Abstract
Recent evidence has shown that long noncoding RNAs (lncRNAs) play major roles in tumorigenesis and cancer progression. The cancer genome atlas program (TCGA) database was used to screen colon adenocarcinoma (COAD)-related differentially expressed lncRNAs, which revealed that lncRNA ELFN1-AS1 was highly expressed in COAD. This study aimed to explore the regulatory role of ELFN1-AS1 in COAD and construct a gene delivery system based on extracellular vesicles (EVs). We found that ELFN1-AS1 levels were obviously increased in COAD patients and COAD tumor cells. Knockdown of ELFN1-AS1 expression by siRNA inhibited COAD cell proliferation and migration. Moreover, silencing ELFN1-AS1 significantly reduced the activation of extracellular signal-regulated protein kinase (Erk), up-regulated the protein expression of E-cadherin and down-regulated vimentin. In addition, we treated human umbilical cord mesenchymal stem cells (hUCMSCs) with siRNA-ELFN1-AS1 and found that EVs from siRNA-ELFN1-AS1-treated hUCMSCs could inhibit COAD cell proliferation and migration in vitro. These findings suggested that ELFN1-AS1 could promote the progression of COAD and that hUCMSC-EVs might be an attractive vehicle for the clinical administration of lncRNA-specific siRNAs in patients with COAD.
Keywords: ELFN1-AS1, colon adenocarcinoma, human umbilical cord mesenchymal stem cells, extracellular vesicles
Introduction
Colon adenocarcinoma (COAD) is one of the main causes of cancer-associated deaths worldwide, and in China, its incidence has shown an increasing trend every year [1,2]. Due to the high frequency of recurrence and metastasis, the 5-year survival rate for patients with COAD is only 30% [3]. Therefore, deeper investigation should be carried out to determine the underlying mechanisms of COAD and to explore more novel potential targets for effective therapies against COAD.
Long noncoding RNAs (lncRNAs) are a class of RNA molecules that exist widely in the mammalian genome, are between 200-100,000 nt long, and have little or no protein-coding function [4]. Accumulating evidence has revealed that lncRNAs may function in diverse crucial physiological and pathological processes [5] and play vital regulatory roles in tumorigenesis and cancer progression [6]. Although some key lncRNAs, such as HOTAIR [7], H19 [8], MALAT1 [9] and UCA1 [10], have been shown to be associated with the progression of COAD, there are still other new key lncRNAs that need to be explored. LncRNA ELFN1-AS1, located antisense to ELFN1, is a novel primate gene with possible microRNA function expressed predominantly in human tumors [11]. A recent study reported that as a diagnostic target for COAD diagnosis, ELFN1-AS1 had a specificity and sensitivity of 100.0 and 87.6%, respectively [12]. Notably, analysis of ELFN1-AS1 genomic profiles from 270 COAD patients in the GEPIA (http://gepia.cancer-pku.cn) online database revealed that the high expression of ELFN1-AS1 in COAD patients was closely associated with lower overall survival [13]. ELFN1-AS1 might be a new key lncRNA in COAD, but its role and the underlying mechanisms in COAD are unclear.
Although lncRNAs provide new opportunities for COAD diagnosis and therapy, the successful delivery of lncRNA-targeting drugs (such as lncRNA-specific siRNAs) to cancer cells has been hampered by difficulties in developing a sustainable and effective transportation system. Extracellular vesicles (EVs) are small membrane vesicles that mediate communication between cells by carrying nucleic acids or proteins [14,15]. Human mesenchymal stem cells (MSCs) are considered to be the ideal source of EVs for drug delivery because MSCs possess lower immunogenicity and are readily available and highly proliferative [16]. Our previous research has demonstrated that human umbilical cord MSC (hUCMSC)-derived EVs (hUCMSC-EVs) are safe for use in animal models and exhibit intrinsic therapeutic effects in sciatic nerve transection [17]. Furthermore, recent studies have revealed that EVs can deliver tumor-related siRNAs or lncRNAs into cancer cells [18,19]. However, the MSC-EV-based lncRNA strategy for COAD treatment has not been explored until now.
In this study, based on the TCGA database, we identified the expression and role of EFLN1-AS1 in COAD. ELFN1-AS1 is upregulated in COAD patients and COAD cells. Knockdown of ELFN1-AS1 inhibited COAD cell proliferation and migration, which was accompanied by changes in Erk activation and epithelial-mesenchymal transition (EMT)-related proteins. In addition, we utilized hUCMSC-EVs to deliver siRNA-ELFN1-AS1 (siRNA-EVs) to COAD cells and found that siRNA-EV treatment decreased COAD cell proliferation and migration. These results highlight the potential role of ELFN1-AS1 in COAD and indicate that hUCMSC-EVs might be a promising vehicle for the clinical administration of lncRNAs-specific siRNAs in COAD treatment.
