Abstract
P4HA2 is one of collagen prolyl 4-hydroxylase (P4H) isoform and increased in several types of human cancer. However, the role of P4HA2 during cervical tumorigenesis remains largely unknown. Here, we report that the protein level of P4HA2 is significantly increased in cervical cancer tissues. Silencing of P4HA2 inhibits cervical cancer cell proliferation, colony formation and migration. We also demonstrate decreased glucose uptake and lactate production in P4HA2 knockdown cells. Mechanistically, P4HA2 promotes cervical cancer cell glycolysis through upregulation of PGK1 and LDHA. We find a positive correlation between P4HA2 and PGK1/LDHA expression in cervical cancer tissues. Importantly, high expression of P4HA2, PGK1 or LDHA has a significantly shorter overall survival period and the survival prediction is enhanced by using combination of P4HA2 and PGK1/LDHA expression. Collectively, we identify P4HA2 as a regulator of glycolysis through PGK1 and LDHA, which may serve as a potential therapeutic target for cervical cancer.
Keywords: P4HA2, glycolysis, PGK1, LDHA, cervical cancer
Introduction
Cervical cancer is one of the most common gynecological malignancies with an estimated global incidence of 570,000 cases and nearly 311,000 deaths in 2018 [1]. Advanced and recurrent cervical cancer is generally considered incurable, and the current therapeutic options show limited effectiveness. Thus, there is an urgent need for identifying valuable predictive biomarkers and developing novel effective therapeutic strategies for cervical cancer.
Mounting evidence has demonstrated that reprogrammed energy metabolism, an emerging hallmark of cancer, facilitates cell growth and proliferation [2]. Cancer cells preferentially utilize glucose for energy production and have higher rates of glycolysis followed by lactic acid fermentation under conditions of normal oxygen tension, which is referred to as the Warburg effect [3]. Previous studies have shown that proliferation and therapeutic resistance of cervical cancer cells are partially dependent on their glycolytic phenotype and several enzymes of the glycolytic pathway have been identified as potential therapeutic targets [4-8]. Therefore, targeting the glycolytic pathway is a promising strategy to suppress cervical cancer progression.
Increased collagen deposition is associated with cervical cancer progression and poor patient survival [9,10]. Collagen prolyl 4-hydroxylase (P4H) catalyzes the hydroxylation of proline to promote the formation of collagen triple helix [11]. Prolyl 4-hydroxylase subunit α2 (P4HA2) is a type of P4H isoform, which is increased in several tumors, including hepatocellular carcinoma [12], breast cancer [13,14], lymphoma [15], oral carcinoma [16] and papillary thyroid cancer [17]. However, the significance of P4HA2 in tumorigenicity and glycolysis of cervical cancer remains largely unknown.
In this research, we aim to explore the role of P4HA2 on the biological behavior of cervical cancer. Our study reveals that the protein level of P4HA2 is significantly increased in cervical cancer tissues and P4HA2 is a predictor of poor patient outcome. Silencing P4HA2 inhibits malignant behavior and glycolysis of cervical cancer cells. Moreover, we have also identified a link between P4HA2 and two glycolytic enzymes (PGK1 and LDHA). Our finding provides a novel perspective on how P4HA2 promotes cervical cancer and indicates that P4HA2 could serve as a potential therapeutic target.
Materials and methods
Human tissue microarray and immunohistochemical analysis
Paired tumor and adjacent non-tumor paraffin tissue microarrays of cervical cancer were purchased from Shanghai Zuocheng Biotech (Shanghai, China). Immunohistochemical (IHC) analysis was performed as previously reported [18]. The protein levels of P4HA2, PGK1 and LDHA were assessed using a semiquantitative method, as previously described [18].
Cell lines
The cervical cancer cell lines Hela and SiHa were purchased from the American Type Culture Collection (ATCC). The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; high glucose, Invitrogen) with 10% fetal bovine serum (FBS, HyClone) and 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Reagents
P4HA2 rabbit polyclonal antibody (13759-1-AP), PGK1 rabbit polyclonal antibody (17811-1-AP), LDHA rabbit polyclonal antibody (21799-1-AP), GAPDH rabbit polyclonal antibody (10494-1-AP) and secondary antibodies were purchased from Proteintech (Wuhan, China). MTT Cell Proliferation and Cytotoxicity Assay Kit (C0009) were obtained from Beyotime Biotechnology (Shanghai, China). Transwell® Polycarbonate Membrane (#3422) was obtained from Corning (New York, USA).
