Abstract
Alterations of the lung microbiota (LM) are associated with clinical features in chronic lung diseases (CLDs) with growing evidence that an altered LM contributes to the pathogenesis of such disorders. The common use of antimicrobial drugs in the management of CLDs likely represents a confounding factor in the study of the LM. The aim of the present study was to assess the effect of oral administration of amoxicillin/clavulanic acid (AC) on the LM in healthy dogs (n = 6) at short (immediately after stopping AC [D10]) and medium-term (16 days after stopping AC [D26]). Metagenetic analyses were performed on the V1–V3 hypervariable region of 16S rDNA after extraction of total bacterial DNA from samples of bronchoalveolar lavage fluid (BALF). AC did not induce significant changes in BALF cellular counts or in the bacterial load or microbial richness, evenness and α-diversity, while the β-diversity was clearly modified at D10 compared with D0 (before AC administration) and D26 (P < 0.01). The relative abundance of Bacteroidetes and Proteobacteria increased at D10 (P < 0.01) in comparison with D0 and D26 (P < 0.01). The relative abundance of Firmicutes decreased from D0 to D10 (P < 0.01) and increased from D10 to D26 (P < 0.01), but was still lower than at D0 (P < 0.01). The proportion of Actinobacteria increased at D26 compared with D0 and D10 (P < 0.01). Significant differences between timepoints at the level of family, genus or species were not found. In conclusion, in healthy dogs, oral administration of AC induces significant changes in LM at the phyla level and in the β-diversity. Most changes normalize within 2 weeks after discontinuation of AC.
Keywords: Microbiology, Zoology, Veterinary medicine, Health sciences, Respiratory system, Antimicrobial, Bacteriology, Clinical research, Lung microbiota, Antimicrobial drug, Dog
Microbiology; Zoology; Veterinary Medicine; Health Sciences; Respiratory System; Antimicrobial; Bacteriology; Clinical Research; Lung microbiota; Antimicrobial drug; Dog
1. Introduction
The lung microbiota (LM) represents the collection of microbes from the lung [1]. In healthy people, the lung microbiota closely resembles that of the oral cavity, although the bacterial biomass is lower [2]. In order to study the LM, at least in healthy individuals, bronchoalveolar lavage (BAL) is considered to be an acceptable sampling method [3]. In the majority of human chronic lung diseases (CLDs), LM alterations have been associated with the disease [4]. However, whether the LM alterations represent a cause or a consequence of the disease is still not clear [5, 6]. In dogs, the LM has been studied much more recently than in man and the literature is sparse [7, 8]. Ericsson et al. (2016) assessed the LM from samples of bronchoalveolar lavage fluid (BALF) in healthy adult experimental beagle dogs [7]. They found that the LM was dominated by the phylum Proteobacteria with a relative abundance of >80%. In parallel, the LM from BALF obtained in healthy adult experimental beagle dogs and client-owned dogs from another breed was studied [8]; results suggest a possible effect on the LM of breed and/or living conditions [7, 8].
In man, the effect of antimicrobial drugs on the gut microbiota has been investigated and a decrease in richness, diversity and modification in up to 30% of the relative abundance of the taxa was shown [9, 10]. Recently, the LM has been shown to be altered by antimicrobial treatment in mice [11]. To our knowledge, the effect of antimicrobial drugs on the LM in healthy individuals has not yet been investigated neither in man nor in dogs.
In the context of the study of the LM alterations in CLD, and since canine patients with CLD often have been treated or are being treated with antimicrobial treatment at the time of referral, there is a need to know how antimicrobial drugs interfere with the LM. Moreover, the time delay needed after cessation of treatment, in order to avoid any interaction of the drug with the LM, has not yet been studied. Therefore, the aim of the present study was to assess the short- and medium-term effect of a widely used oral antimicrobial drug on the LM in healthy adult dogs. The results of the study should provide key information for further investigations of the role of the LM in canine CLDs.
2. Materials and methods
2.1. Dog population
Six healthy experimental beagle dogs (four females and two males) aged between 1 and 11 years (mean 4.4 years), with a mean ± standard deviation body weight of 13.6 ± 1.3 kg, were included in the experimental study approved by the Ethical Committee of the University of Liège (protocol #1910). The dogs were housed on woodchip litter with outdoor access for 3–6 h each day. They had access to clean drinking water ad libitum and were fed with premium commercial dry food. There was no modification in the diet or the living conditions during the study period. The dogs did not receive any antimicrobial drug for at least 1 year prior to the study. At inclusion, the dogs were confirmed to be healthy, based on absence of clinical signs, normal physical examination, normal haematology and serum biochemistry analysis, normal gross appearance during bronchoscopy, and absence of abnormalities in the BALF analysis.
2.2. Protocol
For each dog, 20 mg/kg of amoxicillin/clavulanic acid (AC) (Amoxiclav-VMD, VMD, Arendonk, Belgium) was administered orally twice daily for 10 days. BALF sampling was repeated on each dog at three different timepoints: before AC administration (D0) and immediately (D10) as well as 16 days (D26) after discontinuation of AC.
