Abstract
Patients diagnosed with hepatocellular carcinoma (HCC) suffered a high risk of recurrence and poor prognosis. Identification of differentially expressed genes (DEGs) in HCC provides potential biomarkers for evaluating prognosis and specific therapeutic treatments. In this study, DEGs over-expressed in HCC specimens with a fold change over 2.0 were collected through integrative bioinformatics analysis from GEO datasets. Gene ontology and KEGG pathway enrichment were conducted by applying DAVID database. We noticed Secreted phosphoprotein 1 (SPP1) as one of the signature genes up-regulated in HCC tissues with a close relation to the tumor process. Eighty-seven paired HCC specimens from our medical center were explored to verify the aberrant expression of SPP1 by IHC and qRT-PCR assay. Depletion of SPP1 in HCC Hep3B cells was established. The cell proliferation was impaired in SPP1 depleted cells, along with a resistance of cell apoptosis by down-regulating SPP1. Intriguingly, we further validated a direct interaction between miR-181c and SPP1, which indicated a post-transcriptional regulation mechanism of SPP1 in HCC. Thus, our results suggest that SPP1 may function as an enhancer of HCC growth targeted by miR-181c, and probably provide us an innovational target for HCC diagnose and therapeutic treatment.
Keywords: Hepatocellular carcinoma, secreted phosphoprotein 1, miR-181c, cell growth
Introduction
Hepatocellular carcinoma (HCC) is the most common liver malignancy and the main treatment of this fatal disease is radical resection [1]. In spite of exciting leap in diagnostic techniques and therapeutic treatment including targeted therapy and immunity therapy during the past decades, high rate of recurrence and mortality indicates the unsatisfactory outcome of HCC patients [2,3].
Mining differentially expressed genes (DEGs) from NCBI GEO database provide a possibility of integrating potential biomarkers related to HCC progress and prognosis [4,5]. Yet, the specific evaluation for signature genes from the DEGs screened out from datasets is inadequate. In our previous study, we screened out several genes over-expressed especially in HCC tumor tissues, such as AKR1B10 and ROBO1, which were associated with HCC tumorigenesis respectively [6]. In this study, we set an absolute value of fold-change (FC) of gene mRNA expression with threshold criteria of log2FC ≥ 2.0 and P value <1.0E-04, by which we clustered seven DEGs including Secreted phosphoprotein 1 (SPP1).
We conducted the Gene Ontology (GO) and KEGG pathway enrichment, and found that SPP1 presents critical relationship with signature tumorigenesis process and pathway directly or indirectly, including PI3K/AKT signaling pathway, proteoglycans in cancer and ECM-receptor interaction. Further exploration in either real patients’ specimens or HCC cell lines indicates highly expressed SPP1 in tumor tissues or cells compared with the normal controls. To investigate the bio-function of SPP1 in HCC cells, depletion of SPP1 through sh-RNA method was carried out. As we supposed, down-regulation of SPP1 significantly impaired the cell proliferation of HCC Hep3B cells and arrested the cell cycle in G0/G1 phase. And, the cell apoptosis was enhanced. Noticably, we found microRNA-181c (miR-181c), one of the aberrantly expressed microRNAs exerting differentiated function in multiple tumors like leukemia, lung cancer and gastric cancer [7-9], is the direct regulator up-streaming SPP1 mRNA post-transcriptionally. We suppose SPP1 is a critical regulator participating in HCC tumorigenesis and process, and could probably become a new target for HCC prevention, diagnose and therapeutic treatment.
Materials and methods
Surgical specimens and cell lines
HCC cancer specimens were collected paired with non-cancerous liver tissues from 87 patients performed partial hepatectomy without any preoperative therapy 2013 to 2016 at the Department of Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Informed consent was obtained and the study was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiaotong University School of Medicine. Clinicopathologic features of the patients including gender, age, tumor size, number of lesions, grades et al. were collected.
HCC cell lines Hep3B, HepG2 and Hu7u were purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Science (Shanghai, China), and the normal human hepatic cell line L02 was used as control. Cells above were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), incubator at 37°C, with 100 ug/ml streptomycin and 100 U/ml Penicillin in a humidified cell and an atmosphere of 5% CO2.
Gene expression data process
HCC related Datasets GSE6764, GSE14520 and GSE14323 were downloaded from GEO database. Platforms of these datasets are GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array) for GSE6764, GPL3921 (Affymetrix HT Human Genome U133A Array) for GSE14520, and GPL571 (Affymetrix Human Genome U133A 2.0 Array) for GSE14323.Totally, we enrolled 718 samples from these three datasets for DEGs screening. Dateset GSE6857 containing miRNA expression data was downloaded simultaneously with platform of GPL4700 OSU-CCC MicroRNA Microarray Version 2.0.
Data were preprocessed and normalized by two professional bioinformatics analysts, and then were screened for DEGs according to an absolute value of fold-change (FC) of gene expression with threshold criteria of log2FC ≥ 2.0 and P value <1.0E-04. Funrich Software (Version 3.0, http://funrich.org/index.html) was introduced to analysis the co-expression characteristic of genes detected from the datasets.
GO and KEGG pathway enrichment analysis was conducted by using online tools of the Database for Annotation Visualization and Integrated Discovery (Version 6.7, https://david.ncifcrf.gov/). The cut-off value for significant function and pathway screening was set as P<0.01. GeneMANIA (http://genemania.org/) and STRING database were used and Cytoscape software was applied for the establishment of the DEGs’ network.
KMplot tool (http://www.kmplot.com) was used to evaluate the 5 year over-all survival rate of SPP1 as described in the ‘Statistical analysis’ section below.
qRT-PCR assay, western blot analysis and immunohistochemistry assay
RNA isolation in tissue and cells were conducted according to the instruction of TRIzol reagent (Invitrogen, USA). The first-strand cDNA was synthesized by using High-Capacity cDNA Reverse Transcription Kit (ABI, USA). RT-primers of the mRNAs were synthesized by Sangon Biotech Company (Shanghai, China) as follows: 5’-TCCTAGCCCCACAGACCCTT-3’ (forward) and 5’-CTGTGGAATTCACGGCTGAC-3’ (reverse). Real-time quantitative polymerase chain reaction (qRT-PCR) was conducted following the TaqMan Gene Expression Assays protocol (ABI, USA).
Antibodies against SPP1 respectively were applied (Abcam, USA) following the manufactory instruction. The Western blot analysis and immunohistochemistry assay were performed as previously described [6]. The protein expression levels detected by IHC were blindly assigned to two professional pathologists for examination, and were subjectively set into two groups as staining intensity graded: no to low staining (0~1+) and moderate to high staining (2+~3+).