Bioinformatics analysis
The Gene Expression Omnibus (GEO) database and the Oncomine database (https://www.oncomine.org/) were used to compare the mRNA expression of P4HA1, P4HA2 and P4HA3 in normal cervix tissues and cervical cancer tissues. The cBioPortal (http://www.cbioportal.org/) was used to investigate the correlation of P4HA2 expression with collagen-related gene and glycolysis-related gene expression in Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Provisional) database [19,20]. GSE7410 compared the mRNA expression of three P4H isoforms (P4HA1, P4HA2 and P4HA3) in 21 cervical cancer patients without lymph node metastasis and 19 patients with lymph node metastasis. GSE75034 (GPL6884) compared the mRNA expression of P4HA2 in 73 cervical cancer samples with low hypoxia score and 77 cervical cancer samples with high hypoxia score. GSE75034 (GPL10558) also compared the mRNA expression of P4HA2 in several cervical cancer cell lines exposed to normoxic (95% air) and hypoxic (0.2% O2) condition. Biological processes related to the activities of P4HA2 in cervical cancer were evaluated by GSEA v3.0 software [21]. Processing of the data was according to the guidelines of these databases.
Kaplan-meier plotter
Kaplan-Meier curves were created using the online Kaplan-Meier Plotter website (http://kmplot.com/analysis/) to analyze the prognostic values of P4HA2, PGK1 and LDHA expression in 304 patients with cervical cancer [22].
MTT assay
Exponentially growing cells were seeded in 96-well plates at 2,000 cells per well and incubated for 72 hours. 10 μl MTT solution (10 mg/ml MTT in PBS) was added to each well, and the cells were incubated for another 4 hours at 37°C. Subsequently, 200 μl dimethyl sulfoxide (DMSO) was added to dissolve the crystals. Then the absorbance values were measured at a wavelength of 490 nm using microplate spectrophotometry. Each experiment was carried out at least three repeats.
Colony-formation assay
The transfected cells were seeded in 12-well plates at an initial cell density of 1,000 cells per well. The medium was changed at 3-day intervals. After 10-14 days, the cells were washed with PBS, fixed with 4% paraformaldehyde for 30 minutes, stained with crystal violet for 30 minutes at room temperature, and counted under a microscope. Each experiment was carried out at least three repeats.
Cell migration assay
The transfected cells were inoculated on top of Transwell chambers at an initial cell density of 20,000 cells per chamber after incubated in serum-free medium for 24 hours. Medium was supplemented with serum-free DMEM medium in the upper chamber, and the lower chamber was filled with 10% FBS as a chemoattractant. After incubation for 24 hours, cells on the lower side were fixed with 4% paraformaldehyde for 20 minutes, stained with crystal violet for 20 minutes. Chambers were photographed after washing and drying. Each experiment was carried out at least three repeats.
Wound-healing assay
The transfected cells were plated in 6-well plates at an initial cell density of 1×105 cells per well. On the second day after plating, the cells had covered >90% of the bottom of the plates as a monolayer, and a pipette tip was used to scratch the monolayer across the center of the well. After scratching, the plates were replenished with fresh medium and photos taken at 0 hour were used as the control. Then the plates were placed in a 37°C, 5% CO2 incubator for additional 24 hours and photos were captured.
Plasmid, siRNA and transduction
Flag-P4HA2 plasmid was generated by PCR amplification of sequences obtained from a Hela cell cDNA library and cloned into a pcDNA3.1 vector. The following primers were used to generate the P4HA2 plasmids: forward (5’-CGGGATCCATGAAACTCTGGGTGTCTGC-3’) and reverse (5’-CCGCTCGAGTCAGTCAACTTCTGTTGATC-3’). The following oligonucleotide against genes were used: siRNA against P4HA2-1# (5’-CGAGATACTTTCAAGCATTTA-3’), siRNA against P4HA2-2# (5’-GCGGTACTTTGAGCAGTTATT-3’), siRNA against PGK1 (5’-CAAATGGAACACGGAGGATAA-3’) and siRNA against LDHA (5’-CCGAACTGCAAGTTGCTTATT-3’). Transfection was performed using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer’s recommendations.
Real-time quantitative PCR and western blotting
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. RT-qPCR was carried out by CFX Real-Time PCR Detection System (Bio-Rad) using SYBR qPCR Master Mix (TOYOBO). The following primer sequences were used: P4HA2, sense strand 5’-AGTACCAGGCAATGCTGAGT-3’, antisense strand 5’-CCTCTTCTGTCTACGGGGTG-3’; PGK1, sense strand 5’-GAACAAGGTTAAAGCCGAGCC-3’, antisense strand 5’-GTGGCAGATTGACTCCTACCA-3’; LDHA, sense strand 5’-ATTTGGTCCAGCGTAACGTG-3’ and antisense strand 5’-TGTCCCAAAATGCAAGGAACA-3’; β-actin, sense strand 5’-CATGTACGTTGCTATCCAGGC-3’ and antisense strand 5’-CTCCTTAATGTCACGCACGAT-3’. All reactions were performed in three repeats. Western blot analysis was performed as previously described [18].
Measurement of glucose uptake
The glucose analog 2-deoxyglucose (2-DG) uptake was estimated in a 96-well plate by Glucose Uptake Colorimetric Assay Kit (ab136955, Abcam, Cambridge, MA, USA). Briefly, 2,000 cells were cultured in serum free medium overnight. Then the cells were incubated with 100 µl Krebs-Ringer-Phosphate-HEPES (KRPH) buffer containing 2% BSA at 37°C for 40 minutes. 2-DG was added to the plate for 20 minutes to simulate glucose uptake in cells. Recycling amplification reaction method was used to measure the NADPH levels and glucose uptake was estimated based on 2-DG uptake. Samples were measured at wavelength 412 nm using a kinetic plate reader. Results were normalized by the protein content with a BCA protein assay kit in each assay. Each experiment was carried out at least three repeats.
