Abstract
Pigmented viral plaque is most commonly seen in Pug dogs in association with canine papillomavirus (CPV). In the present study, nucleic acid sequence and localization of viral genes were examined in 4 cases of pigmented viral plaque in Pug dogs. The results of polymerase chain reaction and nucleic acid sequence analysis showed that the 3 cases with pigmented viral plaque were infected with CPV4, and 1 case with CPV18. In the case with CPV18-positive viral plaque, CPV18 gene was also detected in a lesion of cytokeratin-14- and P63-positive basal cell tumor that developed adjacent to a pigmented viral plaque. Moreover, CPV gene was detected in the squamous cells of pigmented viral plaques and the neoplastic cells of basal cell tumor by in situ hybridization. This is the first report of basal cell tumor associated with CPV18-infection in the dog. Infection of CPV18 may be associated with development of basal cell tumor.
Keywords: basal cell tumor, canine papillomavirus, in situ hybridization, pigmented viral plaque
Papillomavirus is small, non-enveloped, double-stranded DNA virus with a circular genome of about 8,000 base pairs (bp) that includes the early (E) and late (L) regions. During the process of infection, E6 and E7 proteins play a role in keratinocyte differentiation, and promote replication of suprabasal cells, accordingly resulting in amplification of the viral genome. Papillomavirus frequently infects the squamous epithelium in many mammalian, and some avian and reptilian species [12]. The virus is associated with the occurrence of benign and malignant neoplasia not only in humans but also in animals, although asymptomatic infections are more common [1, 4, 7, 12, 16, 19]. Canine papillomavirus (CPV) is divided into 19 genotypes and classified into three genera as Lambda, Tau and Chi [2, 7, 20]. CPV may be possibly involved in the onset of canine oral papillomatosis, cutaneous papilloma and pigmented viral plaque in dogs [7, 9]. Pigmented viral plaque is a proliferative lesion of the skin, and is histopathologically characterized by acanthosis and hyperkeratosis of the epidermis with hyperpigmentation. The lesion is most commonly observed in Pug dogs in association with CPV4 infection. Other types of CPV (CPV3, 5, 8, 9, 10, 11, 12, 14, 16 and 18) have also been detected in pigmented viral plaque lesions [6, 8, 15, 17,18,19]. The fact that Pug dogs tend to develop pigmented viral plaque implicates two possibilities in its pathogenesis: host-specific response to the viral infection and/or viral tropism to Pug dogs [14, 21].
There has been no report about neoplastic changes originated from pigmented viral plaque in dogs. The localization of viral nucleic acid has neither been studied in the lesion. In the present study, 4 dogs with pigmented viral plaque were examined for CPV genotype and viral localization in the lesions. In one dog, a neoplastic lesion developed in association with pigmented viral plaque.
MATERIALS AND METHODS
Cases and sample collection
Tissue samples from 4 cases with pigmented viral plaque in Pug dogs are summarized in Table 1. The skin lesions were observed in the forelimb (2 cases), lower abdomen (1 case) and nipple (1 case). In case 4, multiple tumor masses between the digits of the left forelimb were also examined. As a negative control, tissue sample of basosquamous carcinoma (Yorkshire Terrier, 11 years old) was used. All the samples were histopathologically examined and diagnosed at the Laboratory of Veterinary Pathology, the University of Tokyo.
Table 1. Case information and results of papillomavirus detection.
| Case no. | Sample no. | Age | Gender | Location of skin lesions | Histopathological diagnosis of lesions | Papillomavirus detection |
Sequence results (Accession #) |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| IHC | PCR | ISH |
||||||||
| CPV4 | CPV18 | |||||||||
| 1 | (15-838) | 11y | SF | Left forelimb | Pigmented viral plaque | + | + | + | - | CPV4 (LC489227) |
| 2 | (16-118) | 7y | CM | Right lower abdomen | Pigmented viral plaque | + | + | + | - | CPV4 (LC489228) |
| 3 | (16-212) | 11y | SF | Nipple | Pigmented viral plaque | + | + | + | - | CPV4 (LC489229) |
| 4 | A (18-283-1) | 11y | CM | Left forelimb | Pigmented viral plaque | + | + | - | + | CPV18 (LC489230) |
| B (18-283-2) | Between digits of left forelimb | Basal cell tumor | + | + | - | + | CPV18 (LC489231) | |||
| Negative control | 12y | CM | Left forelimb | Basosquamous carcinoma | - | - | - | - | NA | |
SF, spayed female; CM, castrated male; IHC, immunohistochemistry; PCR, polymerase chain reaction; ISH, in situ hybridization; CPV, canine papillomavirus; +, positive; -, negative; NA, not applicable.