Cell transfection
Hep3B cells in exponential phase were prepared and transfected with shRNA suppressing SPP1 translation through pGU6/Neo vectors (GenePharma, Shanghai, China) along with the construction of the control ones. Transfected cells were cultured and selected by using medium added G418 (Santa Cruz Biotechnology, Inc; 400 μg/ml).
Recombinant adenovirus Ad5/F35 (Ad5/F35-SPP1) was used to rescue SPP1 depression in Hep3B cells, and Ad5/F35-Null was set as negative control (GenePharma). Hep3B cells overexpressing miR-181c (Hep3B/miR-181c) were constructed along with Ad5/F35-SPP1 or Ad5/F35-Null treatment, and the negative control ones were set (NigmiR).
Cell proliferation assay and cell cycle analysis
Hep3B cells (1 × 106) either stably transfected were cultured in 96-well microtiter plates in triplicate and incubated for 5 days at 37°C with an atmosphere of 5% CO2. Microplate computer software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for measuring the OD following the Cell Counting Kit-8 (CCK-8) assay kit protocol (Dojindo, Tokyo, Japan). The cell proliferation curves were plotted.
The aforementioned cells were treated in steps with ethanol fixation, RNase A treatment and propidium iodide staining. Flow cytometry detection by using FACSCalibur (Becton-Dickinson, Franklin Lakes, NJ, USA) were conducted. Cell populations at the G0/G1, S and G2/M phases were quantified through ModFit software (Becton-Dickinson). Cell debris and fixation artifacts was excluded.
Cell apoptosis analysis
Cell apoptosis rate calculation was conducted by using Annexin V-FITC Apoptosis Detection Kit I (BD Pharmingen, USA) according to the product instructions. Stable transfected Hep3B cells were resuspended in 1 × Binging Buffer with a concentration about 1 × 106 cells/ml. Five microliter of FITC and 5 μl of PI were added into 100 μl of cell suspension, followed by 15 minutes incubation in darkness and 400 μl × Bingding Buffer was added. The analysis of apoptosis by flow cytometry (Becton Dickinson, USA) was conducted. Both Annexin V-FITC-positive and PI-negative cells were considered as apoptosis ones.
Dual-luciferase reporter assay
MiR-181c was predicted as a potential upstreaming regulator of SPP1 by analysis (microcosm, http://mirecords.biolead.org). A 202 bp sequence from the 3’UTR of SPP1 mRNA was containing putative miR-181c binding site was intercepted: 5’-aauacaauuucucacuuugcauuuagucaaaagaaaaaaugcuuuauagcaaaaugaaagagaacaugaaaugcuucuuucucaguuuauugguugaauguguaucuauuugagucuggaaauaacuaauguguuugauaauuaguuuaguuuguggcuucauggaaacucccuguaaacuaaaagcuucaggguuaugucu-3’. The corresponding sequence mutated was set as follow (Sangon Biotech Co.): 5’-auuucuaauacacucauaggaauaacugauauguauauaagguauuuuggauauucauacacaucuucauaagguacauacacugauaaaucgaucauucucuuuguuuaucacugucguauuuaguuaagagauaguuuaauugauaacuaucucgguacuucguaucacgcagaauaguuauaccaugacgcuaaagaca-3’. Sequences above were cloned into pMIR-REPORT luciferase vectors (Promega, Madison, WI, USA), containing Firefly luciferase, and pRL-TK vectors containg Renilla luciferase used as control. The vectors were co-transfected into Hep3b cells overexpressing miR-181c and the control ones. The luciferase activity was measured by using Dual-Glo Luciferase assay system (Promega) 48 hours post-transfection.
Statistical analysis
Statistical analysis was carried out by using SPSS 20.0 and GraphPad Prism 5.0. As for the analysis of the 87 pairs specimens, and the relative clinicopathologic features study, P-values were calculated through paired t-test and Fisher’s exact test, and P-values <0.05 were considered to indicate a statistically significant result.
The datasets (GSE6764, GSE14520 and GSE14323) were analyzed by using R language. DEGs were screened through a t-test linear models for microarray analysis package in R (Version 3.3, http://www.bioconductor.org) [10].
As for the 5 year over-all survival rate of SPP1, KMplot tool (http://www.kmplot.com) was used for the relative evaluation including 364 HCC patients with the follow-up information. For the expression of SPP1, the Univariate Cox regression analysis was conducted according to the best performing threshold after screening the lower and upper expression quartiles. The hazard ratio with 95% confidence and P-value from the log-rank test were calculated.
The expression of microRNA status between normal and tumor tissues was calculated by using dbDEMC2 software.
Results
SPP1 differentially expressed as one of the DEGs in HCC tumor and normal liver tissues
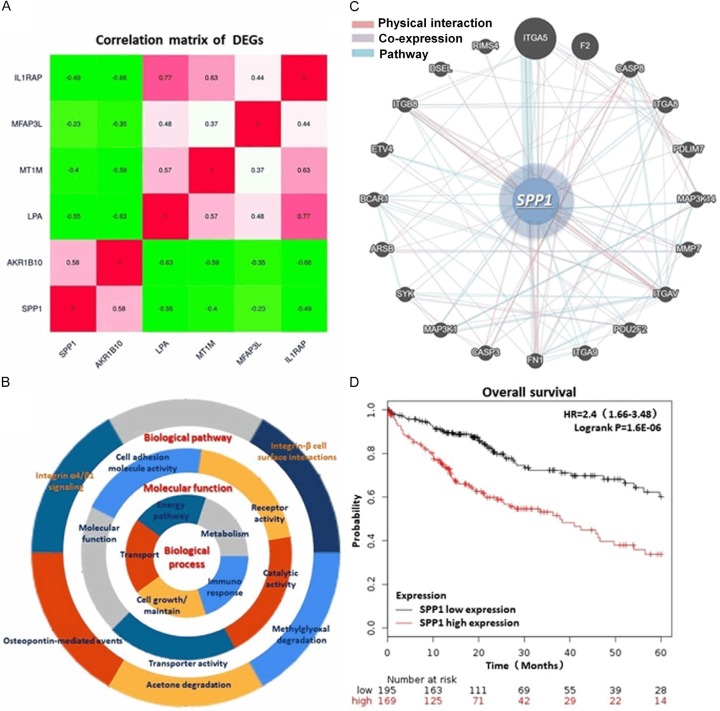
In basis of the criterion of |log2FC| ≥ 2.0 and P value <1.0E-04 for exploring DEGs of HCC through GEO database (https://www.ncbi.nlm.nih.gov/geo/), we totally found 285 genes amplified and 416 genes decreased in HCC tissues compared with the non-cancerous liver tissues. We overlapped these aberrantly expressed genes according to the expression profiles, and finally cohorted 2 up-regulated genes (AKR1B10 and SPP1) and 4 down-regulated ones (LPA, MT1M, MFAP3L and IL1RAP) (Figure 1).