Measurement of lactate production
Lactate production was measured using L-Lactate Assay kit (ab65331, Abcam, Cambridge, MA, USA). Briefly, 2×106 cells were resuspended in lactate assay buffer and the supernatant was collected. Then the reaction mix buffer was added according to the manufacturer’s protocol. Samples were measured at wavelength 450 nm using a microplate reader. Lactate production was calculated with formula provided by the manufacturer. Each experiment was carried out at least three repeats.
Statistical analysis
GraphPad Prism (Version 6.01, GraphPad Software, Inc., USA) was used for all statistical analyses. The data in current study were presented as mean ± SD. Student’s t tests were used to determine statistical significance between experimental groups. The significance of the relationship between P4HA2 and collagen-related or glycolysis-related gene expression was determined using the Pearson correlation coefficient. Significant differences in overall survival (OS) time were assessed with the Cox proportional hazard (log-rank) test. P-value <0.05 was considered significant.
Results
Increased expression of P4HA2 in cervical cancer
In order to address the role of P4HA2 in cervical cancer, we compared the expression of P4HA2 in normal cervix and cervical cancer from the Oncomine database. It revealed that the mRNA expression of P4HA2 in cervical cancer was higher than that in normal cervical tissues (Figure 1A). Immunohistochemistry analysis using the tissue microarray (including 62 paired cervical cancer and adjacent normal cervical tissues) showed that the protein level of P4HA2 was significantly upregulated in cervical cancer compared to adjacent normal tissues (Figure 1B and 1C). According to GSE7410 database, P4HA2 expression was also significantly upregulated in cervical cancer with lymph node metastasis, compared to cervical cancer without lymph node metastasis (Figure 1D). Although the expression of the other P4H isoform P4HA1 was increased in cervical cancer (Figure S1A), there was no difference in cervical cancer with and without lymph node metastasis (Figure S1B). Hypoxia has been reported to induce the expression of P4HA2 [23]. In this study, we confirmed that P4HA2 mRNA expression was elevated in hypoxic conditions of cervical cancer cell lines (GSE75034/GPL10558, Figure S1C) and upregulated in cervical cancer patients with high degree of hypoxia score (GSE75034/GPL6884, Figure S1D). Moreover, Kaplan-Meier log rank analysis showed significant correlation between high expression of P4HA2 and poor OS in cervical cancer patients (Figure 1E).
Figure 1.
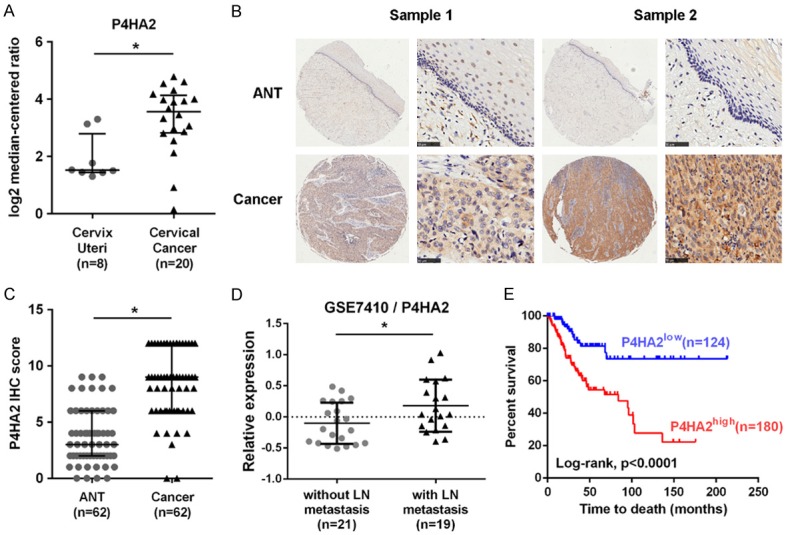
Increased expression of P4HA2 in cervical cancer. A. Analysis of P4HA2 expression in normal cervix and cervical cancer from GSE6791 dataset available at Oncomine (https://www.oncomine.org/), *P<0.05. B. Immunohistochemistry analysis of P4HA2 protein in cervical cancer and adjacent normal tissues. Scale bar =50 μm. C. IHC score of PAHA2 in cervical cancer and adjacent normal tissues, *P<0.05. D. P4HA2 mRNA expression in cervical cancer with and without lymph node metastasis from GSE7410 dataset, *P<0.05. E. Kaplan-Meier log rank analysis in cervical cancer patients with low or high P4HA2 levels from TCGA database, *P<0.01. ANT, adjacent normal tissue; LN, lymph node.