2.3. Samples collection and processing
Dogs were anaesthetised without intubation. The bronchoscope was cleaned and disinfected before each use. A procedural control specimen (PCS) was obtained prior to each BAL procedure by injection of 10 mL of sterile saline solution through the bronchoscope channel followed by aspiration through the same channel into a sterile container using a low-power suction pump. The bronchoscope was then inserted through the oral cavity of the dog. The BAL was performed by injecting 3–4 mL/kg of sterile saline solution divided into three aliquots, including two aliquots with the endoscope inserted into the right diaphragmatic lobe, followed by a third aliquot placed into the left diaphragmatic lobe. Each aliquot was directly aspirated by gentle suction and the fluids recovered from the three aliquots were pooled. After sampling, both PCS and BALF were transferred into cryotubes and stored at -80°C until analysis. A small amount of BALF was used for calculation of the total cell count (TCC) as well as for cytospin preparation (centrifugation at 221 g, for 4 min at 20°C, Thermo Shandon Cytospin©4). Cytosopin preparations were stained by Diff Quick and were used for differential cell count (DCC) determination by counting a minimum of 100 cells.
2.4. 16S rDNA extraction and high throughput sequencing
The analysis of the LM for all dogs and for all 3 timepoints was performed on a single occasion for each step of the LM analysis which included DNA extraction, polymerase chain reactions (PCRs), sequencing and post-sequencing analysis. As required, strict laboratory controls were done to avoid contamination from the PCR reagents and laboratory materials.
Total DNA was extracted from BALFs and PCSs using the DNEasy Blood and Tissue kit (QIAGEN Benelux BV, Antwerp, Belgium) according to the manufacturer's instructions. DNA was eluted into DNase/RNase-free water for a total volume of 30μL and the concentration and purity were evaluated using an ND-1000 spectrophotometer (NanoDrop ND-1000, Isogen, De Meern, The Netherlands).
The bacterial load was assessed by quantitative PCRs (qPCRs) targeting the V2–V3 region of the 16S rDNA. Duplicate qPCRs were conducted in a final volume of 20 μL containing 2.5 μl of template DNA, 0.5 μl of forward primer (5′-ACTCCTACGGGAGGCAGCAG-3’; 0.5 μM), 0.5 μl of reverse primer (5′-ATTACCGCGGCTGCTGG-3’; 0.5 μM) [12], 10 μl of No Rox SYBR 2x MasterMix (Eurogentec, Seraing, Belgium), and 6.5 μl of water. The run also contained non-template controls and a 10-fold dilution series of a V2–V3 purified (Wizard® SV Gel and PCR Clean-Up System, Promega, Leiden, The Netherlands) PCR product quantified by PicoGreen targeting double-stranded DNA (Promega, Leiden, The Netherlands) and used to build the standard curve. Data acquisition was obtained using an ABI 7300 real-time PCR system, with the following cycling sequence: 1 cycle of 50°C for 2 min; 1 cycle of 95°C for 10 min; 40 cycles of 94°C for 15 s; and 1 cycle of 60°C for 1 min. After the PCR, a melting curve was constructed in the range of 64–99°C. Results were expressed in logarithm base 10 copy numbers per milliliter.
For bacterial identification, PCR targeting the V1–V3 region of the 16S rDNA was performed with the following primers: forward (5′-GAGAGTTTGATYMTGGCTCAG-3′) and reverse (5′-ACCGCGGCTGCTGGCAC-3′) and Illumina overhand adapters [13]. Amplicons were purified with the Agencourt AMPure XP beads kit (Beckman Coulter, Villepinte, France) and submitted to a second PCR for indexing using the Nextera XT index primers 1 and 2. After purification, amplicons were quantified by PicoGreen (ThermoFisher Scientific, Waltham, MA, USA) before normalization and pooling. PCSs and the negative control from the extraction and the PCR steps were not sequenced as their PCR products after amplification were <1 ng/μL. Bacterial 16S rDNA amplicon libraries were then sequenced on a MiSeq Illumina sequencer using V3 reagents. A positive control using 20 defined bacterial species DNA was included in the run. Sequence read processing including a first cleaning step for length and sequence quality and a screening for chimera with UCHIME algorithm was made using, respectively, MOTHUR v1.39 and Vsearch [14, 15]. 16S rDNA reference alignment and taxonomical assignation with an operational taxonomic unit (OTU) clustering distance of 0.03 were based on the SILVA database v1.32 using the cluster. split command in MOTHUR v1.39 [16]. A final subsampling was performed to have an identical mean of reads per samples at 5,400 reads.
2.5. Data analysis
Comparisons between events for TCC, DCC and the bacterial load were made using Friedman tests in XLStat (Addinsoft, Paris, France).
Good's coverage index and ecological indicators, including the bacterial richness (Chao1 index), evenness (Simpson index-based measure) and α-diversity (inverse Simpson's index) were calculated with MOTHUR v1.39 and compared between timepoints using Friedman tests in XLStat.
