Abstract
Bladder cancer (BC) is one of the most common cancers in male patients, and the leading cause of cancer-related death in men. Hypoxia plays a critical role in carcinoma biology, including in bladder cancer. However, whether circular RNAs are associated with hypoxia-mediated progression of bladder cancer remain unknown. In this study, our aim was to investigate the role of circular RNA on the hypoxic adaptive response in bladder cancer. Here, we identified a hypoxia-inducible circular RNA, has-circRNA-403658 that contributes to bladder cancer progression. Has-circRNA-403658 is spliced from its host gene, ZNF292, through back-splicing between the 1st and 4th exon. We demonstrated that has-circRNA-403658 was an important circRNA that upregulated in bladder cancer cells under hypoxia, and higher has-circRNA-403658 levels were associated with poorer survival outcome. Silencing has-circRNA-403658 in bladder cancer cells inhibited cell growth and induced cell apoptosis. In addition, has-circRNA-403658 was induced by HIF1α and silencing has-circRNA-403658 inhibited LDHA-mediated aerobic glycolysis, inhibiting bladder cancer cell growth. Thus, our results suggest that has-circRNA-403658 may function as a novel therapeutic target in human bladder cancer.
Keywords: Bladder cancer, circular RNA, HIF1α, LDHA, aerobic glycolysis
Introduction
Bladder cancer (BC) is one of the most common cancers in male patients, and the leading cause of cancer-related deaths in men [1]. Hypoxia plays a critical role in carcinoma biology, including in bladder cancer [2]. BCs achieve a more advanced stage and more distant metastasis with a presentation of hypoxia [3]. A 28-gene hypoxia signature has been shown to predict benefit from hypoxia-modifying therapy in BC patients [4]. HIF1α promotes BC lung metastasis in an animal model by increasing zinc finger E-box-binding homeobox 1 expression and epithelial-to-mesenchymal transition [5]. In addition, hypoxic BC cells facilitate tumor growth and development though the secretion of oncogenic non-coding RNAs [6]. Increasing evidence indicates that aberrantly expressed non-coding RNAs, including circular RNAs (circRNAs), are responsible for BC development [7]. However, the potential pathways or mechanisms underlying BC is not well known.
CircRNAs constitute a spectrum of conserved endogenous RNAs that are formed by exon skipping or back-splicing events. It has been reported that circ-ITCH, circ-HIPK3, circ-MTO1 and circ-MYLK contribute to human bladder cancer [8-11], highlighting the importance of circRNAs in BC development. Given the promising role of hypoxia and circRNAs in BC, we are interested in whether they are associated with hypoxia-mediated progression of BC.
In this study, our aim was to investigate the role of circRNA on the hypoxic adaptive response in BC. We identified a hypoxia-inducible circRNA, has-circRNA-403658 that contributes to BC progression. Has-circRNA-403658 is spliced from its host gene, ZNF292, through back-splicing between the 1sh and 4th exon, and is consequently also named cZNF292. has-circRNA-403658 is induced by hypoxia in cultured endothelial cells and silencing it reduces tube formation and spheroid sprouting of endothelial cells in vitro [12]. Considering the promising role has-circRNA-403658 plays hypoxia-induced angiogenesis, we investigated its role in the adaptive response to hypoxia BC.
Materials and methods
Cell culture
The cell lines (normal bladder epithelial cells CCC-HB-2 and bladder cancer cell lines: (SW780, 5637, T24, J82 and RT4) were purchased from ATCC. These cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Cromwell, CT, USA) under 37°C condition with 5% CO2. For hypoxia culture, the cells were cultured in RPMI 1640 medium supplemented with 10% FBS under 37°C condition with 5% O2 (Biospherix Oxycycler C42, Parish, NY, USA) for 48 h.
CircRNAs array
The normal cultured and hypoxia cultured cells were collected and used for RNA extracted with TRIzol. Total RNAs were digested with Rnase R to remove linear RNAs and enrich circular RNAs. The enriched circular RNAs were used for circRNA microarray (Arraystar Human circRNA Array V2 analysis (Arraystar). Significant differentially expressed lncRNAs were then identified as fold change >2.0 and P<0.05.
Quantitative PCR analysis
After indicated treatment, cells were collected and used for RNA extraction. The expression of circRNAs and mRNA expression of glycometabolism relevant genes was measured by SuperScriptTM IV One-Step RT-PCR System (cat no. 12594100, ThermoFisher Scientific. Shanghai, China) according to manufacturers’ instructions. Expression of β-actin was used as an endogenous control. QPCR was performed at the condition: 60.0°C for 10 min, 98°C for 2 min, 40 circles of 98.0°C for 10 s, 55°C for 10 s and 72°C for 30 s, final extension 72°C for 5 min. The primers were shown in Table 1.
Table 1.
