Abstract
Aerobic glycolysis is a hallmark of metabolic reprogramming in tumor progression. However, the mechanisms regulating glycolytic gene expression remain elusive in neuroblastoma (NB), the most common extracranial malignancy in childhood. Herein, we identify that CUT‐like homeobox 1 (CUX1) and CUX1‐generated circular RNA (circ‐CUX1) contribute to aerobic glycolysis and NB progression. Mechanistically, p110 CUX1, a transcription factor generated by proteolytic processing of p200 CUX1, promotes the expression of enolase 1, glucose‐6‐phosphate isomerase, and phosphoglycerate kinase 1, while circ‐CUX1 binds to EWS RNA‐binding protein 1 (EWSR1) to facilitate its interaction with MYC‐associated zinc finger protein (MAZ), resulting in transactivation of MAZ and transcriptional alteration of CUX1 and other genes associated with tumor progression. Administration of an inhibitory peptide blocking circ‐CUX1‐EWSR1 interaction or lentivirus mediating circ‐CUX1 knockdown suppresses aerobic glycolysis, growth, and aggressiveness of NB cells. In clinical NB cases, CUX1 is an independent prognostic factor for unfavorable outcome, and patients with high circ‐CUX1 expression have lower survival probability. These results indicate circ‐CUX1/EWSR1/MAZ axis as a therapeutic target for aerobic glycolysis and NB progression.
Keywords: circular RNA, CUT‐like homeobox 1, EWS RNA‐binding protein 1, MYC‐associated zinc finger protein, tumor progression
Subject Categories: Cancer, RNA Biology
Aerobic glycolysis is a hallmark of metabolic reprogramming in tumors. This study shows how CUT‐like homeobox 1 (CUX1) and its derived circular RNA (circ‐CUX1) contribute to aerobic glycolysis. The circ‐CUX1/EWSR1/MAZ axis emerges as a possible therapeutic target for neuroblastoma progression.
Introduction
Neuroblastoma (NB), a malignant tumor arising from primitive neural crest, accounts for 15% of cancer‐related mortality in childhood (Brodeur, 2003). For high‐risk NB, the clinical outcome remains poor in despite of multimodal therapeutic approaches (Brodeur, 2003). To support their tumorigenecity and aggressiveness, tumor cells uptake and convert a large amount of glucose into lactic acid even in the presence of adequate oxygen, which is known as aerobic glycolysis or Warburg effect (Hanahan & Weinberg, 2011). Activation of oncogenes (c‐Myc) or inactivation of tumor suppressors (p53) contributes to aberrant expression of transporters and metabolic enzymes of aerobic glycolysis (Shim et al, 1997; Bensaad et al, 2006; Yang et al, 2014). However, identification of transcriptional regulators for aerobic glycolysis in NB still remains to be determined.
Circular RNAs (circRNAs) are a novel class of endogenous noncoding RNAs that are generated from exons or introns, and may function as microRNA (miRNA) sponges, regulators of transcription and splicing, or partners of RNA‐binding protein (RBP) (Lasda & Parker, 2014; Li et al, 2015b). For example, circRNA antisense to cerebellar‐degeneration‐related protein 1 (Cdr1as) harbors 70 binding sites for miR‐7 to regulate its transport in neurons (Piwecka et al, 2017). A special class of exon–intron circRNAs, such as circEIF3J and circPAIP2, is predominantly localized in the nucleus and enhance transcription of their parental genes (Li et al, 2015b). In addition, intronic circRNAs, such as ci‐ankrd52, are able to regulate transcription efficiency of parental genes by binding to RNA polymerase II (Zhang et al, 2013). However, the roles of circRNAs in aerobic glycolysis during tumor progression remain largely elusive.
In this study, we identify CUT‐like homeobox 1 (CUX1) as a transcription factor facilitating aerobic glycolysis and tumor progression in NB. We also reveal the oncogenic functions of a CUX1‐generated intron‐containing circular RNA (circ‐CUX1) in tumorigenesis and aggressiveness. Elevated circ‐CUX1 promotes the aerobic glycolysis, growth, and aggressiveness of NB cells by binding to EWS RNA‐binding protein 1 (EWSR1) and facilitating its interaction with MYC‐associated zinc finger protein (MAZ), resulting in MAZ transactivation and transcriptional alteration of CUX1 and other genes associated with tumor progression, suggesting circ‐CUX1/EWSR1/MAZ axis as a therapeutic target for aerobic glycolysis and NB progression.
Results
CUX1 facilitates aerobic glycolysis and tumor progression
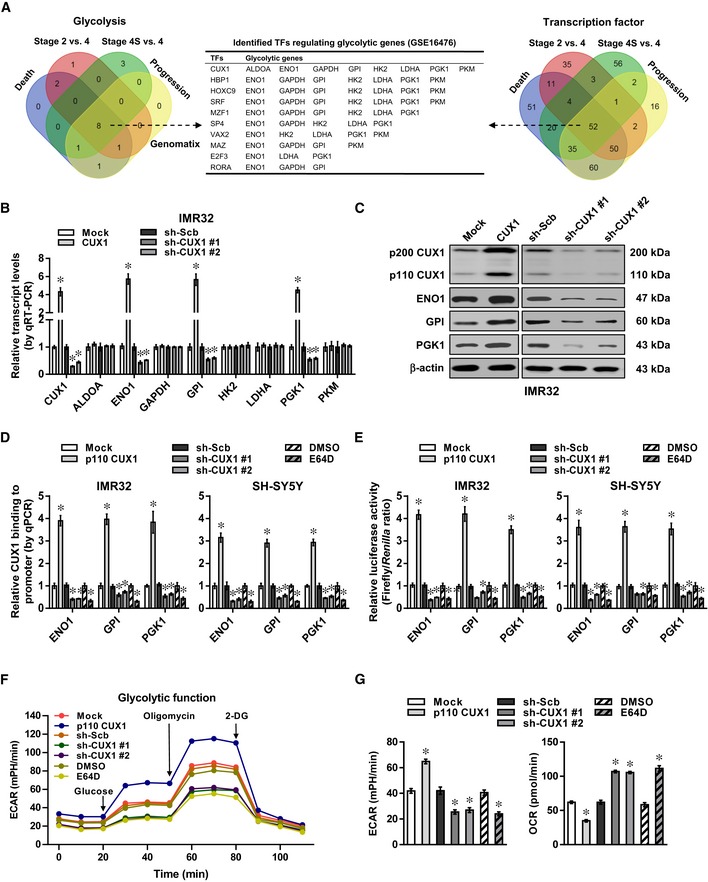
Comprehensive analysis of a microarray dataset (GSE16476) (Molenaar et al, 2012) of 88 NB cases identified 8 differentially expressed glycolytic genes (fold change > 2.0, P < 0.05) that were consistently associated with death, advanced international neuroblastoma staging system (INSS) stages, and clinical progression (Fig 1A). Similarly, we also found 52 transcription factors consistently associated with these clinical features (Fig 1A), which were subjective to further over‐lapping analysis with potential transcription factors regulating all of 8 glycolytic genes revealed by Genomatix program (http://www.genomatix.de/solutions/genomatix-software-suite.html). The results indicated CUX1 as the top transcription factor ranking by number of potential targets (Fig 1A). Higher transcript levels of CUX1 isoform p200 were noted in NB cell lines SH‐SY5Y, IMR32, and SK‐N‐AS, while p75 (Goulet et al, 2002) was expressed at very low levels (Appendix Fig S1A). Consistently, elevated levels of p200 CUX1 and its proteolytically processed isoform p110 were noted in these NB cells, cervical cancer HeLa cells, colon cancer LoVo cells, and prostate cancer PC‐3 cells, than those of non‐transformed normal MCF 10A cells (Appendix Fig S1A). However, both transcript and protein levels of CDP/cut alternatively spliced cDNA (CASP) (Gillingham et al, 2002) were not differently expressed between normal and tumor cells (Appendix Fig S1A). In an independent cohort of 54 primary NB tissues, the transcript levels of p200 CUX1, but not of CASP, were higher than those in normal fetal adrenal medulla (P < 0.05, Appendix Fig S1A), especially in cases with poor stroma (P = 0.0002) or advanced INSS stages (P = 0.007), without association with MYCN amplification (P = 0.56, Appendix Fig S1B).
Figure 1. CUX1 facilitates aerobic glycolysis and tumor progression in NB.
-
AVenn diagram indicating the identification of differentially expressed glycolytic genes and transcription factors (fold change > 2.0, Student's t‐test, P < 0.05) in 88 NB cases (GSE16476), and over‐lapping analysis with potential transcription factors regulating glycolytic genes revealed by Genomatix program.
-
B, CReal‐time qRT–PCR (B, normalized to β‐actin, n = 5) and Western blot (C) assays revealing the expression of CUX1 and glycolytic genes in IMR32 cells stably transfected with empty vector (mock), p200 CUX1, scramble shRNA (sh‐Scb), sh‐CUX1 #1, or sh‐CUX1 #2. Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
-
D, EChIP and qPCR using Flag and CUX1 antibodies (D) and dual‐luciferase (E) assays indicating the p110 CUX1 enrichment and promoter activity of ENO1, GPI, and PGK1 in IMR32 and SH‐SY5Y cells stably transfected with mock, Flag‐tagged p110 CUX1, sh‐Scb, sh‐CUX1 #1, or sh‐CUX1 #2, and those treated with E64D (10 μM) for 24 h (n = 5). Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock, sh‐Scb, or DMSO.
-
F, GSeahorse tracing curves (F), ECAR and OCR (G) of IMR32 cells stably transfected with mock, p110 CUX1, sh‐Scb, sh‐CUX1 #1, or sh‐CUX1 #2, and those treated with E64D (10 μM) for 24 h (n = 5). Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock, sh‐Scb, or DMSO.
