Abstract
Allopolyploids are often faced with the challenge of maintaining well-coordination between nuclear and cytoplasmic genes inherited from different species. The synthetic allotetraploid Cucumis × hytivus is a useful model to explore cytonuclear coevolution. In this study, the sequences and expression of cytonuclear enzyme complex RuBisCO as well as its content and activity in C. × hytivus were compared to its parents to explore plastid–nuclear coevolution. The plastome-coded rbcL gene sequence was confirmed to be stable maternal inheritance, and parental copy of nuclear rbcS genes were both preserved in C. × hytivus. Thus, the maternal plastid may interact with the biparentally inherited rbcS alleles. The expression of the rbcS gene of C-homoeologs (paternal) was significantly higher than that of H-homoeologs (maternal) in C. × hytivus (HHCC). Protein interaction prediction analysis showed that the rbcL protein has stronger binding affinity to the paternal copy of rbcS protein than that of maternal copy in C. × hytivus, which might explain the transcriptional bias of the rbcS homoeologs. Moreover, both the activity and content of RuBisCO in C. × hytivus showed mid-parent heterosis. In summary, our results indicate a paternal transcriptional bias of the rbcS genes in C. × hytivus, and we found new nuclear–cytoplasmic combination may be one of the reasons for allopolyploids heterosis.
Keywords: allopolyploid, plastid–nuclear evolution, RuBisCO, Cucumis
1. Introduction
Allopolyploidization, including interspecific hybridization and whole-genome duplication, has played a key role in the evolution of plant species [1,2,3]. Previous studies have indicated that almost all ancestors of seed plants have experienced at least one round of genome doubling [4,5]. Allopolyploidization often results in complex changes at different aspects, such as genomic changes including homoeologous exchanges and loss of genes [6,7,8], nonadditive gene expression [9,10], and changes in epigenetic modifications [5,11]. During allopolyploidization, not merely two different nuclear genomes combine, but also different sources of chloroplast and mitochondrial genomes interact in the same cell, in view of the complex interactions between the nuclear and organelle genomes [12]. Cytonuclear interaction had been considered as an effective mean for eukaryotes to produce diverse phenotypes, and to improve the competitiveness of survival and reproduction [13,14]. Allopolyploidization disrupts nuclear–cytoplasmic interactions, including changes in gene copy number and stoichiometry [2]. For most angiosperms, organelles (plastid and mitochondrial) are maternally inherited [15]; to date, only very low-frequent biparental or paternal transmission of plastid DNA has been reported in few plant species [16,17,18]. Therefore, in the case of the universal uniparental maternal inheritance of plastid genome in plants, allopolyploids are expected to be more closely matched to maternal homoeologs of plastid targeted genes than to paternal homoeologs [19]. Ribulose-1, 5-bisphosphate carboxylase/oxygenase (RuBisCO), which is localized to chloroplast stroma, is composed of small subunits encoded by the nucleus and large subunits encoded by plastid, and is an ideal system to study the evolution process of plastid–nuclear interactions. Gong et al. [20] found a consistent transcriptional bias of the maternal rbcS homoeologs in four natural allopolyploids, Arabidopsis, Arachis, Brassica, and Nicotiana. Nevertheless, study in synthesized rice allotetraploids has shown there was no consistent pattern of biased expression of maternal-like rbcS homoeologs [21]. A recent study revealed subgenome dominance towards the A subgenome (paternal progenitor) of nuclear genes involved in plastid protein complexes in resynthesized and natural Brassica napus [22]. Hence, cytonuclear coevolution appears to be a complicated but underexplored aspect of allopolyploidization, the genetic and evolutionary forces that allopolyploidization imposes upon cytonuclear interaction still need more comprehensive and systematic study to be well understood.
Synthesized allopolyploids can be very useful models to study cytonuclear interaction since it presents a new nuclear–cytoplasmic combination and can be tested on a known genetic background. An interspecific cross was successfully made in Cucumis between wild Cucumis species, C. hystrix Chakr. (HH, 2n = 2x = 24) and cultivated cucumber, C. sativus L. ‘BeijingJietou’ (CC, 2n = 2x = 14) [23]. The chromosome numbers of C. hystrix × C. sativus F1 interspecific hybrid (HC, 2n = 19) were then doubled through somaclonal variation using embryo culture technique, and a synthesized allotetraploid species, C. × hytivus Chen and Kirkbride (HHCC, 2n = 4x = 38), was obtained [24]. This allotetraploid can self-pollinate to produce viable seeds, providing a unique system to reveal the complicated processes during allopolyploid evolution and speciation.