The primers used in qPCR
| Genes | Forward 5’-3’ | Reverse 5’-3’ |
|---|---|---|
| hsa_circ_000542 | ATGATGAAGATGGCCCAGCT | TGGAAGCTGGTCAAGTGTCT |
| hsa_circ_403658 | GATGGAGAATGGCAGCTGTG | AGCCACTGTGTATACCTCCA |
| hsa_circ_405348 | GAAGAGGAGGAGTGGCTACA | TGTTGGGAATACTCGAGGGG |
| hsa_circ_104082 | TGGCTGTTCTCCATTGTCTG | CAGATCCTTCAGCTGGTAAAGT |
| hsa_circ_001747 | CGCAGGGGAAACTTGAACAT | GCTGGCAAGGTACAACTCAG |
| hsa_circ_039783 | TGAAGGCTCCCATGATTCTGA | GTTTGTCGCTGTTCTCCCTG |
| hsa_circ_103868 | ACGGCTGGACTCTTTTAAGGA | TGGACCCAGGAATTCACAAAA |
| hsa_circ_030448 | ACAGGAGGCAGAGAGAGAGA | GCTGGGCTTCTTTCAATCCA |
| hsa_circ_050054 | TCCAAGATCAAGTTCCCGCT | AGCCAGATCCCGAAGTTCTT |
| hsa_circ_104081 | TGTGCCTTCTTTTCCGTCAC | TACATGCACGCCCTCAAAAG |
| GLUT1 | GAACTCTTCAGCCAGGGTCC | TCACACTTGGGAATCAGCCC |
| GLUT4 | TATCGGCATTCTGATCGCCC | CAACACCGAGACCAAGGTGA |
| HK-II | GTGAATCGGAGAGGTCCCAC | CAAGCAGATGCGAGGCAATC |
| G6P1 | GCAGGTGTATACTACGTGATGGT | AGTCAAAGACGTGCAGGAGG |
| PFK-M | GGCGGAGGAGAGCTAAGACT | TCAGAACGGAAGGTGTCAGC |
| GAPDH | GAAAGCCTGCCGGTGACTAA | TTCCCGTTCTCAGCCTTGAC |
| PGK-1 | GCTGGACAAGCTGGACGTTA | TCTGGGCCTACACAGTCCTT |
| PKM-1 | AGAAAGGTGCCGACTTCCTG | AGGCTCGCACAAGTTCTTCA |
| PKM-2 | TCTCTTCGTCTTTGCAGCGT | AGATCTTGCTGCCCACTTCC |
| LDHA | AGGCTATTCTTGGGCAACCC | TGAGTAGACATCCACCAAGGTT |
| LDHB | CCTGGTAGGTTTCGGCTCAG | CAAGGACAAGTAGGGCCTGG |
Bladder cancer samples
This study was approved by the Ethic Committee of The First Affiliated Hospital of Zhengzhou University. Written informed consents were obtained from all patients. Bladder cancer tissues (N=123) and the matched adjacent tissues were collected from The First Affiliated Hospital of Zhengzhou University. The medical records of patients were collected, including age, sex, clinical TNM staging and survival information.
Lentivirus infection
Has-circRNA-403658 siRNA sequences (si1#: CAGAACACACACUAUAGAG [dT][dT]; si2#: AUCUAGUAGCUCCCUUAA A[dT][dT]), HIF1α and LDHA expressed plasmids were contracted and packaged in lentivirus. The cells were infected with lentivirus (MOI=50) for 48 h and used for further analysis.
BrdU assay
BrdU Cell Proliferation ELISA kit (Abcam, Shanghai, China) was used in this experiment. After indicated treatment, the cells were added BrdU and incubate for 12 hours to incorporate BrdU into their DNA. The cells then were fixed with Fixing Solution and incubate at room temperature. Add the TMB Solution to each well. After color develops and then add the Stop Solution. Immediately begin recording the color development.
Apoptosis analysis
MEBCYTO Apoptosis Kit (MBL BEIJING BIOTECH CO., LTD, Beijing, China) was used in this experiment. After treatment, cells were collected and then resuspended in binding buffer. The cells were incubated with mixed well Annexin V-FITC and Propidium Iodide for 15 min. Finally, Flow cytometric analysis were performed in Attune NxT (Thermofisher, Shanghai, China).
Invasion assays
Cells were cultured in serum-free medium for 12 h and seeded in cell culture inserts. The bottom chamber was filled with 1640 medium containing 10% FBS. After 24 h culture, the cells on the lower surface were fixed with 70% ethyl alcohol, stained with crystal violet. The invasive cell number was counted under microscope.
Immunohistochemistry
The expression of Ki67, c-PARP and LDHA in tumor tissues was analyzed by immunohistochemical analysis. The tumor tissues were fixed and embedded in paraffin and cut into 4-μm-thick sections. The sections were treated with 10 mmol/l sodium citrate buffer at 100°C for 20 min. After blocked with goat serum, the sections were incubated with anti-Ki67 antibody (1:200 dilution) overnight at 4°C. The sections were incubated with secondary antibody for 2 h. Finally, the sections were stained with DAB and counterstained with hematoxylin. The images were obtained by using microscope (Nikon).
Western blot
Protein was extracted using RIPA (Beyotime, Hangzhou, China) and their concentrations were determined by Enhanced BCA Protein Assay Kit (Beyotime, Hangzhou, China). Proteins were separated in SDS/PAG and transferred on PVDF membrane. The membranes were incubated with primary antibody: anti-HIF1α antibody (diluted at 1:1000), anti-LDHA antibody (diluted at 1:1000), anti-VEGFR antibody (diluted at 1:1000), anti-VEGF antibody (diluted at 1:1000). GAPDH was used as control. All the antibodies were purchased from Cell Signaling Technology.