Materials and methods
Ethics statement
COAD samples were collected from 22 patients who provided informed consent, which was in accordance with the ethical standards of the ethics committee of the Affiliated Hospital of Jiangsu University (Zhenjiang, China). All samples were reviewed by a pathologist and were diagnosed as COAD based on histopathological evaluation. These patients had not received radiotherapy or chemotherapy prior to surgical resection.
Cell culture
The human COAD cell lines (CaCO-2, SW-480 and HCT-116) and normal colorectal epithelial cells FHC were purchased from ATCC (Manassas, VA). Cells were not revalidated for this work. All of the cell lines were grown in DMEM medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco) and 1% pen/strep (Gibco). Human umbilical cord mesenchymal stem cells (hUCMSC) were cultured in stem cell culture medium (Cyagen, Guangzhou, China). All the cells were incubated under 5% CO2 and 37°C conditions.
Cell transfection
ELFN1-AS1 siRNAs (si-ELFN1-AS1#1 and si-ELFN1-AS1#2) were designed and synthesized by GenePharma (Shanghai, China). The siRNAs were transfected into COAD cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s procedures.
RNA isolation and quantitative real-time PCR
RNA was extracted from the cell preparations using Trizol (Invitrogen) according to the manufacturer’s protocol. All of the primers (human ELFN1-AS1 and GAPDH) for real-time PCR were purchased from Genecopoeia (Germantown, MD). Real-time PCR was performed with All-in-one™ qPCR Mix (Genecopoeia) in a CFX96™ Real-Time system (Bio-Rad, Hercules, CA). The relative expression of ELFN1-AS1 mRNA was evaluated by the 2-ΔΔCt method and normalized to GAPDH, based on our previous description [20].
CCK-8 proliferation assay
COAD Cells (CaCO-2 and HCT-116) were collected 24 h after transfection by siRNAs, and then the transfected cells were seeded in 96-well plates (2×103/well). Ten microliters of the CCK-8 solution (Beyotime, Nantong, China) was added to each well and incubated for 2 h. The absorbance of the cells in each well was measured at 450 nm using a microplate reader (Synergy HT, BioTek, Biotek Winooski, VT).
HUCMSC extracellular vesicles (hUCMSC-EVs) engineered by si-ELFN1-AS1 (siRNA-EVs) and siRNA-EVs uptake
For the preparation of siRNA-EVs, hUCMSCs were transfected with si-ELFN1-AS1#1 (100 nM) using Lipofectamine 2000, then the EVs were collected [14,17]. The transmission electron microscopy and the particle size distribution of the siRNA-EVs were detected as our previous description [14,17].
For siRNA-EVs uptake, CaCO-2 cells were labeled with 3, 3-Dioctadecyloxacarbocyanine perchlorate (DIO, green) and siRNA-EVs was labeled with CM-Dil (red). After washed twice with PBS, the CM-Dil-labeled siRNA-EVs were incubated with the DIO-labeled CaCO-2 for 2 h. After that, the cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.5% Triton-X 100. Then, the cell nuclei were stained using 4, 6 diamidino-2-phenylindole (DAPI). All reagents were purchased from Invitrogen. Images of siRNA-EVs uptake were obtained using a Nikon Eclipse Ti confocal laser scanning microscope.
Colony forming assay
With respect to the colony formation assay, CaCO-2 and HCT-116 cell lines transfected with si-NC or siRNAs were seeded in six-well plates (500/well) and cultured for two weeks in culture medium with 10% FBS.
To investigate the effect of siRNA-EVs on COAD cell colony formation, CaCO-2 or HCT-116 cells were plated in six-well plates (10% EVs-free FBS complete medium) and treated with hUCMSC-EVs, NC-EVs, siRNA-EVs (200 μg/ml) for 72 h (EVs-treated cells). After that, COAD cells were collected and cultured in another six-well plates (500/well) respectively, and maintained in medium with 10% FBS for 14 days. Then, cells were fixed with methanol and stained with 0.4% crystal violet solution, finally photographed.
Wound healing assay
After 24 h of transfection, 1×106 cells per well were cultured in six-well plates. After the cells reached 80% confluence, scratches were performed by a 200-μl pipette tip. Separated cells were washed out using PBS, and images of the same fields were taken at 48 h after the scratch.
Cell migration assay
SiRNA-transfected COAD cells (CaCO-2 and HCT-116) or siRNA-EV treated cells were harvested and plated into the upper Transwell chamber (CaCO-2: 5×104; HCT-116: 1×105 in 200 μl serum-free meida) for migration assay. The cells were allowed to migrate for 24 h toward the lower chamber that contained 500 μl of medium supplemented with 10% FBS. After incubation, the membranes were fixed with methanol and stained with 0.4% crystal violet solution, imaged using a Carl Zeiss microscope (Jena, Germany), and counted by Image J software.