The genome of cucumber has three different genetic patterns; the chloroplast, mitochondrial, and nuclear genomes are maternally, paternally, and biparentally transmitted, respectively [25,26]. For C. × hytivus, Shen et al. [27] concluded that mitochondrial DNA was paternally inherited, while chloroplast DNA was maternally inherited between Cucumis species. Thus, the newly synthetic allotetraploid C. × hytivus combines two nuclear genomes but inherits only one set of progenitor organellar genomes, respectively, providing a unique model system for analyzing early cytonuclear evolution in allopolyploidization. In this study, the plastid-nuclear coevolution of RuBisCO encoding genes, nuclear rbcS and plastid rbcL genes, as well as RuBisCO content and activity in allotetraploid C. × hytivus and its diploid parents, were characterized. We aimed to explore the RuBisCO in the early stages of allopolyploidization to gain insight into the pace of cytonuclear coevolution of C. × hytivus.
2. Materials and Methods
2.1. Plant Materials, DNA, and RNA Extraction
Three species of Cucumis were used for this study: the cultivated cucumber C. sativus ‘BeijingJietou’ (2n = 14, genome CC), the self-cross plants (S14) of the synthesized new allotetraploid species C. × hytivus (2n = 38, genome HHCC), and the wild species C. hystrix (2n = 24, genome HH). The two diploid plants used in this experiment are the same inbred lines used in the original interspecific cross that generated the interspecific F1. The initial allotetraploid was obtained by chromosome doubling through somaclonal variation using embryo culture technique. The allotetraploid C. × hytivus used in this study was the fourteenth generation (S14) of self-cross plants.
The extraction of total genomic DNA was done using a modified cetyltrimethylammonium bromide (CTAB) method [28]. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) and digested with DNase I for 30 min at 25 °C to remove DNA. Then, 2 µg of total RNA was used to synthesize complementary DNA (cDNA) using a cDNA Synthesis Kit (Fermentas, York, UK).
2.2. Gene Cloning and Quantitative Real-Time PCR Analysis
Primers used to amplify the full-length genomic and CDS sequence of rbcL and rbcS genes in three Cucumis species are listed in Table S1. Polymerase chain reactions (PCRs) were performed as described in Gong et al. [29], and the resulting clones were sequenced (Tsingke, Beijing, China). When amplifying the rbcS genes in allotetraploid C. × hytivus, three parallel independent PCRs for each primer sample were performed, the PCR products were cloned into the pMD-19T vector (Takara, Shiga, Japan) and the resulting all clones were sequenced. To exclude the possibility of PCR recombination and sequencing errors, each sequence was tested at least twice, and only the rbcS copies that sequenced at least 25% supportive clones in each independent PCR experiment were accepted as true copies. The sequences in this study have been uploaded to the GenBank database (accession numbers MK948862–MK948868; https://www.ncbi.nlm.nih.gov/genbank/). Species-specific single-nucleotide polymorphisms (SNPs) were deduced from alignments of the full-length sequence of genomic rbcS orthologs in parental diploids and that of homoeologs in allopolyploid.
The expression of rbcL genes was analyzed by quantitative real-time PCR (qRT-PCR) using the SYBR Premix Ex TaqTM Kit (Takara) as described by Li et al. [30]. The β-Actin gene and F-box gene were used to quantify the relative transcript levels. The relative expression levels of rbcL genes were calculated following the 2−ΔΔCT method [31] with the normalization of data to the geometric average of the internal control genes [32]. The gene-specific primers used for expression analysis were listed in Table S1.
2.3. Quantification and Comparison of rbcS Aallelic and Homeologous Expression Based on RNA-Sequencing
Three biological replicates of next-generation RNA-sequencing (RNA-seq) data of the allotetraploid and diploid parents were downloaded from the SRA database at NCBI (https://www.ncbi.nlm.nih.gov/sra) with accession number SRP155470 (Table S2). The samples of RNA-seq data were the leaves of seedlings of three species under normal growth conditions (14/10 h day/night, 28 ºC/20 ºC day/night, light intensity 500 μmol/m2s1 and air humidity controlled around 70%) at the same developmental stage. Raw data (raw reads) were filtered to produce clean data with high quality. Transcriptome assembly was accomplished based on the ‘left.fq’ and ‘right.fq’ using Trinity [33] fragments per kilobase of transcript per million fragments (FPKM) was used for the evaluation of expression of the rbcS transcripts in three species of C. hystrix (HH), C. × hytivus (HHCC) and C. sativus (CC).
The H- and C-rbcS homoeolog expression levels in allotetraploid C. × hytivus were analyzed according to the method of Gong et al. [29]. The raw reads of each replicate from C. × hytivus RNA-seq data were mapped onto the rbcS cDNA sequence using the Burrows–Wheeler alignment tool [34]. The GATK2 software was used to call SNPs [35]. Raw vcf files were filtered using the GATK standard filter method, and only SNPs with distance > 5 bp were retained. The read number based on specific nucleotide sites of homoeo-SNPs detected in the allotetraploid C. × hytivus was searched. In this way, we can calculate the readings number covering the H-genome homoeo-SNPs and the C-genome homoeo-SNPs, which represented the H- and C-rbcS homoeolog expression levels in C. × hytivus, respectively.
