Abstract
The keratin-associated proteins (KAPs) are structural components of hair/wool fibres. All of the KAPs identified to date contain cysteine, which is thought to form disulphide bonds cross-linking the keratin intermediate filaments. Here, we report the identification of a KAP gene in sheep that would produce a protein that contains a high proportion (63.2 mol%) of glycine and tyrosine, but would not contain any cysteine. This suggests that other forms of intra- and inter-strand interaction may occur with this KAP, such as interactions via ring-stacking and hydrogen-bonding. The gene was dissimilar to any previously reported KAP gene, and was therefore assigned to a new family, and named KRTAP36-1. The KRTAP36-1 genome sequence was almost identical to some EST sequences from sheep and goat skin follicles, suggesting that it is present and expressed in sheep and goats. A BLAST search of the human genome assembly sequence did not reveal any human homologue. Three variant sequences (named A to C) of ovine KRTAP36-1 were identified and four single nucleotide polymorphisms (SNPs) were detected. One SNP was located 32 bp upstream of the coding region, and all of the others were in the coding region and were nonsynonymous. After correcting for potential linkage to the proximal KRTAP20-1, variant B of KRTAP36-1 was found to be associated with increased prickle factor (PF) in wool, suggesting that variation in the gene may have the potential to be used as gene marker for breeding sheep with lower PF.
Keywords: Keratin-associated protein KAP36-1 gene (KRTAP36-1), variation, wool traits, prickle factor, sheep
1. Introduction
Wool fibre is primarily composed of hard α-keratins. These are cysteine-rich, particularly in their head and tail domains [1]. These α-keratins are assembled into keratin intermediate filaments (KIFs), and then embedded in an inter-filamentous matrix comprised of small proteins called the keratin-associated proteins (KAPs). Three broad groups of KAPs have been defined: the high sulphur (HS) KAPs with less than 30 mol% of cysteine, the ultrahigh sulphur (UHS) KAPs with more than 30 mol% of cysteine and the high glycine-tyrosine (HGT) KAPs with 35–60 mol% glycine and tyrosine [2].
The KAP proteins are thought to cross-link KIFs via disulphide bonds [3], but the precise mechanism of linking is still poorly understood. The argument for disulphide cross-linking is in agreement with the observation that the majority (approximately 97.5%) of cysteines in wool are found to be part of disulphide bridges [4] and that most hard α-keratins and KAPs are cysteine rich, even the HGT-KAPs. Despite being more typically rich in glycine and tyrosine, all of the HGT-KAPs identified to date contain cysteine, ranging from 3.2 mol% in ovine KAP8-2 [5] to 14.9 mol% in ovine KAP20-2 [6]. Little is known about whether the HGT-KAPs contribute to cross-linking via cysteine-based disulphide bonding, as the HS- and UHS- KAPs appear to.
There are 17 known KAP gene families (designated as KRTAPs), including seven families (KAP6-KAP8 and KAP19-KAP22) that encode the HGT-KAP proteins in humans [7]. Many of the human KRTAP orthologs remain unidentified in sheep, with only eight having been characterised to date [2,6,8,9]. Three additional HGT-KRTAPs (KRTAP6-4, KRTAP6-5 and KRTAP8-2), which are absent in humans, have been identified in sheep [5,10]. This suggests that sheep have more HGT-KRTAPs than humans, and support the idea that the HGT-KRTAPs play an important role in determining some of the characteristics of the wool fibre.
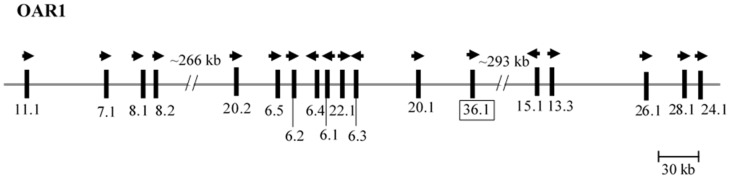
In sheep, the HGT-KRTAPs are clustered on chromosome 1, in a region that is approximately 723-kb in size, and that is between two HS-KRTAPs (KRTAP11-1 and KRTAP15-1) [8]. Bioinformatics analysis of this region led us to identify an open reading frame (ORF) that would encode a glycine and tyrosine-rich protein. This is located near to HGT-KRTAP20-1, and, in this study, the identity of this ORF was investigated, sequence variation in this ORF was described and its effect on some wool traits is reported.