Figure 1.
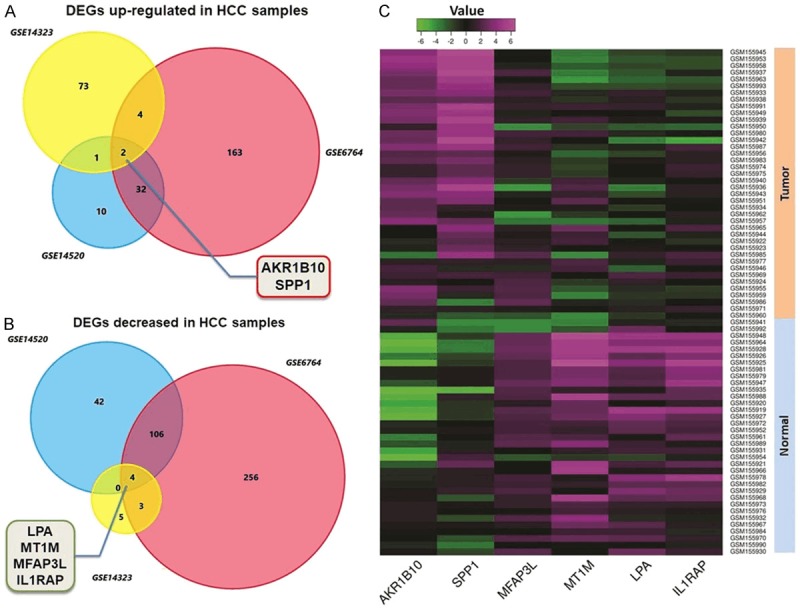
DEGs identified through analysis NCBI GEO datasets. A. Venn chart of the significant up-regulated genes in three HCC datasets (GSE6764, GSE14520 and GSE14323) compared with the non-cancerous liver tissues. AKR1B10 and SPP1 were screened out according to the overlapped results. B. Venn chart of the decreased genes among the three datasets. LPA, MT1M, MFAP3L and IL1RAP were collected finally. C. Representative heatmap generated through GEO datasets illustrates the comprehensive expression profiles of the six significant genes including SPP1. SPP1 presents a relatively higher expression in HCC tissues than in the normal liver tissues.
By conducting GO and KEGG pathway enrichment, we integrated the most significant pathways of these DEGs in HCC, including PI3K/AKT signaling pathway, ECM-receptor interaction, NF-κB signaling pathway and Toll-like receptor signaling pathway. The main gene set enrichment results with a P-value cutoff as 0.05 were shown in Figure 2A-C and Table 1. Protein-protein interaction (PPI) network of these genes was generated through the Search Tool for the Retrieval of Interacting Genes (STRING) database (Version 10.0, http://string-db.org), GeneMANIA (http://genemania.org/) and Cytoscape software (Version 3.4.0, http://www.cytoscape.org/). Analysis comprehensively according to these findings above, we noticed that SPP1 is a critical regulator in the biological process including post-translational protein modification, cell developmental process and cell death process, MAPK cascade and ERK cascade regulation and anginogenesis, which probably participating in the tumorigenesis and progress of HCC (Table 1). Additionally, by analyzing and calculating the 5 year OS rate according to the 364 HCC patient’s follow-up data, we observed a significantly low OS of the patients with a relatively higher SPP1 expression (HR=2.4, logrank P=1.6e-06) (Figure 2D).
Figure 2.
Correlation and interaction between DEGs and the particular analysis of SPP1. A. Correlation matrix of the DEGs. SPP1 presents a highly expression characteristic in HCC tissues sharing co-expression with another HCC promotor, AKR1B10. B. Presentation of the GO and KEGG enrichment analysis indicating the biological process, molecular functions and biological pathways of the DEGs. C. Network that SPP1 involved in including PPI, co-expression and pathway. D. KM plot of the 5 year OS rate generated from 364 HCC cases demonstraed a significant poor outcome of the patients with relatively higher SPP1 expression (HR=2.4, logrank P=1.6e-06).
Table 1.
A presentation of part of the GO and KEGG pathway enrichment analysis for the DEGs associated with cancer genesis and progress
| Category | ID | Term | P-value |
|---|---|---|---|
| BP | GO:0043062 | Regulation of heterotypic cell-cell adhesion | 3.0E-04 |
| BP | GO:0043062 | Extracellular structure organization | 3.0E-04 |
| BP | GO:0006954 | Inflammatory response | 2.9E-03 |
| BP | GO:0019221 | Cytokine-mediated signaling pathway | 5.8E-03 |
| BP | GO:0045595 | Regulation of cell differentiation | 1.3E-02 |
| MF | GO:0004908 | Interleukin-1 receptor activity | 1.1E-03 |
| MF | GO:0070851 | growth factor receptor binding | 2.5E-02 |
| MF | GO:0005102 | Signaling receptor binding | 1.5E-02 |
| MF | GO:0048018 | Receptor ligand activity | 2.4E-02 |
| MF | GO:0070851 | growth factor receptor binding | 2.5E-02 |
| CC | GO:0034358 | Plasma lipoprotein particle | 1.8E-04 |
| CC | GO:0005576 | Extracellular region | 2.3E-04 |
| CC | GO:0071682 | Endocytic vesicle lumen | 8.5E-04 |
| CC | GO:0005788 | Endoplasmic reticulum lumen | 5.2E-03 |
| CC | GO:0005764 | Lysosome | 3.0E-02 |
| KEGG pathway | hsa04512 | ECM-receptor interaction | 9.8E-03 |
| KEGG pathway | hsa04064 | NF-kappa B signaling pathway | 1.0E-02 |
| KEGG pathway | hsa04620 | Toll-like receptor signaling pathway | 1.0E-02 |
| KEGG pathway | hsa04060 | Cytokine-cytokine receptor interaction | 3.7E-02 |
| KEGG pathway | hsa04010 | MAPK signaling pathway | 4.5E-02 |
SPP1 is up-regulated in either tumor tissues of real HCC patients or multiple HCC cell lines
Considering the expression characteristic of SPP1 in HCC observed from biostatistics analysis, we further explored the expression profile of SPP1 in real HCC patients and HCC cell lines. Eighty-seven paired HCC tumor specimens and the adjacent non-cancer tissues were examined through IHC. As differentiating by the intensity of SPP1, these cases were divided into two groups: SPP1 high expressed group and SPP1 low expressed group. For tumor specimens, 70.1% (61/87) of the cases presented high expression status of SPP1, and the rest 29.9% (26/87) cases presented relatively lower expression of SPP1. On the contrary, there is only 26.4% (23/87) non-cancerous liver tissues present a relatively higher SPP1 expression compared with the other 73.6% (64/87).