Tumor xenograft mice
Animal experiments were approved by the Ethics Committee for Animal Research of The First Affiliated Hospital of Zhengzhou University. T24 cells were infected with negative control lentivirus or has-circRNA-403658 si2# lentivirus. Nude mice were randomly divided into two groups (N=5/each group) and subcutaneously injected with transfected T24 cells. Tumor sizes (0.5 × L × W2, L and W are long and short diameters of the tumor mass, respectively) were recorded periodically.
Luciferase reporter assay
LDHA promoter was cloned into the pGL3-Basic vector. Luciferase reporter constructs were co-transfected with an internal control plasmid, pRL-TK (Renilla luciferase reporter plasmid, Promega), into T24 cells, followed by infection with Lv-has-circRNA-403658. The luciferase activity was determined with the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instruction.
RNA pulldown assay
RIP assay was conducted using Magna RIP Kit (EMD Millipore, Billerica, MA). Cells were lysed in RIP lysis buffer, and the cell lysate was treated with magnetic beads conjugated to human anti-Ago2 antibody (Millipore) or isotype-matched control antibody (normal mouse IgG; Millipore). qRT-PCR was performed to detect LDHA and has-circRNA-403658 expression.
Measurement of lactate levels, LDH activity, glucose uptake and ATP content
Lactate ELISA kit was purchased from Cwbiotech (Beijing, China). The cell cultured medium was used for lactate levels measurement by using ELISA kit according to manufacturer’s protocol. The cells were lysed in isolation buffer provided in the LDH activity assay kit (Beyotime, Hangzhou, China), and the LDH activity was determined according to the manufacturer’s instructions. The ATP content was measured by ATP assay kit (Beyotime) following the manufacturer’s protocol. Glucose uptake was measured by glucose uptake colorimetric assay kit (Biovision) following the manufacturer’s protocol.
Statistical analysis
All data from 3 independent experiments were expressed as mean ± SD and processed using SPSS17.0 statistical software. The expression of has_circRNA_403658 greater or equal to the mean was defined as high expression, otherwise were defined as low expression. The overall survival rate estimates over time were calculated using the Kaplan-Meier method with log-rank test. The clinical association between has-circRNA-403658 expression and clinicopathological variables in bladder cancer patients was evaluated by chi-square test. The correlation between LDHA and circRNA_403658 was analyzed by Pearson correlation analysis. The difference between two groups was estimated by Student’s t-test, and three or more groups is estimated by one-way ANOVA with hoc post Turkey test. P<0.05 was statistically significant.
Results
Has_circRNA_403658 is upregulated in patients with bladder cancer
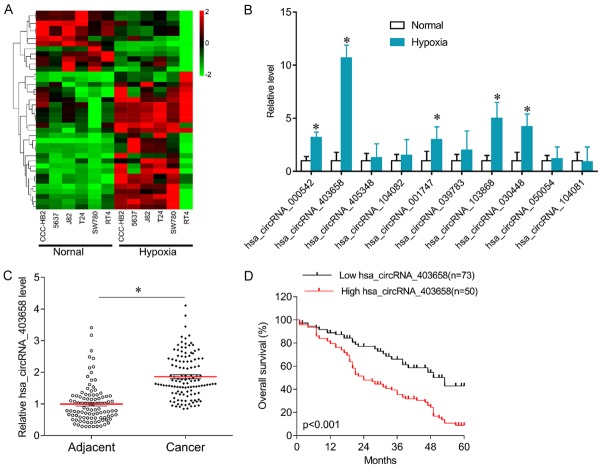
To investigate the role of circRNA in hypoxia-mediated bladder cancer cell growth, circRNA microarray was performed in normal cultured and hypoxia cultured bladder cancer cells. Many differentially expressed circRNAs were screened (Figure 1A). Further qPCR results confirmed that has_circRNA_000542, has_circRNA_403658, has_circRNA_001747, has_circRNA_103868, and has_circRNA_030448 were significantly increased in hypoxia-cultured 5637 cells compared with normal-cultured cells (Figure 1B). As has_circRNA_403658 was the circRNA most increased, we selected it for further investigation. Interestingly, has_circRNA_403658 expression was higher in bladder cancer tissues than in the matched adjacent tissues (Figure 1C). Further statistical analyses showed that patients with bladder cancer who had high has_circRNA_403658 expression displayed poorer overall survival than patients in which expression was low (P<0.001, Figure 1D). In addition, higher has_circRNA_403658 expression was associated with tumor size (P=0.004), distant metastasis (P<0.001) and TNM stage (P<0.001) (Table 2). Univariate and multivariate analyses revealed that has_circRNA_403658 was an independent factor for predicating the prognosis of patients with bladder cancer (Tables 3 and 4). These results indicate that has_circRNA_403658 is implicated in BC growth.
Figure 1.
Expression of has_circRNA_403658 in bladder cancer. A. Heatmap of cluster analysis in bladder cancer cells (ccc-HB2, 5637, J82, T24, SW780 and RT4) under hypoxic and normal condition. B. qPCR was performed to confirm the differentially expressed circRNAs in 5637 cells under hypoxic and normal condition. C. Expression of has_circRNA_403658 in bladder cancer tissues and the matched adjacent tissues. D. Kaplan-Meier curves of overall survival in patients with bladder cancer with low or high has_circRNA_403658 expression. *P<0.05.
Table 2.