2.4. The Prediction of RuBisCO Protein–Protein Complex Binding Affinity
The predicted dissociation constant (Kd) and binding free energy (ΔG) of RuBisCO protein–protein complex were obtained from PPA-Pred2 [36] based on the protein sequences of rbcL and rbcS in C. sativus, C. × hytivus, and C. hystrix.
2.5. Measurement of RuBisCO Activity and Content
RuBisCO activity and content were measured by enzyme-linked immunosorbent assay (ELISA) using Plant RuBisCO ELISA Kit (SenBeiJia Biotechnology Co., Ltd. Nanjing, China) of the SBJ-P1004-48T and SBJ-Pl031-48T, respectively. About 1 g leaves were ground to a fine powder, and 9 mL extraction solution was added. The sample was then obtained by centrifugation at 4 ºC, 8000 g for 10 min, and the supernatant was taken for the next steps according to the manufacturer’s instructions. The kits use the purified plant RuBisCO antibody to coat the microtiter plate, to prepare solid-phase antibody, then add RuBisCO to the microwell of the coated monoclonal antibody, followed by HRP labeling. The antibody binds to form an antibody-antigen-enzyme antibody complex, which is thoroughly washed and then added to the substrate TMB for color development. Finally, the optical delnsity (OD) value of absorbance was measured with a microplate reader, and the activity and content of RuBisCO in the sample were calculated from the corresponding standard curves. All the samples were performed in three biological replicates. The mid-parent heterosis (MPH) was calculated using the following formula: MPH = (allotetraploid value − mid-parental value)/mid-parental value in %, where allotetraploid value is the average value of C. × hytivus, and the mid-parental value is the average value of the two parents (C. hystrix and C. sativus).
2.6. Statistical Analysis
The qRT-PCR results were expressed as the mean ± standard deviation (SE) of three experimental replicates. The values were subjected to Duncan’s multiple range testing with the SPSS 16.0 (SPSS Inc., Chicago, IL, USA). The binomial test was used to assess the bias in rbcS homeologous expression in C. × hytivus, in which the expression of the parental rbcS homeologous gene was set to the success and failure of the ‘binom.test’ function in R programming language (https://www.r-project.org/) (with the hypothesized probability of success being p = 0.5), respectively.
3. Results
3.1. Sequence Variation of RuBisCO Encoding Genes in Cucumis Allopolyploid
Plastome-encoded rbcL gene and nuclear-encoded rbcS gene in C. × hytivus and parental diploids were cloned and sequenced. There was only one copy of the rbcL ortholog and rbcS ortholog gene in diploid parents, C. hystrix (HH) and C. sativus (CC).
Three SNPs were detected between the parental rbcL orthologs (Figure 1), among the three SNP, one is synonymous, and the other two are non-synonymous (Figure S1). Parental rbcS genes consist of three exons and two introns, and 33 SNPs and three indels were detected between the parental rbcS orthologs, of which 8 SNPs (8/33) and all indels (3/3) occurred within the two introns (Figure 2). Corresponding rbcS amino acid sequence alignment shows 15 amino acid differences (Figure S2).
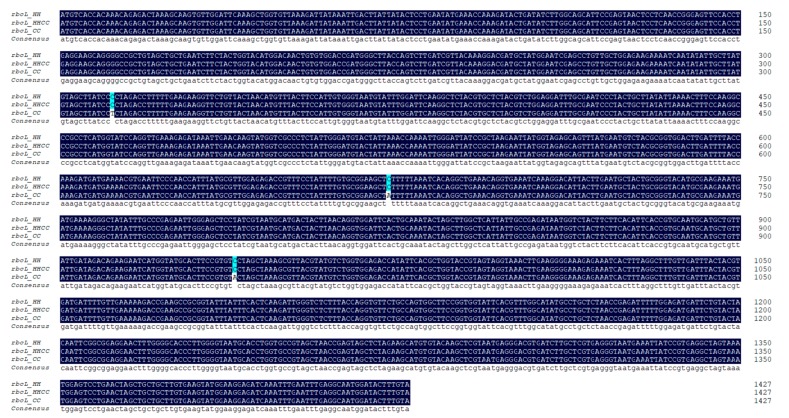
Figure 1.