Simultaneously, we measured the expression of SPP1 at both transcriptional and translational status in three HCC cell lines (Hep3B, HepG2 and Hu7u) compared with the control L02 cells. Consisting with IHC assay in tissues, SPP1 presents a highly expression in both mRNA and protein level than that in L02 cells, especially in Hep3B cells (Figures 3, S1).
Figure 3.
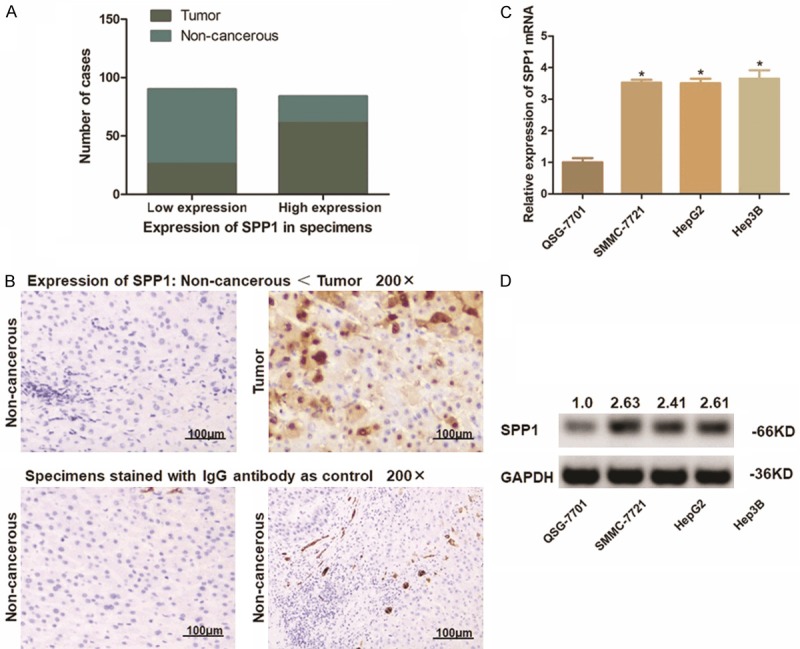
Expression of SPP1 in either HCC patient’s specimens and the HCC cell lines. A. Statistic of number of cases with higher or lower expression of SPP1 in 87 paired HCC specimens. SPP1 was up-regulated in most of the tumor tissues (61/87), and was expressed at a lower level in most of the adjacent non-cancerous tissues (64/87). SPP1 was frequently and significantly higher expressed in tumor tissues (P<0.01). B. Representative graph of immunohistochemistry analysis on tissue microarray (100 ×). Specimens stained IgG anti-body were applied as control. SPP1 in HCC tumor tissues was significantly higher expressed than that in adjacent non-cancerous tissues. C. Transcription level of SPP1 in cell lines was detected through qRT-PCR assay. The SPP1 mRNA expression is significantly higher in HCC cells than that in L02 cells as control (*P<0.05). D. Detection of protein expression of SPP1 in cell lines by Western-blot analysis. SPP1 protein was significantly up-regulated in HCC cells compared with L02 cells. The numbers above the blot indicate normalized protein amounts relative to the negative control, as determined by densitometry.
Highly expressed SPP1 is correlated with HCC clinicopathologic features
The correlation between SPP1 and the clinicopathological features of the 87 HCC patients was studied. As Table 2 shown, there was no significant correlation between SPP1 expression and the patient’s age, gende, virus control status or liver cirrhosis stages. While, higher SPP1 presented a significant positive trend towards, higher serum Alpha-fetoprotein (AFP) level, larger tumor size, advanced TNM stages, more incidence of tumor microsatellite formation and venous invasion.
Table 2.
Correlation between SPP1 and clinicopathologic features in 87 HCC specimens
| Clinicopathologic parameters | SPP1 expression | P * | |
|---|---|---|---|
|
| |||
| Low (n=26) | High (n=61) | ||
| Age (years) | |||
| ≤50 | 12 | 34 | 0.485 |
| >50 | 14 | 27 | |
| Gender | |||
| Male | 17 | 31 | 0.245 |
| Female | 9 | 30 | |
| Diameter (cm) | |||
| ≤5 | 18 | 25 | 0.020 |
| >5 | 8 | 36 | |
| TNM stage | |||
| I~II | 16 | 20 | 0.018 |
| III~IV | 10 | 41 | |
| Tumor encapsulation | |||
| Absent | 8 | 27 | 0.340 |
| Present | 18 | 34 | |
| Tumor microsatellite formation | |||
| Absent | 16 | 22 | 0.035 |
| Present | 10 | 39 | |
| Venous invasion | |||
| No | 15 | 19 | 0.030 |
| Yes | 11 | 42 | |
| HBsAg | |||
| Negative | 3 | 8 | >0.05 |
| Positve | 23 | 53 | |
| AFP (ng/ml) | |||
| ≤400 | 10 | 10 | 0.0478 |
| >400 | 16 | 51 | |
| Cirrhosis | |||
| Absent | 2 | 8 | 0.717 |
| Present | 24 | 53 | |
SPP1 expression level associated with clinicopathologic features in 87 HCC patients, including age, gender, tumor size, tumor stage (AJCC), tumor encapsulation, tumor microsatellite formation, vein invasion, HBsAg status, AFP level, and liver cirrhosis. Statistically significance was assessed by Fish’s exact test.
P<0.05.