Clinical association between Low hsa_circRNA_403658 levels and clinicopathological variables of patients with bladder cancer
| Variable | hsa_circRNA_403658 | χ2 test p value | |
|---|---|---|---|
|
| |||
| Low expression (n=73) | High expression (n=50) | ||
| Age | 0.999 | ||
| <60 | 20 | 13 | |
| ≥60 | 53 | 37 | |
| Gender | 0.233 | ||
| Male | 47 | 38 | |
| Female | 26 | 12 | |
| Tumor size | 0.004 | ||
| <3 cm | 46 | 18 | |
| ≥3 cm | 27 | 32 | |
| Lymph node metastasis | 0.065 | ||
| N0-1 | 36 | 16 | |
| N2-4 | 37 | 34 | |
| Distant metastasis | <0.001 | ||
| No | 60 | 23 | |
| Yes | 13 | 27 | |
| TNM stage | <0.001 | ||
| I-II | 54 | 18 | |
| III-IV | 19 | 32 | |
Table 3.
Univariate analysis of prognostic factors of patients with bladder cancer
| Variable | Hazard ratio | p value |
|---|---|---|
| Age (≥60/<60) | 1.68 | 0.055 |
| Gender (Male/Female) | 1.52 | 0.086 |
| Tumor size (≥3 cm/<3 cm) | 3.45 | 0.036 |
| Lymph node metastasis (N2-4/N0-1) | 4.16 | 0.025 |
| Distant metastasis (Yes/No) | 5.42 | 0.023 |
| TNM stage (III-IV/I-II) | 4.07 | 0.034 |
| hsa_circRNA_403658 (High/Low) | 3.21 | 0.014 |
Table 4.
Multivariate analysis of independent prognostic factors of patients with bladder cancer
| Variable | Hazard ratio | p value |
|---|---|---|
| Tumor size | 2.26 | 0.016 |
| Lymph node metastasis | 3.59 | 0.023 |
| Distant metastasis | 4.08 | 0.017 |
| TNM stage | 3.85 | 0.028 |
| hsa_circRNA_403658 | 4.04 | 0.022 |
Silencing has_circRNA_403658 inhibits bladder cancer cell growth
Has_circRNA_403658 expression was upregulated in bladder cancer cells, especially in 5637 and T24 cells (Figure 2A). 5637 and T24 cells were used for further investigation. Has_circRNA_403658 was knocked down by siRNA in 5637 and T24 cells (Figure 2B). Has_circRNA_403658 knockdown significantly inhibited 5637 and T24 cell proliferation (Figure 2C, 2D), induced cell apoptosis (Figure 2E, 2F) and suppressed cell invasion capacity (Figure 2G, 2H). Furthermore, in a xenograft tumor model, the tumor volume was smaller in mice injected with 5637 cells with has_circRNA_403658 knockdown than in control mice (Figure 2I). Immunohistostaining results showed that Ki67 expression was reduced, while c-PARP expression was upregulated in mice injected with 5637 cells with has_circRNA_403658 knockdown versus in control mice (Figure 2J). These results suggest that silencing has_circRNA_403658 inhibits bladder cancer cell growth in vivo and in vitro.
Figure 2.
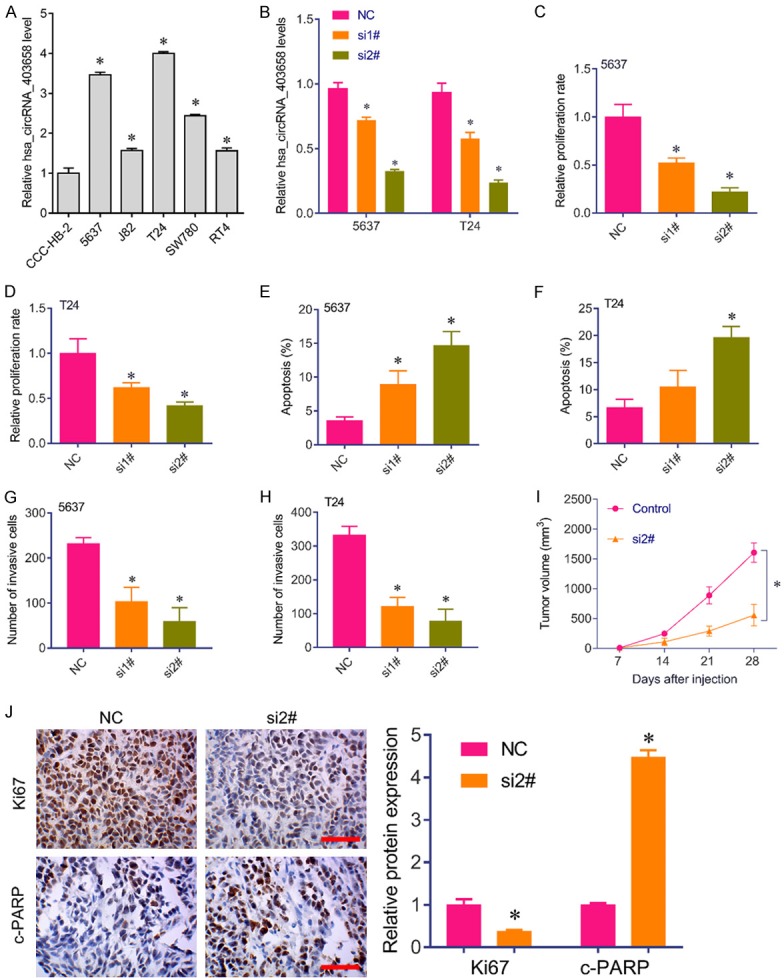
Silencing has_circRNA_403658 inhibited bladder cell growth and invasion. (A) qRT-PCR analysis for has_circRNA_403658 in bladder cancer cell lines, including ccc-HB2, 5637, J82, T24, SW780 and RT4. (B) qRT-PCR analysis for has_circRNA_403658 in 5637 and T24 cells after has_circRNA_403658 siRNA 1# and siRNA 2# transfection. (C, D) Brdu assay was performed to measure the proliferation rate of 5637 (C) and T24 (D) cells after has_circRNA_403658 siRNA transfection. (E, F) Apoptosis analysis for 5637 (E) and T24 (F) cells after has_circRNA_403658 siRNA transfection. (G, H) Transwell assay for 5637 (G) and T24 (H) cells after has_circRNA_403658 siRNA transfection. (I) T24 cells transfected with has_circRNA_403658 siRNA were injected into the back of BALB/c nude mice. The tumor volumes were measured weekly. (J) Immunohistochemistry analysis for Ki67 and c-PARP in xenografted tumor tissues (left), and quantification by imageJ (right). scan bar, 200 μm. *P<0.05 vs control.
