Abstract
Objective: Mechanism by which CCNB1 regulates the cell cycle progression and its prognostic function in non-squamous non-small cell lung cancer (NSCLC) are necessary to be further elucidated. Methods: Data retrieved from gene expression omnibus (GEO) and cancer genome atlas (TCGA) combined with clinical data were used. Survival analysis was conducted in public datasets. Proteomics and co-immunoprecipation assays were designed to unravel proteins with interaction with CCNB1. Short hairpin RNA and small interfering RNA as well as overexpressing genes of interest were used. Results: CCNB1 was not implicated in apoptosis, migration and invasion of NSCLC cells. After either knockdown or overexpression of CCNB1, the occurrence of cell cycle arrest in G2/M phase, fewer cloning formation and diminished dimension of xenograft tumors were observed. CCNB1 expression level was clinically associated with several clinicopathological parameters including gender, smoking, T stage and N stage. Survival analysis showed that the higher level of CCNB1, the more dismal outcome in overall survival as well as in disease-free survival. Mechanistically, we confirmed that the role of CCNB1 on cell cycle and cloning formation was dependent on UBA52, which was able to promote degradation of CCNB1; nevertheless, this consequence relied on APC11. Knockdown of APC11 led to cell cycle arrest in G2/M and less cloning formation even in the presence of overexpressed UBA52. Following upregulation of APC11, the protein of CCNB1 degraded with resultant cell cycle progression and more cloning formation. Conclusion: Degradation of CCNB1 by APC11 via UBA52 ubiquitylation was critical in cell cycle progression and proliferation of NSCLC cell lines.
Keywords: CCNB1, UBA52, APC11, cell cycle, cloning formation
Introduction
Lung cancer is the leading cause of cancer-related death worldwide [1,2]. Although remarkable advances have been made in the treatment of this disease, the 5-year overall survival rate is still only 19% based on the data of patients definitively diagnosed between 2008 and 2014 as lung cancer [1]. Therefore, lung cancer remains a great challenge with respect to the improvement of prognosis within this patient population. Owing to this unsatisfactory status quo, further study on lung cancer is warranted.
Recently, studies on the mechanism of tumorigenesis have revealed that tumors are as a result of various gene mutations followed by abnormality of cellular signal transduction pathways, which are almost usually associated with the signals stimulating the cellular proliferation. It is the fact that cell cycle is the integration of all upstream signal pathways determining whether a cell with tumorigenic mutations is able to develop into cancer; thus the relationship between cell cycle and tumorigenesis has been emphasized [3,4]. In human body, the events of cell turnover are relevant to cell cycle, with most cells as in quiescent state, and the only cells being necessary for proliferation respond to growth signals, whereas they are under strict regulations through a variety of ways. Dysregulation of cell cycle of cancer cells is characterized as one of the critical properties evidenced by previous reports [4-6].
Cell cycle can be divided into several sequential phases as follows: G0/G1, S and G2/M phases with the transition between phases occurring to ensure the completion of cell cycle. The successful transition from one prior phase to the subsequent one is controlled by many specific factors termed cyclins and cyclin-dependent kinases (CDKs) [7]. CCNB1 as one of the essential cyclins plays important roles in G2/M transition and ensuring completion of M phase, with the former effect achieved in conjunction with CCNA; whereas the latter one is only accomplished by sole CCNB1 based on current evidences [8-10]. To our knowledge, the goal of cell cycle of mitosis is to form two daughter cells, which are the consequent of successful completion of M phase of cell cycle. Therefore, the role of CCNB1 to play in M phase is of extreme importance, thereby the exit of cell cycle is implemented. To ensure the completion of cell cycle, the levels of CCNB1 must be decreased in metaphase through the process called ubiquitin-mediated degradation.
Ubiquitin is a small molecular weight protein composed of 76 amino acids, which are edited by four types of genes UBA52, UBA80, UBB and UBC [11,12]. The products of UBB and UBC are polyubiquitin chains [13] and those of UBA52 and UBA80 are fusion proteins containing ubiquitin at the N terminus [13,14]. The aforementioned forms of products of ubiquitin-related genes are sources of ubiquitin in addition to free ubiquitin in cytoplasm. Ubiquitin functions through a process called ubiquitination to induce its substrates to degrade [15,16]. There are three key ubiquitin-related enzymes involving this process detailed as follows: firstly, ubiquitin activating enzyme (E1) via the assistance of adenosine triphosphate (ATP) providing energy activates ubiquitin by formation of thiol ester bond between its active cysteine sites and glycine residual sites; subsequently, activated ubiquitin is transferred to ubiquitin conjugated enzyme (E2) through thiol ester bond formed between activated ubiquitin and E2; eventually, ubiquitin ligase E3 recognizes its substrates and transfers the E2-ubiquitin complex to their substrate to complete the process ubiquitination of specific targets [17-19]. Ubiquitinated substrates are able to be degraded by 26S proteasome [20]. This model is the major approach to promote degradation of proteins in cells [21]. The specificity of ubiquitination is defined by E3 [22]. In mitosis, anaphase promoting complex/cyclosome (APC/C) complex as an E3 ligase plays key roles in the ubiquitination of substrates and is extensively studied. The core structure of APC/C is composed of APC11, APC2 and other eleven components, with APC11 functioning as E3 ligase and APC2 as the skeleton subunit [23,24]. Ubiquitination of CCNB1 in cell cycle via APC/C followed by its degradation is capable of promoting the transition from metaphase to anaphase and the mitotic exit to form two daughter cells [25]. Based on these evidences, ubiquitin mediated degradation of CCNB1 is the essential step to ensure the completion of mitosis.
However, there are four distinct genes associated with origin of ubiquitin, one of which is UBA52 as described above. The expression levels of UBA52 in porcine blastocyst is 6 fold that in metaphase follicles [26]. Similar phenomenon was found in both rhesus and mouse, indicating its importance in the early development of embryo [27,28]. After inhibiting the expression of UBA52, the deceleration of embryo development of rhesus signified that UBA52 is able to promote embryo development [27,29]. It is in fact that the embryo is featured by rapid development, notable cell proliferation and accelerated cell cycle progression; additionally, the level of UBA52 is higher in normal developing embryos than that in stunted embryos. Hence we hypothesized that UBA52 might positively stimulate the proliferation of cells and this hypothesis have proven to be true in a previous study, in which knocking out UBA52 resulted in lethal arrest of porcine oocyte development as well as cell cycle arrest [28,29]. In human tumors, UBA52 has been shown to be able to promote tumorigenesis of colorectal cancer [28]. Nonetheless, whether UBA52 is capable of promoting M phase of cell cycle to progress and proliferation of lung cancer cells through APC11 is unclear. In addition, CCNB1 has been shown to be highly expressed in lung cancer and associated with clinicopathological parameters [30,31]. However, these results were mostly derived from squamous cell carcinoma of non-small cell lung cancer (NSCLC) with a lack of special address of the relationship between CCNB1 and non-squamous NSCLC.
For the sake of further validating the roles of CCNB1 in NSCLC specifically in terms of non-squamous NSCLC histology and probing whether ubiquitination of CCNB1 by APC11 via UBA52 is able to promote cell cycle progression in NSCLC cells, we conducted this study. Our results showed that CCNB1 was associated with clinicopathological parameters of patients with lung cancer and could promote NSCLC cells proliferation and cell cycle progression. Its effect on proliferation and cell cycle progression was accomplished through the process of ubiquitination-mediated degradation which was controlled by APC11 with UBA52 as the ubiquitin source.