Sequence alignment of allopolyploid Cucumis × hytivus (HHCC) RuBisCO coding gene rbcL with those from the progenitor diploid (C. hystrix, HH and C. sativus, CC) rbcL genes. The cloned rbcL gene in sampled allotetraploid C. × hytivus is identical to the rbcL gene in its maternal subspecies Cucumis hystrix (HH).
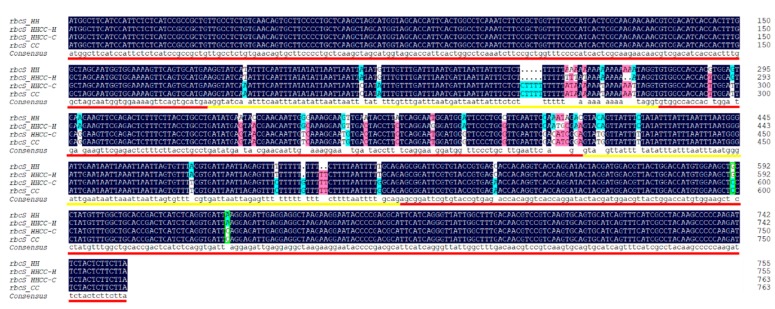
Figure 2.
Sequence alignment of allopolyploid C. × hytivus (HHCC) RuBisCO coding gene rbcS with those from the progenitor diploid (HH and CC) rbcS genes. The red and yellow lines represent the three exons and two introns, respectively. Homoeo-SNPs (homoeo-single nucleotide polymorphisms) at positions 599 (where HH orthologs had a G, but CC orthologs had a C) and 636 (where a HH orthologs had an A, but CC orthologs had a C) are annotated in green boxes.
The rbcL gene in C. × hytivus was confirmed to show maternal inheritance (Figure 1). Sequence alignments of the rbcS gene showed there were two types of rbcS genes in C. × hytivus (HHCC), predicted CC allele was identical with CC rbcS, whereas the HH allele has some mutations compared to the HH rbcS gene, including eight free non-directional mutations and 12 mutations that tend to CC rbcS (Figure 2). Sequence alignment combined with homoeo-SNPs detected in C. × hytivus identified two species-specific SNPs (Figure 2): one of which is located at position 599 in the rbcS genomic sequence (corresponding to position 393 in the cDNA alignment), wherein the HH orthologs have a G but CC orthologs have a C, and the other is located at position 636 in the rbcS genomic sequence (corresponding to position 430 in the cDNA alignment), wherein HH orthologs have an A, but CC orthologs have a C. These SNPs distinguished between paralogs from the H and C genomes in allotetraploid, therefore, these two species-specific SNPs can be used to estimate homoeolog expression in C. × hytivus (HHCC).
3.2. Expression of Maternal Inheritance of the rbcL Gene and Duplicated rbcS Genes in Allotetraploid C. × hytivus
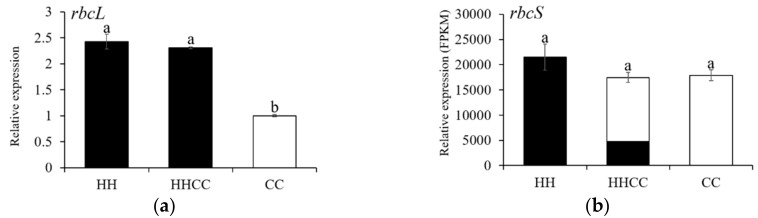
In order to understand how RuBisCO encoded genes perform following genome merger and doubling, qRT-PCR was performed to analyze the expression of rbcL genes in three species of C. hystrix (HH), C. ×hytivus (HHCC), and C. sativus (CC). The rbcL transcriptional level in CC was significantly lower than that in HH and HHCC, and there was no significant difference between HH and HHCC (Figure 3). The expression level of the rbcS gene was analyzed using the method of fragments per kilobase of transcript per million fragments (FPKM) based on the databases of genomic RNAseq, and no significant difference in the expression of the rbcS gene was detected between the two parental diploids HH and CC, suggesting that there is an equal amount of transcription of the rbcS genes relative to the total transcriptome in H- and C-genome progenitors (Figure 3).
Figure 3.
(a) rbcL homoeolog expression and (b) rbcS homoeolog expression in C. hystrix (HH), C. × hytivus (HHCC), and Cucumis sativus (CC). The expression of rbcL and rbcS genes was analyzed in three biological replicates by qRT-PCR and RNA-seq, respectively. H subgenome (black, female parent), C subgenome (white, male parent). Data represents the mean ± standard deviation (SE) (n = 3). The different lower-case letters above the bars show the means are significantly different according to Duncan’s multiple range test at 5% level.