Non-metric multidimensional scaling (NMDS) graph was performed based on a Bray-Curtis dissimilarity matrix at the species level to assess the global bacterial composition (β-diversity) between timepoints (R vegan package). Significant differences between timepoints were calculated with MOTHUR v1.39 using AMOVA and HOMOVA tests. The AMOVA test is a non-parametric analysis for testing the hypothesis that genetic diversity within each timepoint is not significantly different from the genetic diversity in all timepoints together [17]. The HOMOVA test is a nonparametric analysis used to test the hypothesis that the genetic diversity within the different timepoints is homogeneous [18].
Differences in bacterial relative abundances between timepoints were assessed in R using a mixed linear model with Benjamini Hotchberg FDR correction for multiple comparisons.
Results were expressed as median and interquartile range.
All sample raw reads were deposited at the National Center for Biotechnology Information (NCBI) and are available under Bioproject ID PRJNA507075.
3. Results
3.1. BALF cell analysis
There were no significant differences between timepoints for TCC and DCC (Table 1).
Table 1.
Median and interquartile range of the total and differential bronchoalveolar lavage fluid cell count between timepoints.
| Timepoints | Total cell count cells/μL | Macrophages % | Neutrophils % | Lymphocytes % | Eosinophils % |
|---|---|---|---|---|---|
| D0 | 800.0 (702.5–890.0) | 82.5 (80.0–86.5) | 5.5 (4.2–7.5) | 6.0 (0.2–17.0) | 2.0 (1.2–2.8) |
| D10 | 470.0 (275.0–635.0) | 92.0 (90.2–98.2) | 3.0 (0–6.0) | 0.5 (0–2.5) | 1.5 (0.2–2.0) |
| D26 | 220.0 (125.0–330.0) | 78.0 (74.8–81.2) | 5.0 (4.2–9.5) | 9.0 (7.0–11.8) | 0.5 (0–4.0) |
| P-value between the 3 timepoints | 0.07 | 0.07 | 0.31 | 0.40 | 0.25 |
D0, before antimicrobial drug administration; D10, just after antimicrobial drug discontinuation; D26, 16 days after antimicrobial drug discontinuation.
3.2. BALF microbiota analysis
Good's coverage index was >95.91% in all samples (98.74% (97.22–99.04)) and was not different between timepoints (P = 0.31) indicating the same sampling effort per timepoint. A total of 2,236,209 reads were recovered with a median length of 498 nucleotides. After the first cleaning step, 1,691,396 reads were kept and screened for chimera. 1,607,398 reads per samples were retained and used for OTU clustering before the final subsampling.
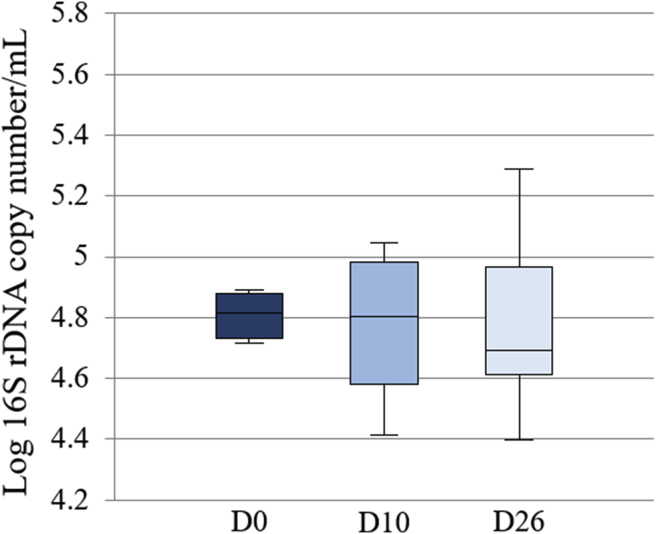
The differences in the bacterial load between timepoints were not significant (P = 0.51) (Fig. 1). In PCSs, the bacterial load was 2.46 (2.41–2.65) copies per millilitre; about 100 times lower than in the BALF samples.
Fig. 1.
Box plot representing the logarithm of the number of 16S rDNA copies per microliter (bacterial load) between timepoints. The medians are represented by the central horizontal bars. The lower and upper limits of the box are the first and third quartiles, respectively. There were no significant differences between timepoints.
Phyla, families, genera and species making up the dog's LM, before AC administration, with a relative abundance of >1.00%, are presented in Table 2.
Table 2.
The top 25 most abundant taxa present in the lung microbiota at the level of phyla, families, genera and species in healthy dogs before administration of the antimicrobial drug.