Material and methods
Antibodies
Antibodies against MMP9 (ab228402), CCNB1 (ab32053), CDK1 (ab131450), CCNA1 (ab133183), CDK2 (ab235941), Bcl-2 (ab196495), UBA52 (ab109227), rabbit IgG (ab172730) and anti-FLAG tag (ab125243) were purchased from Abcam (UK). Antibodies against CCNB1 (for Co-IP, sc-7393), APC11 (sc-517142) and CCNB1 (sc-166757) were purchased from Santa Cruz Biotechnology (USA). Antibodies against ERK1 (#4372), ubiquitin (#3933), mouse IgG (#3420) and APC11 (14090s) were purchased from Cell Signaling Technology (USA). Rabbit anti-FLAG tag antibody (SAB4301135) was purchased from Sigma-Aldrich (USA). Mouse anti-GAPDH antibody was purchased from Bioss (China). Mouse antibody against GAPDH (bsm-0978M) was purchased from BIOSS (China). HRP conjugated goat-anti-mouse IgG (A0216) and HRP conjugated goat-anti-rabbit IgG (A0208) antibodies were purchased from Beyotime Biotechnology (China).
Cell lines and culture
The human NSCLC cell lines (H460 and PLA-801D) were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). A549 and H1299 were kindly presented by Haiyang Hu (Shanghai General Hospital), with 293T and H1581 provided by Haixu Chen (Novobio Scientific, Shanghai, China). Cell lines of 293T, A549, H460 and PLA-801D were cultured in Dulbecco’s modified Eagle’s medium (high glucose, HyClone, USA) with H1299 in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, USA). All culture media were supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin/streptomycin (Life Technologies). Cells were cultured at 37°C under a humidified atmosphere with 5% carbon dioxide.
Patients and tissue samples
A total of 83 patients with NSCLC were recruited between October 2018 and December 2018 in this study. All patients underwent lobectomy for lung cancer with adenocarcinoma in histology as the selection criteria. Fresh samples representing cancerous tissues and paired adjacent normal tissues were obtained and frozen in liquid nitrogen immediately after the completion of procedure of lobectomy. The clinicopathological parameters including age at surgery, sex and pathological information were collected from the medical history. All patients provided written informed consent. This study was approved by the ethics committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China) (No. 2018KY253) and complied with the World Medical Association Declaration of Helsinki.
Quantitative real-time PCR
Total RNA of tissue specimens and cells were extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. M-MLV reverse transcriptase (Themo Fisher Scientific, USA) was used to reversely transcribe total RNA. Quantitative real-time polymerase chain reaction (qPCR) was performed with Maxima SYBR Green qPCR Master Mixes (Thermo Fisher Scientific) in a CFX96TM Real-Time System (Bio-Rad, CA, USA). The corresponding primers used in qPCR were shown as follows: GAPDH, forward 5’-CCCATGTTCGTCATGGGTGT-3’ and reverse 5’-GATGGCATGGACTGTGGTCA-3’; CCNB1, forward 5’-GCAGCACCTGGCTAAGAATG-3’ and reverse 5’-TGCCACAGCCTTGGCTAAAT-3’; UBA52, forward 5’-TGACCAGCAGCGTCTGATATT-3’ and reverse 5’-GGAGCACACATACTTGCGG-3’; APC11, forward 5’-CCCTGATGTCTAGGGAAGAGTC-3’ and reverse 5’-ACACTTGATCTGTGATGCCA-3’; Ubiquitin, forward 5’-CCTGAGGGGTGGCTGTTAAT-3’ and reverse 5’-ATGCTACCATGCAACGAAACC-3’. Each assay was performed in triplicate.
Western blot analysis and CO-IP
Total tissue and cell lysates were extracted using RIPA lysis buffer with the protease inhibitor phenylmethanesuffonyl fluoride (Beyotime Biotechnology, Jiangsu, China). Protein concentration was determined with a BCA protein assay kit (Beyotime Biotechnology) according to the manufacturer’s instructions. The same amounts of total protein 20 ug per lane were electrophoresed via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred to polyvinylidene fluoride (PVDF, Millipore, USA) membranes, which was then blocked with 5% skim milk for one hour at room temperature and followed by incubation with primary antibodies at 4°C overnight. After incubation with a secondary antibody for 1 hour at room temperature, the relative levels of proteins were detected with enhanced chemiluminescence system (Beyotime Biotechnology, Jiangsu, China).
Co-IP assays were conducted using the PierceTM Classic Magnetic IP/Co-IP Kit (Thermofisher, USA) according to the manufacturer’s instruction.
Bioinformatics analysis
Seven microarray datasets regarding lung adenocarcinoma [accession number: GSE19804 [32], GSE18842 [33], GSE191889 [34], GSE31552 [35], GSE43458 [36], GSE75037 [37], GSE31210 [38] were retrieved from gene expression omnibus (GEO) (https://www.ncbi.nlm.nih.gov/gds) database and lung adenocarcinoma dataset in the cancer genome atlas (TCGA) (https://cancergenome.nih.gov/) database was also obtained. The survival data were available in both GSE31210 [38] and TCGA datasets. The differently expressed genes (DEGs) between cancerous and non-cancerous tissue specimens in each dataset were identified by limma package under R language environment (R-3.4.2) from the Bioconductor project with logFoldChang = 2 [39], which was also used to detect DEGs in TCGA dataset. The expression level of gene CCNB1 was validated in these findings to confirm whether its expression levels were upregulated or downregulated in cancerous tissues by comparison with normal lung tissues. Then the patients with lung cancer in GSE31210 and TCGA datasets were categorized into two groups according the median expression levels of CCNB1 as either high or low expression group. Subsequently, the comparison of survival rate between these two groups was conducted.
Flow cytometry to evaluate apoptosis and cell cycle
Apoptosis assays were performed using PE Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s instruction. To assess the cell cycle distribution, cells were trypsinized, washed in PBS and fixed with ice-cold 70% ethanol at 4°C overnight. Prior to being stained, cells were washed with PBS. The staining solution was composed of 50 ug/ml Propidium Iodide (PI) (Beyotime Biotechnology, China), 100 ug/ml ribonuclease-A (RNaseA) (Beyotime Biotechnology, China) and 0.2% Triton X-100 (Invitrogen, USA). The cell pellets were resuspended with the staining solution and stained in the dark for 30 min at room temperature. Cell cycle was evaluated by a flow cytometer (FACSCalibur; BD Biosciences). The data associated with flow cytometry were dealt with FlowJo software.