Notably, ectopic expression or knockdown of p200 CUX1 (referred as CUX1) increased and decreased the levels of p110 CUX1, enolase 1 (ENO1), glucose‐6‐phosphate isomerase (GPI), or phosphoglycerate kinase 1 (PGK1), but not of aldolase, fructose‐bisphosphate A (ALDOA), glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), hexokinase 2 (HK2), lactate dehydrogenase A (LDHA), or pyruvate kinase M (PKM), in IMR32 and SH‐SY5Y cells (Fig 1B and C, Appendix Fig S1C and D). Treatment with E64D, an inhibitor of cathepsin L (Goulet et al, 2004), abolished the up‐regulation of p110 CUX1, ENO1, GPI, and PGK1 induced by CUX1 over‐expression (Appendix Fig S1E). Ectopic expression of p110 CUX1 increased the levels of ENO1, GPI, or PGK1 in IMR32 cells (Appendix Fig S1E). Meanwhile, knockdown of CASP, a Golgi‐localized CUX1 isoform (Gillingham et al, 2002), did not affect the transcript and protein levels of these glycolytic genes in SH‐SY5Y cells (Appendix Fig S1F and G). The CUX1 enrichment and promoter activity of ENO1, GPI, and PGK1 were increased and decreased by p110 CUX1 over‐expression, CUX1 knockdown, or E64D treatment in IMR32 and SH‐SY5Y cells, respectively (Appendix Fig S1H, Fig 1D and E). Over‐expression of p110 CUX1 increased the extracellular acidification rate (ECAR) and reduced the oxygen consumption rate (OCR) in IMR32 cells, while CUX1 knockdown or E64D treatment significantly attenuated the glycolytic process (Fig 1F and G). Accordingly, p110 CUX1 over‐expression, CUX1 knockdown, or E64D treatment increased and decreased the glucose uptake, lactate production, ATP levels, anchorage‐independent growth, and invasion of IMR32 cells, respectively (Appendix Fig S2A–D). Treatment with 2‐deoxyglucose (2‐DG), an established glycolysis inhibitor (Zhang et al, 2014), abolished the increase in these features of IMR32 cells induced by p110 CUX1 over‐expression (Appendix Fig S2A–D). In public datasets, there was positive expression correlation between CUX1 and ENO1, GPI, or PGK1 in NB, colon cancer, or prostate cancer tissues (Appendix Fig S2E), and their levels were associated with poor survival of tumor patients (Appendix Fig S3). Multivariate Cox regression analysis revealed CUX1 as an independent prognostic factor [hazard ratio = 2.105, 95% confidence interval = 1.087–3.243, P = 0.038] for poor survival of NB patients. These findings indicated that CUX1 was a transcription factor facilitating aerobic glycolysis and tumor progression.
Circ‐CUX1 is up‐regulated in NB tissues and cell lines
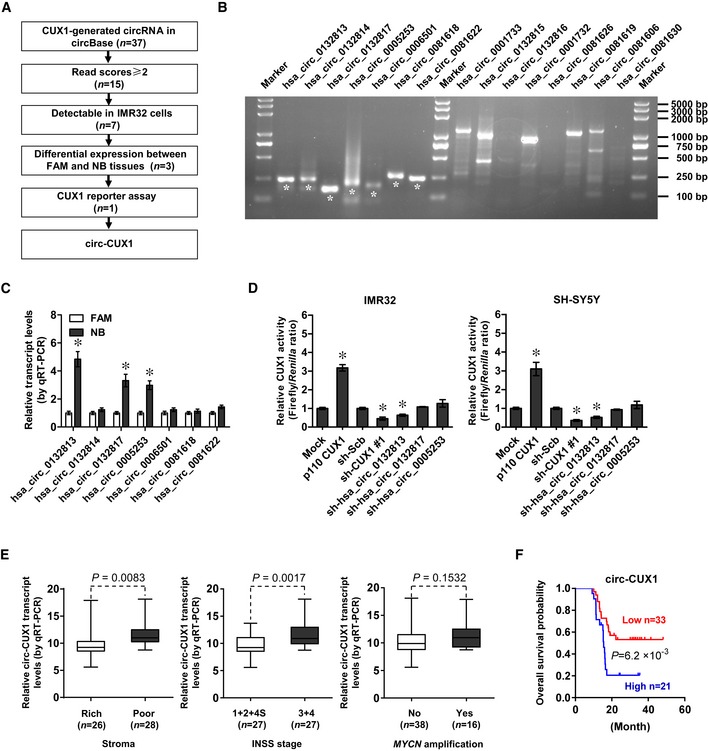
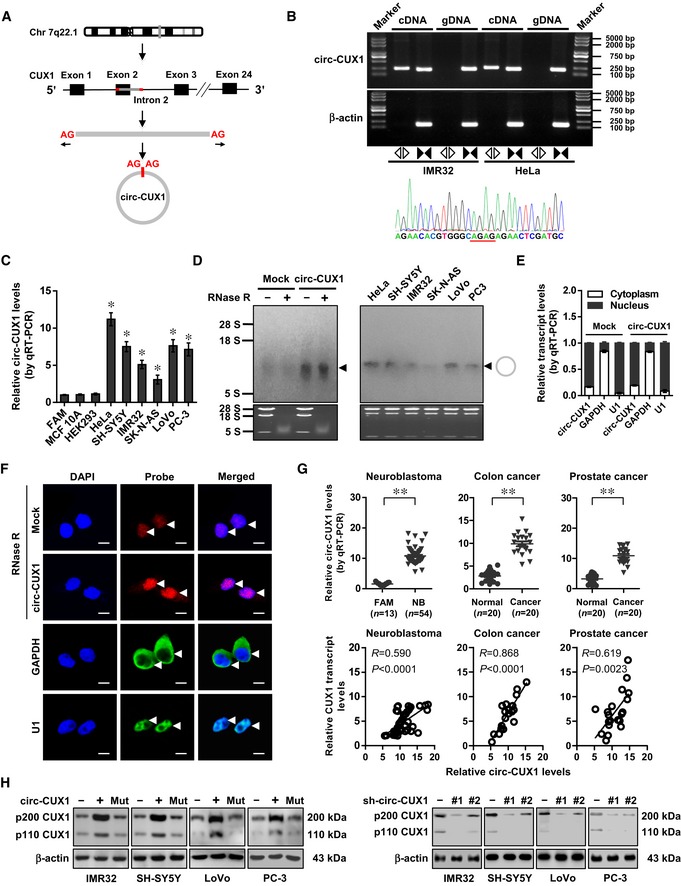
The copy number of CUX1 gene, locating at chr7: 101460882‐101901513, was neither significantly altered in NB (Appendix Fig S4A) nor associated with death, MYCN amplification, INSS stages, or survival of NB cases derived from Oncogenomics database (Appendix Fig S4A and B). There were no genetic variants of CUX1 gene in 563 NB cases of public datasets (Appendix Fig S4C). Among 37 potential circRNAs generated from CUX1 gene in circBase (Glazar et al, 2014), 15 had more than 2 read scores, while further RT–PCR validation revealed 7 detectable circRNAs in IMR32 cells (Fig EV1A and B). Three of them were differentially expressed between normal fetal adrenal medulla and NB tissues (Fig EV1C). Knockdown of hsa_circ_0132813 (termed as circ‐CUX1), but not of hsa_circ_0132817 or hsa_circ_0005253, attenuated the CUX1 transactivation in IMR32 and SH‐SY5Y cells (Fig EV1D). The 393‐nt circ‐CUX1, consisting of exon 2 and partial intron 2 of CUX1 (Fig 2A), was detected by RT–PCR with divergent primers and Sanger sequencing (Fig 2B), and its expression levels were significantly elevated in many tumor cell lines (Fig 2C and D). Endogenous circ‐CUX1 was resistant to RNase R digestion (Fig 2D) and localized within nucleus of IMR32 cells, which was further confirmed by ectopic expression of circ‐CUX1 (Fig 2D–F). Notably, circ‐CUX1 levels were higher in tissues of NB, colon cancer, and prostate cancer, than normal fetal adrenal medulla or adjacent normal tissues (Fig 2G). In addition, circ‐CUX1 levels were positively correlated with those of CUX1 in tissues of NB (R = 0.590, P < 0.0001), colon cancer (R = 0.868, P < 0.0001), or prostate cancer (R = 0.619, P = 0.0023; Fig 2G). In 54 primary NB tumors, higher circ‐CUX1 levels were observed in cases with poor stroma (P = 0.0083) or advanced INSS stages (P = 0.0017), without association with MYCN amplification (P = 0.1532; Fig EV1E). Patients with high circ‐CUX1 expression had lower survival probability (Fig EV1F). These results indicated that circ‐CUX1 was up‐regulated in NB tissues and cell lines.
Figure EV1. Identification and expression levels of circ‐CUX1 .
-
AFlowchart delineating the discovery of circ‐CUX1 from tumor tissues and cell lines. FAM, normal fetal adrenal medulla.
-
BRT–PCR assay with divergent primers showing the detectable circRNAs of correct size (asterisks) in IMR32 cells.
-
CReal‐time qRT–PCR assay indicating the circRNA levels in FAM (n = 13) and NB tissues (n = 20). Student's t‐test, *P < 0.05 versus FAM.
-
DDual‐luciferase assay showing transactivation activity of CUX1 in IMR32 and SH‐SY5Y cells transfected as indicated (n = 5). Student's t‐test, one‐way ANOVA, *P < 0.05 versus empty vector (mock) or scramble shRNA (sh‐Scb).
-
EReal‐time qRT–PCR assay indicating circ‐CUX1 levels (normalized to β‐actin) in NB tissues (n = 54). Bars are means and whiskers show min to max. Student's t‐test.
-
FKaplan–Meier curve showing overall survival of 54 NB patients with low or high circ‐CUX1 levels (cutoff value = 10.77). Log‐rank test.
Figure 2. Circ‐CUX1 is up‐regulated and enhances CUX1 expression in NB.
-
ASchematic illustration showing the generation of circ‐CUX1 from CUX1.
-
BRT–PCR or PCR assay revealing the amplification of circ‐CUX1 from cDNA or genomic DNA (gDNA) of IMR32 and HeLa cells, with validation by Sanger sequencing.
-
C, DReal‐time qRT–PCR (C, normalized to β‐actin, n = 6) and Northern blot (D) indicating the circ‐CUX1 levels in cell lines and IMR32 cells transfected with empty vector (mock) or circ‐CUX1 and treated with RNase R (3 U μg−1). One‐way ANOVA, *P < 0.05 versus HEK293.
-
E, FReal‐time qRT–PCR (E, normalized to β‐actin) and RNA‐FISH with antisense junction probe and RNase R (3 U μg−1) treatment (F) showing the distribution and localization (arrowheads) of circ‐CUX1 in IMR32 cells stably transfected with mock or circ‐CUX1 (n = 5), using GAPDH and U1 as controls. Scale bar: 10 μm.
-
GReal‐time qRT–PCR assay indicating circ‐CUX1 expression (normalized to β‐actin) and its correlation with CUX1 levels (Pearson's correlation coefficient) in tumor tissues, normal fetal adrenal medulla (FAM), or normal counterparts. Student's t‐test, **P < 0.01 versus FAM or normal.
-
HWestern blot showing the CUX1 levels in tumor cells stably transfected as indicated.