2. Materials and Methods
This research was undertaken in accordance with the Animal Welfare Act 1999 (New Zealand Government) and the collection of sheep blood drops by the nicking of their ears was covered by Section 7.5 Animal Identification, in: Code of Welfare: Sheep and Beef Cattle (2016); a code of welfare issued under the Animal Welfare Act 1999 (New Zealand Government).
2.1. Sheep Investigated and Wool Samples
A total of 415 sheep were investigated. These included 46 New Zealand (NZ) Romney sheep (sourced from five farms that are not believed to be connected genetically), 48 Merino sheep (sourced from five farms that are not believed to be connected genetically) and 321 Southdown × Merino-cross lambs (sourced from the same farm, but from six sire-lines).
The association studies were carried out on the 321 Southdown × Merino-cross lambs. All these lambs were ear-tagged with an identification number at birth, and their birth dates, birth weights, birth ranks (i.e., whether they were a single, twin or triplet), gender and dam identity were recorded. All the lambs were managed as a single mob on the same farm up to weaning, when they were separated to two mobs based on their gender. They were shorn at twelve months of age. At shearing, the greasy fleece weight (GFW) was measured for each lamb, and a wool sample was collected from the mid-side region for wool trait measurement using International Wool Textile Organisation (IWTO) standardised methods, at the New Zealand Wool Testing Authority Ltd. (NZWTA, Napier, NZ). This included measurement of wool yield (Yield), mean staple length (MSL), mean staple strength (MSS), mean fibre diameter (MFD), fibre diameter standard deviation (FDSD), coefficient of variation of fibre diameter (CVFD), mean fibre curvature (MFC) and prickle factor (PF; the percentage of fibres of diameter greater than 30 microns). Lamb clean fleece weights (CFWs) were calculated from the GFW and Yield measurements.
A sample of blood from each sheep was collected onto TFN paper (Munktell Filter AB, Sweden) and genomic DNA was purified using a two-step washing techniques detailed in Zhou et al. [11].
2.2. PCR Amplification of the Newly Identified Open Reading Frame
A 174-bp ORF that appeared to encode a glycine and tyrosine-rich protein was identified near ovine KRTAP20-1, at position nt123318135–123318308 (NC_019458.2) on chromosome 1. Sequences flanking this ORF were used to design two PCR primers to amplify a 367-bp fragment spanning the entire ORF. These primers were 5’-GGTTTACCACACCCACAATG-3’ and 5’-GTAGCATAGCAAGAGTGAAG-3’, and they were synthesised by Integrated DNA Technologies (Coralville, IA, USA).
PCR amplification was performed in a 15-μL reaction containing the genomic DNA on one 1.2-mm punch of TFN paper, 0.25 μM of each primer, 150 μM of each dNTP (Eppendorf, Hamburg, Germany), 2.5 mM of Mg2+, 0.5 U of Taq DNA polymerase (Qiagen, Hilden, Germany) and 1× the reaction buffer supplied with the enzyme. The thermal profile consisted of an initial denaturation for 2 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C, and with a final extension of 5 min at 72 °C. Amplification was carried out in S1000 thermal cyclers (Bio-Rad, Hercules, CA, USA).
2.3. Screening for Sequence Variation and Variant Sequencing
PCR amplicons were subject to SSCP analysis to screen for sequence variation. A 0.7 μL aliquot of each amplicon was mixed with 7 μL of loading dye (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene-cyanol). After denaturation at 95 °C for 5 min, samples were placed rapidly on wet ice and then loaded on 16 cm × 18 cm, 14% acrylamide: bisacrylamide (37.5:1) (Bio-Rad) gels. Electrophoresis was performed using Protean II xi cells (Bio-Rad), at 300 V for 18 h at 11 °C in 0.5 × TBE buffer. The gels were silver-stained by the method described by Byun et al. [12].