| Phylum | Family | Genus | Species | Median relative abundance, % |
|---|---|---|---|---|
| Firmicutes | Streptococcaceae | Streptococcus | Streptococcus mitis | 3.5 (2.1–4.1) |
| Streptococcus cristatus | 0.5 (0.2–0.8) | |||
| Streptococcus salivarius | 0.4 (0–1.0) | |||
| Staphylococcaceae | Staphylococcus | Staphylococcus epidermidis | 1.8 (1.7–2.7) | |
| Staphylococcus warneri | 0.8 (0.4–2.2) | |||
| Staphylococcus xylosus | 0.5 (0.1–0.7) | |||
| Erysipelotrichaceae | Allobaculum | Allobaculum HM124340 | 1.3 (0.1–3.5) | |
| Allobaculum DQ113686 | 0.5 (0.2–1.3) | |||
| Allobaculum 16S_OTU48 | 0.3 (0–1.3) | |||
| Turicibacter | Turicibacter FJ880353 | 0.3 (0.1–0.6) | ||
| Veillonellaceae | Veillonella | Veillonella JQ449520 | 1.1 (0.8–1.4) | |
| Bacillales Family XI | Gemella | Gemella haemolysans | 0.9 (0.4–1.0) | |
| Actinobacteria | Propionibacteriaceae | Propionibacterium | Propionibacterium acnes | 7.0 (5.2–17.0) |
| Corynebacteriaceae | Corynebacterium_1 | Corynebacterium_1 tuberculostearicum | 1.1 (0.4–1.5) | |
| Micrococcaceae | Micrococcus | Micrococcus luteus | 0.6 (0.4–0.8) | |
| Rothia | Rothia mucilaginosa | 0.6 (0.1–1.0) | ||
| Rothia dentocariosa | 0.4 (0–0.9) | |||
| Proteobacteria | Moraxellaceae | Acinetobacter | Acinetobacter_johnsonii | 0.4 (0.1–2.5) |
| Enhydrabacter | Enhydrobacter_osloensis | 0.4 (0.1–1.0) | ||
| Bacteroidetes | Flavobacteriaceae | Flavobacterium | Flavobacterium EU802240 | 0.7 (0.1–1.1) |
| Elizabethkingia | Elizabethkingia miricola | 0.6 (0.1–1.0) | ||
| Chryseobacterium | Chryseobacterium haifense | 0.4 (0–1.0) | ||
| Fusobacteria | Fusobacteriaceae | Fusobacterium | Fusobacterium AJ867041 | 0.6 (0–1.6) |
| Fusobacterium nucleatum | 0.4 (0.1–0.9) | |||
| Verrucomicrobia | Verrucomicrobiaceae | Verrucomicrobiaceae_ge | Verrucomicrobiaceae_ge 16S_OTU17 | 0.8 (0.4–2.2) |
The relative abundances are presented in median and interquartile range.
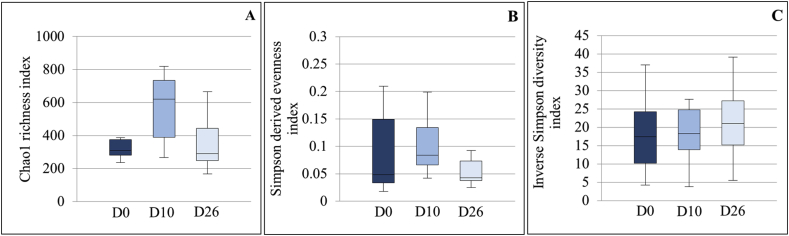
The bacterial richness, evenness and α-diversity were not significantly modified between timepoints (P = 0.31, 0.61 and 0.85 respectively) (Fig. 2).
Fig. 2.
Box plot graphs representing the bacterial richness (A), evenness (B) and alpha diversity (C) at the 3 timepoints. The medians are represented by the central horizontal bars. The lower and upper limits of the box are the first and third quartiles, respectively.
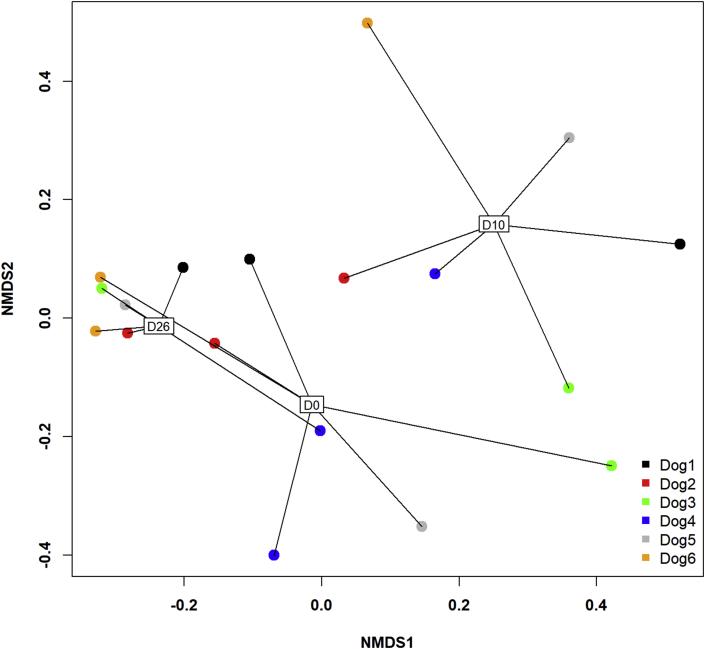
The NMDS graph of the β-diversity showed clear differences between D10 and the other timepoints (Fig. 3). Significant differences were found between timepoints with the AMOVA test (P < 0.001) with significant differences in the post-hoc tests between D0 and D10 (P = 0.002) and D10 and D26 (P < 0.001), but not between D0 and D26. The HOMOVA test showed significant differences between timepoints (P = 0.009) with significant difference in the post-hoc tests only between D10 and D26 (P = 0.004).