Circ‐CUX1 enhances CUX1 expression at transcriptional level
To investigate the effects of circ‐CUX1 on expression of parental gene CUX1, circ‐CUX1 or two independent short hairpin RNAs (shRNAs) targeting junction site of circ‐CUX1 (sh‐circ‐CUX1) were stably transfected into tumor cell lines. Transfection of circ‐CUX1 vector, but not of that with mutant back‐splicing elements (circ‐CUX1‐Mut), resulted in obvious circ‐CUX1 production in IMR32 cells, which was resistant to RNase R digestion (Appendix Fig S5A). Transfection of sh‐circ‐CUX1 #1 and sh‐circ‐CUX1 #2 increased the enrichment of Argonaute 2 (AGO2) on circ‐CUX1, but not on CUX1 mRNA, in IMR32, SH‐SY5Y, LoVo, and PC‐3 cells (Appendix Fig S5B). In a luciferase reporter‐based assay monitoring shRNA specificity (Bramsen et al, 2010), transfection of sh‐circ‐CUX1 #1 or sh‐circ‐CUX1 #2 decreased the activity of circ‐CUX1 reporter, without impact on that of CUX1 reporter (Appendix Fig S5C). Notably, stable transfection of circ‐CUX1, but not of circ‐CUX1‐Mut, into IMR32, SH‐SY5Y, LoVo, and PC‐3 cells resulted in its over‐expression (Fig EV2A), increased CUX1 promoter activity (Fig EV2B), and elevated levels of CUX1 isoforms p200 and p110, but not of CASP (Figs EV2C and 2H). The stability of CUX1 mRNA was not altered in IMR32 cells stably transfected with circ‐CUX1 (Fig EV2D). Meanwhile, stable knockdown of circ‐CUX1 led to decrease in CUX1 promoter activity and expression of p200 and p110 in tumor cells, without effects on CUX1 mRNA stability or CASP levels (Figs EV2A–D and 2H). These results illustrated that circ‐CUX1 enhanced CUX1 expression at transcriptional level in tumor cells.
Figure EV2. Circ‐CUX1 facilitates CUX1 transcription and aerobic glycolysis of tumor cells.
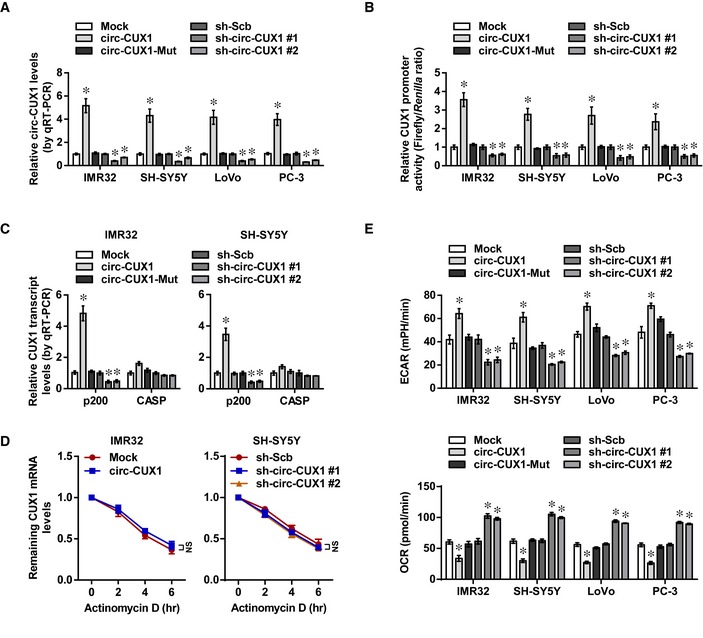
-
AReal‐time qRT–PCR assay indicating expression of circ‐CUX1 (normalized to β‐actin) in IMR32, SH‐SY5Y, LoVo, or PC‐3 cells stably transfected as indicated (n = 5). One‐way ANOVA, *P < 0.05 versus empty vector (mock) or scramble shRNA (sh‐Scb).
-
BDual‐luciferase assay showing the activity of CUX1 promoter in IMR32, SH‐SY5Y, LoVo, or PC‐3 cells stably transfected as indicated (n = 5). One‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
-
CReal‐time qRT–PCR assay indicating expression of CUX1 isoforms (normalized to β‐actin) in IMR32 and SH‐SY5Y cells stably transfected as indicated (n = 5). One‐way ANOVA, *P < 0.05 versus empty vector (mock) or scramble shRNA (sh‐Scb).
-
DReal‐time qRT–PCR assay indicating CUX1 mRNA levels in IMR32 and SH‐SY5Y cells stably transfected as indicated (n = 5), and those treated with actinomycin D (5 μg ml−1). One‐way ANOVA, NS: non‐significant.
-
ESeahorse extracellular flux assay showing ECAR and OCR in IMR32, SH‐SY5Y, LoVo, or PC‐3 cells stably transfected as indicated (n = 6). One‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
Circ‐CUX1 exerts an oncogenic role in tumor progression
We further observed the potential effects of circ‐CUX1 on biological features of tumor cells. The ECAR was increased and decreased in IMR32, SH‐SY5Y, LoVo, and PC‐3 cells stably transfected with circ‐CUX1 or sh‐circ‐CUX1, along with reduced and enhanced OCR, while transfection of circ‐CUX1‐Mut did not affect these features (Fig EV2E). Notably, ectopic expression of circ‐CUX1 increased the glucose uptake, lactate production, and ATP levels of IMR32 cells, which was attenuated by 2‐DG treatment (Appendix Fig S6A). Stable over‐expression or knockdown of circ‐CUX1 increased and decreased the anchorage‐independent growth and invasion of IMR32 and SH‐SY5Y cells, respectively (Fig 3A and B). Consistently, stable transfection of circ‐CUX1 or sh‐circ‐CUX1 #1 into IMR32 cells resulted in a significant increase or decrease in growth, tumor weight, Ki‐67 proliferation index, CD31‐positive microvessels, glucose uptake, lactate production, and ATP levels of subcutaneous xenograft tumors in nude mice (Fig 3C–E). Athymic nude mice treated with tail vein injection of IMR32 cells with stable over‐expression or knockdown of circ‐CUX1 displayed more or less lung metastatic colonies, with lower or greater survival probability, respectively (Fig 3F). These results indicated that circ‐CUX1 exerted an oncogenic role in tumorigenesis and aggressiveness.
Figure 3. Circ‐CUX1 exerts an oncogenic role in tumorigenesis and aggressiveness.
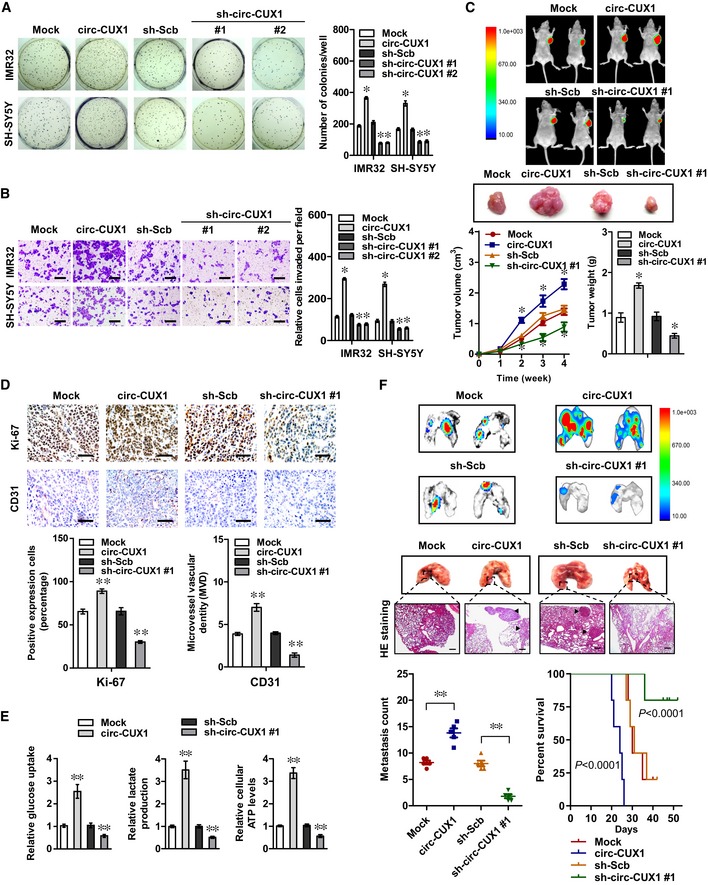
-
A, BSoft agar (A) and Matrigel invasion (B) assays showing the anchorage‐independent growth and invasion capability of IMR32 and SH‐SY5Y cells stably transfected with empty vector (mock), circ‐CUX1, scramble shRNA (sh‐Scb), or sh‐circ‐CUX1 (n = 5). Scale bars: 100 μm. Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
-
CRepresentative fluorescence images, in vivo growth curve, and weight at the end points of subcutaneous xenograft tumors formed by IMR32 cells stably transfected as indicated in nude mice (n = 5 for each group). Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
-
D, EImmunohistochemical staining showing the expression of Ki‐67 and CD31 (D) and glucose uptake, lactate production, and ATP levels (E) within subcutaneous xenograft tumors formed by IMR32 cells stably transfected as indicated (n = 5 for each group). Scale bars: 100 μm. Student's t‐test, **P < 0.01 versus mock or sh‐Scb.
-
FRepresentative images, HE staining (arrowheads), quantification of lung metastatic colonization, and Kaplan–Meier curves of nude mice treated with tail vein injection of IMR32 cells stably transfected as indicated (n = 5 for each group). Scale bar: 100 μm. Student's t‐test, **P < 0.01 versus mock or sh‐Scb. Log‐rank test for survival comparison.
Circ‐CUX1 directly interacts with EWSR1 protein in NB cells
To explore the protein partner of circ‐CUX1, RNA pull‐down was performed using biotin‐labeled probes generated by ligation of linear transcript in vitro (Petkovic & Muller, 2015) or synthesized as oligonucleotides targeting junction site (Fig 4A). Mass spectrometry revealed 47 proteins consistently pulled down by exogenous circ‐CUX1 and antisense probe targeting endogenous circ‐CUX1, but not by linear transcript or sense probe, and 18 of them were RBPs defined by RBPDB (http://rbpdb.ccbr.utoronto.ca). Further comprehensive analysis of protein interacting with transcription factors of CUX1 promoter revealed by Genomatix and BioGRID database (https://thebiogrid.org) indicated three potential circ‐CUX1‐interacting partners (Fig 4A), including EWSR1, ELAV‐like RNA‐binding protein 1 (ELAVL1), and synaptotagmin‐binding cytoplasmic RNA‐interacting protein (SYNCRIP). Further validating RNA pull‐down assay using circ‐CUX1 probes revealed the specific enrichment of endogenous or exogenous circ‐CUX1, but not p200 CUX1 or CASP transcript, and the physical interaction of circ‐CUX1 with EWSR1, but not with ELAVL1 or SYNCRIP, in non‐transfected IMR32 cells (Fig 4B). Endogenous binding of EWSR1 protein to circ‐CUX1, but not to p200 CUX1 or CASP transcript, was also observed in SH‐SY5Y cells, which was facilitated by transfection of circ‐CUX1 (Fig 4C). In NB cell lines SH‐SY5Y, SK‐N‐AS, BE(2)‐C, and IMR32, PCR assay indicated no fusion of EWSR1 with Fli1 gene (Appendix Fig S6B). Three‐dimensional (3D) confocal images of dual RNA fluorescence in situ hybridization (RNA‐FISH) and immunofluorescence assay confirmed endogenous co‐localization of circ‐CUX1 and EWSR1 in IMR32 cells, which was facilitated by transfection of circ‐CUX1 (Fig 4D and Movie EV1). Consistently, RNA electrophoretic mobility shift assay (EMSA) showed that circ‐CUX1 interacted with EWSR1 protein within nuclear extracts of SH‐SY5Y cells (Fig 4E). The RNA recognition motif (RRM) domain [361–447 amino acids (aa)], but not amino‐ or carboxyl‐terminus, of glutathione S‐transferase (GST)‐tagged EWSR1 protein was necessary for its interaction with circ‐CUX1, but not with p200 CUX1 or CASP transcript (Fig 4F). Mutation of 394–397 or 406–410 aa of RRM domain, potential interacting regions analyzed by catRAPID (Agostini et al, 2013), abolished the interaction of EWSR1 with circ‐CUX1 (Fig 4F). These results suggested that circ‐CUX1 directly interacted with EWSR1 protein in NB cells.