Transfection, infection and establishment of stable cell lines
The short-hairpin RNA (shRNA) interference and overexpression assays were conducted on the lentivirus-based plasmid pL6.3-shRNA-NoGFP and pL6.3/MCS-BSD, respectively, both of which were purchased from Novobio Scientific (Shanghai, China). The restriction sites BsmBI were selected to construct the interference vector with NheI and AscI as the restriction enzymatic sites for overexpression vector. The shRNAs were synthesized by GENEray (Shanghai) and after successfully annealed into interfering vector, lentivirus-packing plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen, USA). The primary lentivirus were harvested at 48 hour or 72 hours after transfection, which were used to infect A549 and H1299 cells. The sequences of corresponding genes for overexpression experiments were synthesized by GENEray (Shanghai). The shRNAs used were shown as follows: CCNB1-shRNA2 forward: 5’-CACCGGTTGTTGCA-GGAGACCATGTCGAAACATGGTCTCCTGCAACAACC-3’, reverse: 5’-AAAA-GGTTGTTGCAGGAGACCATGTTTCGACATGGTCTCCTGCAACAACC-3’; CCNB1-shRNA3 forward: 5’-CACCGCAACATACTTTGGCCAAATACGAATAT-TTGGCCAAAGTATGTTGC-3’, reverse: 5’-AAAAGCAACATACTTTGGCCAA-ATATTCGTATTTGGCCAAAGTATGTTGC-3’; UBA52-shRNA2 forward: 5’-CAC-CGGTGGCATTATTGAGCCTTCTCGAAAGAAGGCTCAATAATGCCACC-3’, reverse: 5’-AAAAGGTGGCATTATTGAGCCTTCTTTCGAGAAGGCTCAATAA-TGCCACC-3’; UBA52-shRNA3 forward: 5’-CACCGCTTGCCCAGAAATACAA-CTGCGAACAGTTGTATTTCTGGGCAAGC-3’, reverse: 5’-AAAAGCTTGCCC-AGAAATACAACTGTTCGCAGTTGTATTTCTGGGCAAGC-3’; APC11-siRNA1, sense: 5’-GCCCUUGAUCAAGAGACCATT-3’, anti-sense: 5’-UGGUCUCUUGA-UCAAGGGCTT-3’; APC11-siRNA2, sense: 5’-CCUUGGUGCCUUGACCAUU-TT-3’, anti-sense: AAUGGUCAAGGCACCAAGGTT-3’.
Transwell migration and Matrigel invasion assays
Tumor cell migration and invasion assays were performed using Boyden chamber (Corning, USA) containing 8 μm pore polycarbonate membranes. Additional Matrigel (BD Biosciences, USA) was used in invasion assay compared with migration one. A total of 200 μl of cell suspension (1 × 104 cells) free of FBS was added to the upper chamber and the lower chamber was filled with 600 μl of medium supplemented with 10% FBS. After incubation for 24 hours, the cells in the upper chamber were removed with those on the lower side of the membrane fixed with 4% paraformaldehyde and subsequently stained with 1% crystal violet for 5 minutes. Cells were counted in five random fields.
Cloning formation assays
A amount of 200 cells were seeded in each well of 6-well plate and incubated for 8-12 days until the cloning was apparent. Prior to evaluation, the cells were fixed by 4% paraformaldehyde and then stained with 1% crystal violet for 10 minutes. The number of colonies was calculated with the naked eye.
In vivo xenograft assay
The animal experiments were approved by the Institutional Animal Care and Use Committee of Experimental Animal Center of Shanghai Tongji University. Five-week-old male BALB/c-nu mice were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Science (Shanghai, China). Six in number of mice were assigned into each group. A total of 2 × 106 cells suspended in 100 ul PBS were subcutaneously injected into the right flanks of each nude mouse. Tumor dimension measured every 3 days from 10 days after injection. Tumor volume was calculated as follows: volume = 1/2 × length × width2. Six weeks later, all mice were sacrificed and tumors were removed.
Proteomic analysis by LC-MS/MS
Cell lysates were electrophoresed onto SDS-PADGE, which was subsequently digested by Filter-aided sample preparation (FASP) Protein Digestion Kit (Expedeon, UK) according to the manufacturer’s instruction. Samples were dissolved in solvent A (0.1% formic acid) and loaded onto Zorbax300SB-C18peptidtraps (Agilent Technologies, Wilmington, DE). The peptides were separated by a liquid chromatography column (0.15 mm*150 mm, RP-C18, Column Technology Inc.) with a gradient of 4%-50% solvent B (0.1% formic acid in 84% methyl cyanides) for 50 min, 50%-100% solvent B for 4 min and then holding at 100% solvent B for the ultimate 6 min. The products were analyzed by Q Exactive mass spectrometer (Thermo Fisher).
Statistical analysis
All assays were in triplicate test. The data were expressed as mean ± standard deviation (SD). Student’s t test or Wilcoxon test were used for comparison between groups. The association of CCNB1 mRNA level with clinicopathological parameters was addressed using binary logistic regression analysis. Survival analysis was performed using Kaplan-Meier method with log-rank test in SPSS software (17.0). The bioinformatic analysis method was described above. Except survival test with SPSS, all statistical analysis was conducted in the environment of R language (R-3.4.2). P<0.05 was considered statistically significant. The symbol * denotes P<0.05 with ** for P<0.001.
Results
CCNB1 affected cell cycle and proliferation of NSCLC cells
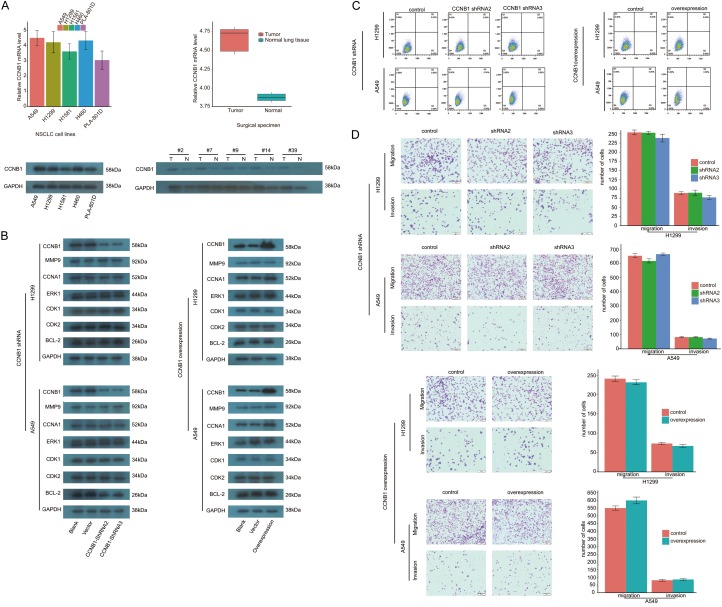
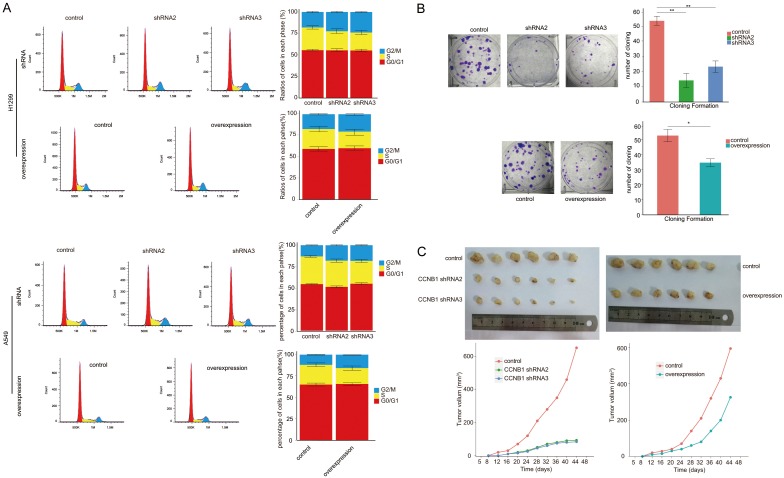
Both of mRNA and protein of CCNB1 were detected in several NSCLC cell lines. The similar manner of expression of CCNB1 mRNA was found in surgical specimens from patients with NSCLC, with the level of CCNB1 protein expression being significantly higher in lung cancer tissues than that in adjacent normal lung tissues (Figure 1A). After the process of inhibiting expression of CCNB1 mRNA using shRNA, the levels of several other proteins were not affected; similarly, the expression of these proteins was not influenced by the upregulation of CCNB1 (Figure 1B). Following the process of both inhibiting and upregulating CCNB1, there was no statistical difference observed in apoptotic assay and migration and invasion study both in A549 and H1299 cells (Figure 1C, 1D). In contrast, significant difference was found both in cell cycle and proliferation assay. In A549, the ratios of cells in G2/M phase to cells of whole cell cycle in shRNA2 and shRNA3 groups were (18±0.73)% and (18.42±0.56)%, respectively. In comparison to control group (13.1±0.47)%, there was a significant difference in each interfering group with both P value less than 0.001. Similar consequence was detected in H1299 in cell cycle assay with cells in G2/M accounting for (22.39±0.91)% in shRNA2 group and (18±0.9)% for shRNA3 group. When compared with their corresponding controls, statistical significance was obtained both in shRNA2 and shRNA3 groups with P value as 0.008 and 0.001, respectively. As to the impact of the upregulation of CCNB1 on cell cycle, flow cytometry study showed that the ratios of cells in G2/M phase both in overexpression group and control were (15.75±0.39)% and (11.9±0.49)%, respectively, with P value less than 0.001 when compared to control group. Upregulating CCNB1 also resulted in G2/M arrest in H1299 with the ratio rising from (11.9±0.49)% in control to (15.75±0.39)% (P less than 0.001). Whereas no difference was noted in the ratio of cells in G0/G1 phase as well as in S phase both in A549 and H1299 cells (Figure 2A).