The effect of P4HA2 on cervical cancer cell malignant behavior
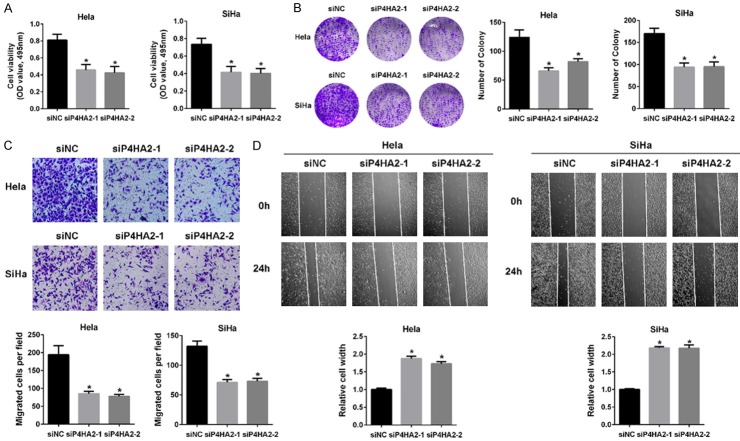
To examine the function of P4HA2 in cervical cancer, we silenced P4HA2 expression in a panel of cervical cancer cell lines (Hela and SiHa) with two different siRNAs (siP4HA2-1# and siP4HA2-2#). Cell proliferation was examined by MTT and colony formation assay. After silencing P4HA2, the growth of Hela and SiHa cell was significantly decreased (Figure 2A). Similarly, significant reduction in the number of colonies of Hela and SiHa cell transfected with P4HA2 siRNA was also observed (Figure 2B). Transwell and wound healing assays further demonstrated that knockdown of P4HA2 significantly reduced cervical cancer cell migration (Figure 2C and 2D). Moreover, the expressions of collagen deposition related genes (Col1A1, Col4A1, Col4A5, Col5A1, Col5A2 and Col7A1) were significantly correlated with P4HA2 in cervical cancer cohort from TCGA (Figure S2). These findings indicate that the development and progression of cervical cancer is accompanied by P4HA2.
Figure 2.
The effect of P4HA2 on cervical cancer cell malignant behavior. A. Cell viability of P4HA2-silenced Hela and SiHa cells with two different siRNAs, *P<0.05 versus siNC group. B. Colony formation was examined in Hela and SiHa cells from siNC, siP4HA2-1# and siP4HA2-2# groups, *P<0.05 versus siNC group. C. Transwell migration assay in Hela and SiHa cells from siNC, siP4HA2-1# and siP4HA2-2# groups. The number of cells per field was counted, *P<0.05 versus siNC group. D. Wound healing assay in Hela and SiHa cells from siNC, siP4HA2-1# and siP4HA2-2# groups, *P<0.05 versus siNC group.
Knockdown of P4HA2 inhibits glycolysis in cervical cancer cells
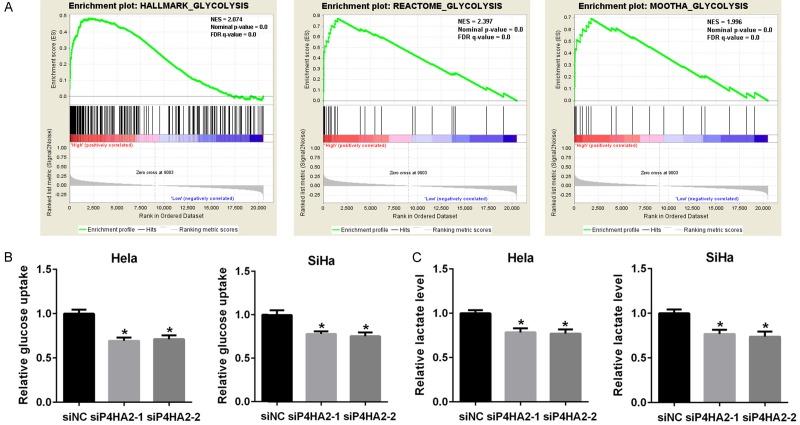
To characterize the function of P4HA2 in glycolysis, GSEA analysis was performed using TCGA data for 309 samples from cervical cancer. Gene sets of HALLMARK_GLYCOLYSIS (NES=2.074, P<0.01), REACTOME_GLYCOLYSIS (NES=2.397, P<0.01) and MOOTHA_GLYCOLYSIS (NES=1.996, P<0.01) were highly enriched in P4HA2-high expression group (Figure 3A). It reveals that the expression of P4HA2 is positively correlated with gene expression signatures involved in response to glycolysis. Then we detected decreased glucose uptake and lactate production in P4HA2 knockdown Hela and SiHa cells (Figure 3B and 3C). These results indicate that P4HA2 affects cervical cancer cell glycolysis.
Figure 3.