Figure 4. Circ‐CUX1 directly interacts with EWSR1 protein in NB cells.
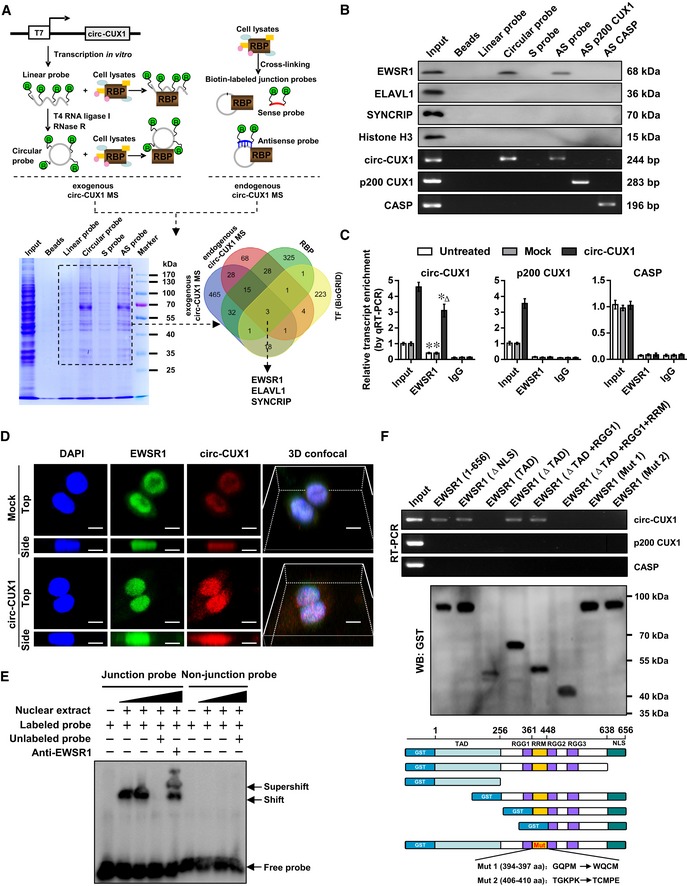
-
ASchematic illustration, Coomassie Blue staining, and Venn diagram showing the differential proteins pulled down by biotin‐labeled linear or circular exogenous circ‐CUX1, sense (S) or antisense (AS) probe targeting junction site of endogenous circ‐CUX1 from IMR32 cells, and over‐lapping analysis with RBP and proteins interacting with potential transcription factors of p200 CUX1 revealed by Genomatix program and BioGRID database.
-
BWestern blot (upper panel) and RT–PCR (lower panel) assays indicating the proteins and transcripts pulled down by biotin‐labeled linear or circular exogenous circ‐CUX1, sense (S) or antisense (AS) probe targeting junction site of endogenous circ‐CUX1 from IMR32 cell lysates, using AS probes of p200 CUX1 or CASP as controls.
-
CRIP and real‐time qRT–PCR assays revealing the interaction of EWSR1 with circ‐CUX1, p200 CUX1, or CASP in SH‐SY5Y cells and those stably transfected with empty vector (mock) or circ‐CUX1 (n = 5). Student's t‐test, *P < 0.05 versus IgG; Δ P < 0.01 versus mock.
-
D3D confocal images of dual RNA‐FISH and immunofluorescence staining assay showing the co‐localization of circ‐CUX1 and EWSR1 in IMR32 cells stably transfected with mock or circ‐CUX1. Scale bar: 10 μm.
-
ERNA EMSA determining the interaction between biotin‐labeled circ‐CUX1 probe and EWSR1 protein within nuclear extracts of SH‐SY5Y cells (arrowheads).
-
FIn vitro binding assay depicting the recovered circ‐CUX1, p200 CUX1, or CASP detected by RT–PCR (upper panel) after incubation with GST‐tagged recombinant EWSR1 protein validated by Western blot (lower panel).
Circ‐CUX1 facilitates EWSR1‐mediated MAZ transactivation
To further investigate target genes of circ‐CUX1, RNA sequencing (RNA‐seq) was performed and revealed 781 up‐regulated and 434 down‐regulated genes (fold change > 1.5, P < 0.05) in IMR32 cells upon circ‐CUX1 over‐expression (Fig 5A). Transcription factors regulating these genes were analyzed by ChIP‐X program (Lachmann et al, 2010), which revealed top five potential ones, including E12, lymphoid enhancer‐binding factor 1 (LEF1), MAZ, nuclear factor of activated T cells (NFAT), and specificity protein 1 (SP1) (Fig 5B). Further over‐lapping analysis with EWSR1‐interacting proteins derived from BioGRID database revealed that MAZ protein was the only transcription factor involved in this process (Fig 5B). Endogenous interaction between EWSR1 and MAZ was observed in IMR32 cells (Appendix Fig S6C). The RRM domain (361–448 aa), but not transactivation domain (TAD), Arg‐Gly‐Gly (RGG) 1, RGG2, or RGG3 domain, of Myc‐tagged EWSR1 was necessary for its interaction with MAZ protein (Appendix Fig S6D). Meanwhile, zinc finger (ZNF) domain (198–477 aa), but not N‐terminus or C‐terminus, of Flag‐tagged MAZ was necessary for its interaction with EWSR1 (Appendix Fig S6E). Ectopic expression or knockdown of circ‐CUX1 increased and decreased the interaction between EWSR1 and MAZ in IMR32 and SH‐SY5Y cells, respectively (Fig 5C and D, and Appendix Fig S6F).
Figure 5. Circ‐CUX1 facilitates EWSR1‐mediated MAZ transactivation in NB cells.
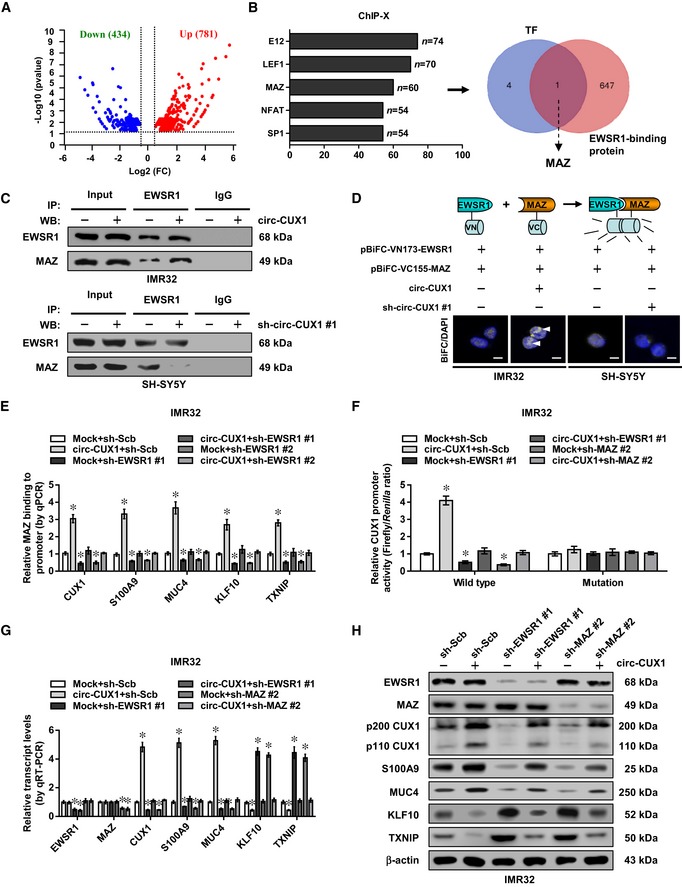
-
AVolcano plots indicating RNA‐seq results of 781 up‐regulated and 434 down‐regulated genes in IMR32 cells upon stable circ‐CUX1 over‐expression (fold change > 1.5, P < 0.05).
-
BChIP‐X analysis (left panel) showing top five transcription factors regulating the altered genes, and Venn diagram (right panel) indicating the identification of MAZ by over‐lapping analysis of five transcription factors and EWSR1‐interacting proteins derived from BioGRID database.
-
CCo‐IP and Western blot assays showing the interaction between EWSR1 and MAZ in IMR32 and SH‐SY5Y cells stably transfected with circ‐CUX1 or sh‐circ‐CUX1 #1, respectively.
-
DBiFC assay revealing the interaction (arrowheads) of EWSR1 and MAZ in IMR32 and SH‐SY5Y cells stably transfected as indicated, with nuclei stained by DAPI. Scale bars: 10 μm.
-
EChIP assay showing the binding of MAZ (normalized to input, n = 5) to target gene promoters in IMR32 cells stably transfected as indicated. One‐way ANOVA, *P < 0.05 versus mock+sh‐Scb.
-
FDual‐luciferase assay revealing the relative activity of p200 CUX1 promoter with wild‐type or mutant MAZ‐binding site in IMR32 cells stably transfected as indicated (n = 5). One‐way ANOVA, *P < 0.05 versus mock+sh‐Scb.
-
G, HReal‐time qRT–PCR (G, normalized to β‐actin, n = 5) and Western blot (H) assays showing the expression of EWSR1, MAZ, and their target genes in IMR32 cells stably transfected as indicated. One‐way ANOVA, *P < 0.05 versus mock+sh‐Scb.