Fig. 3.
Two-dimensional non-parametric representation of the global bacterial composition at the species level between timepoints for each dog based on a Bray-Curtis matrix of dissimilarity. Lung communities are clustered by timepoints. D0: before antimicrobial administration; D10: just after antimicrobial discontinuation; D26: 16 days after antimicrobial discontinuation; NMDS: non-metric multidimensional scaling.
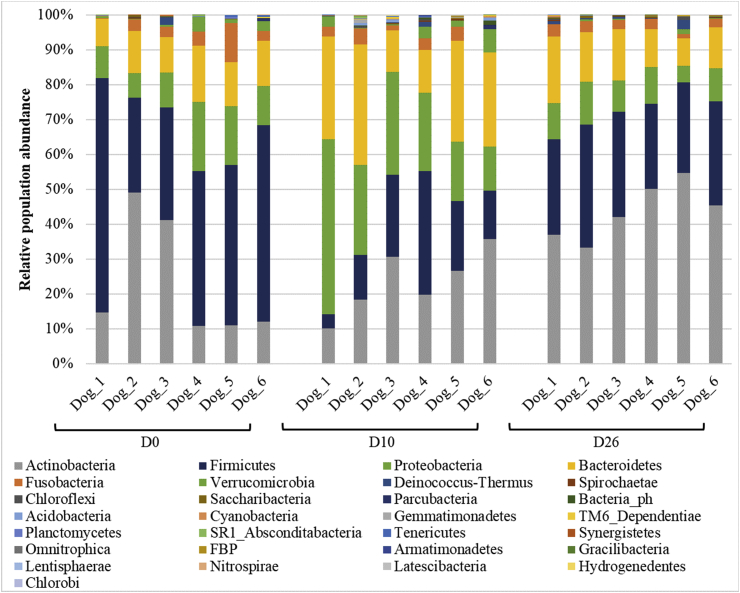
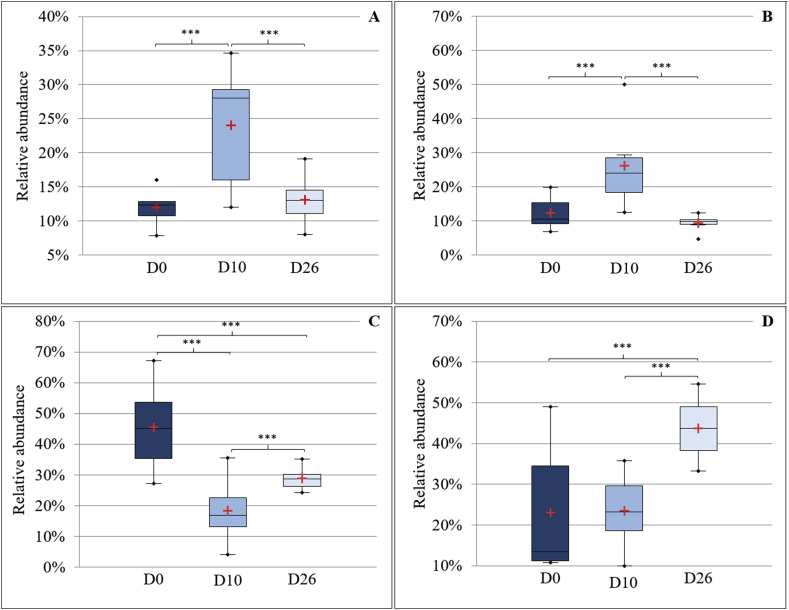
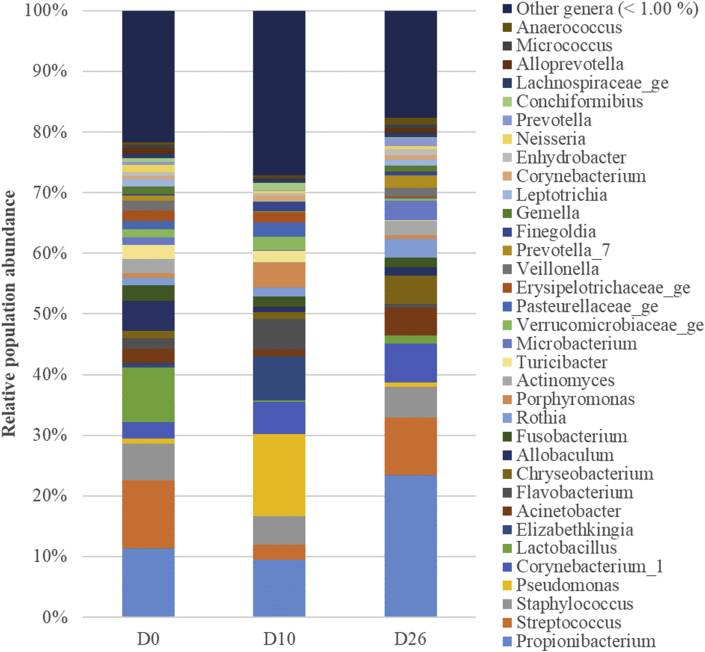
Fig. 4 illustrates the distribution of bacterial relative abundance at the phyla level in all dogs at the different timepoints. The Bacteroidetes (Fig. 5A), the Proteobacteria (Fig. 5B), the Firmicutes (Fig. 5C) and the Actinobacteria (Fig. 5D) were significantly different between timepoints. No significant differences were shown between timepoints at the level of families, genera and species. However, as shown in Fig. 6, at D10, some genera decreased, such as Streptococcus spp. (Firmicutes), Staphylococcus spp. (Firmicutes) and Lactobacillus spp. (Firmicutes), others increased, such as Pseudomonas spp. (Proteobacteria), Flavobacterium spp. (Bacteroidetes) and Chryseobacterium spp. (Bacteroidetes), while some remained stable, such as Propionibacterium spp. (Actinobacteria).