Figure 1.
Significance of CCNB1 for NSCLC. (A) The extensive expression of CCNB1 in NSCLC cells and in NSCLC cancerous tissues. (B) None impact of CCNB1 expression on several other proteins observed. CCNB1 showed effect neither in apoptosis (C) nor in migration and invasion (D) in NSCLC cell lines. NSCLC: non-small cell lung cancer. shRNA: short hairpin RNA.
Figure 2.
CCNB1 influences cell cycle progression and migration and invasion of NSCLC cells. Both shRNA interference and upregulation of CCNB1 induce the arrest of cell cycle of NSCLC cells in G2/M (A) and lessen the ability of cloning formation (B) as well as xenograft tumor growth (C). NSCLC: non-small cell lung cancer. shRNA: short hairpin RNA. amplification × 100 under light microscope. *: P<0.05. **: P<0.001.
To evaluate the role of CCNB1 to play in promoting proliferation of NSCLC cell, cloning formation assay was conducted. The amount of cloning formation for A549 was 13±5.7/well in shRNA2 group and 22±4.9/well for shRNA3; when compared with control (52±4.1/well), the difference was significant with P value of 0.0014 and 0.0027 for shRNA2 and shRNA3 groups, respectively. After the overexpression of CCNB1, the upregulated group conversely demonstrated less cloning formation (34±3.3/well) than control (52±4.9/well) with P value of 0.012 (Figure 2B). The A549 cells receiving the same treatment used in proliferative assay were subcutaneously injected in nude mice, which were sacrificed 44 days after injection. The ultimate volume of the xenograft tumor was significantly less in two CCNB1 knockdown groups than that in control, with both P values of less than 0.001. Like the results observed in proliferation assay, the ability of tumorigenesis of A549 cells after overexpression of CCNB1 also declined as shown in Figure 2C.
Validation of CCNB1 expression and its predictive value of survival in NSCLC patients in public database
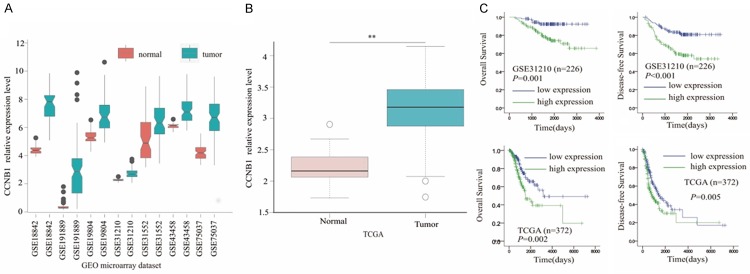
Following the evaluation of the impact of CCNB1 on cell cycle and proliferation in NSCLC cells, we further retrieved seven microarray datasets of lung adenocarcinoma from GEO database to investigate and validate its expression in lung specimens and its prediction of prognosis of NSCLC patients. A total of the seven datasets showed that the expression of CCNB1 was significantly higher in tumors than in adjacent normal lung tissues. Subsequently, the similar result was documented in lung adenocarcinoma dataset from TCGA database. As to its capability of predicting prognosis of patients with lung adenocarcinoma, the survival data from GSE31210 and TCGA were interpreted with log-rank analysis. The overall survival was significantly poorer in CCNB1 high expression group than in low expression group as validated in TCGA dataset with P value of 0.001 and 0.002, respectively. With respect to the disease-free survival, CCNB1 high expression was a dismal indicator in this concern both in GEO and TCGA datasets with P value of less than 0.001 and 0.005, respectively as shown in Figure 3. In addition to the relationship between the level of expression of CCNB1 and its prediction of prognosis of NSCLC patients, the question whether its expression level was related to clinicopathological parameters was further to be elucidated.
Figure 3.
Validation of the expression and the predictive role of CCNB1. (A) Each dataset from gene expression omnibus shows that there is a higher expression level of CCNB1 in tumor tissues than in normal lung tissues. Similar results are found in TCGA dataset (B). (C) Higher level of CCNB1 expression predicts poorer overall survival and disease-free survival in GSE31210 dataset as well as in TCGA dataset. GEO: gene expression omnibus. TCGA: the cancer genome atlas. **: P<0.001.
Association of CCNB1 expression with the clinical information of NSCLC patients
In our 83 patients with adenocarcinoma of the lung in histology, they were categorized into high and low expression groups according to the relative expression level of CCNB1 mRNA in cancerous tissues, with its median value as the threshold value. The use of this criteria lead to 41 patients divided into high-expression group with the remaining 42 into the low-expression group. The age and state of metastases appeared no difference between two groups. In contrast, there was significant difference among gender, smoking, T stage, N stage and TNM stage between the two groups as shown in Table 1. In the univariate analysis, gender, smoking, T stage, N stage and TNM stage were related to the level of CCNB1 mRNA as validated in TCGA dataset. The variables that showed statistical difference in univariate analysis underwent multivariate analysis subsequently, which confirmed the association of the expression level of CCNB1 with gender, smoking, T stage as well as N stage. And this relationship was validated by TCGA dataset (Table 2).
Table 1.