Notably, higher MAZ levels were observed in NB tissues than those in normal fetal adrenal medulla (P < 0.0001), especially in those with poor stroma (P = 0.0205) or advanced INSS stages (P = 0.0097), without association with MYCN amplification (P = 0.6445, Appendix Fig S7A). Among 60 MAZ target genes derived from RNA‐seq results and ChIP‐X analysis, the expression of CUX1, S100 calcium‐binding protein A9 (S100A9), mucin 4 (MUC4), Kruppel‐like factor 10 (KLF10), or thioredoxin‐interacting protein (TXNIP) was significantly correlated with that of MAZ in 54 NB cases (Appendix Fig S7B). In addition, higher expression of EWSR1, MAZ, S100A9, or MUC4 and lower expression of KLF10 or TXN1P were associated with poor survival of NB patients (GSE16476, Appendix Fig S7C). In RNA pull‐down and chromatin isolation by RNA purification (ChIRP) (Chu & Chang, 2016) assays using biotin‐labeled circ‐CUX1 junction probe, circ‐CUX1 was associated with EWSR1 and MAZ protein, and promoters of target genes (CUX1, S100A9, MUC4, KLF10, or TXNIP), but not with transcripts of downstream genes in SH‐SY5Y cells (Appendix Fig S8A). Ectopic expression or knockdown of circ‐CUX1 enhanced and reduced the binding of MAZ to these target gene promoters in IMR32 and SH‐SY5Y cells, while silencing or over‐expression of EWSR1 abolished these effects (Fig 5E and Appendix Fig S8B). The activity of wild‐type CUX1 promoter, but not of that with mutant MAZ‐binding site, was increased and decreased by ectopic expression or knockdown of circ‐CUX1 (Fig 5F and Appendix Fig S8C). In addition, the levels of CUX1, S100A9, MUC4, KLF10, or TXNIP were significantly altered in IMR32 and SH‐SY5Y cells stably transfected with circ‐CUX1 or sh‐circ‐CUX1 #1 (Fig 5G and H, Appendix Fig S8D and E). Knockdown or ectopic expression of EWSR1 or MAZ rescued tumor cells from these changes (Fig 5F–H, Appendix Figs S8C–E and S9A). These data indicated that circ‐CUX1 facilitated EWSR1‐mediated MAZ transactivation and transcriptional alteration of target genes in NB cells.
Therapeutic peptide blocking the circ‐CUX1‐EWSR1 interaction
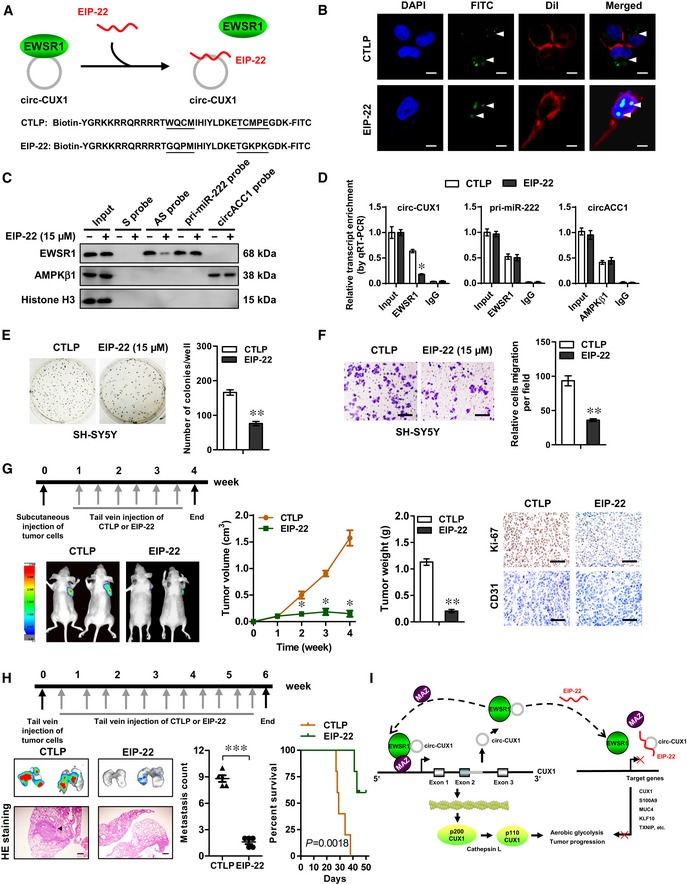
Based on the importance of RRM domain (especially 394–397 or 406–410 aa) of EWSR1 in interacting with circ‐CUX1, we further designed a cell‐penetrating peptide, named as EWSR1 inhibitory peptide of 22 amino acids (EIP‐22), that might potentially block circ‐CUX1‐EWSR1 interaction (Fig 6A). Treatment of SH‐SY5Y cells with EIP‐22 resulted in its obvious aggregation within the nucleus (Fig 6B). Biotin‐labeled peptide pull‐down assay revealed the binding of EIP‐22 to endogenous circ‐CUX1 in SH‐SY5Y cells (Appendix Fig S9B). In addition, EIP‐22 treatment reduced the interaction between circ‐CUX1 and EWSR1, but not that of pri‐miR‐222 and EWSR1 (Ouyang et al, 2017) or circACC1 and AMP‐activated protein kinase beta 1 (AMPKβ1) (Li et al, 2019; Fig 6C and D). Administration of EIP‐22 inhibited the viability, anchorage‐independent growth, and invasion of SH‐SY5Y cells (Appendix Fig S9C, Fig 6E and F), with alteration of circ‐CUX1 downstream gene expression (Appendix Fig S9D). In contrast, EIP‐22 treatment resulted in no significant alteration in the viability of MCF 10A, non‐transformed normal cells with very low circ‐CUX1 expression (Fig 2C and Appendix Fig S9C). Notably, EIP‐22 treatment synergized the suppressing effects of glycolysis inhibitors, 2‐DG and 3‐bromopyruvate (3‐BP) (Cardaci et al, 2012; Zhang et al, 2014), on the viability, growth, and invasion of IMR32 and SH‐SY5Y cells (Appendix Fig S9E–G). Intravenous administration of EIP‐22 significantly reduced the growth, tumor weight, Ki‐67 proliferation index, and CD31‐positive microvessels, altered circ‐CUX1 target gene expression, and decreased the glucose uptake, lactate production, and ATP levels in subcutaneous xenograft tumors formed by injection of SH‐SY5Y cells (Fig 6G and Appendix Fig S10A–C). Moreover, administration of EIP‐22 via tail vein reduced the lung metastatic colonies and prolonged the survival time of athymic nude mice received tail vein injection of SH‐SY5Y cells (Fig 6H). These data suggested that EIP‐22 suppressed NB progression by blocking circ‐CUX1‐EWSR1 interaction.
Figure 6. Inhibitory peptides suppress tumor progression by blocking circ‐CUX1‐EWSR1 interaction.
-
A, BSchematic illustration (A) and distribution (arrowheads) (B) of mutant control (CTLP) or inhibitory (EIP‐22) peptide within SH‐SY5Y cells (at 48 h), with nuclei and cellular membrane stained by DAPI or Dil. Scale bars: 10 μm.
-
C, DWestern blot (C), RIP, and real‐time qRT–PCR (D, n = 5) assays indicating the proteins (EWSR1, AMPKβ1) and transcripts (circ‐CUX1, pri‐miR‐222, circACC1) pulled down by biotin‐labeled sense (S) or antisense (AS) junction probes of circ‐CUX1, and antisense probes of pri‐miR‐222 or circACC1 in SH‐SY5Y cells treated with CTLP or EIP‐22 (15 μM) for 48 h. Student's t‐test, *P < 0.05 versus IgG.
-
E, FSoft agar (E) and Matrigel invasion (F) assays indicating the growth and invasion of SH‐SY5Y cells treated with CTLP or EIP‐22 (15 μM) for 48 h (n = 5). Scale bars: 100 μm. Student's t‐test, **P < 0.01 versus CTLP.
-
G, HRepresentative images, in vivo growth curve, tumor weight, Ki‐67 and CD31 expression of xenograft tumors (G) and lung metastatic colonization, and Kaplan–Meier curves (H) of nude mice (n = 5 for each group) treated with subcutaneous or tail vein injection of SH‐SY5Y cells and CTLP or EIP‐22 (5 mg kg−1). Scale bars: 100 μm. One‐way ANOVA, Student's t‐test, **P < 0.01, ***P < 0.001 versus CTLP. Log‐rank test for survival comparison.
-
ISchematic illustration of circ‐CUX1‐promoted tumor progression: circ‐CUX1 binds to EWSR1 to facilitate its interaction with MAZ, resulting in MAZ transactivation and transcriptional alteration of CUX1 and other genes associated with aerobic glycolysis and tumor progression. An inhibitory peptide blocking circ‐CUX1‐EWSR1 interaction suppresses tumor progression.
Therapeutic lentivirus‐mediated circ‐CUX1 knockdown in vivo
We further explored the therapeutic efficiencies of circ‐CUX1 knockdown on athymic nude mice bearing xenograft tumors formed by subcutaneous or tail vein injection of IMR32 cells. Lentivirus‐mediated knockdown of circ‐CUX1 significantly reduced the growth, tumor weight, Ki‐67 proliferation index, and CD31‐positive microvessels of subcutaneous xenograft tumors (Appendix Fig S11A and B), with altered expression of circ‐CUX1 target genes (Appendix Fig S11C). The glucose uptake, lactate production, and ATP levels were significantly decreased in xenograft tumors of nude mice received tail vein injection of lentivirus carrying sh‐circ‐CUX1 (Appendix Fig S11D). In addition, administration of lentivirus carrying sh‐circ‐CUX1 #1 decreased the lung metastatic colonies and prolonged the survival time of nude mice (Appendix Fig S11E). These results indicated that lentivirus‐mediated circ‐CUX1 knockdown inhibited aerobic glycolysis and NB progression in vivo.
Discussion
Recent studies show that although LDHA and LDHB promote tumorigenicity, they are dispensable for aerobic glycolysis in NB (Dorneburg et al, 2018), suggesting the involvement of other glycolytic genes in this process. So far, interrogative screening of transcriptional regulators of aerobic glycolysis in NB remains unknown. In this study, we identified CUX1 as a transcription factor facilitating the expression of glycolytic genes ENO1, GPI, and PGK1 in NB. We demonstrate that circ‐CUX1 interacts with EWSR1 protein to increase MAZ transactivation, which subsequently regulates the transcription of CUX1 and other genes associated with tumor progression in cis and in trans (Fig 6I), such as S100A9 (Lim et al, 2016), MUC4 (Rowson‐Hodel et al, 2017), KLF10 (Weng et al, 2017), and TXNIP (Shen et al, 2015). The discovery of circ‐CUX1/EWSR1/MAZ axis represents a promising step for therapeutic intervention against tumors.