Western blotting
The proteins in COAD cells were extracted using RIPA buffer (Cell Signaling Technology Inc., Danvers, MA) containing PMSF (Beyotime, Nantong, China). The protein extracts (50 μg) were separated on 10% sodium dodecyl SDS-PAGE and then electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA), as our previous description [20]. The antibodies to human E-cadherin was purchased from cell signaling technology, Erk, phosphorylated-Erk (p-Erk), GAPDH, Vimentin, β-actin, CD63, calreticulin and HRP-linked anti-rabbit/mouse IgG were all purchased from Abcam (Cambridge, MA).
Statistical analysis
GraphPad Prism software (Version 5.0; La Jolla, CA) was used for statistical analyses. The data are expressed as the means ± SD. Statistical significance was determined using Mann-Whitney test or one-way analysis of variance. Results were considered statistically significant at P-values less than 0.05.
Results
LncRNA ELFN1-AS1 was upregulated in human colon adenocarcinoma (COAD)
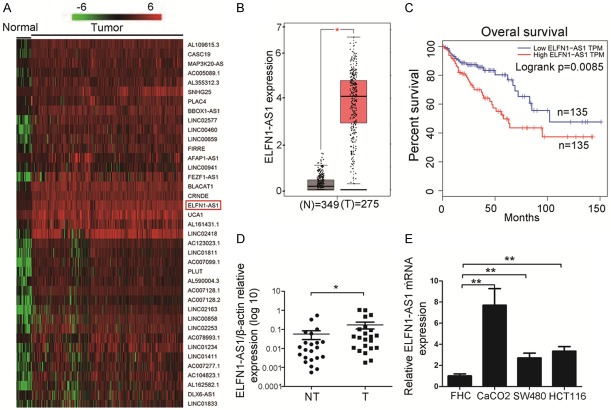
First, we analyzed RNA sequencing data of COAD and para-cancerous tissues downloaded from TCGA (39 normal and 398 cancer specimens). As expected, ELFN1-AS1 was shown to be highly expressed in COAD tissues (Figure 1A). The results shown in the box plots revealed that ELFN1-AS1 expression was dramatically increased in COAD tissues, and the survival curves of COAD patients showed that the expression level of ELFN1-AS1 was significantly associated with the overall survival rate by analyzing the data from the bioinformatic tool GEPIA (Figure 1B and 1C). In addition, we observed that ELFN1-AS1 was markedly upregulated in COAD tissues compared with adjacent nontumor tissues (Figure 1D). Moreover, ELFN1-AS1 was found to be expressed at higher levels in COAD cell lines than in the normal colorectal epithelial cell line FHC (Figure 1E). All of these results supported that ELFN1-AS1 played an oncogenic role in the progression of COAD.
Figure 1.
ELFN1-AS1 expression is up-regulated in colon adenocarcinoma (COAD). A. Hierarchical cluster heat map of differentially expressed lncRNAs in COAD and corresponding normal tissues generated from RNA sequencing data from the TCGA database. Red in the heat map denotes upregulation; green denotes downregulation. The red arrow indicates ELFN1-AS1. B. Expression of ELFN1-AS1 in the GEPIA database. *P<0.05 vs. Normal group (Mann-Whitney test). C. Kaplan-Meier survival analysis of COAD patients’ overall survival based on ELFN1-AS1 expression in GEPIA. D. The mRNA level of ELFN1-AS1 was determined by qRT-PCR in 22 pairs of tumor tissues and adjacent non-tumor tissues. *P<0.05 vs. non-tumor group (Mann-Whitney test). E. ELFN1-AS1 expression in COAD cell lines (CaCO-2, SW-480, HCT-116) compared with normal colorectal epithelial cells FHC detected by qRT-PCR. **P<0.01 vs. FHC group (one-way analysis of variance).
Inhibition of ELFN1-AS1 impeded the proliferation of COAD cells
To study the potential biological functions of ELFN1-AS1, we designed two independent small interfering RNAs (siRNAs) to silence ELFN1-AS1. As shown in Figure 2A and 2B, ELFN1-AS1 expression was strongly reduced 24 h after siRNA transfection of CaCO-2 and HCT-116 cells. Next, CCK8 assays demonstrated that ELFN1-AS1 knockdown dramatically inhibited cell growth (Figure 2C and 2D). A similar effect was also observed in the colony formation assay, where colony numbers were decreased upon knockdown of ELFN1-AS1 (Figure 2E). These results suggested that ELFN1-AS1 inhibition suppressed COAD cell proliferation.
Figure 2.