Histopathological and immunohistochemical analyses
Four µm-thick formalin-fixed, paraffin-embedded (FFPE) sections were deparaffinized, and stained with hematoxylin and eosin (HE). Histopathologic diagnosis was performed by 2 Japanese College of Veterinary Pathologists (JCVP)-certified veterinary pathologists (K.U. and J.K.C.). The neoplastic lesion in case 4 was diagnosed through the Classification of Epithelial Tumors of the Skin, Surgical Pathology of Tumors of Domestic Animals [5]. Primary antibodies used for immunohistochemistry (IHC) were summarized in Table 2. Deparaffinized sections were subjected to antigen retrieval. Namely, non-specific reactions were blocked by immersing the sections in 3% hydrogen peroxide in methanol at room temperature for 5 min, followed by incubation in 8% skimmed milk at 37°C for 40 min. The sections were then incubated at 4°C overnight with one of the primary antibodies. After washed 3 times with Tris-buffered saline (TBS), the sections were incubated with Dako EnVision+System horseradish peroxidase-labedled anti-mouse or anti-rabbit secondary antibody polymers (Dako, Tokyo, Japan) at 37°C for 40 min. To visualize reaction products, sections were subjected to chromogen treatment with 0.05% 3,3′-diaminobenzidine and 0.03% hydrogen peroxide in Tris-hydrochloric buffer, and counterstained with hematoxylin. In the IHC detection of canine papillomavirus, a known CPV-associated cutaneous papilloma section was used as a positive control. Normal skin tissues were used as positive controls for cytokeratin 14 and p63 stainings. For negative controls, primary antibodies were replaced with TBS.
Table 2. Primary monoclonal antibodies used for immunohistochemistry.
| Antibody to | Monoclonal type (clone) | Dillution | Source |
|---|---|---|---|
| Papillomavirus | Mouse (BPV1/1H8+CAMVIR) | 1:80 | Abcam, Cambridge, MA, U.S.A. |
| p63 | Mouse (BC4A4) | 1:100 | Biocare Medical, Concord, CA, U.S.A. |
| Cytokeratin14 | Mouse (LL002) | 1:50 | Leica Biosystems, Milton Keynes, U.K. |
| Ki-67 | Mouse (MIB-1) | Ready to use | Dako, Tokyo, Japan |
PCR amplification and sequence assay
DNA was extracted from FFPE tissues with QIAamp DNA FFPE Tissue Kit (QIAGEN GmbH, Hilden, Germany) [13]. The DNA of canine papillomavirus type 2, extracted from a cutaneous papilloma lesion (Dachshund, 14 years old), was used as a positive control. L1 gene of papillomavirus (389 bp) was amplified using canPVf/FAP64 primer set (5′-CTTCCTGAWCCTAAYMAKTTTGC-3′; 5′-CCWATATCWVHCATNTCNCCATC-3′) [9, 10]. For CPV18 analysis, two more CPV18- specific primer sets to amplify the L1 genome (1F/1R 5′-TCAACCGCCCTTATTGGCTT-3′; 5′-CCTGGAGCCCGATATTCCAC-3′ and 2F/2R 5′-ACTTGGTGAGCACTGGTCTG-3′; 5′-AAGCCAATAAGGGCGGTTGA-3′) were used, and the amplified products of which were 300 and 451 bp, respectively. PCR reactions were performed using C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, U.S.A.). All amplifications were performed using KOD Fx Neo kit (Toyobo, Tokyo, Japan), according to the instructions. Amplification conditions were 94°C for 2 min, 98°C for 10 sec, an appropriate annealing temperature for each primer sets for 30 sec and 68°C for 15 sec by 44 cycles with an infinite holding at 20°C. The annealing temperature for canPVf/FAP64 and CPV18- specific primer sets were 50°C and 60°C, respectively. PCR products were electrophoresed on a 2% agarose gel and detected by UV light irradiation. The extracted products from the gel were cloned using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA, U.S.A.), and sequenced using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, U.S.A.). The obtained sequences were compared with the known sequences in the GenBank through the BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For sequence analysis, SnapGene software (Version 2.6.2; SnapGene, GSL Biotech, Chicago, IL, U.S.A.) was used.