CUX1 is a transcription factor involved in embryonic development (Michl et al, 2005; Harada et al, 2008) and regulates cellular proliferation, migration, and epithelial‐to‐mesenchymal transition, suggesting its emerging roles in tumorigenesis and aggressiveness (Michl et al, 2005). Elevated CUX1 expression has been documented in many tumors and is associated with poor survival of patients (Liu et al, 2013). Full‐length p200 CUX1 binds rapidly but only transiently to DNA (Liu et al, 2013), while its proteolytic product p110 isoform activates gene transcription (Harada et al, 2008; Kedinger et al, 2009). In this study, our results indicated that CUX1 was an independent prognostic marker for progression and poor outcome of NB. In addition, p110 CUX1 promoted the expression of target genes ENO1, GPI, and PGK1 in NB cells. As a glycolytic enzyme, ENO1 acts as a metabolic tumor promoter by contributing to Warburg effect (Chen et al, 2018). GPI is a housekeeping cytosolic enzyme responsible for catalytic interconversion between glucose‐6‐phosphatase and fructose‐6‐phosphate, and plays a key role in glycolytic pathway (Ždralević et al, 2017). During the glycolytic process, PGK1 contributes to ATP generation and participates in tumor progression (Li et al, 2016). Our gain‐ and loss‐of‐function studies indicated that CUX1 promoted the aerobic glycolysis, growth, and invasiveness of NB cells, suggesting its oncogenic roles in NB progression.
Human CUX1 gene locates at chromosome 7q22, a region associated with copy number gain that contributes to multidrug resistance in NB (Mazzocco et al, 2015). However, we found no alteration of copy number or genetic variants of CUX1 in NB cohorts, indicating other mechanisms facilitating its over‐expression. CircRNAs play important roles in regulating gene expression at post‐transcriptional or transcriptional levels (Hansen et al, 2013; Li et al, 2015b). For example, ciRS‐7 and circSry serve as sponges of miR‐7 and miR‐138 in the cytoplasm (Hansen et al, 2013). Exonic circRNAs also exert regulatory functions in the cytoplasm by forming a ribonucleoprotein complex with miRNA and AGO protein (Lasda & Parker, 2014). Meanwhile, exon–intron circRNAs are predominantly localized in the nucleus and regulate their parent gene expression in a cis‐acting manner through specific RNA–protein interaction (Li et al, 2015b). In this study, circ‐CUX1 was identified as an intron‐containing circRNA up‐regulated in tumor tissues and cells. Circ‐CUX1 enhanced the expression of CUX1 at transcriptional level, and tumor‐promoting functions of circ‐CUX1 were mediated, at least in part, through interacting with EWSR1 protein in NB cells.
As one member of EWS family of RNA‐binding proteins, EWSR1 participates in gene transcription, splicing, and miRNA processing (Luo et al, 2015). Chromosomal translocation of ESWR1 has been discovered in Ewing sarcoma (Sohn et al, 2010). However, our results revealed no EWS‐Fli1 gene fusion in NB cells. Due to lack of DNA‐binding domain, EWSR1 usually acts as a potent transcriptional cofactor in tumor progression via interacting with transcription regulatory proteins, such as CREB‐binding protein and p300 (Chakravarti et al, 1996). In this study, we found that RRM domain of EWSR1 was necessary for its interaction with ZNF domain of MAZ. As a ubiquitously expressed transcription factor, MAZ binds to GC‐rich cis‐elements through its C2H2‐type ZNF motif (Parks & Shenk, 1996) and activates transcription of KRAS and vascular endothelial growth factor (VEGF) in pancreatic cancer, cervical cancer, and glioblastoma cells (Smits et al, 2012; Cogoi et al, 2013). Our evidence indicated that through interplay with its cofactor EWSR1, MAZ regulated the transcription of CUX1, S100A9, MUC4, KLF10, or TXNIP in NB cells. Notably, circ‐CUX1 bound to RRM region of EWSR1, resulting in EWSR1‐mediated MAZ transactivation, suggesting the oncogenic roles of circ‐CUX1/EWSR1/MAZ axis in aerobic glycolysis and tumor progression.
In summary, we demonstrate that elevated CUX1 and its generated circ‐CUX1 are associated with poor outcome of NB patients, and exert oncogenic roles in aerobic glycolysis and tumor progression. Mechanistically, circ‐CUX1 binds to EWSR1 protein to facilitate MAZ transactivation, resulting in transcriptional alteration of CUX1 and other genes associated with NB progression. An inhibitory peptide (EIP‐22) blocking circ‐CUX1‐EWSR1 interaction or lentivirus‐mediated circ‐CUX1 knockdown suppresses the aerobic glycolysis, tumorigenesis, and aggressiveness of NB cells. Combinational administration of EIP‐22 and glycolysis inhibitors (2‐DG or 3‐BP) targeting HK2, GPI, or GAPDH (Cardaci et al, 2012; Zhang et al, 2014) exerts synergistic effects in suppressing growth and aggressiveness of NB cells. Due to limited size of cohort, the prognostic value of circ‐CUX1 and CUX1 and their association with MYCN amplification in NB warrant further investigation. This study extends our knowledge about the regulation of aerobic glycolysis by transcription factor and its generated circRNA, and suggests that circ‐CUX1/EWSR1/MAZ axis may be a potential therapeutic target for NB.
Materials and Methods
Cell culture
Human MCF 10A (CRL‐10317), HeLa (CCL‐2), SH‐SY5Y (CRL‐2266), IMR32 (CCL‐127), SK‐N‐AS (CRL‐2137), BE(2)‐C (CRL‐2268), SK‐N‐MC (HTB‐10), LoVo (CCL‐229), PC‐3 (CRL‐1435), HEK293 (CRL‐1573), and HEK293T (CRL‐3216) cells were obtained from American Type Culture Collection (Rockville, MD), authenticated by short tandem repeat profiling, and used within 6 months after resuscitation of frozen aliquots. Mycoplasma contamination was regularly examined using Lookout Mycoplasma PCR Detection Kit (Sigma, St. Louis, MO). Tumor cells, HEK293, and HEK293T cells were cultured in RPMI1640 supplied with 10% fetal bovine serum (Life Technologies, Inc., Gaithersburg, MD), while MCF 10A cells were cultured in DMEM/F12 medium containing 5% horse serum (Invitrogen, Carlsbad, CA) and 20 ng ml−1 epidermal growth factor (PeproTech, Rocky Hill, NJ) at 37°C, and treated with E64D, actinomycin D (ActD), 2‐DG, or 3‐BP (Sigma).
RT–PCR and real‐time quantitative RT–PCR
Nuclear, cytoplasmic, and total RNA was extracted using RNA Subcellular Isolation Kit (Active Motif, Carlsbad, CA) or RNeasy Mini Kit (Qiagen Inc., Redwood City, CA), with or without RNase R (3 U μg−1, Epicenter, Madison, WI) digestion at 37°C for 15 min. Reverse transcription and real‐time PCR were performed using Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN), SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), and primers (Appendix Table S1). The transcript levels were analyzed by 2−ΔΔCt method. De novo RNA synthesis was blocked by ActD (5 μg ml−1) treatment, while mRNA stability was examined by transcript levels at indicated time points.
Northern blot
The non‐junction and junction probes specific for circ‐CUX1 were synthesized and labeled by digoxigenin (DIG, Appendix Table S2). For Northern blot, 20 μg of total RNA was separated on 3‐(N‐morpholino)propanesulfonic acid‐buffered 2% (w/v) agarose gel containing 1.2% (v/v) formaldehyde under denaturing condition at 80 V for 4 h, and transferred to Hybond‐N+ membrane (Pall Corp., Port Washington, NY). Hybridization was performed at 65°C for 16–18 h in DIG Easy Hyb solution (Roche) and detected by anti‐DIG antibody (1:500 dilution) and chemiluminescence substrate CSPD (Roche).
Western blot
Tissue or cellular protein was extracted with 1× cell lysis buffer (Promega, Madison, WI). Western blot was performed as previously described (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018b), with antibodies (1:500 dilution) specific for CUX1 (sc‐514008, Santa Cruz Biotechnology, Santa Cruz, CA), ENO1 (ab155102), GPI (ab66340), PGK1 (ab38007), EWSR1 (ab93837), ELAVL1 (ab136542), SYNCRIP (ab184946), MAZ (ab85725), MUC4 (ab60720), S100A9 (ab92507), KLF10 (ab73537), TXNIP (ab188865), AMPKβ1 (ab32112), β‐actin (ab8227), Flag (ab18230), Myc (ab9106, Abcam Inc., Cambridge, MA), GST (sc‐33614), or histone H3 (sc‐10809, Santa Cruz Biotechnology).
Gene over‐expression and knockdown
Human circ‐CUX1 linear sequence (393 bp) was obtained from NB tissues by PCR (Appendix Table S2) and inserted into pLCDH‐ciR (Geenseed Biotech Co., Guangzhou, China). Human p200 CUX1 construct was provided by Dr. George Stratigopoulos, while p110 CUX1 was released by digestion or amplified using primers (Appendix Table S2), and subcloned into pcDNA3.1 (Invitrogen) or pCMV‐3Tag‐1A (Addgene, Cambridge, MA). Human EWSR1 cDNA (1,971 bp) and MAZ cDNA (1,482 bp) were provided by Dr. Ralf Janknecht or amplified from NB tissues with primers (Appendix Table S2), and their truncations were subcloned into pCMV‐N‐Myc or pCMV‐3Tag‐1A (Addgene). Mutation of circ‐CUX1 or EWSR1 was prepared with GeneTailor™ Site‐Directed Mutagenesis System (Invitrogen) and primers (Appendix Table S2). Oligonucleotides specific for shRNAs (Appendix Table S3) were inserted into GV298 (GeneChem Co., Ltd, Shanghai, China). Stable cells were screened by neomycin or puromycin (Invitrogen).
Rescue of target gene expression
To rescue circ‐CUX1 knockdown‐altered target gene expression, EWSR1 or MAZ was transfected into stable cell lines. To restore target gene expression altered by circ‐CUX1, shRNAs against EWSR1 or MAZ (Appendix Table S3) were transfected into tumor cells using GeneSilencer Transfection Reagent (Genlantis, San Diego, CA).
Lentivirus packaging
Lentiviral vectors were co‐transfected with packaging plasmids psPAX2 and pMD2G (Addgene) into HEK293T cells. Infectious lentivirus was harvested at 36 and 60 h after transfection, and filtered through 0.45 μm PVDF filters. Recombinant lentivirus was concentrated 100‐fold by ultracentrifugation (2 h at 120,000 g), dissolved in phosphate‐buffered saline (PBS), and injected into mice within 48 h.
RNA‐seq assay
Total RNA of tumor cells (1 × 106) was extracted using TRIzol® reagent (Life Technologies, Inc.). Library preparation and transcriptome sequencing on an Illumina HiSeq X Ten platform were carried out at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Fragments per kilobase of transcript per million fragments mapped (FPKM) of each gene were calculated.