Fig. 4.
Phyla-level composition of bronchoalveolar lavage fluid (BALF) microbiota at the 3 timepoints. Bar charts showing relative abundance annotated to the taxonomic level of phylum for all taxa detected in BALF collected from 6 healthy adult beagle dogs, before (D0) and 10 days (D10) as well as 16 days after the discontinuation of the drug.
Fig. 5.
Box plot graphs representing Bacteroidetes (A), Proteobacteria (B), Firmicutes (C) and Actinobacteria (D) relative abundances between timepoints. The means and the medians are represented by the red crosses and the central horizontal bars respectively. The lower and upper limits of the box are the first and third quartiles, respectively. Points are considered as outliers. ***Statistically different (P < 0.001).
Fig. 6.
Genus-level composition of bronchoalveolar lavage fluid (BALF) microbiota at the 3 timepoints. Bar charts showing relative abundance annotated to the taxonomic level of genus of all taxa detected in BALF collected from 6 healthy adult beagle dogs, before (D0) and 10 days (D10) as well as 16 days after the discontinuation of the antimicrobial drug.
4. Discussion
To the best of our knowledge, this is the first study in dogs investigating how an antimicrobial drug interferes with the LM. Since the use of antimicrobial drugs such as oral treatment with AC is common in the management of canine CLDs, this study is a prerequisite before assessing the role of alterations of the LM in the pathogenesis of canine CLDs. In the present study, oral administration of AC to healthy beagles induced an obvious shift in the β-diversity of the LM as well as significant changes in the proportion of the major phyla, and the majority of these changes were no longer present at 2 weeks after drug discontinuation. Furthermore, the bacterial load and the ecological indices of richness, evenness and α-diversity were not significantly modified.
In the study of the LM, avoiding bacterial contamination is crucial, because of the low bacterial biomass of the respiratory tract [2, 19]. Amplification of contaminants could modify the data obtained and provide aberrant results [20]. Origins of contamination can be numerous and may occur at different steps involving the laboratory analyses, the materials, mainly the bronchoscope, and the sampling procedure [20, 21]. In order to minimize contamination from extraction and sequencing reagents, strict laboratory controls of all reagents and machines were performed. PCSs were collected before each sampling in order to detect contamination via the bronchoscope itself, the sterile saline solution and the device used for the lavage. In the present study, analysis of PCSs indicated that this source of contamination could only minimally alter our results [22]. Finally, during the procedure, care was taken to avoid contact with the oropharyngeal, laryngeal and tracheal mucosae during insertion of the bronchoscope. In spite of these handling precautions, some contamination during passage of the bronchoscope through the upper airway cannot be excluded. However, it has been shown that such contamination only minimally alters the LM in bronchoscopically-acquired specimens [3, 21, 22].
In the present study, the four major phyla (Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes) found in the lung of healthy beagles were the same as described in previous studies in beagles, although the abundance order differed [7, 8]. The observed differences in relative abundance of major phyla can be attributed to several factors. Firstly, the LM largely depends on the environmental conditions [2, 23] and differences in housing, type of food, geographical area and dog behaviour may have had an impact on the LM [2, 24]. Secondly, the technique used to sample and analyse BALF in the present study differs from that used by Ericsson et al. (2016), in which a catheter was passed through a sterile endotracheal tube to collect the BALF. In the present study the use of a bronchoscope was chosen, according to a technique that has been approved for investigation of the LM in man [22]. Finally, the relative abundance of Firmicutes in this study compared with others performed on beagles was higher. This elevated percentage of Firmicutes might have been slightly overestimated because bacteria composing the Firmicutes phylum appear to have more 16S rDNA copies in their genome than bacteria in other phyla [25].
In order to limit the variations between samples related to contamination or factors influencing the LM as mentioned above, the dogs in the present study were from the same breed, co-housed in a stable environment and fed with the same standardized diet. The sampling procedure was highly standardized and repeated identically at the three timepoints. Moreover, each step of the LM analysis (DNA extraction, PCRs, sequencing and post sequencing analysis) was performed at a single occasion for all samples together (from all dogs and from all 3 timepoints).