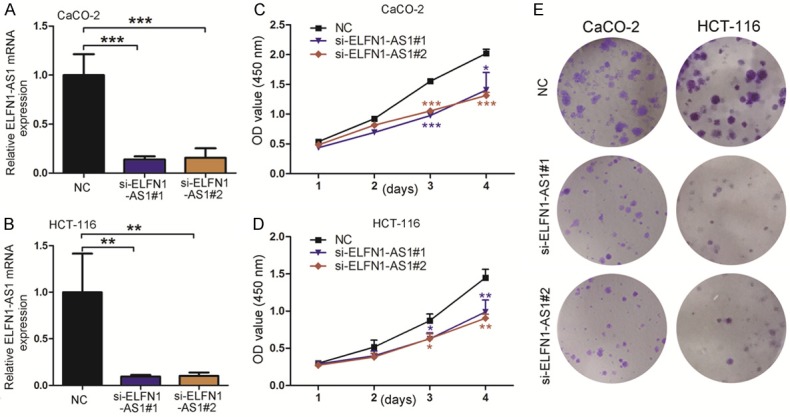
Down-regulation of ELFN1-AS1 expression inhibits COAD cell proliferation. A, B. The expression levels of ELFN1-AS1 mRNA in CaCO-2 and HCT-116 siRNA group were detected by real-time PCR. **P<0.05 and ***P<0.001 vs. NC group (one-way analysis of variance). C, D. CaCO-2 and HCT-116 cell lines transfected with si-RNA or si-NC were cultured in 96-well plates, cell proliferation were measured via CCK-8. *P<0.05, **P<0.05 and ***P<0.001 vs. NC group (one-way analysis of variance). E. The colony forming growth assay was performed to determine the proliferation of siRNA/si-NC transfected COAD cells.
LncRNA ELFN1-AS1 promoted the migration of COAD cells in vitro
To validate the function of ELFN1-AS1 on the migration of COAD cells, we first carried out a scratch wound assay in which silencing ELFN1-AS1 significantly decreased the migration ability of both CaCO-2 and HCT-116 cells (Figure 3A and 3B). The results of the Transwell migration assay revealed that compared with that of the control group, siRNA-ELFN1-AS1 treatment dramatically decreased the number of migrated COAD cells (Figure 3C and 3D). Taken together, these results suggested that ELFN1-AS1 could promote the migration of COAD cells.
Figure 3.
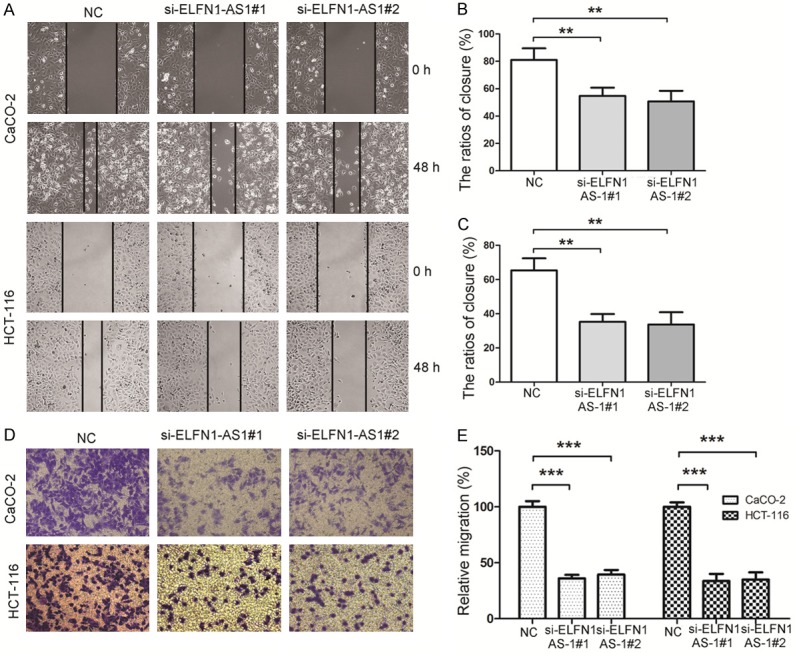
Down-regulation of ELFN1-AS1 gene expression inhibits COAD cell migration. A. A wound healing assay was applied to analyze the migration capacity in COAD cells after transfection with siRNA or NC. B, C. The quantifications of COAD cell migration were presented as percentage the ratios of closure. **P<0.05 vs. NC group (one-way analysis of variance). D. Transwell migration assay in down-regulation of ELFN1-AS1 cells and their corresponding control cells. E. Image J software was used for cell counting and the columns represent the mean of cell numbers from at least three independent experiments. ***P<0.001 vs. NC group (one-way analysis of variance).