The parental origin of each rbcS and homoeolog in the allotetraploid C. × hytivus can be distinguished by using homoeo-SNPs. The aforementioned two species-specific SNPs were used to estimate homoeolog expression to assess whether there was a biased expression of rbcS genes in C. × hytivus. In all three replicates of C. × hytivus RNA-seq libraries, the observed total expression of C-homoeologs (indicated by C subtotal in Table 1) was significantly higher than that of H-homoeologs (indicated by H subtotal in Table 1) in allopolyploid C. × hytivus (HHCC). Therefore, rbcS expression has significant genomic bias in the direction of its paternal genome donor (Figure 3 and Table 1).
Table 1.
rbcS homoeolog expression in Cucumis allopolyploid C. × hytivus.
| Homoeolog | SNP:393 | SNP:430 | |
|---|---|---|---|
| HHCC-1 | H Subtotal | 687 | 687 |
| C Subtotal | 959 | 805 | |
| Total | 1646 | 1445 | |
| HHCC-2 | H Subtotal | 458 | 359 |
| C Subtotal | 603 | 439 | |
| Total | 1061 | 794 | |
| HHCC-3 | H Subtotal | 440 | 369 |
| C Subtotal | 660 | 558 | |
| Total | 1100 | 927 |
3.3. The Prediction of the RuBisCO Protein–Protein Complex Binding Affinity
In order to find out the possible reasons for paternally-biased rbcS gene expression in C. × hytivus.
PPA-Pred2 was used to predict dissociation constants and binding free energy (ΔG) of RuBisCO complex formed by rbcL and rbcS proteins in C. sativus, C. × hytivus, and C. hystrix, respectively. The results showed that diploid parents have higher binding efficiency than tetraploid, CC diploid’s rbcL and rbcS has higher binding free energy than that of HH diploid. Within the allopolyploid C. × hytivus, rbcS C-homolog has a lower dissociation constant and higher binding free energy for rbcL relative to H homolog (Table 2). The values observed suggest that rbcS C-homoeologs have a higher affinity to rbcL than that of H-homoeologs. The result indicates that the paternal copy of rbcS protein had a stronger interaction with rbcL protein, which possibly explained the transcriptional bias of the paternal-like rbcS homoeologs.
Table 2.
Prediction of the binding affinity of protein –protein complex of rbcL and rbcS.
| rbcL | rbcS | Kd (Dissociation Constant) | ΔG (Binding Free Energy) |
|---|---|---|---|
| rbcL HHCC | rbcS HHCC-H | 1.51·10-07 M | −9.30 kcal/mol |
| rbcS HHCC-C | 6.87·10−08 M | −9.77 kcal/mol | |
| rbcL HH | rbcS HH | 9.30·10−09 M | −10.95 kcal/mol |
| rbcL CC | rbcS CC | 8.19·10−09 M | −11.03 kcal/mol |
HHCC-H: HH allele in HHCC, HHCC-C: CC allele in HHCC.
3.4. The Influence of Allopolyploidization on RuBisCO Content and Activity
To investigate how allopolyploidization affects the function of RuBisCO, we measured and compared the RuBisCO content and activity of C. × hytivus with its diploid parents. When normalized for fresh weight, both RuBisCO content and activity in leaves of C. × hytivus showed positive mid-parent heterosis (MPH, mean that heterotic traits outperform to the average of its two parents) (6.9% and 14.32%, respectively) (Table 3). The result indicates that C. × hytivus show hybrid vigor of RuBisCO.
Table 3.
RuBisCO content and activity in C. hystrix (HH), C. × hytivus (HHCC), and C. sativus (CC).
| HH | HHCC | CC | MPH (%) | |
|---|---|---|---|---|
| RuBisCO content (ng/g FW) | 1.41 ± 0.03 | 1.70 ± 0.03 | 1.77 ± 0.02 | 6.90% |
| RuBisCO activity (μmol/min/g FW) | 7.24 ± 0.17 | 6.86 ± 0.10 | 4.76 ± 0.14 | 14.32% |
FW:Fresh weight; MPH: Mid-parent heterosis. Values represent the mean ± SE (n = 3), the experiment was performed in three biological replicates.