Depletion of SPP1 suppresses cell proliferation and arrests the cell cycle of Hep3B cells
Hep3B cells with a high level of SPP1 expression were selected and transfected with pGU6/Neo vectors for depleting SPP1. The significant decrease of SPP1 in Hep3B cells was verified through qRT-PCR assay and Western blot analysis (Figures 4A, 4B, S1). CCK-8 assay on the aforementioned cells indicated an obvious impair of cell proliferation in SPP1 depleted Hep3B cells compared with the control. In detail, the P-value was <0.05 for days 1~2 and <0.01 for days 3~4 (Figure 4C). Flow cytometric analysis was conducted to learn the effect of SPP1 on cell apoptosis. As we observed, when SPP1 depleted, the percentage of Hep3B cells maintaining at G0/G1 phase was increased from 44.37% to 57.01% (P<0.01). Simultaneously, the S phase cells and the G2/M phase cells were respectively declined from 24.58% to 18.57% (P<0.05) and from 31.05% to 23.74% (P<0.05) (Figure 4D, 4E). Meanwhile, we found that the cell apoptosis was significantly enhanced along with SPP1 depletion with an apoptosis rate from 38.19% to 78.91% in average (P<0.01), demonstrating that the resistance of cell apoptosis in Hep3B cells was crippled (Figure 4F, 4G).
Figure 4.
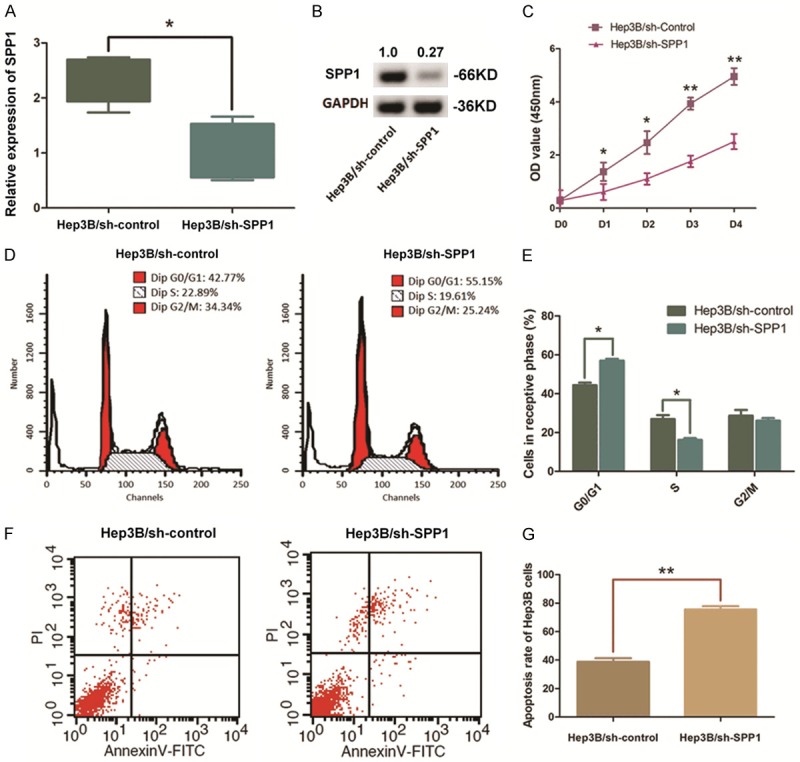
Depletion of SPP1 in Hep3B cells impairs the cell growth and promotes cell apoptosis. A. QRT-PCR assay indicated a significant down-regulation of SPP1 mRNA level in Hep3B cells after pGU6/Neo vectors transfection (**P<0.01). B. Western blot analysis validated the decline of SPP1 after transfection. Numbers above the blot indicate the protein amounts normalized. C. CCK8 assay was conducted to illustrate the effect of SPP1 on cell proliferation. Cell proliferation of Hep3B cells was significantly impaired by depleting SPP1 (**P<0.01, *P<0.05). D, E. Representative histograms describing the cell cycle profiles of Hep3B cells. Significantly, the cell cycle was arrested in the G0/G1 phase following depletion of SPP1 in Hep3B cells (*P<0.05). F, G. Apoptosis rate of was detected by flow cytometry. Representative graph of cell apoptosis rate was demonstrated and statistics analysis of cell apoptosis rate indicates that depletion of SPP1 in Hep3B cells significantly promotes cell apoptosis compared with the control ones (**P<0.01).
SPP1 is directly targeted by miR-181c post-transcriptionally in HCC cells
MiR-181c presents relatively lower expression status in multiple human maligancies, including HCC, calculated by using dbDEMC2 software (http://www.picb.ac.cn/dbDEMC) (Figure 5A). And we further validated the expression decrease of miR-181c in HCC cell lines compared with control L02 cells (Figure 5B). In this study, MiR-181c was predicted in this study through microcosm bioinformatics analysis online software. The potential binding site was visulized, and the minium free energy hybridization of miR-181c and SPP1 mRNA 3’-UTR was presented as Figure 5C shown.
Figure 5.
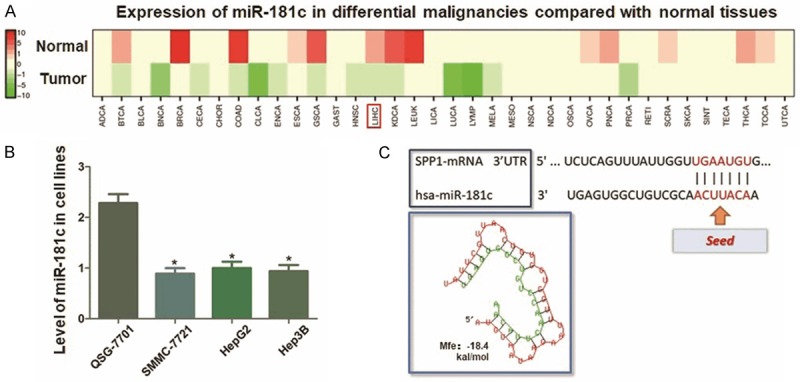
Analysis of miR-181c expression in HCC and the prediction of its targeting effect on SPP1. A. Calculation of miR-181c expression through dbDEMC2 software. MiR-181c is relatively lower expressed in multiple human maligancies, including HCC. B. qRT-PCR assay indicated significantly lower expression of miR-181c in HCC cell lines compared with L02 cells (*P<0.05). C. The predicted miR-126 binding site in the wild-type SPP1 mRNA 3’-UTR (3’UTR-WT) is visulizated, and the minimum free energy hybridization of miR-181c and SPP1 mRNA 3’-UTR is calculated (Mfe: -18.4 kal/mol).