PCR amplicons representative of different SSCP patterns from sheep that appeared to be homozygous were sequenced at the Lincoln University DNA Sequencing Facility. For those variants that were only found in heterozygous sheep, they were sequenced using a rapid approach described previously [13]. In this approach, a band corresponding to the variant was excised as a gel slice from the polyacrylamide gel, macerated and then used as a template for reamplification with the original primers. This second amplicon was then sequenced.
2.4. Sequence Analyses
Sequence alignments, translations, comparisons and the construction of phylogenetic tree were carried out using DNAMAN (version 5.2.10, Lynnon BioSoft, Vaudreuil, Canada). The BLAST algorithm was used to search the NCBI GenBank (www.ncbi.nlm.nih.gov/) databases for homologous sequences.
2.5. Genotyping of KRTAP20-1
The 321 Southdown × Merino-cross lambs used for the association analyses were also genotyped for variation in KRTAP20-1 using a PCR-SSCP technique described previously [8]. Briefly, KRTAP20-1 was amplified using the PCR primers 5’-TCATATTCTGCAAGCAAAGGC-3’and 5’-GCTGATGGGTCTCAGTCAC-3’. After denaturation, amplicons were electrophoresed using 14% acrylamide: bisacrylamide (37.5:1) (Bio-Rad) gels containing 1.0% v/v glycerol, at 8 °C and 390 V for 18 h. Polymerase chain reaction amplicons of the previously described variants [8] were included as references to determine genotypes in the gels.
2.6. Statistical Analyses of Associations
All the statistical analyses were undertaken using Minitab version 16 (Minitab Incorporated, State College, PA, USA).
General linear mixed-effect models (GLMMs) were employed to individually evaluate the effect of the presence or absence (coded as “1” or “0”) of the three variants of the ORF (A, B and C), on the ten wool traits that had been measured or calculated. In these models, gender and sire were included as fixed and random factors respectively, as they affected all of the wool traits. Differences in the marginal means derived from these models were considered to be significant when p < 0.05, and trends were noted when 0.05 ≤ p < 0.10.
As a consequence of variants occurring in genotypes, it is possible that the effect of one variant in the genotype is affected by the presence of the other variant in that genotype. Accordingly, any variant sequence of the ORF in the initial GLMMs, which had an association with a wool trait of p < 0.200 and thus was potentially associated with the trait (albeit at a low threshold), was included as an explanatory factor in a second set of multivariant presence/absence models. Once again, gender and sire were included as fixed and random factors respectively in this second set of models, as they affected all the wool traits. Differences in the marginal means derived from these models were once again considered to be significant when p < 0.05, and trends were noted when 0.05 ≤ p < 0.10.
Finally, given that variation in the nearby gene KRTAP20-1 has been described as affecting wool yield and mean fibre diameter-associated traits [8], and to test whether the associations identified above between the ORF variants and variation in the wool traits was as a consequence of proximity to KRTAP20-1, a third set of GLMMs that included KRTAP20-1 genotype as an explanatory factor, were subsequently undertaken. Once again, gender and sire were included as fixed and random factors respectively in this third set of models, as they affected all the wool traits. Differences in the marginal means derived from these models were once again considered to be significant when p < 0.05, and trends were noted when 0.05 ≤ p < 0.10.
Birth rank was not found to affect the ten wool traits and thus it was not included as an explanatory factor in any of the above models.