4. Discussion
Allopolyploid speciation often faces the challenges of stoichiometric disruption due to the combination of two divergent nuclear genomes and only one parental set of organelle genomes [21]. Changes in one genomic compartment may influence other genomes evolution in a cell [37]. The RuBisCO is a cytonuclear enzyme complex, making it an ideal system to study plastid–nuclear coordination. It is often used as a window to explore the cytonuclear evolutionary features of allopolyploidization. In the case of allopolyploid C. × hytivus, the maternal plastid interacts with the biparentally inherited nuclear rbcS alleles. The predicted CC allele in C. ×hytivus (HHCC) was identical with CC diploid rbcS gene, whereas the HH allele has some mutations compared to HH diploid rbcS gene, including some mutations without direction and some mutations that tend to CC rbcS, presumably due to being affected by subgenomic dominant or subsequent nucleo-cytoplasmic interactions. The observed total expression of C-homoeologs (paternal) was significantly higher than that of H-homoeologs (maternal) in allopolyploid C. × hytivus (HHCC) in its RNA-seq libraries. This result indicates a significant genomic bias in rbcS expression in the direction of its paternal genome donor in C. × hytivus. The ratio of rbcS: rbcL expression is much higher in CC than in HH or HHCC, there is such a possibility that rbcS’s relatively high expression in CC diploid appears to carry over to the CC homoeolog in the allotetraploid, and cis regulation may play a role in regulating gene expression. Beyond that, the protein–protein interaction prediction indicates that the paternal copy of the rbcS proteins had a stronger interaction with rbcL protein, which might explain the transcriptional bias of the paternal-like rbcS homoeologs. We point out that the heterologous expression of chloroplast-associated nuclear genes in allopolyploids may be related to chloroplast function. Although the expression bias of homoeologous genes in allopolyploids has been reported to be related to nuclear genome stability, studies on the relationship between organelles and nuclear stability have been ignored [38]. We hope this research will be a stepping stone to explore the relationship between chloroplasts and mitochondria with the different parental genome behaviors.
Previous studies about cytonuclear coevolution in allopolyploids reported transcriptional bias of the maternal rbcS homoeologs were detected in five natural allopolyploids, including Arabidopsis, Arachis, Brassica, Gossypium, and Nicotiana [20,29]. However, our research and several recent studies of allopolyploids [21,22,39] do not support the hypothesis of preferential expression of maternal transcripts. In addition to the influence of nucleus-organelle interactions, the expression bias of rbcS in C. × hytivus could also be the weak selection pressure from the limited non-synonymous substation between diverged H- and C-genome rbcL or insufficient evolution time allowed for such initial synthesized HHCC to achieve the ultimate cytonuclear coevolution. The expression of individual genes varies greatly [40], and it does not represent the global genomic bias in transcription, but its related studies can supplement the mechanism of subgenome transcriptome asymmetry. Subgenome expression of both synthetic and natural allopolyploids was conditioned by parental legacy and modified by transcriptome shock. However, natural allopolyploids also experience cumulative effects during the evolutionary process under natural and/or human selection [41]. Different regulatory modules may cause inconsistent expression bias in allopolyploids. We conclude that cytonuclear evolution after genome doubling is a necessary and complicated coevolving process, the direction of evolution could be related to different types of species and evolutionary time.
Differential gene expression can play a role in coordinating cytonuclear interactions in allopolyploids [19]. Allopolyploids may have the advantage of integrating more genetic resources through genome reorganization and flexible gene expression [42]. Chen [43] systematically characterized the roles of nonadditive gene expression, epigenetic regulation, and small RNAs in hybrid vigor. And in this study, both the activity and content of RuBisCO in C. × hytivus showed mid-parent heterosis (MPH) (Table 3), indicating C. × hytivus showed hybrid vigor of RuBisCO. We propose that new nuclear–cytoplasmic combination may be one of the reasons for heterosis in allopolyploid. Another possibility of the departure from mid-parent expectations in allopolyploid may be the outcome of separate inheritance of the nuclear vs. cytoplasmic genomes. It may be possible to verify this theory through a synthetic reciprocal combination. A better understanding of the regulatory mechanisms for heterosis in allopolyploid will help us efficiently select the better combinations of parents to produce the best performing hybrids and polyploids. Further study of the accommodation in other cytonuclear coencoded complexes (including mitochondria and chloroplast related proteins), and comprehensive analysis over generations may provide valuable information on the mechanism of heterosis and the pace of cytonuclear co-evolution.
5. Conclusions
Here, synthesized Cucumis allopolyploid was used to explore the cytonuclear interaction of RuBisCO. The nuclear rbcS gene responded to nuclear–cytoplasmic interactions in the new genetic systems, specifically showed transcriptional paternal bias expression, which supports that maternally-biased gene expression of the cytonuclear enzyme complexes does not always occur in allopolyploid. This pattern of transcriptional bias is highly likely to be related to the physical and chemical properties of protein, in other words, the maternal plastid-encoded rbcL can have priority for the biparental nuclear-encoded rbcS with higher affinity. Our results point out the contribution of protein interaction affinity in the evolution of nuclear genes involved in plastid protein complexes, and we suggest that new nuclear–cytoplasmic combination may be one of the reasons for heterosis in allopolyploid. It is necessary to carry out extensive research on other cytonuclear enzyme complexes in allopolyploids.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/11/869/s1, Figure S1: Alignment of translated CDS regions of allopolyploid C. × hytivus (HHCC) with those from the progenitor diploid (HH and CC) rbcL genes, Figure S2: Alignment of translated CDS regions of allopolyploid C. × hytivus (HHCC) with those from the progenitor diploid (HH and CC) rbcS genes, Table S1: The sequence of primers employed in this study, Table S2: The RNA-seq data used in this study.