In situ hybridization
In situ hybridization was performed to detect papillomaviral DNA on tissue sections. Digoxigenin DNA Labeling Kit (Roche Applied Science GmbH, Penzberg, Germany) was used to produce hybridization probes by labeling PCR products (primer pair canPVf/FAP64) of each case (case 1, 2, 3 and 4). ISH HRP detection kit (Biocare medical, Pacheco, CA, U.S.A.) was used to detect the probe as follows. After pronase and autoclave-pretreatment in citrate buffer, DNA was denatured by heating the sections in boiling water for 10 min and quickly chilled at 4°C. Then the sections were hybridized with each one of the Digoxigenin-labeled DNA probes at 37°C for 20 hr. The reaction was stopped by keeping the sections at 65°C for 10 min. After washing, the sections were incubated with mouse anti-digoxigenin antibody at 4°C overnight, and then incubated with Dako EnVision+System horseradish peroxidase-labeled anti-mouse secondary antibody polymer (Dako) at 37°C for 40 min. To visualize the reaction products, sections were subjected to a chromogen treatment with 0.05% 3, 3′-diaminobenzidine and 0.03% hydrogen peroxide in Tris-hydrochloric buffer, and finally counterstained with hematoxylin. Probes produced from the PCR products of case 2 and case 4 did not show any cross reactivity to each other, therefore they were used for detecting CPV4 and CPV18, respectively.
RESULTS
Gross and histopathological findings
Histopathological lesions of pigmented plaque in the present 4 dogs were characterized by acanthosis and hyperkeratosis of the epidermis with hyperpigmentation (Fig. 1A). Koilocytes (cytoplasmic clearing) and nuclear inclusions were sporadically observed (Fig. 1B).
Fig. 1.
(A) Pigmented viral plaque. Acanthosis and hyperkeratosis of the epidermis with hyperpigmentation (case 1). HE. Bar, 50 µm. (B) Pigmented viral plaque. Intranuclear viral inclusion in the epidermis (arrow, inset) (case 2). HE. Bar, 25 µm. (C) Basal cell tumor and pigmented viral plaque. A black nodule (arrow) between digits of the left forelimb adjacent to pigment viral plaque lesion (arrow head) (case 4). (D) Basal cell tumor and pigmented viral plaque. A tumor mass with a distinct margin in the dermis and subcutaneous (basal cell tumor; arrow). Arrowhead indicates adjacent pigmented viral plaque lesion (case 4). HE. Bar, 1 cm. (E) Basal cell tumor. Lobules of round basaloid tumor cells separated by fibrous tissues (case 4). HE. Bar, 50 µm. (F) Basal cell tumor. Mitotic figures (arrows) and pigments within the neoplastic tissue (case 4). HE. Bar, 25 µm.
In one dog (case 4), a black nodule of 2 cm in diameter was found adjacent to pigmented plaque lesions between digits of left forelimb (Fig. 1C). Histopathologically, the nodule had a distinct margin, and were composed of neoplastic epithelial cells which formed cystic and lobular patterns (Fig. 1D). Neoplastic cells were round-shaped and had an indistinct cellular margin, giving a basal cell-appearance without showing any adnexal differentiation (Fig. 1E). The number of mitotic figures was 3 per 10 high power-fields. Melanin pigments were often observed within the neoplastic cells (Fig. 1F). Koilocytes and nuclear inclusions were not observed. Based on these findings, the nodule was diagnosed as basal cell tumor.