Clinicopathological parameters of lung cancer patients between CCNB1 high and low expression groups
| Variables | Number of patients | P | |
|---|---|---|---|
|
| |||
| CCNB1 high-expression group | CCNB1 low-expression group | ||
| Number | 41 | 42 | 0.913 |
| Age (years) | 0.915 | ||
| >70 | 7 | 5 | |
| 55-70 | 20 | 25 | |
| <55 | 14 | 12 | |
| Gender | 0.012 | ||
| Female | 26 | 15 | |
| Male | 15 | 27 | |
| Smoking (pack/year) | 0.002 | ||
| None | 17 | 7 | |
| 1-20 | 3 | 2 | |
| 20-50 | 8 | 6 | |
| >50 | 13 | 27 | |
| T stage | 0.012 | ||
| T1 | 19 | 7 | |
| T2 | 18 | 30 | |
| T3 | 4 | 4 | |
| T4 | 0 | 1 | |
| N stage | 0.019 | ||
| N0 | 29 | 20 | |
| N1 | 10 | 14 | |
| N2 | 2 | 8 | |
| Metastases | 0.986 | ||
| No | 40 | 41 | |
| Yes | 1 | 1 | |
| TNM stage | 0.033 | ||
| I | 18 | 12 | |
| II | 21 | 20 | |
| III | 1 | 9 | |
| IV | 1 | 1 | |
Table 2.
The association of CCNB1 expression level with variables from surgical specimen and TCGA lung adenocarcinoma dataset
| Variable | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Patient data | TCGA | Patient data | TCGA | |||||
|
|
|
|
|
|||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Age | 1.010 (0.523-1.946) | 0.977 | 1.284 (0.940-1.754) | 0.116 | - | - | - | - |
| Gender | 3.120 (1.274-7.642) | 0.013 | 2.065 (1.363-3.128) | 0.001 | 3.149 (1.091-9.092) | 0.034 | 2.051 (1.335-3.149) | 0.001 |
| Smoking | 1.708 (1.188-2.454) | 0.004 | 1.177 (1.006-1.377) | 0.043 | 1.648 (1.091-2.486) | 0.017 | 1.185 (1.007-1.395) | 0.041 |
| T stage | 2.418 (1.146-5.103) | 0.021 | 1.498 (1.099-2.041) | 0.010 | 3.374 (1.304-8.729) | 0.012 | 1.475 (1.007-2.162) | 0.046 |
| N stage | 2.259 (1.144-4.460) | 0.019 | 1.502 (1.120-2.016) | 0.007 | 4.869 (1.302-18.212) | 0.019 | 1.659 (1.069-2.573) | 0.024 |
| Metastases | 0.976 (0.059-16.147) | 0.986 | 3.034 (0.855-10.763) | 0.086 | - | - | - | - |
| TNM stage | 1.959 (1.033-3.721) | 0.039 | 1.372 (1.057-1.780) | 0.017 | 0.433 (0.123-1.527) | 0.193 | 0.837 (0.540-1.298) | 0.837 |
Notes: OR: odds ratio; CI, confidence interval; TCGA: The Cancer Genome Atlas.
Owing to the significance of CCNB1 in NSCLC cell lines and clinical data, the further elucidation of its potential mechanism by which it influences the cell cycle and proliferation of NSCLC cell is warranted.
Identification of proteins interacting with CCNB1 and determination of the gene of interest to study
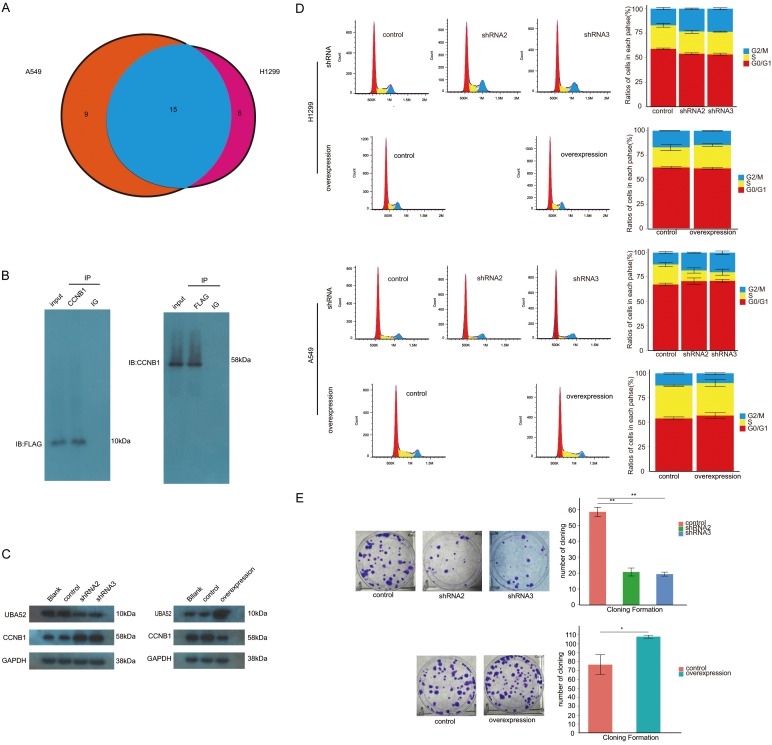
Co-immunoprecipitation assay was conducted using anti-CCNB1 antibody both in A549 and H1299 cells. The immunoprecipitation samples were sent for proteomics study. The findings showed that there were 15 proteins pulled down by anti-CCNB1 antibody in both cells shown in Figure 4A. After a combination of our finding and referring to literatures, we selected UBA52 as the potential gene of interest. Then, we further validated whether there was the presence of interaction between UBA52 and CCNB1 with UBA52 overexpressed and labeled with FLAG tag. We first performed CO-IP using anti-CCNB1 antibody and immunostained samples with anti-FLAG antibody, with the resulting identification of FLAG-UBA52 band. Simultaneously, CO-IP was conducted, whereas with anti-FLAG antibody as the one that pulled down its target protein and with anti-CCNB1 antibody as the antibody that was used in the process of immunostaining blot. The band related to CCNB1 was still present (Figure 4B). Therefore, we deemed that UBA52 might interact with CCNB1 in NSCLC cells.
Figure 4.
Identification of an interaction between UBA52 and CCNB1 and the effect of UBA52 on cell cycle and cloning formation. A. Proteomics analysis detected 15 proteins with possible interaction with CCNB1 including UBA52. B. Co-immuoprecipation confirmed the presence of interaction between CCNB1 and UBA52. C. Both inhibiting and enhancing the expression of UBA52 resulted in a change in the level of CCNB1 protein in Western-blot study. D. Inhibition of UBA52 caused the arrest of cell cycle in G2/M of non-small cell lung cancer cells with its overexpression reversing this impact. E. Both the interference and overexpression of UBA52 affected A549 cloning formation with the former showing inhibitive property, as opposed to the reinforcing potential in the latter group. IP: immunoprecipitation. ov: overexpression. shRNA: short hairpin RNA. *: P<0.05. **: P<0.001.
UBA52 was able to affect cell cycle progression and A549 cell proliferation
Following the knockdown and overexpression of UBA52, the former would lead to the detection of relatively increased level of CCNB1 protein with WB and the latter to a decreased level of CCNB1 protein (Figure 4C). Of concern was whether these alterations could affect the cell cycle and proliferation of NSCLC cells. As to cell cycle, an arrest in G2/M phase secondary to shRNA knockdown of UBA52 was observed, with the ratio of cells in G2/M increasing from (12.06±0.93)% in control to (18.23±0.50)% in shRNA2 group and (20±1.71)% in shRNA3 group in H1299 with each P value of 0.001 and 0.005, respectively; in A549, similar trends were observed with the ratios of cells in G2/M phase being (17.04±0.06)%, (23.36±0.03)% and (23.69±1.62)% in control, shRNA2 and shRNA3 groups, respectively (Figure 4D). In A549 proliferation assay, the count of cloning formation was 20±3.3 per well in number in shRNA2 group and 18±2.4 per well in shRNA3 group, as compared with 61±4.5 per well in control with both P values of less than 0.001. After the overexpression of UBA52 in A549, the amount of cloning formation increased from 61±6.1 per well in control to 96±4.5 per well in overexpressed group, with P value of 0.003 (Figure 4E).