Effects of altered has_circRNA_403658 expression on aerobic glycolysis
Considering has_circRNA_403658 is upregulated under hypoxia, we further investigated the effect of altered has_circRNA_403658 expression on aerobic glycolysis in bladder cancer cells. We found that has_circRNA_403658 significantly inhibited LDHA expression, but not other glycometabolism relevant genes in 5637 and T24 cells (Figure 3). As a result, silencing has_circRNA_403658 decreased lactate production (Figure 4A), LDH activity (Figure 4B), ATP production (Figure 4C) and glucose uptake (Figure 4D) in bladder cancer cells. We examined whether has_circRNA_403658 is correlated with the sensitivity of cells to glycolytic inhibitors. As expected, silencing has_circRNA_403658 enhanced the inhibitory effects of glycolytic inhibitors, oxamate and 2-DG on cell proliferation (Figure 4E, 4F). In addition, we found that overexpression of HIF-1α (Figure 5A) significantly induced has_circRNA_403658 expression (Figure 5B), and silencing has_circRNA_403658 reduced HIF1α-mediated bladder cancer cell proliferation and apoptosis (Figure 5C-F). Therefore, the above results demonstrate that silencing has_circRNA_403658 could reduce aerobic glycolysis and increase the sensitivity of bladder cancer cells to glycolytic inhibitors.
Figure 3.
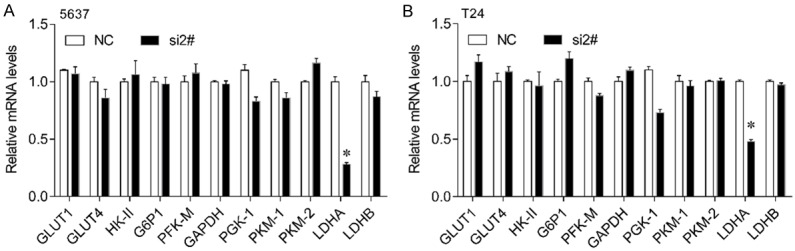
Effects of circRNA_403658 knockdown on glycometabolism relevant gene expression. A, B. 5637 and T24 cells were transfected with circRNA_403658 siRNA 2# for 48 h, and then qPCR was performed to determine the expression of glycometabolism relevant genes. circRNA_403658 knockdown significantly decreased the expression of LDHA. *P<0.05.
Figure 4.
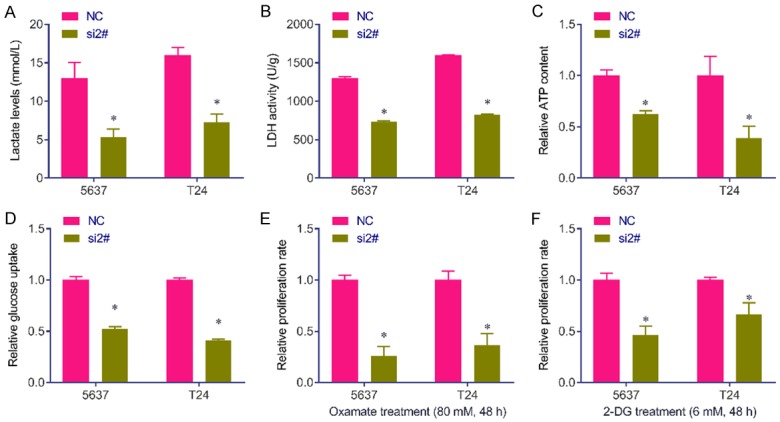
Silencing has_circRNA_403658 inhibits lactate production and LDH activity in bladder cancer cells. A. Lactate concentrations were determined in 5637 and T24 cells after 48 h transfection with has_circRNA_403658 siRNA 2#. B. LDH activity was determined in 5637 and T24 cells after 48 h transfection with has_circRNA_403658 siRNA 2#. C. ATP concentration was determined in 5637 and T24 cells after 48 h transfection with has_circRNA_403658 siRNA 2#. D. Glucose uptake was determined by Glucose Uptake Colorimetric Assay kit in 5637 and T24 cells after 48 h transfection with has_circRNA_403658 siRNA 2#. E. 5637 and T24 cells were transfected with has_circRNA_403658 siRNA and then treated with oxamate sodium (Oxamate, 80 mM) for 48 h. The cell viability was measured using a Brdu assay. F. 5637 and T24 cells were transfected with has_circRNA_403658 siRNA and then were treated with 6 mM 2-DG for 48 h. Cell viability was measured using a Brdu assay. *P<0.05.