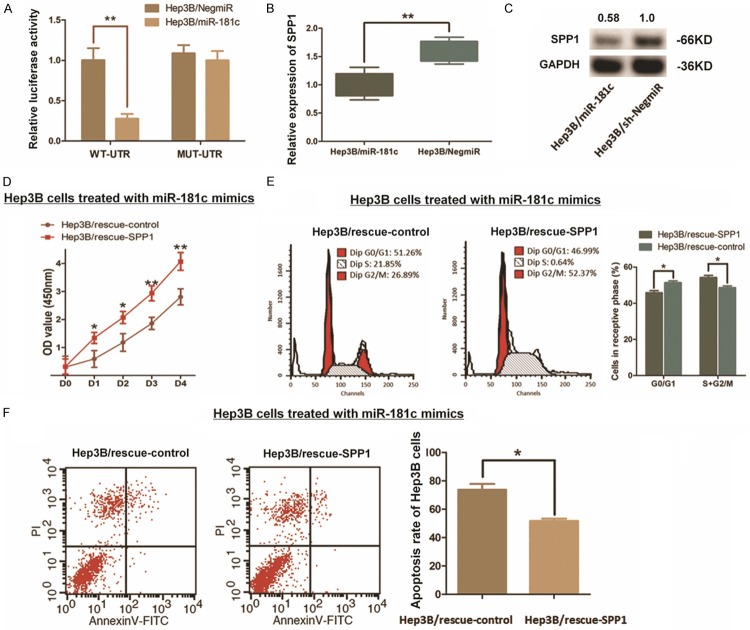
We constructed luciferase reporter vectors containing 202 bp 3’-UTR sequence of SPP1 mRNA (WT-UTR) and the control vectors consist of corresponding mutated sequence (MUT-UTR). Like Figure 6A shown, mimics indued miR-181c up-regulation (Hep3B/miR-181c) in Hep3B cells significantly decreased the luciferase signal of SPP1/pMIR/WT compared with the control ones (Hep3B/NigmiR). As supposed, this suppressing effect induced by miR-181c was abolished in Hep3B cells concerning with MUT-UTR. And the expression of SPP1 was significantly decreased within the miR-181c up-regulated Hep3B cells (Figures 6B, 6C, S1). Intringulingly, we further found that the cell growth impacts and the resistence of cell apoptosis in Hep3B cells induced by miR-181c could be patly rescured when SPP1 was ectopicly up-regulated (Figure 6D-F). All the findings above indicate a direct interaction and post-transcriptional regulation of miR-181c to SPP1.
Figure 6.
Modulation of SPP1 in Hep3B cells induced by miR-181c. A. The direct interaction between SPP1 and miR-181c was detected by Dual-luciferase reporter assay. Up-regulation miR-181c through mimics in Hep3B (Hep3B/miR-181c) decreased the luciferase signal of SPP1/pMIR/WT significantly in cells with compared with the negative control (Hep3B/NigmiR), while mutation of the putative miR-181c-binding site abolished this suppressive effect (**P<0.01). B. qRT-PCR assay demonstrated that the mRNA expression of SPP1 was significantly decreased by introducing miR-181c into Hep3B cells (**P<0.01). C. Western blot analysis indicated that the SPP1 protein was significantly decreased by introducing miR-181c into Hep3B cells. D-F. Rescue experiment was conducted due to re-upregulating SPP1 in Hep3B cells transfected with miR-181c. CCK8 assay and the cytometry analysis respectively indicated a partly rescue effect through up-regulating SPP1, which induced recovery of cell proliferation, along with the promotion of cell cycle process and resistence to cell apoptosis (*P<0.05).
Discussion
Worldwide, HCC presents as one of the most prevalent human maligant neoplasms leading to high mortality [11]. Quite amount of HCC patients were no longer capable for radical resection because of the large tumor size, venous invasion and multi-intrahepatic metastasis when diagnosed at later stages [12].
According to the preclinical an dclinical research, molecule-targeted therapy, like sorafenib and lenvatinib application, has been introduced into advanced HCC therapautic treatment, which has been playing important role in comprehensive treatment of HCC [13,14]. However, as for recent acknowledgement, this innovation treatment is still meeting with the challenge of drug resistance by targeting merely single target [15,16]. Thus, it is important and meaningful to discover new biomarkers and targets for comprehensive and combined stratigies in HCC treatment.
Biostatistic analysis provides the possibility for intensive and integrative insight of DEGs in cancer research, which means by mining the datasets of a certain carcinoma could assist the researchers to find new candidate genes in tumor provention, diagnose, and treatment. On basis of this strategy, we downloaded datasets frome GEO database with either mRNAs or non-coding RNAs (ncRNAs) expression information of HCC patients. The analysis of GSE6764, GSE14520 and GSE14323 respectively gave out 285 genes up-regulated in HCC tumors with a fold change over 2.0, and also 416 decreased ones. Among them, we further clustered SPP1 and AKR1B10 as two significent genes sharing co-expression profile in these three individual datasets, and collected another four decreased genes.
Combining with the GO and KEGG enrichment results, we discovered that these 6 candidate genes were involved in critical process such as PI3K/AKT signaling pathway, ECM-receptor interaction and NF-κB signaling pathway, by which were supposed to participatie in HCC tumorigenesis and process. Intriguingly, we noticed SPP1, also known as osteopontin, is one of the most significant over-expressed genes in HCC present close relationship with HCC progress. SPP1 is a multifunctional genes, which was first reported as one of the biomarkers in cell epitheial transformation process [17]. Further research described that SPP1 was involved in the attachment of osteoclasts to mineralized bone matrix [18]. And also, this gene participates in the up-regulation of IFN-γ and IL-12 as a cytokine [19,20].
In cancer research, literatures ever published have provided a basic imagiation of SPP1’s bio-functions in tumorigensis and process. In glioma, SPP1 plays a role of the ligand of CD44, assisting to enhance oncogene EPAS-1 expression and to promote aggresive glioma growth [21]. In colorectal cancer, high SPP1 expression is correlated with poor survival with positive venous invasion and high TNM stage [22]. And SPP1 was found highly expressed in epithelial ovarian cancer tissues, specifically activating Integrin-β/FAK/AKT pathway, promoting cell growth and mobility in ovarian cancer [23]. In prostate cancer, SPP1 is concerned with the progress of tumor recurrence and metastasis by mediating the bilogical processes of Smad4/PTEN pathway [24].
Reports of SPP1 in liver diseases, especially in liver tumor, is insufficient yet. Accodring to the limited literatures, we suppose SPP1 as the biomarker participating in polymorphisms process, and the pro-inflammatory genes regulation, which associates in liver injury, HBV and HCV infection status and HCC occurence [25-27]. However, execpt for the aberrant expession of SPP1 in liver tissues, the potent mechanisms of this gene affecting HCC tumorigenesis and progress leave us largely unkonwn.