UBA52 exerts its role as a source of ubiquitin [21], which interacts with ubiquitin ligase E3 to promote degradation of specific substrates [40]. In cell cycle regulation, APC/C was reported as one of the major component of the E3 family. Its core structure is APC11, which defines its activity as a ubiquitin ligase E3 [40]. Therefore, the interaction between UBA52 and APC11 and the impact of APC11 on cell cycle and proliferation of NSCLC cells warrants to be studied.
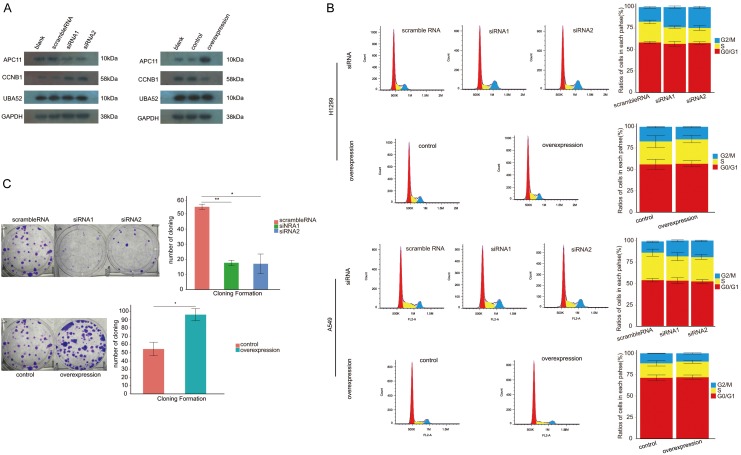
APC11 expression correlates with the expression level of UBA52 and CCNB1 proteins and functions on cell cycle and proliferation of NSCLC cells
After inhibiting expression of APC11 protein using siRNA, the increased level of CCNB1 protein was detected using WB method. In contrast, the level of CCNB1 protein was reduced after overexpression of APC11. In both settings, the UBA52 was rarely affected in the form of protein level in WB study as shown in Figure 5A. With respect to cell cycle, in H1299, the ratios of cells in G2/M were (17.02±0.88)%, (23.64±1.42)% and (24.39±1.40)% in control, siRNA1 and siRNA2 groups respectively, with P value of 0.005 as comparing those ratios in siRNA1 with that in control and 0.003 for siRNA2 group. In A549, the relevant ratios were represented by (13.19±0.74)%, (18.47±1.14)% and (19.45±0.74)% in control, siRNA1 and siRNA2 groups, respectively. The P value for siRNA1 compared with control was 0.005 and 0.001 for siRNA2 (Figure 5B). In A549 proliferative assay, the number of cloning formation was 53±2.4 per well in control compared to 16±1.2 per well in siRNA1 and 14±2.1 per well in siRNA2 with both P values of less than 0.001 for two experimental groups, respectively (Figure 5C).
Figure 5.
The function of APC11. A. APC11 affected the level of CCNB1. B. Suppressing the expression of APC11 by small interference RNA led to cell cycle arrest in G2/M in both A549 and H1299. C. APC11 demonstrated an impact on the capability of cloning formation in A549. ov: overexpression. siRNA: small interfering RNA. *: P<0.05. **: P<0.001.
As the aforementioned statement, the consequence of interfering with the expression of one of the three genes CCNB1, UBA52 and APC11 alone were clarified, especially in the areas of cell cycle and cloning formation. However, whether this process of interfering would affect another one or more interfering process for these genes was needed to further verify their interaction and determine their role in the regulation of cell cycle and proliferation.
UBA52-mediated degradation of CCNB1 via APC11 promotes cell cycle progression and proliferation of NSCLC cells
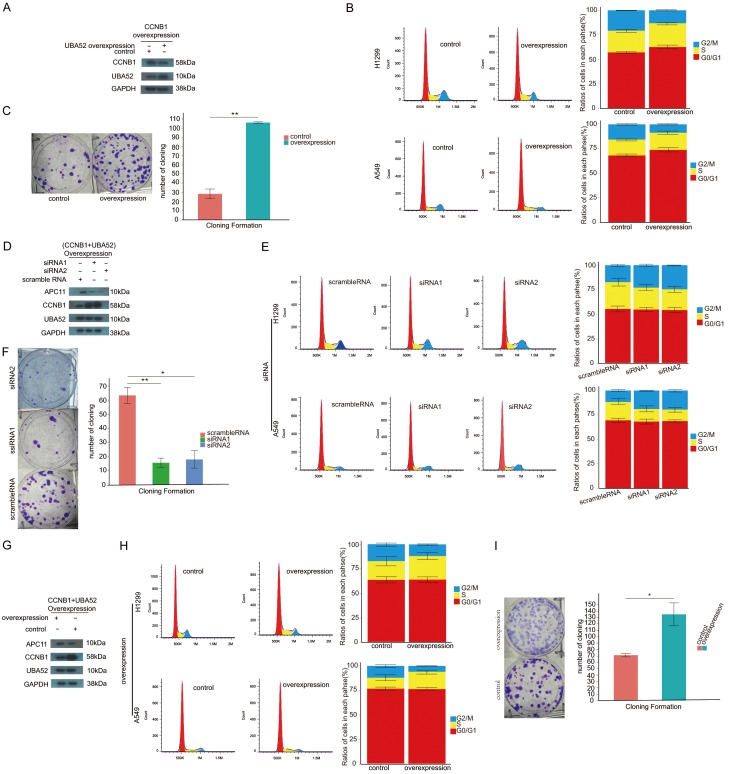
As evidenced in Figures 1D, 2A, 2B, enhanced expression of CCNB1 inhibited the ability of migration and invasion and cell cycle as well as proliferation of NSCLC cells. In comparison with these facts, overexpressed UBA52 counteracted the effects of the overexpression of CCNB1 alone as illustrated in Figure 6A-C as follows: overexpression of UBA52 induced the decreased level of CCNB1 protein, eliminated the inhibition to cell cycle progression and proliferation in CCNB1 overexpressed NSCLC cell line. As shown in Figure 6C, the capability of cloning formation was significantly reinforced with the number of cloning in treated group versus in control being 106±7.8 vs 27±4.5 with P value of less than 0.001. In cell cycle analysis, in H1299, the ratios of cells in G2/M phase were (13.28±0.16)% in both gene overexpressed group compared with (17.34±0.62)% in control with P value of 0.003. No statistical significance of ratios of cells in both G0/G1 and S phases between groups was observed.
Figure 6.
Both APC11 and UBA52 required for degradation of CCNB1. (A) Overexpression of UBA52 reduced the level of protein CCNB1 in CCB1 overexpressed A549 cell. (B) Overexpression of UBA52 facilitated cell cycle progression in CCNB1 overexpressed cells, which showed cell cycle arrest in G2/M without manipulation of UBA52 as shown in Figure 2A. (C) As features in cell cycle, overexpression of UBA52 increased the ability of cloning formation of A549 with overexpression of CCNB1. In the circumstance with both UBA52 and CCNB1 overexpressed in non-small cell lines, inhibiting APC11 by small interference RNA resulted in relatively higher level of CCNB1 protein (D), cell cycle arrest in G2/M (E) and less cloning formation (F); in contrast, overexpression of APC11 led to lower level of CCNB1 protein (G), smooth cell cycle progression (H) and more cloning formation (I). ov: overexpression. siRNA: small interfering RNA. *: P<0.05. **: P<0.001.