Knockdown of ELFN1-AS1 inhibited Erk pathway and epithelial-mesenchymal transition (EMT) process in COAD cells
As lncRNA ELFN1-AS1 is thought to participate in the COAD progression, and Erk pathway been shown to play an important role in promoting proliferation and metastasis [21]. Therefore, we firstly explored the correlation of ELFN1-AS1 and Erk pathway. We found that knockdown of ELFN1-AS1 by siRNA transfection can reduce the protein levels of phosphorylated-Erk (p-Erk) both in CaCO2 and HCT116 cells (Figures 4A, 4B and S1), suggesting that Erk signal acted as a downstream of ELFN1-AS1.
Figure 4.
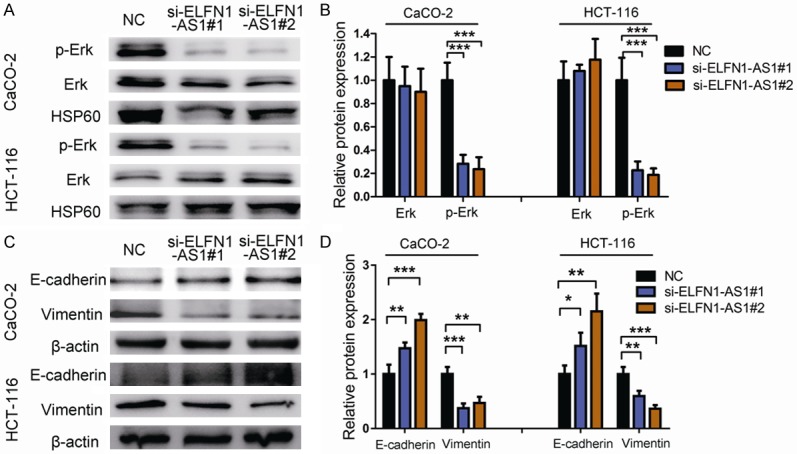
Down-regulation of ELFN1-AS1 regulates the EMT in COAD cell lines. A. The effect of ELFN1-AS1 expression on the levels of Erk and p-Erk. B. Protein expression of Erk and p-Erk. ***P<0.001 vs. NC group (one-way analysis of variance). C. The effect of ELFN1-AS1 expression on the levels of E-cadherin, and Vimentin in COAD cell lines transfected with si-RNA or si-NC by Western blot. D. Protein expression of E-cadherin, and Vimentin were analyzed by Image J software. *P<0.05, **P<0.05 and ***P<0.001 vs. NC group (one-way analysis of variance).
Epithelial-mesenchymal transition (EMT) is a process defined by the loss of tight junctions and an increase in cell motility, which has an important effect on tumor progression [22,23]. Erk pathway is an inducer of EMT, which could upregulate vimentin and downregulate E-cadherin [24]. Next, we investigated whether ELFN1-AS1 had any influence on EMT progression. Our results showed that the downregulation of ELFN1-AS1 significantly induced E-cadherin protein expression but decreased the expression of vimentin (Figures 4C, 4D and S1). These findings suggested that in COAD cells, the knockdown of ELFN1-AS1 could reduce the activity of Erk, inhibit EMT progression and decrease cell proliferation and metastasis, which indicated that silencing ELFN1-AS1 might be a novel effective therapy against COAD.
Extracellular vesicles (EVs) from hUCMSCs siRNA against ELFN1-AS1 modified effectively suppressed the proliferation and migration of COAD in vitro
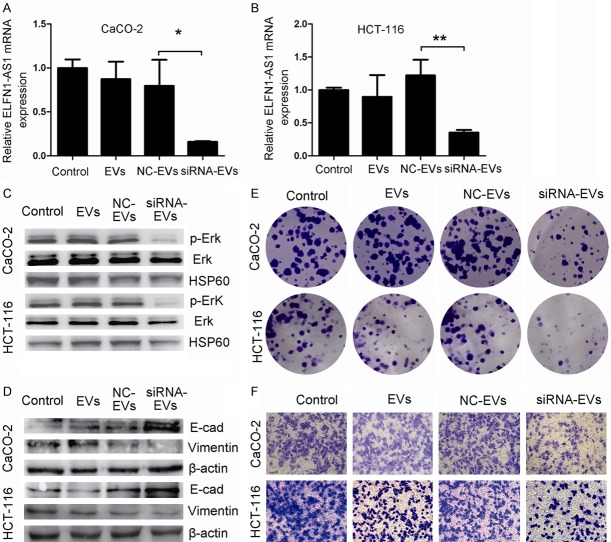
However, an effective targeted gene delivery to tumors is still a challenge. Most recently, several studies indicated that EVs, especially those derived from MSCs, may be used as delivery vehicles for cancer therapeutics (such as the delivery of miRNAs) [25]. Here, we wanted to determine whether MSC-EVs could be loaded with ELFN1-AS1-siRNA to affect COAD progression. To address this issue, we first randomly selected si-ELFN1-AS1#1 to transfect hUCMSCs. Then, we collected EVs from the si-ELFN1-AS1#1-treated hUCMSCs and named them siRNA-EVs. As shown in Figure 5A, siRNA-EVs were round membrane-bound vesicles and had an average diameter of approximately 148 nm with a size distribution of 50 nm to 650 nm (Figure 5B). Western blotting demonstrated that the EV marker protein CD63 was present in these vesicles (Figures 5C and S2). Then, we treated CaCO-2 cells with CM-Dil-labeled siRNA-EVs and found that these vesicles could fuse with the membranes of CaCO-2 cells (Figure 5D), suggesting that the siRNA-EVs could be taken up by COAD.