Figure 5.
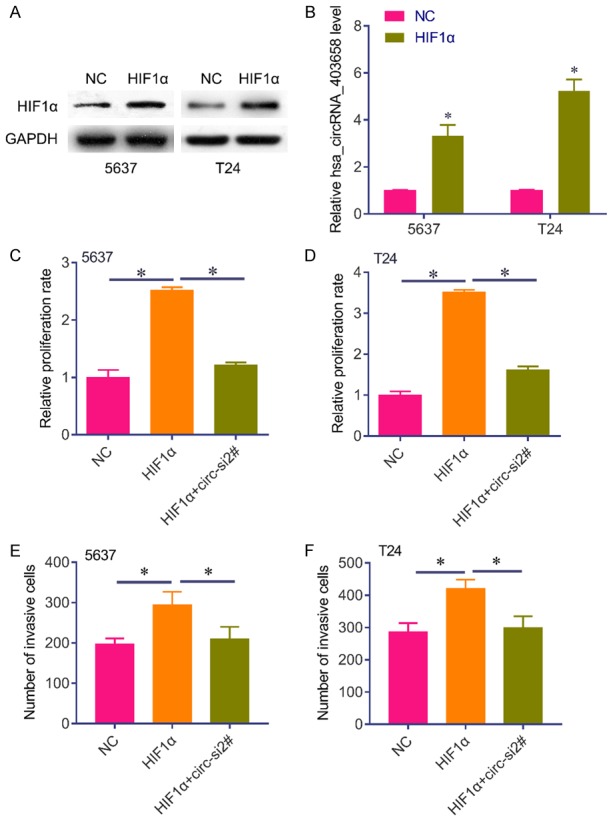
HIF1α induces has_circRNA_403658 expression. (A) Western blot analysis of the expression of has_circRNA_403658 in 5637 and T24 cells after HIF1α expressed plasmid transfection. (B) The expression of has_circRNA_403658 was determined in 5637 and T24 cells after HIF1α expressed plasmid transfection. (C, D) Brdu assay was performed to measure the proliferation rate of 5637 (C) and T24 (D) cells after HIF1α expressed plasmid or plus has_circRNA_403658 siRNA transfection. (E, F) Transwell assay for 5637 (E) and T24 (F) cells after HIF1α expressed plasmids or plus has_circRNA_403658 siRNA transfection. *P<0.05.
Has_circRNA_403658 functions as a positive regulator of LDHA
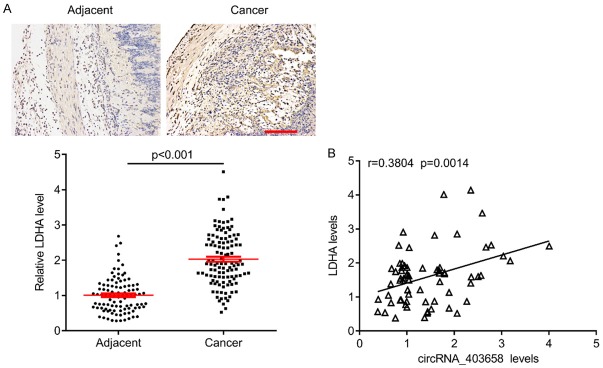
To investigate whether has_circRNA_403658 could impact LDHA, a key checkpoint of glycolysis, we analyzed the protein expression of LDHA after silencing has_circRNA_403658. Results showed that the protein levels of LDHA, VEGFR and VEGF in has_circRNA_403658-knocked down bladder cancer cells were significantly lower than those in control cells (Figure 6A). Thus, has_circRNA_403658 positively regulates LDHA expression. RNA IP assay revealed that has_circRNA_403658 interacted with LDHA (Figure 6B), and luciferase reporter gene assay showed that upregulation of has_circRNA_403658 greatly increased the luciferase activity of the LDHA promoter (Figure 6C). These results indicate that has_circRNA_403658 positively regulates LDHA expression through promoting the activity of the LDHA promoter. Intriguingly, restoring the expression of LDHA in has_circRNA_403658-knocked down bladder cancer cells apparently reversed the inhibitory roles of has_circRNA_403658 downregulation in regulating cell proliferation, and apoptosis compared with the has_circRNA_403658-knocked down 5637 cells (Figure 6D and 6E). In addition, LDHA overexpression reversed has_circRNA_403658 siRNA-inhibited aerobic glycolysis, which was evaluated by increased lactate, LDH activity, ATP content and glucose uptake in 5637 and T24 cells (Figure 7). Furthermore, we measured LDHA expression in human bladder cancer tissues, and found that LDHA was upregulated in bladder cancer tissues compared with the adjacent tissues (Figure 8A). We also observed that LDHA expression was positively correlated with circRNA_403658 in bladder cancer (Figure 8B). These data demonstrate that has_circRNA_403658 functions as an oncogene through an LDHA-dependent mechanism, and might be associated with aerobic glycolysis.