Knockdown of P4HA2 inhibits glycolysis in cervical cancer cells. A. Gene sets enrichment analysis showed that the gene set of HALLMARK_GLYCOLYSIS, REACTOME_GLYCOLYSIS and MOOTHA_GLYCOLYSIS were enriched in the P4HA2-high group from cervical cancer samples. NES, normalized enrichment score; FDR, false discovery rate. B. Glucose uptake in Hela and SiHa cells from siNC, siP4HA2-1# and siP4HA2-2# groups, *P<0.05 versus siNC group. C. Lactate production in Hela and SiHa cells from siNC, siP4HA2-1# and siP4HA2-2# groups, *P<0.05 versus siNC group.
P4HA2 promotes glycolysis through PGK1 and LDHA
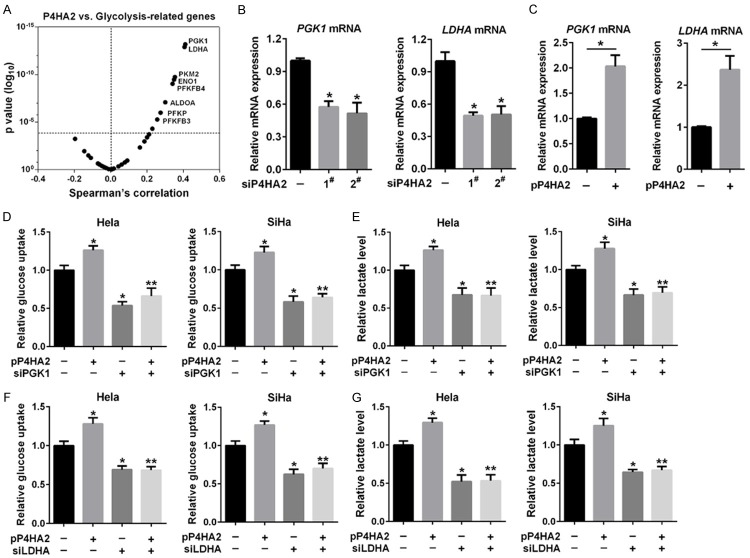
To figure out the mechanism of P4HA2 involved in glycolysis, we analyzed correlation between the expression of P4HA2 and glycolysis-related genes in the TCGA cervical cancer dataset. As shown in Figure 4A, P4HA2 expression was positively correlated with glycolysis-related genes (PGK1, LDHA, PKM2, ENO1, PFKFB4, ALODA, PFKP and PFKFB3). Then we tried to investigate whether P4HA2-enhanced glycolysis in cervical cancer was dependent upon top two related genes PGK1 and LDHA. A real-time PCR analysis showed that knockdown of P4HA2 reduced PGK1 and LDHA mRNA levels (Figures 4B, S3A), whereas overexpression of P4HA2 had the opposite effect (Figure 4C). We also found that PGK1 and LDHA protein levels were reduced in P4HA2-silenced cells (Figure S3B). To complement loss-of-function studies, Hela and SiHa cells were transfected with P4HA2 expression plasmid or with a control empty vector. Overexpression of P4HA2 increased glucose uptake and lactate level, whereas knockdown of PGK1 or LDHA significantly reduced glucose uptake and lactate level induced by P4HA2 (Figure 4D-G). These findings suggest that P4HA2 promotes cervical cancer cell glycolysis through PGK1 and LDHA.
Figure 4.
P4HA2 promotes glycolysis through PGK1 and LDHA. (A) The correlation between the mRNA expression of P4HA2 and glycolysis-related genes based on TCGA cervical cancer dataset. (B) PGK1 and LDHA mRNA expression in Hela cells treated with siP4HA2-1# and siP4HA2-2#, *P<0.05 versus siNC group. (C) PGK1 and LDHA mRNA expression in Hela cells transfected with P4HA2 expression plasmid, *P<0.05. (D and E) Glucose uptake (D) and lactate production (E) were measured in Hela and SiHa cells transfected with P4HA2 expression plasmid and PGK1 siRNA as indicated, *P<0.05 versus siNC group. **P<0.05 versus siPGK1 group. (F and G) Glucose uptake (F) and lactate production (G) were measured in Hela and SiHa cells transfected with P4HA2 plasmid and LDHA siRNA as indicated, *P<0.05 versus siNC group. **P<0.05 versus siLDHA group.
Association of P4HA2 expression with PGK1/LDHA expression in cervical cancer
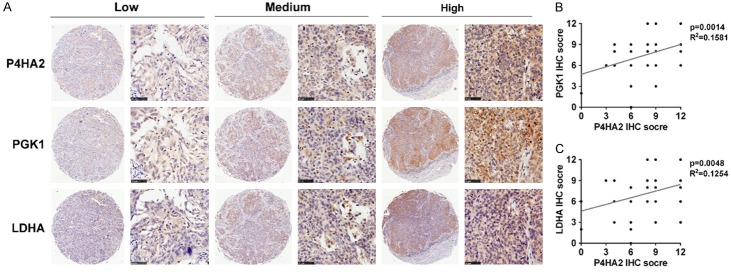
To further evaluate the relationship among P4HA2, PGK1 and LDHA in cervical cancer tissues, IHC analysis were performed. We observed that the protein level of P4HA2 was positively correlated with PGK1 at a statistically significant level (R2=0.1581, P=0.0014; Figure 5A and 5B). Similar results were obtained between P4HA2 and LDHA (R2=0.1254, P=0.0048; Figure 5A and 5C). These data verify the association of P4HA2 expression with PGK1 and LDHA expression from clinical evidence.