The stability of the LM over time is an important source of experimental and clinical variability and might have interfered with our results. Indeed, it has been shown in mice, that the LM is dynamic and rapidly converges in cohoused mice placed in shared cages [11]. Dogs of our study were housed in the same conditions before and during the study period. As a consequence, it is reasonable to expect that such a stable environment helped to reduce time-induced variations.
AC, a beta-lactam antimicrobial drug, is a broad-spectrum antimicrobial drug acting against Gram-positive and to a lesser extent Gram-negative bacteria [26]. AC was chosen as an antimicrobial agent since it is largely used by veterinarians in dogs with lower airway disease. Moreover, AC is a drug recommended in both human and veterinary medicine for the treatment of acute pneumonia, including acute aspiration pneumonia [27, 28]. The dosage currently used in canine practice and recommended for pulmonary infections was used [29].
Significant modifications were found at the phyla level after AC administration. As expected, the Firmicutes phylum mainly represented by bacteria that are sensitive to AC decreased. The increased relative abundance in the phylum Proteobacteria appeared to be related to an increase in the genus Pseudomonas spp., which is known to be resistant to AC [29]. Finally, the main genera composing the phylum Bacteroidetes increased at D10 were Gram-negative bacteria which are less sensitive to AC [26]. According to these results and to the significant modification in the β-diversity shown just after discontinuation of the drug, AC appears to have an effect on the LM in healthy dogs, even if differences were not significant under the phyla level.
Absence of significant differences in the relative abundances under the phyla level and in bacterial load, richness, evenness and α-diversity might be attributed to a possible high resilience of the LM to disturbances, compared with microbiota from other sites of the body. Such a hypothesis is supported by the fact that differences in resilience of microbiota have been shown, according to their niche. For example, the salivary microbiota was shown to be more resilient to disturbance after antimicrobial drug administration compared to that of the gut [9]. Another explanation would be that in healthy individuals, such as the dogs in this study, the permeability of the alveolar-capillary wall is lower than in diseased lungs [30], leading to a limited penetration of AC into the parenchyma and airways and therefore a limited effect on the LM. Indeed it has been shown that amoxicillin concentration in the sputum in man may differ according to different host- and drug-related factors, such as alveolar-capillary permeability [31, 32]. As alveolar-capillary permeability increases in the case of inflammation, the concentration of AC, which passively diffuses in the alveolar space [31], is probably decreased in healthy airways. It should be remembered that the use of another antimicrobial drug with improved airways penetration could have induced different LM modifications.
The inability to highlight significant differences under the phyla level might also be due to the number of data, including about 5,400 sequences per sample, as well as to the limited number of dogs included in the study. This contributed to a lack of power of the statistical tests mainly associated with the corrections for multiple tests more significant with a large dataset [33].
5. Conclusion
In summary, in healthy dogs, oral administration of a commonly used broad-spectrum antimicrobial drug induced significant changes in the pulmonary microbial population and the majority of these changes were no longer present at 2 weeks after the discontinuation of the drug. As a consequence, for investigation of associations between the LM and CLDs in dogs, discontinuance of any antimicrobial medication at a minimum of 2 weeks before sampling is advised. However, further studies are warranted to investigate the effect of other antimicrobial drugs and to identify the optimal delay between antimicrobial drug discontinuation and sampling, in order to avoid any interference with the LM analysis.
Declarations
Author contribution statement
Aline Fastrès, Cécile Clercx: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Bernard Taminiau: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Emilie Vangrinsven: Conceived and designed the experiments; Performed the experiments.
Alexandru-Cosmin Tutunaru: Performed the experiments.
Evelyne Moyse, Frederic Farnir: Analyzed and interpreted the data.
Georges Daube: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at the National Center for Biotechnology Information (NCBI) under the accession number PRJNA507075.
Acknowledgements
We thank Sylvain Romijn and Belinda Albert for their help in the sample collection and processing. We thank Dr. Dickson R.P. and his laboratory team, especially Mr. Brown C. for their help in statistical analyses and in the presentation of data. Finally, we would like to thank Pr. Day M.J. for proofreading the English text.