Figure 5.
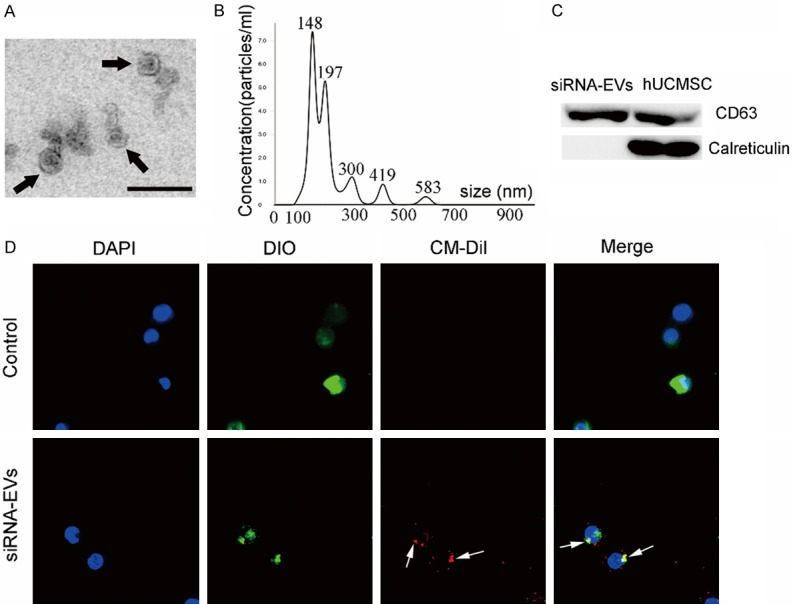
Extracellular Vesicles (EVs) engineered by ELFN1-AS1 inhibitor (siRNA-EVs). A. siRNA-EVs were observed under a transmission electron microscope; some of the siRNA-EVs are indicated by arrows. Scale bar = 200 nm. B. Size distributions of siRNA-EVs were detected using the Nanoparticle Tracking Analysis. C. Western blotting analysis of CD63 and Calreticlin expression in lysates from siRNA-EVs and hUCMSC. D. DIO-labeled CaCO-2 cells (green) were incubated with CM-Dil-labeled siRNA-EVs (red) for 2 h, and the siRNA-EVs uptake by CaCO-2 cells was determined. The arrows indicate the fusion of the membrane. The images are shown at ×600.
Next, we evaluated the roles of siRNA-EVs on COAD cells. There was a significant decrease in ELFN1-AS1 expression in COAD cells treated with siRNA-EVs (Figure 6A and 6B), which was accompanied by the inhibition of p-Erk, upregulation of E-cadherin and downregulation of vimentin (Figures 6C, 6D and S3). Moreover, compared to NC-EV group, siRNA-EV treatment inhibited COAD cell proliferation (Figure 6E) and migration (Figure 6F). Taken together, these results suggested that EVs from hUCMSCs treated with siRNA against ELFN1-AS1 could effectively suppress the proliferation and migration of COAD cells in vitro.
Figure 6.
SiRNA-EVs inhibit COAD cell proliferation and migration in vitro. A, B. The expression of ELFN1-AS1 in COAD cells treated with siRNA-EVs (200 μg/ml for 72 h) was detected by real-time PCR. *P<0.05 and **P<0.01 vs. NC-EV group (one-way analysis of variance). C. Erk and p-Erk protein were detected by western blotting after treatment by siRNA-EVs in COAD cells. D. EMT relevant proteins (E-cadherin and Vimentin) were detected by western blotting analysis after treatment by siRNA-EVs in COAD cells. E. Colon forming assays were used to determine the colony-forming ability of COAD cells after siRNA-EVs treatment. F. Transwell migration assay in siRNA-EVs treated-COAD cells and their corresponding control cells.
Discussion
Colon adenocarcinoma (COAD) occurs increasingly frequently worldwide [1,2]. Although accumulated evidence has shown that the dysregulation of lncRNAs is closely related to tumorigenesis and cancer progression [6], there are still a limited number of reports about the key lncRNAs in COAD. In this study, we analyzed RNA sequencing data from the TCGA database and found that lncRNA ELFN1-AS1 was increased; moreover, upregulation of ELFN1-AS1 in COAD patients was closely associated with lower overall survival. Although Liu et al. [12] reported that ELFN1-AS1 was involved in the early stage of COAD and had potential diagnostic value, the potential roles of ELFN1-AS1 in the progression of COAD are still unclear.