Immunohistochemistry
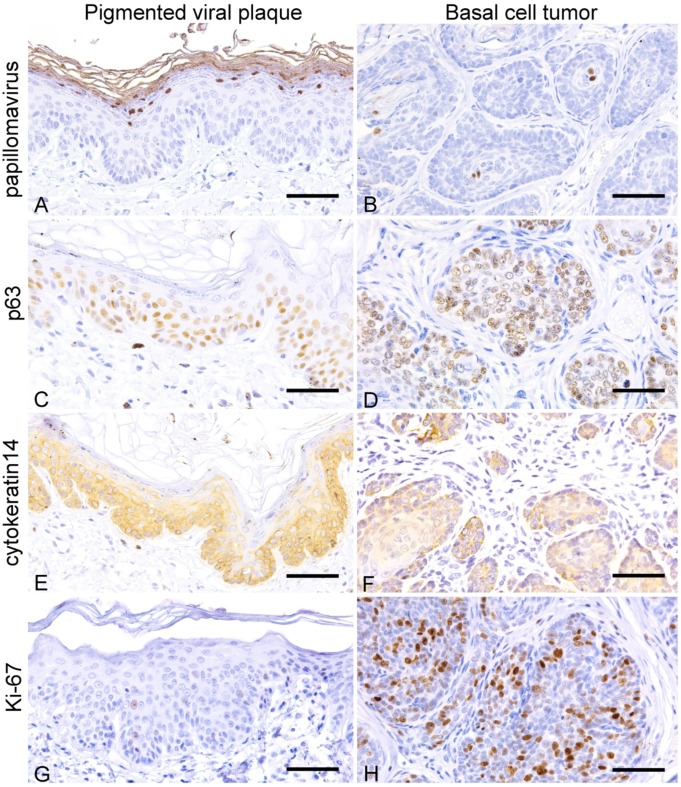
Papillomavirus antigen was immunohistochemically detected in the nucleus of epithelial cells in the granular layer of pigmented viral plaque lesions (Table 1, Fig. 2A). In the lesion of basal cell tumor, papillomavirus antigen was detected in the nucleus of epithelial tumor cells in the center of the neoplastic lobules (Fig. 2B). Thickened basal cell layer of the pigmented viral plaque lesions of all cases and neoplastic cells of basal cell tumor lesion in case 4 were positive for p63 and cytokeratin 14 (Fig. 2C–F). The number of Ki-67-positivite cells was higher in the neoplastic lesion compared to pigmented viral plaque lesions of cases 1, 2, 3 and 4 (Fig. 2G and 2H).
Fig. 2.
Pigmented viral plaque: A, C, E and G; basal cell tumor: B, D, F and H. (A) Nuclei of epithelial cells in the stratum granulosum are positive for papillomavirus antigen (case 2). IHC. (B) Tumor cells in the center of neoplastic lobules are positive for canine papillomavirus antigen (case 4). IHC. (C) The basal layer is thickened with the proliferation of p63-positive basal cells (case 2). IHC. (D) Nuclei of tumor cells are positive for p63 (case 4). IHC. (E) The basal layer is thickened with the proliferation of cytokeratin14-positive basal cells (case 2). IHC. (F) Tumor cells are positive for cytokeratin 14 (case 4). IHC. (G) Few Ki-67-positive cells in the basal layer (case 2). IHC. (H) Many tumor cells are positive for ki-67 (case 4). IHC. Bars, 50 µm.
PCR and sequence analyses
By using the primer sets for detecting canine papilloma virus (canPVf/FAP64), a single positive band was observed through electrophoresis of PCR products, in all ceses (Table 1). The sequences of the PCR products amplified with the canPVf/FAP64 primer set were 99% identical in case 1, 100% in cases 2 and 3, to the reference sequence of CPV4 (GenBank accession number: EF584537.1), and 100% in case 4, to the reference sequence of CPV18 (GenBank accession number: KT326919.1). Moreover, in case 4, the sequencing results were also confirmed with the CPV18- specific primer set 1F/R and 2F/R, were 99 and 100% identical to the reference sequence of CPV18.
In situ hybridization
By in situ hybridization, CPV4 gene was detected in the nucleus of epithelial cells in the suprabasal spinous layer and granular layer of pigmented viral plaque lesions in cases 1, 2 and 3 (Table 1, Fig. 3A), while not in case 4. In contrast, CPV18 gene was detected in the nucleus of epithelial cells in the spinous layer and granular layer of the pigmented viral plaque as well as in that of neoplastic lesions in case 4 (Fig. 3B). CPV18 gene was not detected in the lesions of cases 1, 2 and 3. The number of CPV gene-positive cells was comparable between pigmented viral plaque lesions of cases 1, 2, 3 and 4, however it was lower in basal cell tumor lesion (case 4) compared to that of pigmented viral plaque lesions.
Fig. 3.
(A) Pigmented viral plaque. Nuclei of epithelial cells in the spinous layer and granular layer are positive for CPV4 gene (case 2). ISH. (B) Basal cell tumor. Nuclei of tumor cells are positive for CPV18 gene (arrow) (case 4). ISH. Bars, 50 µm.