Nevertheless, these findings relied on APC11; after interfering with the expression of APC11 with siRNA in both CCNB1 and UBA52 overexpressed A549 cell, the level of CCNB1 seemed to slightly rise (Figure 6D). The ability of cloning formation in CCNB1 and UBA52 overexpressed A549 was significantly suppressed in APC11 interfered groups with 62±3.7 per well of cloning in number in control, when compared with 15±2.9 in siRNA1 and 17±2.9 in siRNA2, resulting in both P values of less than 0.001 (Figure 6F). These suppressed trends were also detected in cell cycle assay. In H1299, the ratios of cells in G2/M were (22.99±1.07)%, (24.67±0.51)% and (17.35±1.02)% in siRNA1, siRNA2 and control groups, respectively, with P value of siRNA1 vs control being 0.006 and less than 0.001 for siRNA2 vs control. Similarly, these inhibited events were present in A549, in which the ratios of cells increased from (11.72±0.96)% in control to (18.9±0.49)% in siRNA1 and (19.48±1.20)% in siRNA2 with the P values of latter two groups compared with the control being less than 0.001 and 0.002, respectively (Figure 6E). In consistent with exception of resulting events after upregulating APC11 in both CCNB1 and UBA52 overexpressed NSCLC cells, the level of CCNB1 protein was reduced (Figure 6G) and the ability of cloning formation in A549 was enhanced from 71±6.9 per well in control to 135±13.9 per well in triple gene overexpressed group with P value of 0.004 (Figure 6I). As for the cell cycle of NSCLC cells, the ratios of cells in G2/M phase in H1299 decreased from (17.23±1.61)% in control to (11.97±0.76)% and in A549 decreased from (12.1±1.47)% to (5.97+0.37)% with P values in both H1299 and A549 cell lines being 0.014 and 0.005, respectively. The ratios of cells in G0/G1 and S phases in both H1299 and A549 cell lines were comparable (Figure 6H).
Discussion
CCNB1 was able to be predictive of the prognosis of patients with lung cancer, with the higher level of CCNB1 resulting in the poorer outcome [30,31]. However, these findings are mainly stemmed from the studies on squamous cell histology in NSCLC; thus its role in non-squamous NSCLC warrants further investigation. Moreover, in previous studies, the level of expression of CCNB1 was predominantly determined using immunohistochemistry approach. As the advent of microarray technology and the establishment of public database such as TCGA and GEO, a vast amount of gene-associated expression data in the form of RNA sequencing were available for assessing the prognostic significance of gene expression [32-38]. Therefore, our present study utilized the median levels of CCNB1 mRNA as the criteria for dividing the patients with NSCLC into high expression group and low expression group, resulting in nearly each 50% of patients in both groups. Based on previous reports, the ratios of CCNB1 overexpression in NSCLC ranged from 40.9%-88% [41,42]; accordingly, the median of CCNB1 mRNA as cutoff value leading to nearly equal number of patients in two groups is reasonable. Yet the optimal method to evaluate the expression state of CCNB1 warrants further study. In our clinical data, the level of CCNB1 mRNA correlated with gender, smoking status, T stage and N stage and this association was validated via TCGA lung adenocarcinoma dataset as reported by Yoshida T. and associates [31]. Owing to our lack of follow-up material, we used the survival data in GSE31210 [38] from GEO to evaluate the prognosis of CCNB1 level in the fashion of RNA seq with the dataset from TCGA as the validation set. After survival analysis, we revealed that the higher level of CCNB1 RNA seq indicates a more dismal outcome both in disease-free survival and in overall survival as well as in TCGA validation dataset. In NSCLC cell lines, CCNB1 had no role to play on apoptosis, migration, invasion and interference with the expression of a variety of other proteins. In the cell cycle assay, inhibiting the expression of CCNB1 led to the cell cycle arrest in G2/M phase, which was consistent with the previous evidence [43,44]. This event of cell cycle arrest in G2/M might result from the dominant role of CCNB1 in promoting cell cycle progression to accomplish the mitosis [45-49]. To our knowledge, the goal of mitosis is to produce two daughter cells to induce tumor cell proliferation. Arrest in cell cycle inevitably causes suppression of tumor cell growth. Therefore, the ability of cloning formation in A549 reduced following inhibition of the expression of CCNB1. Of interest, overexpression of CCNB1 also resulted in cell cycle arrest in G2/M and inhibition of A549 cell proliferation. This finding might be explained by the study of Chang D.C et al that introduction of undegradable CCNB1, even in a relatively low level, into cells would block cell cycle progression [45]. The means of overexpressing CCNB1 in the present study was in a lentivirus-mediated fashion, which might produce the amount of CCNB1 protein exceeding the level of its degradation.
During the cell cycle progression, degradation of CCNB1 enables the transition from metaphase to anaphase in mitosis. The process involving the degradation of CCNB1 is termed ubiquitination-mediated degradation, which is accomplished under the sequential interaction of ubiquitin-associated enzymes such as E1, E2 and E3 with its substrates [17,19,50,51]. Ubiquitin is the initiator of this process. In human body, there are several genes being able to provide ubiquitin, one of which is UBA52 [11,12]. UBA52 functioned essentially in early embryogenesis [52,53]. After knockout of UBA52, fatal arrest of development of porcine oocytes occurred [27]. Although UBA52 might play an important role in embryogenesis, its roles on cell cycle and proliferation of NSCLC cell lines are unknown. In our proteomics analysis, UBA52 was identified as one of the proteins with potential interaction with CCNB1, and then this probable interaction was verified by co-immunoprecipitation assay. Upregulation of UBA52 resulted in both a higher amount of cloning formation and an increase in cell cycle progression as well as in combined CCNB1 overexpressed cell lines. In contrast, the reverse events were observed following knock-down of UBA52. As to how UBA52 influences these processes, this is our concern aimed at exploring its mechanism by which it conducts its role in NSCLC cell lines.
In WB assay, we detected that the level of CCNB1 protein was relatively reduced after overexpression of UBA52 even in CCNB1 overexpressed cell lines and was increased after knock-down of UBA52. Based on the fact that UBA52 is a source of ubiquitin, we hypothesized that the impact on CCNB1 by manipulation of UBA52 be a process depended on degradation mediated by UBA52 in NSCLC cell lines. As we described above, ubiquitination is related to a variety of enzymes, including E3 which determines the specificity of substrates recognized [54]. In cell cycle, E3 is represented by APC/C [55], which consists of 19 subunits with APC11 as its enzymatic unit [56]. Our findings were agreement with the previous reports, which showed that inhibiting the expression of APC11 induced the cell cycle arrest in G2/M [56,57]. Additionally, APC11 contributed to the A549 cell proliferation in cloning formation assay. As to its impact on CCNB1, overexpression of APC11 also led to decreased level of CCNB1 protein and its downregulation resulted in higher level of CCNB1 in WB study with no change in the level of UBA52 protein observed. These findings encouraged us to speculate that in NSCLC cell lines degradation of CCNB1 by UBA52 requiring the participation of APC11. In our further investigation, with the level of APC11 expression being altered in CCNB1 and UBA52 combined overexpressed NSCLC cell lines, knock-down of APC11 lead to the cell cycle arrest in G2/M in NSCLC cell lines and decreased the ability of cloning formation in A549; whereas the facilitating effects in both cell cycle and cloning formation were detected after upregulation of APC11.In protein level of both CCNB1 and UBA52 overexpressed A549 cell, overexpression and inhibition of APC11 caused the level of CCNB1 to be relatively declined and remain in high level, respectively.