In this study, we first examined the expression of ELFN1-AS1 in COAD patients and COAD cancer cell lines by real-time PCR and found that the expression of ELFN1-AS1 was not only significantly upregulated in COAD patients but was also increased in COAD cell lines. In addition, silencing ELFN1-AS1 significantly inhibited the proliferation, colony formation and migration of COAD cell lines in vitro. These findings indicate that ELFN1-AS1 may be involved in promoting COAD progression.
Epithelial-mesenchymal transition (EMT) is a process in which epithelial cells lose cell polarity and gain the ability metastasize [22], and it has been confirmed that EMT is closely associated with cancer progression [23,26]. Jiang et al. [27] found that lncRNA SNHG15 could promote colon cancer progression by modulating EMT. Similar to this report, we found that knockdown of ELFN1-AS1 could downregulate the expression of vimentin, while E-cadherin was upregulated, suggesting that the function of ELFN1-AS1 in COAD might be associated with EMT. In addition, we found that silencing ELFN1-AS1 dramatically inhibited the activation of p-Erk. Erk pathway plays an important role in promoting COAD proliferation and metastasis and is a common inducer of EMT [21,24]. Thus, our results indicated that the inhibition of the COAD progression induced by the knockdown of ELFN1-AS1 might due to the decreased activation of the Erk pathway. While, the potential mechanism of ELFN1-AS1 in COAD still needs to be elucidated. In addition, it is also worthwhile to explore the role of ELFN1-AS1 in other kinds of tumors.
ELFN1-AS1 may be a potential target for tumor treatment. However, it is still a challenge to efficiently deliver lncRNA-targeting drugs (for example, lncRNA-specific siRNAs) to tumors due to degradation of the delivered gene, poor cellular uptake, and lack of tumor targeting ability [28]. Although liposomes and viral-based delivery systems have been assessed, all of these approaches exhibit low efficiency [29]. Extracellular vesicles (EVs) are nanoscale membranous vesicles that can serve as a novel gene drug delivery mechanism that combines high drug carrying capacity and targeting specificity, making them useful in tumor treatment [30]. Thus, using EVs as biological vehicles to deliver tumor suppressor siRNAs or lncRNAs is a promising approach. In addition, MSCs are an efficient mass producer of EVs for drug delivery [16], and MSC-EVs have been shown to be safe in some clinical trials [31,32]. Recently, Li et al. [33] found that EVs derived from bone-marrow-derived MSCs (BMSCs) treated with siRNA against GRP78 suppress sorafenib resistance in hepatocellular carcinoma. In this study, we found that EVs from siRNA-ELFN1-AS1-treated hUCMSCs (siRNA-EVs) could significantly decrease ELFN1-AS1 expression in COAD cells and effectively inhibit COAD cell growth and migration. Thus, MSC-EV-based lncRNA-specific siRNA therapy may be a new strategy for the treatment of COAD. Interestingly, a previous study reported that human BMSC-EVs could promote SW480 growth [34]. While, in our study, we found that hUCMSC-EV treatment alone could not significantly influence COAD growth and migration. This discrepancy may be attributed to the possibility that the quantity of EVs was insufficient to affect tumor growth or to the different sources of MSC-EVs.