DISCUSSION
The locations, dog breeds and histopathological findings, of the lesions of pigmented viral plaque in the present study were consistent with those in the previous studies [21]. CPV types detected, vary according to the proliferative lesions including pigmented viral plaque, and oral or cutaneous papilloma and squamous cell carcinoma [6]. A novel type CPV (CPV18) was recently identified from pigmented viral plaque lesions of a dog in the United States [6, 14]. In the present study, DNA sequence of CPV4 was detected in pigmented viral plaque lesions of 3 dogs investigated (case 1, 2 and 3), while CPV18 gene was detected in lesions of pigmented viral plaque and basal cell tumor in case 4. This is the first report of CPV18 detection in a neoplastic lesion in the dog.
Though the number and size of pigmented viral plaque lesions in a dog may increase or decrease spontaneously, malignant transformation of the lesions has never been reported. In the present study, basal cell tumor was found adjacent to the lesion of pigmented viral plaque and the tumor cells were immunohistochemically positive for papillomavirus antigen (case 4). CPV18 gene was detected by PCR and in situ hybridization in the epithelial cells of pigmented viral plaque as well as in the neoplastic cells.
Pigmented viral plaque lesions were characterized by the thickened basal cell layer of the epidermis, small projections consisting of basal cells into the dermis, and the increased number of p63- and cytokeratin 14-positive basal cells (Fig. 2C and 2E). The neoplastic cells in case 4 were diffusely positive for p63 and cytokeratin 14, indicating a basal cell phenotype (Fig. 2D and 2F). Moreover, the number of Ki-67-positive cells was increased in the tumor tissue compared to that in the pigmented viral plaque. Therefore, basal cells in the pigmented viral plaque may had undergone neoplastic change and developed basal cell tumor in case 4.
Papillomavirus infects basal cells, and viral gene can be maintained in the cells at as low as 100 copies, causing an asymptomatic infection [3]. Infected basal cells proliferate and induce the keratinocyte differentiation [11, 12, 18]. In the present study, papillomavirus antigen was detected by immunohistochemistry only in the granular layer of pigmented viral plaque lesions and in the center of basal cell tumor lobules. Also, by in situ hybridization, CPV4 and CPV18 DNAs were detected in the suprabasal cells of the pigmented viral plaque lesions and in the center of basal cell tumor lobule. This staining pattern is likely to be associated with cell differentiation as the hybridization probe detects the L1 gene of the virus genome, which is expressed in differentiated epithelial cells. Replication of CPV18 gene may be associated with the proliferation of the tumor cells.
In conclusion, CPV4 and CPV18 genes were detected in pigmented viral plaque lesions in the dog. In the CPV18-positive case, basal cell tumor developed together with pigmented viral plaque, and the virus gene was detected also in the tumor tissue. The present study shows that CPV18 infection may be associated with neoplastic proliferation of epidermal basal cells.
Acknowledgments
The authors are grateful to Dr. Hidekatsu Shimakura for his technical assistance. The GenBank/EMBL/DDBJ accession numbers of the nucleotide sequences are LC489227 (case 1, 15-838), LC489228 (case 2, 16-118), LC489229 (case 3, 16-212), LC489230 (case 4A, 18-283-1), and LC489231 (case 4B, 18-283-2).