Author Contributions
Conceptualization, J.C., X.Y. and Y.Z.; methodology, Y.Z.; software, P.W.; validation, Y.Z., Z.Z. and Y.M.; resources, Q.Z.; data curation, Y.Z. and J.L.; writing—original draft preparation, Y.Z.; writing—review and editing, J.C. and X.Y.; project administration, X.Y. and J.L.; funding acquisition, J.C.
Funding
This research was partially supported by the key program from the National Natural Science Foundation of China (No. 31430075), Natural Science Foundation of Jiangsu Province, China (No. BK20180536), China Postdoctoral Science Foundation (No. 2017M611834), National Key Research and Development Program of China (No. 2016YFD0100204-25) and National Key Research and Development Program of China (No. 2016YFD0101705-5).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Leitch I.J., Bennett M.D. Polyploidy in angiosperms. Trends Plant Sci. 1997;2:470–476. doi: 10.1016/S1360-1385(97)01154-0. [DOI] [Google Scholar]
- 2.Wendel J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000;42:225–249. doi: 10.1023/A:1006392424384. [DOI] [PubMed] [Google Scholar]
- 3.Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C.F., Sankoff D., de Pamphilis C.W., Wall P.K., Soltis P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y.N., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H.Y., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–U113. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 5.Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant. Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Lim K.Y., Kovarik A., Matyasek R., Chase M.W., Clarkson J.J., Grandbastien M.A., Leitch A.R. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- 7.Buggs R.J.A., Chamala S., Wu W., Tate J.A., Schnable P.S., Soltis D.E., Soltis P.S., Barbazuk W.B. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 2012;22:248–252. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Yu X.Q., Wang X.X., Hyldgaard B., Zhu Z.B., Zhou R., Kjaer K.H., Ouzounis T., Lou Q.F., Li J., Cai Q., et al. Allopolyploidization in Cucumis contributes to delayed leaf maturation with repression of redundant homoeologous genes. Plant J. 2018;94:393–404. doi: 10.1111/tpj.13865. [DOI] [PubMed] [Google Scholar]
- 9.Flagel L.E., Wendel J.F. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010;186:184–193. doi: 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- 10.Bottani S., Zabet N.R., Wendel J.F., Veitia R.A. Gene expression dominance in allopolyploids: Hypotheses and models. Trends Plant Sci. 2018;23:393–402. doi: 10.1016/j.tplants.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Paun O., Bateman R.M., Fay M.F., Hedren M., Civeyrel L., Chase M.W. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae) Mol. Biol. Evol. 2010;27:2465–2473. doi: 10.1093/molbev/msq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor W.C. Regulatory interactions between nuclear and plastid genomes. Annu. Rev. Plant Phys. 1989;40:211–233. doi: 10.1146/annurev.pp.40.060189.001235. [DOI] [Google Scholar]
- 13.Crespi B., Nosil P. Conflictual speciation: Species formation via genomic conflict. Trends Ecol. Evol. 2013;28:48–57. doi: 10.1016/j.tree.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Campbell D.R., Waser N.M., Aldridge G., Wu C.A. Lifetime fitness in two generations of ipomopsis hybrids. Evolution. 2008;62:2616–2627. doi: 10.1111/j.1558-5646.2008.00460.x. [DOI] [PubMed] [Google Scholar]
- 15.Reboud X., Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. doi: 10.1038/hdy.1994.19. [DOI] [Google Scholar]
- 16.Wang T., Li Y., Shi Y., Reboud X., Darmency H., Gressel J. Low frequency transmission of a plastid-encoded trait in Setaria italica. Theor. Appl. Genet. 2004;108:315–320. doi: 10.1007/s00122-003-1424-8. [DOI] [PubMed] [Google Scholar]
- 17.Azhagiri A.K., Maliga P. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 2007;52:817–823. doi: 10.1111/j.1365-313X.2007.03278.x. [DOI] [PubMed] [Google Scholar]
- 18.Svab Z., Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc. Natl. Acad. Sci. USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharbrough J., Conover J.L., Tate J.A., Wendel J.F., Sloan D.B. Cytonuclear responses to genome doubling. Am. J. Bot. 2017;104:1277–1280. doi: 10.3732/ajb.1700293. [DOI] [PubMed] [Google Scholar]