In summary, we confirmed that in COAD cells, lncRNA ELFN1-AS1 was upregulated and promoted tumor proliferation and migration. ELFN1-AS1 functions in the tumorigenicity of COAD cells at least in part by regulating p-Erk and EMT, suggesting that ELFN1-AS1 might be a potential molecular target for COAD treatment. In addition, to our knowledge, this study provides the first evidence that hUCMSC-EVs might be a promising vehicle to deliver lncRNA-specific siRNAs to COAD cells to inhibit tumor progression.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (81900562 and 81871243), key research and development plan of Zhenjiang city (SH2019047), the key research and development programs of Jiangsu Province (BE2017697), the Six Talent Peaks of Jiangsu Province (WSN-009), “LiuGeYi” Projects of Jiangsu Province (LGY2016055), “XueDiJiFang” Projects of Jiangsu Province (x201812) and the Science and Technology Plan Project of Changzhou (CJ20180001).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Chen VW, Hsieh MC, Charlton ME, Ruiz BA, Karlitz J, Altekruse SF, Ries LA, Jessup JM. Analysis of stage and clinical/prognostic factors for colon and rectal cancer from SEER registries: AJCC and collaborative stage data collection system. Cancer. 2014;120(Suppl 23):3793–3806. doi: 10.1002/cncr.29056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-kappaB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, Gao X, Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, Li G, Wang G, Song J, Li Z, Zhang Z, Yuan W, Li X. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627–2644. doi: 10.1038/s41388-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, Basile F, Di Pietro C, Purrello M, Ragusa M. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–241. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polev DE, Karnaukhova IK, Krukovskaya LL, Kozlov AP. ELFN1-AS1: a novel primate gene with possible microRNA function expressed predominantly in human tumors. Biomed Res Int. 2014;2014:398097. doi: 10.1155/2014/398097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JX, Li W, Li JT, Liu F, Zhou L. Screening key long non-coding RNAs in early-stage colon adenocarcinoma by RNA-sequencing. Epigenomics. 2018;10:1215–1228. doi: 10.2217/epi-2017-0155. [DOI] [PubMed] [Google Scholar]
- 13.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L, Pu Y, Zhang L, Qi Q, Xu L, Li W, Wei C, Wang X, Zhou S, Zhu J, Wang X, Liu F, Chen X, Su C. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles promote lung adenocarcinoma growth by transferring miR-410. Cell Death Dis. 2018;9:218. doi: 10.1038/s41419-018-0323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Dong L, Zhong H, Yang L, Li Q, Su C, Gu W, Qian Y. Extracellular vesicles (EVs) from lung adenocarcinoma cells promote human umbilical vein endothelial cell (HUVEC) angiogenesis through yes kinase-associated protein (YAP) transport. Int J Biol Sci. 2019;15:2110–2118. doi: 10.7150/ijbs.31605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, Lim SK. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Dong L, Zhou D, Li L, Zhang W, Zhen Y, Wang T, Su J, Chen D, Mao C, Wang X. Extracellular vesicles from human umbilical cord mesenchymal stem cells improve nerve regeneration after sciatic nerve transection in rats. J Cell Mol Med. 2019;23:2822–2835. doi: 10.1111/jcmm.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, Lv Q, Qin C, Chu H, Wang M, Yuan L, Qian J, Zhang Z. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143. doi: 10.1186/s12943-018-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong L, Wang X, Tan J, Li H, Qian W, Chen J, Chen Q, Wang J, Xu W, Tao C, Wang S. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med. 2014;18:2213–2224. doi: 10.1111/jcmm.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Lee JY, Kong G. Roles and epigenetic regulation of epithelial-mesenchymal transition and its transcription factors in cancer initiation and progression. Cell Mol Life Sci. 2016;73:4643–4660. doi: 10.1007/s00018-016-2313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding C, Luo J, Li L, Li S, Yang L, Pan H, Liu Q, Qin H, Chen C, Feng J. Gab2 facilitates epithelial-to-mesenchymal transition via the MEK/ERK/MMP signaling in colorectal cancer. J Exp Clin Cancer Res. 2016;35:5. doi: 10.1186/s13046-015-0280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, Shahar T, Yamashita S, Parker Kerrigan B, Fueyo J, Sawaya R, Lang FF. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro Oncol. 2018;20:380–390. doi: 10.1093/neuonc/nox152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong AJ. Epithelial-mesenchymal transition in cancer progression. Clin Adv Hematol Oncol. 2011;9:941–943. [PubMed] [Google Scholar]
- 27.Jiang H, Li T, Qu Y, Wang X, Li B, Song J, Sun X, Tang Y, Wan J, Yu Y, Zhan J, Zhang H. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87. doi: 10.1016/j.canlet.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 28.Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review) Mol Med Rep. 2012;6:9–15. doi: 10.3892/mmr.2012.871. [DOI] [PubMed] [Google Scholar]
- 29.van der Meel R, Fens MH, Vader P, van Solinge WW, Eniola-Adefeso O, Schiffelers RM. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release. 2014;195:72–85. doi: 10.1016/j.jconrel.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 30.Sun D, Zhuang X, Zhang S, Deng ZB, Grizzle W, Miller D, Zhang HG. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Deliv Rev. 2013;65:342–347. doi: 10.1016/j.addr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Kordelas L, Rebmann V, Ludwig AK, Radtke S, Ruesing J, Doeppner TR, Epple M, Horn PA, Beelen DW, Giebel B. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973. doi: 10.1038/leu.2014.41. [DOI] [PubMed] [Google Scholar]
- 32.Nassar W, El-Ansary M, Sabry D, Mostafa MA, Fayad T, Kotb E, Temraz M, Saad AN, Essa W, Adel H. Umbilical cord mesenchymal stem cells derived extracellular vesicles can safely ameliorate the progression of chronic kidney diseases. Biomater Res. 2016;20:21. doi: 10.1186/s40824-016-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Yang C, Shi Y, Zhao L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnology. 2018;16:103. doi: 10.1186/s12951-018-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W, Huang L, Li Y, Zhang X, Gu J, Yan Y, Xu X, Wang M, Qian H, Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315:28–37. doi: 10.1016/j.canlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.