REFERENCES
- 1.Antonsson A., Forslund O., Ekberg H., Sterner G., Hansson B. G.2000. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 74: 11636–11641. doi: 10.1128/JVI.74.24.11636-11641.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard H. U., Burk R. D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E. M.2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401: 70–79. doi: 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doorbar J.2005. The papillomavirus life cycle. J. Clin. Virol. 32 Suppl 1: S7–S15. doi: 10.1016/j.jcv.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 4.Howley P. M.2013. Papillomaviruses. pp. 1662–1703. In: Fields Virology (Howley, P. M., Schiller, J. S. and Lowy, D. R. eds.), Lippincott Williams & Wikins, Philadelphia. [Google Scholar]
- 5.Kiupel M.2018. Surgical pathology of tumors of domestic animals. pp. 36–74. In: Volume 1: Epithelial Tumor of the Skin, 3rd ed. (Kiupel, M., Goldschmidt, M. H. and Munday, J. S. eds.), Davis-Thompson Foundation, Gurnee. [Google Scholar]
- 6.Lange C. E., Diallo A., Zewe C., Ferrer L.2016. Novel canine papillomavirus type 18 found in pigmented plaques. Papillomavirus Res 2: 159–163. doi: 10.1016/j.pvr.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange C. E., Favrot C.2011. Canine papillomaviruses. Vet. Clin. North Am. Small Anim. Pract. 41: 1183–1195. doi: 10.1016/j.cvsm.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 8.Lange C. E., Tobler K., Schraner E. M., Vetsch E., Fischer N. M., Ackermann M., Favrot C.2013. Complete canine papillomavirus life cycle in pigmented lesions. Vet. Microbiol. 162: 388–395. doi: 10.1016/j.vetmic.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 9.Lange C. E., Zollinger S., Tobler K., Ackermann M., Favrot C.2011. Clinically healthy skin of dogs is a potential reservoir for canine papillomaviruses. J. Clin. Microbiol. 49: 707–709. doi: 10.1128/JCM.02047-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luff J. A., Affolter V. K., Yeargan B., Moore P. F.2012. Detection of six novel papillomavirus sequences within canine pigmented plaques. J. Vet. Diagn. Invest. 24: 576–580. doi: 10.1177/1040638712443360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maglennon G. A., McIntosh P., Doorbar J.2011. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology 414: 153–163. doi: 10.1016/j.virol.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munday J. S.2014. Bovine and human papillomaviruses: a comparative review. Vet. Pathol. 51: 1063–1075. doi: 10.1177/0300985814537837 [DOI] [PubMed] [Google Scholar]
- 13.Munday J. S., Kiupel M., French A. F., Howe L., Squires R. A.2007. Detection of papillomaviral sequences in feline Bowenoid in situ carcinoma using consensus primers. Vet. Dermatol. 18: 241–245. doi: 10.1111/j.1365-3164.2007.00600.x [DOI] [PubMed] [Google Scholar]
- 14.Munday J. S., Pasavento P.2017. Papillomaviridae and Polyomaviridae. pp. 229–243. In: Fenner’s Veterinary Virology, 5th ed. (Munday, J. S., Pasavento, P., MacLachlan, N. J. eds.), Academic Press, London. [Google Scholar]
- 15.Munday J. S., Thomson N. A., Luff J. A.2017. Papillomaviruses in dogs and cats. Vet. J. 225: 23–31. doi: 10.1016/j.tvjl.2017.04.018 [DOI] [PubMed] [Google Scholar]
- 16.Munday J. S., Tucker R. S., Kiupel M., Harvey C. J.2015. Multiple oral carcinomas associated with a novel papillomavirus in a dog. J. Vet. Diagn. Invest. 27: 221–225. doi: 10.1177/1040638714567191 [DOI] [PubMed] [Google Scholar]
- 17.Nagata M., Nanko H., Moriyama A., Washizu T., Ishida T.1995. Pigmented plaques associated with papillomavirus infection in dogs: is this epidermodysplasia verruciformis? Vet. Dermatol. 6: 179–186. doi: 10.1111/j.1365-3164.1995.tb00063.x [DOI] [PubMed] [Google Scholar]
- 18.Narama I., Kobayashi Y., Yamagami T., Ozaki K., Ueda Y.2005. Pigmented cutaneous papillomatosis (pigmented epidermal nevus) in three pug dogs; histopathology, electron microscopy and analysis of viral DNA by the polymerase chain reaction. J. Comp. Pathol. 132: 132–138. doi: 10.1016/j.jcpa.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 19.Stokking L. B., Ehrhart E. J., Lichtensteiger C. A., Campbell K. L.2004. Pigmented epidermal plaques in three dogs. J. Am. Anim. Hosp. Assoc. 40: 411–417. doi: 10.5326/0400411 [DOI] [PubMed] [Google Scholar]
- 20.Tisza M. J., Yuan H., Schlegel R., Buck C. B.2016. Genomic sequence of canine papillomavirus 19. Genome Announc. 4: e01380–e16. doi: 10.1128/genomeA.01380-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobler K., Lange C., Carlotti D. N., Ackermann M., Favrot C.2008. Detection of a novel papillomavirus in pigmented plaques of four pugs. Vet. Dermatol. 19: 21–25. [DOI] [PubMed] [Google Scholar]