Dual‐luciferase reporter assay
Complementary oligonucleotides containing four canonical binding sites of CUX1 or MAZ, and promoter fragments of ENO1 (−1,880/+301), GPI (−1,854/+247), PGK1 (−882/+246), or p200 CUX1 (−2,084/+106) amplified from genomic DNA (Appendix Table S2) were subcloned into pGL3‐Basic (Promega). Mutation of MAZ‐binding site was performed with GeneTailor™ Site‐Directed Mutagenesis System (Invitrogen) and primers (Appendix Table S2). To test specificity of shRNA, target sequences of circ‐CUX1 and CUX1 were amplified using primers (Appendix Table S2) and subcloned into 3′‐untranslated region of Renilla luciferase within psiCHECK2 (Promega). Dual‐luciferase assay was performed as previously described (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018b).
RNA pull‐down and mass spectrometry
Biotin‐labeled oligonucleotide probes targeting junction sites of circRNAs were synthesized (Invitrogen). Linear circ‐CUX1 was in vitro transcribed using Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase, incubated with guide oligonucleotides (Appendix Table S2), circularized using T4 RNA ligase I, treated with RNase R, and purified with RNeasy Mini Kit (Qiagen Inc.). RNA pull‐down was performed as previously described (Li et al, 2018b). Retrieved protein was detected by Western blot or mass spectrometry (Wuhan Institute of Biotechnology, Wuhan, China), while recovered transcripts were measured by RT–PCR using primers (Appendix Table S1). In ChIRP assay, cells were harvested, cross‐linked, sonicated, hybridized with probes, and mixed with streptavidin magnetic beads (Chu & Chang, 2016). The retrieved DNA was detected by PCR using primers (Appendix Table S1).
RNA‐FISH
Biotin‐labeled antisense or sense probe for circ‐CUX1 junction was synthesized (Appendix Table S2). The probes for GAPDH and U1 were generated by in vitro transcription of PCR products (Appendix Table S1) using DIG Labeling Kit (MyLab Corporation, Beijing, China). Cells were incubated with 40 nM FISH probe in hybridization buffer (100 mg ml−1 dextran sulfate, 10% formamide in 2 × SSC) at 37°C for 16 h, with or without RNase R (3 U μg−1) treatment. The signals of circ‐CUX1 were detected by Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China), with nuclei staining by 4′,6‐diamidino‐2‐phenylindole (DAPI).
Fluorescence immunocytochemical staining
Tumor cells were grown on coverslips, incubated with antibodies specific for EWSR1 (ab93837; Abcam Inc.; 1:100 dilution) at 4°C overnight, and treated with Alexa Fluor 594 goat anti‐rabbit IgG (1:1,000 dilution) and DAPI (300 nM) staining. The images were photographed under a Nikon A1Si Laser Scanning Confocal Microscope and applied for 3D reconstruction using NIS‐Elements Viewer (Nikon Instruments Inc., Japan).
Co‐immunoprecipitation (co‐IP)
Co‐IP was performed as previously described (Jiao et al, 2018; Li et al, 2018b), with antibodies (1:200 dilution) specific for EWSR1 (sc‐28327, Santa Cruz Biotechnology), MAZ (ab85725), Flag (ab18230), or Myc (ab9106, Abcam Inc.). Bead‐bound proteins were released and analyzed by Western blot.
Bimolecular fluorescence complementation system (BiFC)
Human EWSR1 cDNA (1,971 bp) and MAZ cDNA (1,482 bp) were subcloned into BiFC vectors pBiFC‐VN173 and pBiFC‐VC155 (Addgene), and co‐transfected into tumor cells for 24 h. The fluorescence emission was observed under a confocal microscope, with excitation and emission wavelengths of 488 and 500 nm, respectively (Kerppola, 2008).
Chromatin immunoprecipitation (ChIP)
ChIP assay was undertaken using EZ‐ChIP kit (Upstate Biotechnology, Temecula, CA) (Zhao et al, 2016; Li et al, 2018b), with antibodies (1:100 dilution) specific for CUX1 (#81557, Cell Signaling Technology, Inc., Danvers, MA) or MAZ (ab85725, Abcam Inc.) and primers targeting gene promoters (Appendix Table S1).
Cross‐linking RNA immunoprecipitation (RIP)
Cells (1 × 108) were ultraviolet light cross‐linked at 254 nm (200 J cm−2). RIP assay was performed using Magna RIP™ RNA‐Binding Protein Immunoprecipitation Kit (Millipore) (Zhao et al, 2016; Li et al, 2018b), with antibodies (1:100 dilution) specific for AGO2 (ab186733, Abcam Inc.), EWSR1 (#11910, Cell Signaling Technology, Inc.), GST (sc‐33614, Santa Cruz Biotechnology), or AMPKβ1 (ab32112, Abcam Inc.). Co‐precipitated RNAs were detected by RT–PCR or real‐time qRT–PCR with specific primers (Appendix Table S1).
In vitro binding assay
A series of EWSR1 truncations were amplified with primers (Appendix Table S2), subcloned into pGEX‐6P‐1 (Addgene), and transformed into E. coli to produce GST‐tagged EWSR1 protein (Zhao et al, 2016; Li et al, 2018b). The EWSR1‐circRNA complexes were pulled down using GST beads (Sigma). Protein was detected by SDS–PAGE and Western blot, while circRNA was measured by RT–PCR with specific primers (Appendix Table S1).
RNA EMSA
Biotin‐labeled circ‐CUX1 probe was synthesized as described above. RNA EMSA using nuclear extracts was performed using LightShift Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific, Inc., Waltham, MA).
Design and synthesis of inhibitory peptides
Based on interacting region of EWSR1 revealed by mutagenesis and in vitro binding assays, wild‐type and mutant inhibitory peptides blocking circ‐CUX1 and EWSR1 interaction were designed and synthesized by linking with biotin‐labeled 11 amino acid cell‐penetrating peptide (YGRKKRRQRRR) from Tat protein transduction domain at the N‐terminus and conjugating with fluorescein isothiocyanate (FITC) at the C‐terminus (ChinaPeptides Co. Ltd, Shanghai, China), with purity larger than 95%.
Biotin‐labeled peptide pull‐down
Total RNA was isolated using RNeasy Mini Kit (Qiagen Inc.) and incubated with biotin‐labeled peptide at 4°C overnight. Then, incubation of RNA‐peptide complex with streptavidin‐agarose was undertaken at 4°C for 2 h. Beads were extensively washed, and circRNAs pulled down were measured by real‐time qRT–PCR.
Aerobic glycolysis and seahorse extracellular flux assays
Cellular glucose uptake, lactate production, and ATP levels were detected as previously described (Ma et al, 2014). ECAR and OCR were measured in XF media under basal conditions and in response to glucose (10 mM), oligomycin (2 μM), and 2‐deoxyglucose (50 mM) using a Seahorse Biosciences XFe24 Flux Analyzer (North Billerica, MA).
In vitro cell viability, growth, and invasion assays
The 2‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT; Sigma) colorimetric (Li et al, 2015a, 2018b), soft agar (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018a,b), and Matrigel invasion (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018a,b) assays were undertaken to measure in vitro viability, growth, and invasive capabilities of tumor cells.
In vivo growth, metastasis, and therapeutic assays
All animal experiments were carried out in accordance with NIH Guidelines for the Care and Use of Laboratory Animals, and approved by the Animal Care Committee of Tongji Medical College (approval number: Y20080290). For in vivo tumor growth and experimental metastasis studies, tumor cells (1 × 106 or 0.4 × 106) were injected into dorsal flanks or tail vein of blindly randomized 4‐week‐old female BALB/c nude mice (National Rodent Seeds Center, Shanghai, China) breeding at specific pathogen free (SPF) condition (n = 5 per group) (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018a,b). For in vivo therapeutic studies, tumor cells (1 × 106 or 0.4 × 106) were injected into dorsal flanks or tail vein of nude mice, respectively. One week later, mice were blindly randomized and treated by tail vein injection of synthesized cell‐penetrating peptide (ChinaPeptides, Shanghai, China) or lentivirus (1 × 107 plaque‐forming units) as indicated. The in Vivo Optical Imaging System (In‐Vivo FX PRO, Bruker Corporation, Billerica, MA) was applied to acquire fluorescent images of xenograft tumors in nude mice.
Patient tissue samples
Human tissue study was approved by the Institutional Review Board of Tongji Medical College (approval number: 2011‐S085). All procedures were conformed to principles set forth by Declaration of Helsinki and Department of Health and Human Services Belmont Report. Written informed consent was obtained from all patients without preoperative chemotherapy or other treatment. Fresh tumor tissues were collected at surgery, validated by pathological diagnosis, and stored at −80°C. Total RNAs of normal fetal adrenal medulla were purchased from Clontech (Mountain View, CA).
Immunohistochemistry
Immunohistochemical staining and quantitative evaluation were performed as previously described (Zhang et al, 2012; Zhao et al, 2016; Li et al, 2018a,b), with antibodies specific for Ki‐67 (1:500, sc‐23900, Santa Cruz Biotechnology) or CD31 (1:500, ARG52748, Arigo, Hsinchu City, Taiwan). The degree of positivity was blindly assessed by at least two pathologists.
Statistical analysis
All data were shown as mean ± standard error of the mean (SEM). Cutoff of gene expression was defined by average values. Two‐sided unpaired Student's t‐test and one‐way ANOVA were used to compare difference. Pearson's correlation coefficient assay was used to analyze expression correlation. Log‐rank test and Cox regression models were used to assess survival difference and hazard ratio. All statistical tests were considered significant when P < 0.05. Randomization and blinding strategies were used whenever possible. Experimental sample size was determined on the basis of power analysis assuming a significance level (alpha) of 0.05 and a power of 80%. Animal cohort sizes were determined on the basis of similar studies. The exact P‐values and number of replicates were indicated in Appendix Table S4.
Author contributions
Conception and design: QT and LZ; Methodology and resources: HL, FY, XW, EF, and DL; Acquisition of data: HL, FY, AH, XW, EF, YC, DL, HS, JW, YG, YL, and HJL; Supervision: QT, LZ, and KH; Writing the manuscript: HL, FY, QT, and LZ.
Conflict of interest
The authors declare that they have no conflict of interest.
The paper explained.
Problem
Neuroblastoma (NB) is the most common extracranial tumor in childhood. Although countless efforts have been made to improve the therapeutic efficiency, the outcome of patients suffering from high‐risk NB still remains poor. Aerobic glycolysis is a hallmark of metabolic reprogramming that contributes to tumor progression. Further in‐depth investigation of mechanisms regulating aerobic glycolysis during NB progression is vital for resolution of these issues.
Results
By integrating analysis of public datasets, we identify that CUX1 and CUX1‐generated circular RNA (circ‐CUX1) facilitate aerobic glycolysis and NB progression. Mechanistically, transcription factor p110 CUX1, a proteolytic product of p200 CUX1, promotes expression of glycolytic genes, while circ‐CUX1 facilitates EWSR1‐mediated MAZ transactivation to alter expression of p200 CUX1 and other genes associated with tumor progression. High expression of circ‐CUX1 or p200 CUX1 is associated with poor outcome of NB patients. Administration of an inhibitory peptide blocking circ‐CUX1‐EWSR1 interaction or lentivirus mediating circ‐CUX1 knockdown suppresses aerobic glycolysis, growth, and aggressiveness of NB cells.