Figure 6.
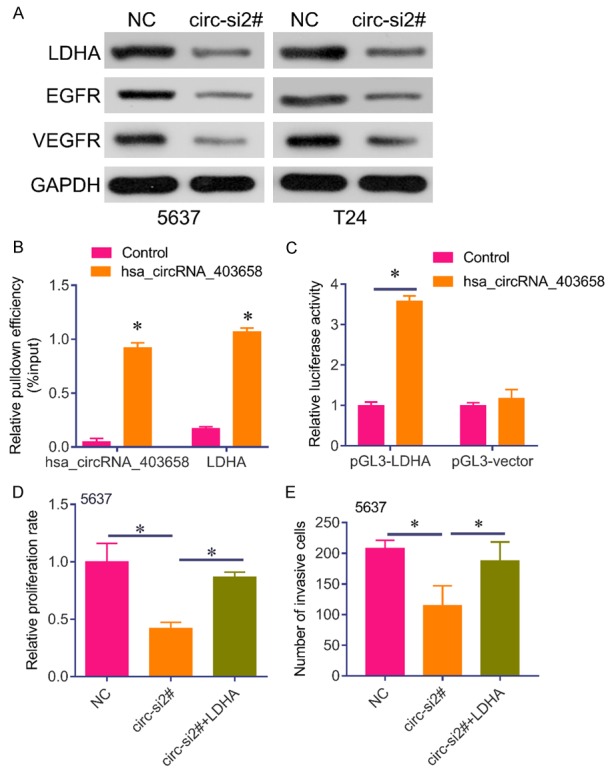
LDHA blocked has_circRNA_403658 siRNA-mediated tumor-suppressive effects in bladder cancer cells. A. Western blot analysis of LDHA, EGFR, VEGFR in 5637 and T24 cells after has_circRNA_403658 siRNA transfection. B. RNA pull-down assay for the amount of has_circRNA_403658 and LDHA with either has_circRNA_403658 or control probe in 5637 cells. C. Luciferase reporter assay for the luciferase activity of pGL3-LDHA in 5637 cells co-transfected with has_circRNA_403658. D. Brdu assay was performed to measure the proliferation rate of 5637 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. E. Transwell assay for 5637 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. *P<0.05.
Figure 7.
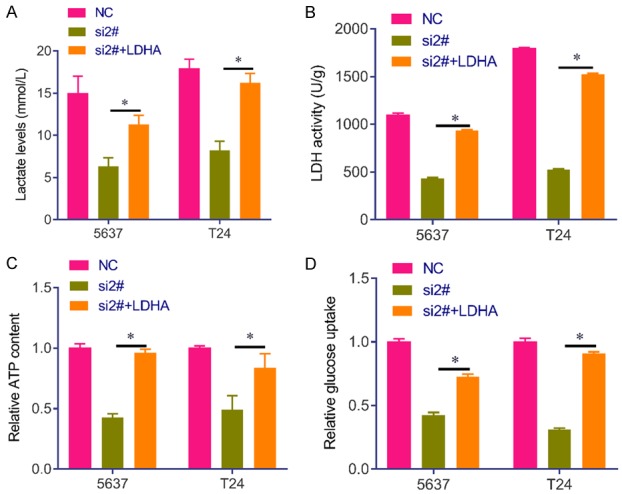
LDHA reverses has_circRNA_403658 siRNA-inhibited aerobic glycolysis in bladder cancer cells. A. Lactate concentrations were determined in 5637 and T24 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. B. LDH activity was determined in 5637 and T24 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. C. ATP concentration was determined in 5637 and T24 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. D. Glucose uptake was determined by Glucose Uptake Colorimetric Assay kit in 5637 and T24 cells after LDHA expressed plasmid or plus has_circRNA_403658 siRNA transfection. *P<0.05.
Figure 8.
Correlation between LDHA and circRNA_403658 in bladder cancer tissues. A. Representative IHC images for LDHA in bladder tissues (upper), and quantification by ImageJ (lower). Bar=200 μm. B. Pearson correlation analysis for LDHA and circRNA_403658 in bladder cancer tissues.
Discussion
In this study, we first demonstrated that has-circRNA-403658, an important circRNA is upregulated in bladder cancer cells under hypoxia, and that higher has-circRNA-403658 levels are associated with poorer survival outcome. In addition, we also revealed that has-circRNA-403658 is induced by HIF1α, and that silencing has-circRNA-403658 inhibits LDHA-mediated aerobic glycolysis, inhibiting bladder cancer cell growth. The Warburg effect plays an important role in bladder cancer development. Thus, our results suggest that has-circRNA-403658 may function as a novel therapeutic target in human bladder cancer.
CircRNAs were initially considered as junk products. However, there is increasing evidence to suggest an important regulatory role for circRNAs in cancer biology [11]. Dysregulated circRNAs have been reported in bladder cancer [13]. For example, hsa_circ_0000285 may be an independent prognostic factor for bladder cancer patients [14]. circ0001429 negatively regulates miR-205-5p to increase VEGFA and promote the development of bladder cancer [15]. CircFNDC3B serves as a tumor suppressor by sponging miR-1178 in human bladder cancer [16]. CircPRMT5 acts as an oncogene by promoting urothelial carcinoma of EMT in bladder cells [17].