Our study showed that CCNB1 in NSCLC cell lines was essential to the cell cycle progression as well as cloning formation and these roles were dependent on the its interaction with UBA52, which is a source of ubiquitin [11-14] and induced the degradation of CCNB1 to accomplish the above function. However, this interaction was under the regulation of APC11, a subunit of E3 ligase in ubiquitin-mediated degradation of various substrates [23,24]. In combined overexpression of CCNB1 and UBA52, knock-down of APC11 resulted in cell cycle arrest in G2/M and limited amount of cloning formation, both of which were reversed in NSCLC cells following upregulation of APC11. In addition, reduced level of CCNB1 protein after overexpressing APC11 was observed even in CCNB1 overexpressed cells. In summary, based on the previous evidence that UBA52 and APC11 participate in ubiquitin-mediated degradation of substrates, our study showed that, in non-squamous NSCLC cells, degradation of CCNB1 by APC11 via UBA52 ubiquitination induced cell cycle progression and enhanced cellular proliferation.
Acknowledgements
We are grateful to Feng Huihui, Ye Jun and Chen Haixu for their instruction to our certain laboratory procedures.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci U S A. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams GH, Stoeber K. Cell cycle markers in clinical oncology. Curr Opin Cell Biol. 2007;19:672–679. doi: 10.1016/j.ceb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 7.Malumbres M, Barbacid M. Is cyclin D1-CDK4 kinase a bona fide cancer target? Cancer Cell. 2006;9:2–4. doi: 10.1016/j.ccr.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker RT, Board PG. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 1991;19:1035–1040. doi: 10.1093/nar/19.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 13.Redman KL, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–440. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- 14.Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, Tanaka K. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell. 2009;137:549–559. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 15.Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol. 2012;47:17–28. doi: 10.3109/10409238.2011.620943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinngrebe J, Montinaro A, Peltzer N, Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014;15:28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 20.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 21.Wiborg O, Pedersen MS, Wind A, Berglund LE, Marcker KA, Vuust J. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985;4:755–759. doi: 10.1002/j.1460-2075.1985.tb03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamenz J, Ferrell JE Jr. The temporal ordering of cell-cycle phosphorylation. Mol Cell. 2017;65:371–373. doi: 10.1016/j.molcel.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 25.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 26.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Mao J, O’Gorman C, Sutovsky M, Zigo M, Wells KD, Sutovsky P. Ubiquitin A-52 residue ribosomal protein fusion product 1 (Uba52) is essential for preimplantation embryo development. Biol Open. 2018;7 doi: 10.1242/bio.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503:2095–2100. doi: 10.1016/j.bbrc.2018.07.165. [DOI] [PubMed] [Google Scholar]
- 29.Sadritdinova AF, Dmitriev AA, Snezhkina AV, Belenikin MS, Krasnov GS, Manylov OG, Kudryavtsev AA, Melnikova NV, Speranskaya AS, Darii MV, Lakunina VA, Uroshlev LA, Smurov AO, Stepanov OA, Kudryavtseva AV. A new reliable reference gene UBA52 for quantitative real-time polymerase chain reaction studies in pyloric cecal tissues of the starfish asterias rubens. Genet Mol Res. 2014;13:3972–3980. doi: 10.4238/2014.May.23.8. [DOI] [PubMed] [Google Scholar]
- 30.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 31.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15:252–256. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 32.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, Chuang EY. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19:2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Farez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 34.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J, Marquardt G, Mullapudi N, Wang T, Han W, Shi M, Keller S, Zhu C, Locker J, Spivack SD. Lung cancer transcriptomes refined with laser capture microdissection. Am J Pathol. 2014;184:2868–2884. doi: 10.1016/j.ajpath.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabbout M, Garcia MM, Fujimoto J, Liu DD, Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, Behrens C, Lee JJ, Wistuba II, Kadara H. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II, Minna JD, Gazdar AF. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22:4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–325. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 41.Cooper WA, Kohonen-Corish MR, McCaughan B, Kennedy C, Sutherland RL, Lee CS. Expression and prognostic significance of cyclin B1 and cyclin A in non-small cell lung cancer. Histopathology. 2009;55:28–36. doi: 10.1111/j.1365-2559.2009.03331.x. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez Salmon A, Garrido M, Abdulkader I, Gude F, Leon L, Ruibal A. The immunohistochemical expression of cyclin B1 is associated with higher SUV in 18F-FDG-PET in non-small cell lung cancer patients. Initial results. Rev Esp Med Nucl. 2009;28:63–65. [PubMed] [Google Scholar]
- 43.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang LL, Feng ZL, Su MX, Jiang XM, Chen X, Wang Y, Li A, Lin LG, Lu JJ. Downregulation of cyclin B1 mediates nagilactone E-induced G2 phase cell cycle arrest in non-small cell lung cancer cells. Eur J Pharmacol. 2018;830:17–25. doi: 10.1016/j.ejphar.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Chang DC, Xu N, Luo KQ. Degradation of cyclin B is required for the onset of anaphase in mammalian cells. J Biol Chem. 2003;278:37865–37873. doi: 10.1074/jbc.M306376200. [DOI] [PubMed] [Google Scholar]
- 46.Dunphy WG. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y, Sramkoski RM, Jacobberger JW. The kinetics of G2 and M transitions regulated by B cyclins. PLoS One. 2013;8:e80861. doi: 10.1371/journal.pone.0080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter T, Pines J. Cyclins and cancer. Cell. 1991;66:1071–1074. doi: 10.1016/0092-8674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 49.Soni DV, Sramkoski RM, Lam M, Stefan T, Jacobberger JW. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle. 2008;7:1285–1300. doi: 10.4161/cc.7.9.5711. [DOI] [PubMed] [Google Scholar]
- 50.Iconomou M, Saunders DN. Systematic approaches to identify E3 ligase substrates. Biochem J. 2016;473:4083–4101. doi: 10.1042/BCJ20160719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi M, Oshima S, Maeyashiki C, Nibe Y, Otsubo K, Matsuzawa Y, Nemoto Y, Nagaishi T, Okamoto R, Tsuchiya K, Nakamura T, Watanabe M. The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci Rep. 2016;6:36780. doi: 10.1038/srep36780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mtango NR, Latham KE. Ubiquitin proteasome pathway gene expression varies in rhesus monkey oocytes and embryos of different developmental potential. Physiol Genomics. 2007;31:1–14. doi: 10.1152/physiolgenomics.00040.2007. [DOI] [PubMed] [Google Scholar]
- 54.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 55.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 56.Chang TS, Jeong W, Lee DY, Cho CS, Rhee SG. The RING-H2-finger protein APC11 as a target of hydrogen peroxide. Free Radic Biol Med. 2004;37:521–530. doi: 10.1016/j.freeradbiomed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Moore R, Boyd L. Analysis of RING finger genes required for embryogenesis in C. elegans. Genesis. 2004;38:1–12. doi: 10.1002/gene.10243. [DOI] [PubMed] [Google Scholar]