Impact
Our results extend the knowledge about regulation of aerobic glycolysis by transcription factor and its generated circRNA. First of all, transcription factor CUX1 is essential for glycolytic gene expression during NB progression. Secondly, EWSR1 interacts with MAZ to facilitate its transactivation and regulate downstream gene expression. Thirdly, circ‐CUX1 facilitates the interaction of EWSR1 with MAZ. Finally, blocking circ‐CUX1‐EWSR1 interaction might be a novel therapeutic strategy for NB and other tumors.
For more information
Publically available datasets can be found
(i)GEO database (GSE16476, GSE62564, GSE41258, GSE5851, GSE6956, https://www.ncbi.nlm.nih.gov/geo).
(ii)The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov).
(iii) Kaplan–Meier plotter (http://kmplot.com).
(iv)Oncogenomics (https://pob.abcc.ncifcrf.gov/cgi-bin/JK).
(v)cBioPortal for Cancer Genomics (http://cbioportal.org).
(vi)Tumor alterations relevant for genomics‐driven therapy (TARGET, https://software.broadinstitute.org/cancer/cga/target).
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Acknowledgements
We appreciate Drs. George Stratigopoulos and Ralf Janknecht for providing vectors. This work was granted by the National Natural Science Foundation of China (Grants # 81472363, 81572423, 81672500, 81773094, 81772967, 81874085, 81874066, 81802925, 81903011, 81903008), and Fundamental Research Funds for the Central Universities (Grant # 2019kfyRCPY032).
EMBO Mol Med (2019) 11: e10835
Contributor Information
Liduan Zheng, Email: ld_zheng@hotmail.com.
Qiangsong Tong, Email: qs_tong@hotmail.com.
Data availability
RNA‐seq data have been deposited in Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo), under accession number GSE136135.
References
- Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG (2013) catRAPID omics: a web server for large‐scale prediction of protein‐RNA interactions. Bioinformatics 29: 2928–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak M, Vidal M, Nakano K, Bartrons R, Gottlieb E, Vousden K (2006) TIGAR, a p53‐inducible regulator of glycolysis and apoptosis. Cell 126: 107–120 [DOI] [PubMed] [Google Scholar]
- Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW et al (2010) A screen of chemical modifications identifies position‐specific modification by UNA to most potently reduce siRNA off‐target effects. Nucleic Acids Res 38: 5761–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM (2003) Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 3: 203–216 [DOI] [PubMed] [Google Scholar]
- Cardaci S, Desideri E, Ciriolo MR (2012) Targeting aerobic glycolysis: 3‐bromopyruvate as a promising anticancer drug. J Bioenerg Biomembr 44: 17–29 [DOI] [PubMed] [Google Scholar]
- Chakravarti DLV, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM (1996) Role of CBP/P300 in nuclear receptor signalling. Nature 383: 99–103 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang Y, Wang H, Zeng YY, Li Z, Li ML, Li FF, You J, Zhang ZM, Tzeng CM (2018) WW domain‐binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma. Cell Death Dis 9: 347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Chang HY (2016) Understanding RNA‐chromatin interactions using chromatin isolation by RNA purification (ChIRP). Methods Mol Biol 1480: 115–123 [DOI] [PubMed] [Google Scholar]
- Cogoi S, Zorzet S, Rapozzi V, Geci I, Pedersen EB, Xodo LE (2013) MAZ‐binding G4‐decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res 41: 4049–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorneburg C, Fischer M, Barth TFE, Mueller‐Klieser W, Hero B, Gecht J, Carter DR, de Preter K, Mayer B, Christner L et al (2018) LDHA in neuroblastoma is associated with poor outcome and its depletion decreases neuroblastoma growth independent of aerobic glycolysis. Clin Cancer Res 24: 5772–5783 [DOI] [PubMed] [Google Scholar]
- Gillingham A, Pfeifer A, Munro S (2002) CASP, the alternatively spliced product of the gene encoding the CCAAT‐displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol Biol Cell 13: 3761–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20: 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet B, Watson P, Poirier M, Leduy L, Bérubé G, Meterissian S, Jolicoeur P, Nepveu A (2002) Characterization of a tissue‐specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res 62: 6625–6633 [PubMed] [Google Scholar]
- Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A (2004) A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell 14: 207–219 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- Harada R, Vadnais C, Sansregret L, Leduy L, Berube G, Robert F, Nepveu A (2008) Genome‐wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res 36: 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W, Chen Y, Song H, Li D, Mei H, Yang F, Fang E, Wang X, Huang K, Zheng L et al (2018) HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene 37: 2728–2745 [DOI] [PubMed] [Google Scholar]
- Kedinger V, Sansregret L, Harada R, Vadnais C, Cadieux C, Fathers K, Park M, Nepveu A (2009) p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E‐cadherin and occludin. J Biol Chem 284: 27701–27711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK (2008) Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol 85: 431–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A (2010) ChEA: transcription factor regulation inferred from integrating genome‐wide ChIP‐X experiments. Bioinformatics 26: 2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mei H, Pu J, Xiang X, Zhao X, Qu H, Huang K, Zheng L, Tong Q (2015a) Intelectin 1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up‐regulation of N‐myc downstream regulated gene 2. Mol Cancer 14: 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L et al (2015b) Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264 [DOI] [PubMed] [Google Scholar]
- Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T et al (2016) Mitochondria‐translocated phosphoglycerate kinase 1 functions as a protein kinase to coordinate glycolysis and TCA cycle in tumorigenesis. Mol Cell 61: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Song H, Mei H, Fang E, Wang X, Yang F, Li H, Chen Y, Huang K, Zheng L et al (2018a) Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat Commun 9: 2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang X, Mei H, Fang E, Ye L, Song H, Yang F, Li H, Huang K, Zheng L et al (2018b) Long noncoding RNA pancEts‐1 promotes neuroblastoma progression through hnRNPK‐mediated β‐catenin stabilization. Cancer Res 78: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, Thorne RF, Zhang XD, Hu W, Wu M (2019) CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab 30: 157–173 [DOI] [PubMed] [Google Scholar]
- Lim SY, Yuzhalin AE, Gordon‐Weeks AN, Muschel RJ (2016) Tumor‐infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene 35: 5735–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KC, Lin BS, Zhao M, Wang KY, Lan XP (2013) Cutl1: a potential target for cancer therapy. Cell Signal 25: 349–354 [DOI] [PubMed] [Google Scholar]
- Luo Y, Blechingberg J, Fernandes AM, Li S, Fryland T, Børglum AD, Bolund L, Nielsen AL (2015) EWS and FUS bind a subset of transcribed genes encoding proteins enriched in RNA regulatory functions. BMC Genom 16: 929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Li C, Sun L, Huang D, Li T, He X, Wu G, Yang Z, Zhong X, Song L et al (2014) Lin28/let‐7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun 5: 5212 [DOI] [PubMed] [Google Scholar]
- Mazzocco K, Defferrari R, Sementa AR, Garaventa A, Longo L, De Mariano M, Esposito MR, Negri F, Ircolò D, Viscardi E et al (2015) Genetic abnormalities in adolescents and young adults with neuroblastoma: a report from the Italian Neuroblastoma Group. Pediatr Blood Cancer 62: 1725–1732 [DOI] [PubMed] [Google Scholar]
- Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D'Arrigo C, Ryder K, Menke A, Gress T et al (2005) CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell 7: 521–532 [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J et al (2012) Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483: 589–593 [DOI] [PubMed] [Google Scholar]
- Ouyang H, Zhang K, Fox‐Walsh K, Yang Y, Zhang C, Huang J, Li H, Zhou Y, Fu XD (2017) The RNA binding protein EWS is broadly involved in the regulation of pri‐miRNA processing in mammalian cells. Nucleic Acids Res 45: 12481–12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks CL, Shenk T (1996) The serotonin 1a receptor gene contains a TATA‐less promoter that responds to MAZ and Sp1. J Biol Chem 271: 4417–4430 [DOI] [PubMed] [Google Scholar]
- Petkovic S, Muller S (2015) RNA circularization strategies in vivo and in vitro . Nucleic Acids Res 43: 2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez‐Miranda LR, Memczak S, Wolf SA, Rybak‐Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P et al (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: eaam8526 [DOI] [PubMed] [Google Scholar]
- Rowson‐Hodel AR, Wald JH, Hatakeyama J, O'Neal WK, Stonebraker JR, VanderVorst K, Saldana MJ, Borowsky AD, Sweeney C, Carraway KL III (2017) Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene 37: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, O'Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL, Welm AL, Ayer DE (2015) Metabolic reprogramming in triple‐negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci USA 112: 5425–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla‐Favera R, Dang CV (1997) c‐Myc transactivation of LDH‐A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits M, Wurdinger T, van het Hof B, Drexhage JA, Geerts D, Wesseling P, Noske DP, Vandertop WP, de Vries HE, Reijerkerk A (2012) Myc‐associated zinc finger protein (MAZ) is regulated by miR‐125b and mediates VEGF‐induced angiogenesis in glioblastoma. FASEB J 26: 2639–2647 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Li H, Reidy K, Beers LF, Christensen BL, Lee SB (2010) EWS/FLI1 oncogene activates caspase 3 transcription and triggers apoptosis in vivo . Cancer Res 70: 1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng CC, Hawse JR, Subramaniam M, Chang VHS, Yu WCY, Hung WC, Chen LT, Cheng KH (2017) KLF10 loss in the pancreas provokes activation of SDF‐1 and induces distant metastases of pancreatic ductal adenocarcinoma in the KrasG12D p53flox/flox model. Oncogene 36: 5532–5543 [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y, Wu M (2014) Reciprocal regulation of HIF‐1α and lincRNA‐p21 modulates the Warburg effect. Mol Cell 53: 88–100 [DOI] [PubMed] [Google Scholar]
- Ždralević M, Marchiq I, de Padua MMC, Parks SK, Pouysségur J (2017) Metabolic plasiticy in cancers‐distinct role of glycolytic enzymes GPI, LDHs or membrane transporters MCTs. Front Oncol 7: 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, Huang K, Zheng L, Tong Q (2012) MicroRNA‐145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia‐inducible factor 2 alpha. Oncogene 33: 387–397 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51: 792–806 [DOI] [PubMed] [Google Scholar]
- Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y (2014) 2‐Deoxy‐D‐glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett 355: 176–183 [DOI] [PubMed] [Google Scholar]
- Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, Tong Q (2016) CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene 35: 3565–3576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Data Availability Statement
RNA‐seq data have been deposited in Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo), under accession number GSE136135.