Herein, our results demonstrate that has-circRNA-403658 is an oncogene in bladder cancer under hypoxia. Importantly, further functional studies showed that silencing has-circRNA-403658 in bladder cancer cells inhibited cell growth and induced cell apoptosis. The oncogene role for has-circRNA-403658 under hypoxia was strongly supported by our clinical in vitro and in vivo studies.
The common mechanism by which circRNAs function is via sponging miRNAs [18]. For example, circBCRC4 sponges miR-101 [19], circCEP128 sponges miR-145-5p and circ-VANGL1 sponges miR-605 in bladder cancer [20,21]. However, has-circRNA-403658 has no validated microRNA-binding sites (as demonstrated through high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation data sets), suggesting that it does not act as a microRNA sponge [12]. It has been reported that cZNF292 initially exhibits proangiogenic activities in cultured endothelial cells under hypoxia [12]. Further investigation revealed that cZNF292 silencing suppresses tube formation by inhibiting glioma cell proliferation and cell-cycle progression via the Wnt/β-catenin signaling pathway and related genes such as VEGFR and EGFR [22]. In this study, we revealed that in has-circRNA-403658 silencing-inhibited bladder cancer cells, the Warburg effect could be reversed by enforced overexpression of LDHA, a key enzyme that catalyzes the conversion of pyruvate to lactate. has-circRNA-403658 silencing also reduced the expression of VEGFR and EGFR, two important genes for tumor angiogenesis [23]. These results suggest a critical regulatory role in hypoxia and angiogenesis by the has-circRNA-403658-LDHA regulatory axis. We have confirmed the interaction between has-circRNA-403658 and LDHA. However, the regulatory mechanism by which has-circRNA-403658 induces LDHA needs further investigation. Collectively, these data suggested a critical role of the has-circRNA-403658-LDHA pathway in the control of bladder cancer cell growth and invasion.
In summary, our findings provide comprehensive evidence that raised has-circRNA-403658 is a biomarker for poor prognostic in bladder cancer. has-circRNA-403658 exerts a regulatory role in promoting LDHA and the Warburg effect to facilitate bladder cancer aggressive nature. Our findings may also have highlighted a novel therapeutic target, has-circRNA-403658, for human bladder cancer.
Disclosure of conflict of interest
None.
References
- 1.Massari F, Ciccarese C, Santoni M, Iacovelli R, Mazzucchelli R, Piva F, Scarpelli M, Berardi R, Tortora G, Lopez-Beltran A, Cheng L, Montironi R. Metabolic phenotype of bladder cancer. Cancer Treat Rev. 2016;45:46–57. doi: 10.1016/j.ctrv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 2.McConkey DJ, Choi W. Molecular subtypes of bladder cancer. Curr Oncol Rep. 2018;20:77. doi: 10.1007/s11912-018-0727-5. [DOI] [PubMed] [Google Scholar]
- 3.Tsui KH, Hou CP, Chang KS, Lin YH, Feng TH, Chen CC, Shin YS, Juang HH. Metallothionein 3 is a hypoxia-upregulated oncogene enhancing cell invasion and tumorigenesis in human bladder carcinoma cells. Int J Mol Sci. 2019;20:E980. doi: 10.3390/ijms20040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Roberts D, Takhar M, Erho N, Bibby B, Thiruthaneeswaran N, Bhandari V, Cheng WC, Haider S, McCorry A, McArt D, Jain S, Alshalalfa M, Ross A, Schaffer E, Den RB, Jeffrey KR, Klein E, Hoskin PJ, Freedland SJ, Lamb AD, Neal DE, Buffa FM, Bristow RG, Boutros PC, Davicioni E, Choudhury A, West CML. Development and validation of a 28-gene hypoxia-related prognostic signature for localized prostate cancer. EBioMedicine. 2018;31:182–189. doi: 10.1016/j.ebiom.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Huang Z, Zhang M, Wang W, Liang H, Zeng J, Wu K, Wang X, Hsieh JT, Guo P, Fan J. HIF-1alpha promotes ZEB1 expression and EMT in a human bladder cancer lung metastasis animal model. Oncol Lett. 2018;15:3482–3489. doi: 10.3892/ol.2018.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, Li X. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Liu Y, Zhang X, Liu J, Wang P. Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene. 2018;677:189–197. doi: 10.1016/j.gene.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991–996. doi: 10.1016/j.bbrc.2018.12.046. [DOI] [PubMed] [Google Scholar]
- 12.Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang X, Chen W, Han Y, Shao L, Gao Y, He C. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18:35. doi: 10.1186/s12943-019-0996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, Gui SL, Zhou SB, Qin WB, Wu DM, Wang SQ. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66:197–202. doi: 10.4149/neo_2018_180318N185. [DOI] [PubMed] [Google Scholar]
- 15.Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. doi: 10.3233/CBM-182380. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, Lin T, Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, Xiao KH, Liu ZW, Luo JH, Zhou FJ, Xie D. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 18.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ, Xiao XY. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2017;37:886–890. doi: 10.1007/s11596-017-1822-9. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Huang W, Wang X, Wang T, Chen Y, Chen B, Liu R, Bai P, Xing J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24:40. doi: 10.1186/s10020-018-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 22.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvorak HF. Tumor stroma, tumor blood vessels, and antiangiogenesis therapy. Cancer J. 2015;21:237–243. doi: 10.1097/PPO.0000000000000124. [DOI] [PubMed] [Google Scholar]