Figure 5.
Association of P4HA2 expression with PGK1/LDHA expression in cervical cancer. A. IHC analysis of P4HA2, PGK1 and LDHA in cervical cancer tissues. Scale bar =50 μm. B. Positive correlation of P4HA2 IHC score with PGK1 IHC score (R2=0.1581, P=0.0014). C. Positive correlation of P4HA2 IHC score with LDHA IHC score (R2=0.1254, P=0.0048).
Increased P4HA2/PGK1/LDHA expression is associated with poor survival in cervical cancer patients
To emphasize the prognostic role of P4HA2-induced glycolysis in cervical cancer patients, the online Kaplan-Meier Plotter website was used to analyze the prognostic values of P4HA2, PGK1 and LDHA expression in 304 patients with cervical cancer. Kaplan-Meier log rank analysis showed that patients whose tumors with high PGK1 or LDHA expression levels had a significantly shorter OS period (Figure 6A and 6B), consistent with results of P4HA2. We further analyzed prognosis of patients with different expression levels of P4HA2 and PGK1 or LDHA. High P4HA2/PGK1 and P4HA2/LDHA groups showed shorter survival times compared to low P4HA2/PGK1 and P4HA2/LDHA expression groups (Figure 6C and 6D). Moreover, P4HA2high/PGK1high/LDHAhigh group had a poorer prognosis compared to P4HA2low/PGK1low/LDHAlow group (Figure 6E). The survival prediction was enhanced by using both P4HA2 and PGK1/LDHA expression. Overall, these results indicate that P4HA2 and PGK1/LDHA are poor prognostic factors in cervical cancer patients.
Figure 6.

P4HA2/PGK1/LDHA expression is associated with poor survival in cervical cancer patients. A and B. Kaplan-Meier analysis of patient survival stratified by P4HA2 and PGK1 or LDHA level from TCGA database. C and D. Kaplan-Meier analysis of patient survival stratified according to the combination of P4HA2 and PGK1 or LDHA. E. Kaplan-Meier analysis of patient with high or low P4HA2/PGK1/LDHA expression. High/low, gene expression above/below the median level.
Discussion
In the current study, we revealed that P4HA2 promoted glycolysis in cervical cancer cells via its downstream effecters PGK1 and LDHA. Importantly, our results further demonstrated that increased expression of P4HA2/PGK1/LDHA was associated with poor OS in cervical cancer patients. Therefore, we provided a previously unappreciated mechanism to explain the role of P4HA2 in glycolysis. Together with previous results in hepatocellular and breast ductal carcinoma [12-14], our data extended the prognostic value of P4HA2 in patients with cervical cancer.
Collagen is a main component of extracellular matrices (ECM) and usually enhances the growth and migration of cancer cells [24,25]. As one of the key regulator enzymes for collagen homeostasis, P4HA2 has been reported to be overexpressed in several malignancies [12-17]. Consistent with these studies, we showed that the expression of P4HA2 was increased in cervical cancer based on the Oncomine database. Immunohistochemical staining also revealed elevated protein level of P4HA2 in cervical cancer tissues compared to adjacent non-tumor tissues. Moreover, we also found the correlation between lymph node metastasis and high P4HA2 expression, while the association with other members of P4H (P4HA1 and P4HA3) were not found. These data suggest that P4HA2 may play important roles in cervical cancer.
Numerous studies suggest that malignant tumor is not only a genetic disease, but also an energy metabolic disease, and oncogenic alterations actively reprogram metabolism in cancer cells [2]. Warburg effect, which is a phenomenon that cancer cells heavily rely on glycolysis for energy metabolism even when the oxygen supply is sufficient, is considered a metabolic signature related to the occurrence and development of cancer [3]. This abnormal glycolysis of cancer cells promotes glucose uptake and lactate production. A recent study implicates that the expression of P4HA1 contributes to glycolysis and hypoxic response during cancer development [26]. This prompted us to explore whether P4HA2 could regulate glycolysis in cervical cancer. In current study, GSEA analysis demonstrated that the glycolysis pathway was significantly enriched in response to high expression of P4HA2 in cervical cancer patients. The hypothesis was also supported by the fact that silencing P4HA2 was accompanied by reduced glucose uptake and lactate level in cervical cancer cells. In this context, we identified a novel role of P4HA2 in promoting cervical carcinogenesis through glycolysis.