In this study, again, we noticed a remarkable up-regulation of SPP1 in HCC tissues compared with the non-cancerous liver tissues through data mining and IHC assay in 87 cases of the real patients. And also, we validated the significant high expression characteristic of SPP1 at both transcriptional and translational status in multiple HCC cell lines according to the baseline of L02 cells. Further analysis combined with the clinicopathologic features from these real patients provided us a clear imagination of the correlationship between SPP1 expression status and serum APF level, tumor size, tumor invasion degrees and the total evaluation of TNM stages.
Along with the findings above, the 5 year over-all survival rate generated from 364 HCC cases demonstraed a poor outcome of the patients with relatively higher SPP1 expression. This finding clearly elucidates a trend of SPP1 up-regulation in HCC patients with poor prognosis, which suggests SPP1 as a probable target for further study in HCC prevention and diagnosis.
Depletion of SPP1 and the followed rescue experiment were carried out in Hep3B cells by using either pGU6/Neo vectors or recombinant adenovirus Ad5/F35. As we showed in our results, depleting SPP1 not only impaired the Hep3B cells’ proliferation, but also arrested the cell cycle in G0/G1 phase instead of maintaining at the G2/M stages. Simultaneously, apoptosis rate in Hep3B with SPP1 depletion was significantly enhanced. On the contrary, rescue experiment by re-expressing SPP1 in Hep3B cells reversed the changings induced by SPP1 depletion, including accelerating the cell proliferation, and inhibiting the cell apoptosis. All these above indicate that targeting SPP1 could effectively suppress the cell growth and promote programmed cell death in HCC.
Being members of microRNAs, miRNA-181 family function as post-transcriptional regulators effecting on gene expression [28,29]. As acknowledeged, family of miRNA-181 consists of four homological molecules respectively named miR-181a, miR-181b, miR-181c and miR-181d [30]. Accumulating evidence demonstrates that miR-181 members are associated with human cancer prognosis and survival, and involved in tumor invasion and metastasis in different ways [31-33]. For example, miR-181a and miR-181b act as promotors enhancing breast cancer cell proliferation through regulating PI3K/AKT signaling pathway [34]. And, miR-181d presents different functions in diverse human maligancies, like suppressing cell proliferation in glioblastoma via regulating NF-κB signaling pathway [35], or promoting osteosarcoma and colon cancer metastasis through either regulating FOXP1 feedback loop or modulating the process of glycolysis [36,37]. Noticeably, miR-181c commonly presents to be suppressing factor in diverse malignancies. In gasrtic cancer, miR-181c is significantly down-regulated as a potential biomarker indicating apoptosis resistence in tumor cells and relatively poor prognosis [9]. In breast cancer, miR-181c is the up-streaming regulator of PPAR-α involved in epithelial-mesenchymal transition, and is remarkably decreased in cancer cells [38]. Expression profile of miR-181 family in HCC and the correlated bio-functions are not distinguished between the four members, and the definite research on miR-181c is limited and not updated yet. Thus, whether miR-181c plays promotional effects like miR-181b or acts as inhibitor in HCC process need to be further elucidated.
In this study, miR-181c was predicted as the up-streaming regulator of SPP1 according to bioinformatics analysis. The predicted binding site demonstrated a strong potential interaction between miR-181c seed sequence and 3’-UTR of SPP1 mRNA. Detection in HCC cells indicated a down-regulation of miR-181c in tumor cells compared with L02 cells. And the direct interaction between miR-181c and SPP1 mRNA was further validated through dual-luciferase reporter assay. Interestingly, up-regulation of miR-181c in Hep3B cells sequentially decreased the expression of SPP1, and demonstrated us a clear image of the modulation and regulation mechanism involved with SPP1 in HCC.
In summary, we suppose SPP1 as one of the probable genes participating in the enhance of HCC cell growth, which provide us new potenital target for HCC prevention and treatment. Moreover, miR-181c presents direct interaction characteristic in HCC cells with SPP1 as an up-streaming inhibitor, confidently and hopefully suggesting new stratiges in HCC research and treatment for establishment of interventional practice at molecule level.
Acknowledgements
The authors thank Shen Chen, Di Ma, Ye Lu, Xiaoyong Gong, Jiajun Ren and Yuchen Yang for providing valuable technical supports and assistance. This study was kindly supported by grants from the following: National Natural Science Foundation of China (No. 81602544); Shanghai Pujiang Talent Project (No. 18PJD029).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Reig M, Mariño Z, Perelló C, Iñarrairaegui M, Ribeiro A, Lens S, Díaz A, Vilana R, Darnell A, Varela M, Sangro B, Calleja JL, Forns X, Bruix J. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol. 2016;65:719–726. doi: 10.1016/j.jhep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Nishibatake Kinoshita M, Minami T, Tateishi R, Wake T, Nakagomi R, Fujiwara N, Sato M, Uchino K, Enooku K, Nakagawa H, Asaoka Y, Shiina S, Koike K. Impact of direct-acting antivirals on early recurrence of HCV-related HCC: comparison with interferon-based therapy. J Hepatol. 2019;70:78–86. doi: 10.1016/j.jhep.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 4.Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L. Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem. 2019;120:10069–10081. doi: 10.1002/jcb.28290. [DOI] [PubMed] [Google Scholar]