- 20.Gong L., Olson M., Wendel J.F. Cytonuclear evolution of rubisco in four allopolyploid lineages. Mol. Biol. Evol. 2014;31:2624–2636. doi: 10.1093/molbev/msu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X.F., Dong Q.L., Li X.C., Yuliang A.Z., Yu Y.N., Li N., Liu B., Gong L. cytonuclear variation of rubisco in synthesized rice hybrids and allotetraploids. Plant. Genome Us. 2017:10. doi: 10.3835/plantgenome2017.05.0041. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira de Carvalho J., Lucas J., Deniot G., Falentin C., Filangi O., Gilet M., Legeai F., Lode M., Morice J., Trotoux G., et al. Cytonuclear interactions remain stable during allopolyploid evolution despite repeated whole-genome duplications in Brassica. Plant J. 2019;98:434–447. doi: 10.1111/tpj.14228. [DOI] [PubMed] [Google Scholar]
- 23.Chen J.F., Staub J.E., Tashiro Y., Isshiki S., Miyazaki S. Successful interspecific hybridization between Cucumis sativus L. and C. C-hystrix Chakr. Euphytica. 1997;96:413–419. doi: 10.1023/A:1003017702385. [DOI] [Google Scholar]
- 24.Chen J.F., Kirkbride J.H. A new synthetic species of Cucumis (Cucurbitaceae) from interspecific hybridization and chromosome doubling. Brittonia. 2000;52:315–319. doi: 10.2307/2666583. [DOI] [Google Scholar]
- 25.Havey M.J., McCreight J.D., Rhodes B., Taurick G. Differential transmission of the Cucumis organellar genomes. Theor Appl Genet. 1998;97:122–128. doi: 10.1007/s001220050875. [DOI] [Google Scholar]
- 26.Shen J., Zhao J., Bartoszewski G., Malepszy S., Havey M., Chen J.F. Persistence and protection of mitochondrial DNA in the generative cell of cucumber is consistent with its paternal transmission. Plant. Cell Physiol. 2015;56:2271–2282. doi: 10.1093/pcp/pcv140. [DOI] [PubMed] [Google Scholar]
- 27.Shen J., Kere M.G., Chen J.F. Mitochondrial genome is paternally inherited in Cucumis allotetraploid (C. x hytivus) derived by interspecific hybridization. Sci. Hortic-Amsterdam. 2013;155:39–42. doi: 10.1016/j.scienta.2013.03.009. [DOI] [Google Scholar]
- 28.Murraary M.G., Thompsom W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong L., Salmon A., Yoo M.J., Grupp K.K., Wang Z.N., Paterson A.H., Wendel J.F. The cytonuclear dimension of allopolyploid evolution: An example from cotton using rubisco. Mol. Biol. Evol. 2012;29:3023–3036. doi: 10.1093/molbev/mss110. [DOI] [PubMed] [Google Scholar]
- 30.Li J., Li Z.G., Tang L., Yang Y.W., Zouine M., Bouzayen M. A conserved phosphorylation site regulates the transcriptional function of ETHYLENE-INSENSITIVE3-like1 in tomato. J. Exp. Bot. 2012;63:427–439. doi: 10.1093/jxb/err289. [DOI] [PubMed] [Google Scholar]
- 31.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q.D., et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yugandhar K., Gromiha M.M. Protein-protein binding affinity prediction from amino acid sequence. Bioinformatics. 2014;30:3583–3589. doi: 10.1093/bioinformatics/btu580. [DOI] [PubMed] [Google Scholar]
- 37.Sloan D.B. Using plants to elucidate the mechanisms of cytonuclear co-evolution. New Phytol. 2015;205:1040–1046. doi: 10.1111/nph.12835. [DOI] [PubMed] [Google Scholar]
- 38.Ge X.H., Ding L., Li Z.Y. Nucleolar dominance and different genome behaviors in hybrids and allopolyploids. Plant Cell Rep. 2013;32:1661–1673. doi: 10.1007/s00299-013-1475-5. [DOI] [PubMed] [Google Scholar]
- 39.Sehrish T., Symonds V.V., Soltis D.E., Soltis P.S., Tate J.A. Cytonuclear coordination is not immediate upon allopolyploid formation in Tragopogon miscellus (Asteraceae) allopolyploids. Plos ONE. 2015;10:e0144339. doi: 10.1371/journal.pone.0144339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams K.L., Cronn R., Percifield R., Wendel J.F. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X.T., Zhang H.K., Li Y.L., Zhang Z.B., Li L.F., Liu B. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 2016;209:1264–1277. doi: 10.1111/nph.13678. [DOI] [PubMed] [Google Scholar]
- 42.Barker M.S., Arrigo N., Baniaga A.E., Li Z., Levin D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016;210:391–398. doi: 10.1111/nph.13698. [DOI] [PubMed] [Google Scholar]
- 43.Chen Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant. Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.