The coding genes of key glycolysis-related enzymes are directly responsible for the regulation of glycolysis, including ALDOA, ENO1, HK2, LDHA, PFKP, PGK1, PKM2, PDK1, PFKFB2, PFKFB3, et al. To figure out the mechanism by which P4HA2 regulates glycolysis, we utilized TCGA databases for bioinformatics analysis to examine the correlation between P4HA2 and glycolysis-related genes and confirmed that LDHA and PGK1 were the potential downstream targets of P4HA2. PGK1, one of two ATP-generating enzymes in glycolysis, has been reported overexpressed in numerous malignancies. PGK1 is a promoter of metastasis in colon cancer and gastric cancer [27,28]. It is also a potential survival biomarker in breast cancer and endometrial cancer [29,30]. PGK1 regulates energy homeostasis and is regulated by multiple mechanisms [31]. The protein modification of PGK1 is essential for its function. For example, the acetylation of PGK1 enhances its enzymatic activity and promotes liver cancer development [32]. Moreover, phosphorylated PGK1 promotes its mitochondrial translocation and regulates mitochondrial pyruvate utilization [33]. LDHA, the other glycolysis-related enzyme, catalyzes the conversion of pyruvate to lactate in the final step of glycolysis. Cumulative studies demonstrate LDHA is aberrantly highly expressed in several cancer types, which is associated with the progression of cancer [34]. In cervical cancer, HPV E6/p53 induces down-regulation of miR-34a/LDHA axis and facilitates tumor growth and invasion [35]. A recent study shows that nuclear LDHA senses intracellular reactive oxygen species (ROS) to produce α-hydroxybutyrate for HPV16 E7-induced cervical cancer development [36]. Herein we provide evidence that P4HA2 affects glycolysis in cervical cancer cells through PGK1 and LDHA. We show that silencing P4HA2 decreased expression of PGK1 and LDHA. In addition, knockdown of PGK1 or LDHA reversed P4HA2-induced glycolysis. Furthermore, we also observed the association of P4HA2 with PGK1/LDHA protein levels in cervical cancer tissues. However, additional studies are needed to explore the mechanism by which P4HA2 regulates PGK1 and LDHA.
Two recent studies have demonstrated that P4HA2 is a predictor of poor outcome in patients with hepatocellular and breast ductal carcinoma [12,13]. Our data also showed that high expression of P4HA2, PGK1 or LDHA had a significantly shorter OS period. Moreover, the survival prediction was enhanced by using combination indicates of P4HA2, PGK1 and LDHA. These results indicate that glycolysis induced by P4HA2 plays a critical role in cervical cancer progression. Further studies are recommended to investigation the underlying mechanisms.
In summary, we have identified a novel function of P4HA2 in regulating glycolysis in cervical cancer. The observations also show the important role of P4HA2 in cervical cancer malignant behavior. Moreover, our findings suggest that combination of P4HA2 and PGK1/LDHA is a promising biological marker for prognosis of cervical cancer patients. Taken together, these findings indicate that P4HA2 is a promising therapeutic target for cervical cancer.
Acknowledgements
This study was supported by Grants from the National Natural Science Foundation of China (81502230 and 81802589), Grants from Shanghai Municipal Commission of Health and Family Planning (20164Y0190).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W, He T, Liu S, Zheng Y, Xiang L, Pei X, Wang Z, Yang H. The PIK3CA E542K and E545K mutations promote glycolysis and proliferation via induction of the β-catenin/SIRT3 signaling pathway in cervical cancer. J Hematol Oncol. 2018;11:139. doi: 10.1186/s13045-018-0674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Zhai H, Ouyang Y, Lu Z, Chu C, He Q, Cao X. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019;19:129. doi: 10.1186/s12935-019-0845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashmi R, Huang X, Floberg JM, Elhammali AE, McCormick ML, Patti GJ, Spitz DR, Schwarz JK. Radioresistant cervical cancers are sensitive to inhibition of glycolysis and redox metabolism. Cancer Res. 2018;78:1392–1403. doi: 10.1158/0008-5472.CAN-17-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji Y, Wu KH, Zhang YW, Zhang YX, Hu JF, Yu MM. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am J Cancer Res. 2016;6:312–320. [PMC free article] [PubMed] [Google Scholar]
- 8.Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, Zhang S, Wang M, Xiao G, Liao H. Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-α-mediated signaling. Cell Death Dis. 2014;5:e1367. doi: 10.1038/cddis.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17:73. doi: 10.1186/s12935-017-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou T, Tong C, Kazobinka G, Zhang W, Huang X, Huang Y, Zhang Y. Expression of COL6A1 predicts prognosis in cervical cancer patients. Am J Transl Res. 2016;8:2838–2844. [PMC free article] [PubMed] [Google Scholar]