References
- 1.Segal L.N., Rom W.N., Weiden M.D. Lung microbiome for clinicians: new discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc. 2014;11(1):108–116. doi: 10.1513/AnnalsATS.201310-339FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The microbiome and the respiratory tract. Annu. Rev. Physiol. 2016;78(1):481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Beck J.M., Huffnagle G.B. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa A.N., Costa FM da, Campos S.V., Salles R.K., Athanazio R.A., Costa A.N. The pulmonary microbiome: challenges of a new paradigm. J. Bras. Pneumol. 2018;44(5):424–432. doi: 10.1590/S1806-37562017000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fastrès A., Felice F., Roels E., Moermans C., Corhay J.-L., Bureau F. The lung microbiome in idiopathic pulmonary fibrosis: a promising approach for targeted therapies. Int. J. Mol. Sci. 2017;18(12):2735. doi: 10.3390/ijms18122735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y., Ma S.-F., Espindola M.S., Vij R., Oldham J.M., Huffnagle G.B. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017;196(2):208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ericsson A.C., Personett A.R., Grobman M.E., Rindt H., Reinero C.R. Composition and predicted metabolic capacity of upper and lower airway microbiota of healthy dogs in relation to the fecal microbiota. Wilson B.A., editor. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0154646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roels E., Taminiau B., Darnis E., Neveu F., Daube G., Clercx C. Comparative analysis of the respiratory microbiota of healthy dogs and dogs. J. Vet. Intern. Med. 2017;31(1):230–231. [Google Scholar]
- 9.Zaura E., Brandt B., Teixeira de Mattos M.J., Buijs M., Caspers M., Rashid M. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio. 2015;6(6) doi: 10.1128/mBio.01693-15. e01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiemann S., Smit N., Strowig T. Antibiotics and the intestinal microbiome: individual responses, resilience of the ecosystem, and the susceptibility to infections. Curr. Top. Microbiol. Immunol. 2016;398:123–146. doi: 10.1007/82_2016_504. [DOI] [PubMed] [Google Scholar]
- 11.Dickson R.P., Erb-Downward J.R., Falkowski N.R., Hunter E.M., Ashley S.L., Huffnagle G.B. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Respir. Crit. Care Med. 2018;198(4):497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bindels L.B., Neyrinck A.M., Salazar N., Taminiau B., Druart C., Muccioli G.G. Non digestible oligosaccharides modulate the gut microbiota to control the development of leukemia and associated cachexia in mice. PLoS One. 2015;10(6):1–16. doi: 10.1371/journal.pone.0131009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngo J., Taminiau B., Fall P.A., Daube G., Fontaine J. Ear canal microbiota – a comparison between healthy dogs and atopic dogs without clinical signs of otitis externa. Vet. Dermatol. 2018;29(5) doi: 10.1111/vde.12674. 425-e140. [DOI] [PubMed] [Google Scholar]
- 14.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq. Appl Env Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss P. 2013. Amova [Internet]https://www.mothur.org/wiki/Amova [cited 2019 Oct 21]. Available from. [Google Scholar]
- 18.Schloss P. 2018. Homova [Internet]https://www.mothur.org/wiki/Homova [cited 2019 Oct 21]. Available from. [Google Scholar]
- 19.Marsh R.L., Nelson M.T., Pope C.E., Leach A.J., Hoffman L.R., Chang A.B. How low can we go? The implications of low bacterial load in respiratory microbiota studies. Pneumonia. 2018;10(1):7. doi: 10.1186/s41479-018-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Falkowski N.R., Huffnagle G.B. Bacterial topography of the healthy human lower respiratory tract. mBio. 2017;8(1) doi: 10.1128/mBio.02287-16. e02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassis C.M., Erb-Downward J.R., Dickson R.P., Freeman C.M., Schmidt T.M., Young V.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6(2):1–10. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd C.M., Marsland B.J. Lung homeostasis: influence of age, microbes, and the immune system. Immunity. 2017;46(4):549–561. doi: 10.1016/j.immuni.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Dickson R.P., Huffnagle G.B. The lung microbiome: new principles for respiratory Bacteriology in Health and disease. PLoS Pathog. 2015;11(7) doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Větrovský T., Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8(2):1–10. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur S.P., Rao R., Nanda S. Amoxicillin: a broad spectrum antibiotic. Int. J. Pharm. Pharm. Sci. 2011;3(3):30–37. [Google Scholar]
- 27.File T.M. The development of pharmacokinetically enhanced amoxicillin/clavulanate for the management of respiratory tract infections in adults. Int. J. Antimicrob. Agents. 2007;30:131–134. doi: 10.1016/j.ijantimicag.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 28.Lappin M.R., Blondeau J., Boothe D., Breitschwerdt E.B., Guardabassi L., Lloyd D.H. Antimicrobial use guidelines for treatment of respiratory tract disease in dogs and cats: antimicrobial guidelines working group of the international society for companion animal infectious diseases. J. Vet. Intern. Med. 2017;31:279–294. doi: 10.1111/jvim.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plumb D.C. eighth ed. Wiley-Blackwell; Ames, Iowa: 2015. pp. 80–84. (Plumb’s Veterinary Drug Handbook). [Google Scholar]
- 30.Brusse-Keizer M., Valk P Vander, van der Zanden R.W., Nijdam L., van der Palen J., Hendrix R. Amoxicillin concentrations in relation to beta-lactamase activity in sputum during exacerbations of chronic obstructive pulmonary disease. Int J COPD. 2015;10:455–461. doi: 10.2147/COPD.S70355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honeybourne D. Antibiotic penetration into lung tissues. Thorax. 1994;49(2):104–106. doi: 10.1136/thx.49.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pennington J. Penetration of antibiotics into respiratory secretions. Rev. Infect. Dis. 1981;3(1):67–73. doi: 10.1093/clinids/3.1.67. [DOI] [PubMed] [Google Scholar]
- 33.Desquilbet L. 2015. Puissance Statistique D’une Étude Version 3 [Internet]https://eve.vet-alfort.fr/course/view.php?id=353 [cited 2018 May 3]. Available from. [Google Scholar]