsis.
Supplemental Table 3. Hazard ratios and associated 95% confidence intervals for the association between G1 dominant APOL1 risk genotype group and incident sepsis among blacks overall and stratified by diabetes and CKD.
Supplemental Table 4. Hazard ratios and associated 95% confidence intervals for the association between G2 dominant APOL1 risk genotype group and incident sepsis among blacks overall and stratified by diabetes and CKD.
Supplemental Table 5. Hazard ratios and 95% confidence intervals for the association between G1 and G2 additive models of APOL1 genotype and incident sepsis among blacks overall and stratified by diabetes and CKD.
Supplemental Table 6. Hazard ratios and associated 95% confidence intervals for the association between G1 recessive APOL1 risk genotype group and incident sepsis among blacks overall and stratified by diabetes and CKD.
Supplemental Table 7. Hazard ratios and associated 95% confidence intervals for the association between G2 recessive APOL1 risk genotype group and incident sepsis among blacks overall and stratified by diabetes and CKD.
Supplemental Table 8. Type of organ dysfunction across APOL1 models among sepsis participants.
Supplemental Table 9. Number of organ dysfunctions across APOL1 models among sepsis participants.
References
- 1.Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homblé F, Vanhamme L, Tebabi P, Pays A, Poelvoorde P, Jacquet A, Brasseur R, Pays E: Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science 309: 469–472, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limou S, Dummer PD, Nelson GW, Kopp JB, Winkler CA: APOL1 toxin, innate immunity, and kidney injury. Kidney Int 88: 28–34, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brauweiler AM, Goleva E, Leung DYM: Interferon-γ protects from staphylococcal alpha toxin-induced keratinocyte death through apolipoprotein L1. J Invest Dermatol 136: 658–664, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samanovic M, Molina-Portela MP, Chessler AD, Burleigh BA, Raper J: Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog 5: e1000276, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor HE, Khatua AK, Popik W: The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol 88: 592–603, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Götz N, Heilmann J, Mack K, Sauter D, Thompson D, Perreaud J, Rausell A, Munoz M, Ciuffi A, Kirchhoff F, Telenti A: Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 12: 41, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikulak J, Oriolo F, Portale F, Tentorio P, Lan X, Saleem MA, Skorecki K, Singhal PC, Mavilio D: Impact of APOL1 polymorphism and IL-1β priming in the entry and persistence of HIV-1 in human podocytes. Retrovirology 13: 63, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard VJ, Cushman M, Pulley L, Gomez CR, Go RC, Prineas RJ, Graham A, Moy CS, Howard G: The reasons for geographic and racial differences in stroke study: Objectives and design. Neuroepidemiology 25: 135–143, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Wang HE, Shapiro NI, Griffin R, Safford MM, Judd S, Howard G: Chronic medical conditions and risk of sepsis. PLoS One 7: e48307, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson N, Price AL, Reich D: Population structure and eigenanalysis. PLoS Genet 2: e190, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR: Epidemiology of severe sepsis in the United States: Analysis of incidence, outcome, and associated costs of care. Crit Care Med 29: 1303–1310, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Mayr FB, Yende S, Angus DC: Epidemiology of severe sepsis. Virulence 5: 4–11, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC: The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315: 801–810, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutiérrez OM, Irvin MR, Chaudhary NS, Cushman M, Zakai NA, David VA, Limou S, Pamir N, Reiner AP, Naik RP, Sale MM, Safford MM, Hyacinth HI, Judd SE, Kopp JB, Winkler CA: APOL1 nephropathy risk variants and incident cardiovascular disease events in community-dwelling black adults. Circ Genom Precis Med 11: e002098, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper A, Ilboudo H, Alibu VP, Ravel S, Enyaru J, Weir W, Noyes H, Capewell P, Camara M, Milet J, Jamonneau V, Camara O, Matovu E, Bucheton B, MacLeod A: APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. Elife 6: e25461, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, Doumatey A, Ayodo G, Alper SL, Pollak MR, Friedman DJ, Raper J: Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A 111: E2130–E2139, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An P, Kirk GD, Limou S, Binns-Roemer E, Kopp JB, Winkler CA: Impact of APOL1 genetic variants on HIV-1 infection and disease progression. Front Immunol 10: 53, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heymann J, Winkler CA, Hoek M, Susztak K, Kopp JB: Therapeutics for APOL1 nephropathies: Putting out the fire in the podocyte. Nephrol Dial Transplant 32[Suppl 1]: i65–i70, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA: Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem 283: 21540–21549, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA: ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy 4: 1079–1082, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taneja R, Parodo J, Jia SH, Kapus A, Rotstein OD, Marshall JC: Delayed neutrophil apoptosis in sepsis is associated with maintenance of mitochondrial transmembrane potential and reduced caspase-9 activity. Crit Care Med 32: 1460–1469, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Akl I, Lelubre C, Uzureau P, Piagnerelli M, Biston P, Rousseau A, Badran B, Fayyad-Kazan H, Ezedine M, Vincent JL, Boudjeltia KZ, Vanhamme L: Apolipoprotein L expression correlates with neutrophil cell death in critically ill patients. Shock 47: 111–118, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Sharma NK, Ferreira BL, Tashima AK, Brunialti MKC, Torquato RJS, Bafi A, Assuncao M, Azevedo LCP, Salomao R: Lipid metabolism impairment in patients with sepsis secondary to hospital acquired pneumonia, a proteomic analysis. Clin Proteomics 16: 29, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.