- 5.Yin F, Shu L, Liu X, Li T, Peng T, Nan Y, Li S, Zeng X, Qiu X. Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:127. doi: 10.1186/s13046-016-0403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Zhou Y, Fei X, Chen X, Chen Y. Biostatistics mining associated method identifies AKR1B10 enhancing hepatocellular carcinoma cell growth and degenerated by miR-383-5p. Sci Rep. 2018;8:11094. doi: 10.1038/s41598-018-29271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C, Song L, Chen J, Wang F, Zhao HL, Ma YN, Yu J, Zhang JW. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2015;34:3226–3239. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Xing Y, Rong L. miR-181 regulates cisplatin-resistant non-small cell lung cancer via downregulation of autophagy through the PTEN/PI3K/AKT pathway. Oncol Rep. 2018;39:1631–1639. doi: 10.3892/or.2018.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zabaglia LM, Bartolomeu NC, Dos Santos MP, Peruquetti RL, Chen E, de Arruda Cardoso Smith M, Payão SLM, Rasmussen LT. Decreased microRNA miR-181c expression associated with gastric cancer. J Gastrointest Cancer. 2018;49:97–101. doi: 10.1007/s12029-017-0042-7. [DOI] [PubMed] [Google Scholar]
- 10.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong MC, Jiang JY, Goggins WB, Liang M, Fang Y, Fung FD, Leung C, Wang HH, Wong GL, Wong VW, Chan HL. International incidence and mortality trends of liver cancer: a global profile. Sci Rep. 2017;7:45846. doi: 10.1038/srep45846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, Baron A, Park JW, Han G, Jassem J, Blanc JF, Vogel A, Komov D, Evans TRJ, Lopez C, Dutcus C, Guo M, Saito K, Kraljevic S, Tamai T, Ren M, Cheng AL. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 13.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 14.Thomas H. Liver cancer: lenvatinib non-inferior to sorafenib for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2018;15:190. doi: 10.1038/nrgastro.2018.20. [DOI] [PubMed] [Google Scholar]
- 15.Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: etiology and current and future drugs. J Clin Exp Hepatol. 2019;9:221–232. doi: 10.1016/j.jceh.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudo M. Systemic therapy for hepatocellular carcinoma: latest advances. Cancers (Basel) 2018;10 doi: 10.3390/cancers10110412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Wang W, He J, Jiang L, Li X. Osteopontin as a biomarker for osteosarcoma therapy and prognosis. Oncol Lett. 2019;17:2592–2598. doi: 10.3892/ol.2019.9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraher D, Hodge JM, Collier FM, McMillan JS, Kennedy RL, Ellis M, Nicholson GC, Walder K, Dodd S, Berk M, Pasco JA, Williams LJ, Gibert Y. Citalopram and sertraline exposure compromises embryonic bone development. Mol Psychiatry. 2016;21:656–664. doi: 10.1038/mp.2015.135. [DOI] [PubMed] [Google Scholar]
- 19.Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 20.Kashani-Sabet M, Nosrati M, Miller JR 3rd, Sagebiel RW, Leong SPL, Lesniak A, Tong S, Lee SJ, Kirkwood JM. Prospective validation of molecular prognostic markers in cutaneous melanoma: a correlative analysis of E1690. Clin Cancer Res. 2017;23:6888–6892. doi: 10.1158/1078-0432.CCR-17-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe EK, Yi JW, Chai YJ, Park KJ. Upregulation of the adipokine genes ADIPOR1 and SPP1 is related to poor survival outcomes in colorectal cancer. J Surg Oncol. 2018;117:1833–1840. doi: 10.1002/jso.25078. [DOI] [PubMed] [Google Scholar]
- 23.Zeng B, Zhou M, Wu H, Xiong Z. SPP1 promotes ovarian cancer progression via integrin beta1/FAK/AKT signaling pathway. Onco Targets Ther. 2018;11:1333–1343. doi: 10.2147/OTT.S154215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong QZ, Zhang XF, Zhao Y, Jia HL, Zhou HJ, Dai C, Sun HJ, Qin Y, Zhang WD, Ren N, Ye QH, Qin LX. Osteopontin promoter polymorphisms at locus -443 significantly affect the metastasis and prognosis of human hepatocellular carcinoma. Hepatology. 2013;57:1024–1034. doi: 10.1002/hep.26103. [DOI] [PubMed] [Google Scholar]
- 26.Shin HD, Park BL, Cheong HS, Yoon JH, Kim YJ, Lee HS. SPP1 polymorphisms associated with HBV clearance and HCC occurrence. Int J Epidemiol. 2007;36:1001–1008. doi: 10.1093/ije/dym093. [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Hafiz SM, Hamdy HE, Khorshed FM, Aboushousha TS, Safwat G, Saber MA, Seleem M, Soliman AH. Evaluation of osteopontin as a biomarker in hepatocellular carcinomas in Egyptian patients with chronic HCV cirrhosis. Asian Pac J Cancer Prev. 2018;19:1021–1027. doi: 10.22034/APJCP.2018.19.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 29.Pop-Bica C, Pintea S, Cojocneanu-Petric R, Del Sal G, Piazza S, Wu ZH, Alencar AJ, Lossos IS, Berindan-Neagoe I, Calin GA. MiR-181 family-specific behavior in different cancers: a meta-analysis view. Cancer Metastasis Rev. 2018;37:17–32. doi: 10.1007/s10555-017-9714-9. [DOI] [PubMed] [Google Scholar]
- 30.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, Dohner K, Neilly MB, Price C, Lussier YA, Zhang Y, Larson RA, Le Beau MM, Caligiuri MA, Bullinger L, Valk PJ, Delwel R, Lowenberg B, Liu PP, Marcucci G, Bloomfield CD, Rowley JD, Chen J. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, Lax S, Samonigg H, Hoefler G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67:198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 33.Liu YS, Lin HY, Lai SW, Huang CY, Huang BR, Chen PY, Wei KC, Lu DY. MiR-181b modulates EGFR-dependent VCAM-1 expression and monocyte adhesion in glioblastoma. Oncogene. 2017;36:5006–5022. doi: 10.1038/onc.2017.129. [DOI] [PubMed] [Google Scholar]
- 34.Strotbek M, Schmid S, Sánchez-González I, Boerries M, Busch H, Olayioye MA. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int J Cancer. 2017;140:2310–2320. doi: 10.1002/ijc.30661. [DOI] [PubMed] [Google Scholar]
- 35.Yang F, Liu X, Liu Y, Liu Y, Zhang C, Wang Z, Jiang T, Wang Y. miR-181d/MALT1 regulatory axis attenuates mesenchymal phenotype through NF-kappaB pathways in glioblastoma. Cancer Lett. 2017;396:1–9. doi: 10.1016/j.canlet.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Guo X, Zhu Y, Hong X, Zhang M, Qiu X, Wang Z, Qi Z, Hong X. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017;8:e2958. doi: 10.1038/cddis.2017.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Xiao Z, Yu R, Wang Y, Xu R, Zhu X. miR-181d-5p-FOXP1 feedback loop modulates the progression of osteosarcoma. Biochem Biophys Res Commun. 2018;503:1434–1441. doi: 10.1016/j.bbrc.2018.07.060. [DOI] [PubMed] [Google Scholar]
- 38.Rajarajan D, Selvarajan S, Charan Raja MR, Kar Mahapatra S, Kasiappan R. Genome-wide analysis reveals miR-3184-5p and miR-181c-3p as a critical regulator for adipocytes-associated breast cancer. J Cell Physiol. 2019;234:17959–17974. doi: 10.1002/jcp.28428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.