- 11.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von hippel-lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 12.Wang T, Fu X, Jin T, Zhang L, Liu B, Wu Y, Xu F, Wang X, Ye K, Zhang W, Ye L. Aspirin targets P4HA2 through inhibiting NF-κB and LMCD1-AS1/let-7g to inhibit tumour growth and collagen deposition in hepatocellular carcinoma. EBioMedicine. 2019;45:168–180. doi: 10.1016/j.ebiom.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toss MS, Miligy IM, Gorringe KL, AlKawaz A, Khout H, Ellis IO, Green AR, Rakha EA. Prolyl-4-hydroxylase a subunit 2 (P4HA2) expression is a predictor of poor outcome in breast ductal carcinoma in situ (DCIS) Br J Cancer. 2018;119:1518–1526. doi: 10.1038/s41416-018-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong G, Deng L, Zhu J, Rychahou PG, Xu R. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer. 2014;14:1. doi: 10.1186/1471-2407-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Zhou X, Li Z, Liu K, Wang W, Tan R, Cong X, Shan J, Zhan Y, Cui Z, Jiang L, Li Q, Shen S, Bai M, Cheng Y, Li B, Tan M, Ma DK, Liu JO, Dang Y. Prolyl 4-hydroxylase 2 promotes B-cell lymphoma progression via hydroxylation of Carabin. Blood. 2018;131:1325–1336. doi: 10.1182/blood-2017-07-794875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis PP, Waldron L, Perez-Ordonez B, Pintilie M, Galloni NN, Xuan Y, Cervigne NK, Warner GC, Makitie AA, Simpson C, Goldstein D, Brown D, Gilbert R, Gullane P, Irish J, Jurisica I, Kamel-Reid S. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer. 2011;11:437. doi: 10.1186/1471-2407-11-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarzab B, Wiench M, Fujarewicz K, Simek K, Jarzab M, Oczko-Wojciechowska M, Wloch J, Czarniecka A, Chmielik E, Lange D, Pawlaczek A, Szpak S, Gubala E, Swierniak A. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Wang L, Qiu M, Liu Z, Qian W, Yang Y, Wu S, Feng Y. The protein levels of MCM7 and p63 in evaluating lesion severity of cervical disease. Int J Gynecol Cancer. 2013;23:318–324. doi: 10.1097/IGC.0b013e31827f6f06. [DOI] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Vasta JD, Raines RT. Collagen prolyl 4-Hydroxylase as a therapeutic target. J Med Chem. 2018;61:10403–10411. doi: 10.1021/acs.jmedchem.8b00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilkes DM, Chaturvedi P, Bajpai S, Wong CC, Wei H, Pitcairn S, Hubbi ME, Wirtz D, Semenza GL. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73:3285–3296. doi: 10.1158/0008-5472.CAN-12-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong G, Stewart RL, Chen J, Gao T, Scott TL, Samayoa LM, O’Connor K, Lane AN, Xu R. Collagen prolyl 4-hydroxylase 1 is essential for HIF-1α stabilization and TNBC chemoresistance. Nat Commun. 2018;9:4456. doi: 10.1038/s41467-018-06893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad SS, Glatzle J, Bajaeifer K, Bühler S, Lehmann T, Königsrainer I, Vollmer JP, Sipos B, Ahmad SS, Northoff H, Königsrainer A, Zieker D. Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int J Oncol. 2013;43:586–590. doi: 10.3892/ijo.2013.1971. [DOI] [PubMed] [Google Scholar]
- 28.Zieker D, Königsrainer I, Tritschler I, Löffler M, Beckert S, Traub F, Nieselt K, Bühler S, Weller M, Gaedcke J, Taichman RS, Northoff H, Brücher BL, Königsrainer A. Phosphoglycerate kinase 1 a promoting enzyme for peritoneal dissemination in gastric cancer. Int J Cancer. 2010;126:1513–1520. doi: 10.1002/ijc.24835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu D, He C, Wei J, Zhang Z, Luo Y, Tan H, Ren C. PGK1 is a potential survival biomarker and invasion promoter by regulating the HIF-1α-mediated epithelial-mesenchymal transition process in breast cancer. Cell Physiol Biochem. 2018;51:2434–2444. doi: 10.1159/000495900. [DOI] [PubMed] [Google Scholar]
- 30.Guo S, Xiao Y, Li D, Jiang Q, Zhu L, Lin D, Jiang H, Chen W, Wang L, Liu C, Fang W, Lin L. PGK1 and GRP78 overexpression correlates with clinical significance and poor prognosis in Chinese endometrial cancer patients. Oncotarget. 2017;9:680–690. doi: 10.18632/oncotarget.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43:301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, Liu X, Tao Y, Yin H, Zhou H, Zhou L, Ye D, Ye Q, Gao D. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology. 2017;65:515–528. doi: 10.1002/hep.28887. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61:705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase a: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7:6124–6136. doi: 10.1002/cam4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji Y, Wu KH, Zhang YW, Zhang YX, Hu JF, Yu MM. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am J Cancer Res. 2016;6:312–320. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Guo JZ, Liu Y, Wang K, Ding W, Wang H, Liu X, Zhou S, Lu XC, Yang HB, Xu C, Gao W, Zhou L, Wang YP, Hu W, Wei Y, Huang C, Lei QY. Nuclear lactate dehydrogenase a senses ros to produce α-hydroxybutyrate for HPV-induced cervical tumor growth. Nat Commun. 2018;9:4429. doi: 10.1038/s41467-018-06841-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.