3. Results
3.1. Identification of KRTAP36-1 in Sheep and the Absence of a Homologue in the Human Genome
The ORF at nt123318135–123318308 (NC_019458.2) was located between KRTAP20-1 and KRTAP15-1 on sheep chromosome 1 (Figure 1). This ORF had a nucleotide sequence that was different to all of the ovine KRTAPs identified to date, but shared 99% identity to two GenBank sequences labelled as ovine KRTAP16-1 (KF543056.1) and caprine KRTAP16-1 (AY502950.1). A BLAST search of the NCBI Expressed Sequence Tag (EST) database, revealed that this ORF sequence had 99% identity to eight ovine mRNA sequences (JK724590.1, GO705930.1, EE851605.1, EE847453.1, EE753136.1, GO779858.1, EE848868.1 and EE848117.1), and 98% identity to one caprine mRNA sequence (CD052106.1) derived from skin tissue/wool follicle. This suggests that this ORF is expressed in the wool follicle and the sequence differences between this ORF and ovine ESTs may reflect sequence variation, or errors in RT-PCR and/or genomic sequencing.
Figure 1.
KRTAPs identified on the sheep chromosome 1 region that contains a newly identified KRTAP36-1. The newly identified gene is shown in a box, and the 17 previously identified KRTAPs are also shown. Vertical bars represent the location of different KRTAPs and the arrowheads indicate the direction of transcription. The numbers below the bars indicate the name of the respective KAP genes (i.e., 11.1 is KRTAP11-1). The nucleotide distances are approximately and refer to NC_019458.2.
The ORF was predicted to encode a protein of 57 amino acid residues. Five amino acids were common (totalling 93 mol%) in this protein, with the most common being glycine (35.1 mol%), followed by tyrosine (28.1 mol%), serine (14.0 mol%), leucine (8.8 mol%) and phenylalanine (7.0 mol%). The protein would not contain any cysteine, which excludes it from being assigned to either the HS- or UHS-KAP groups.
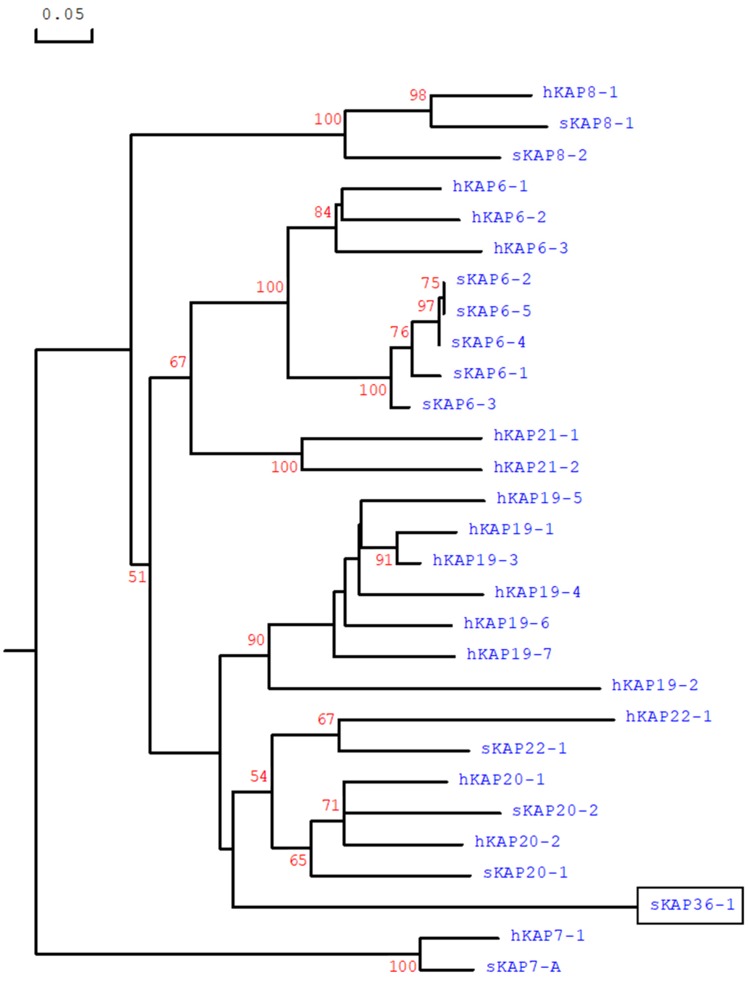
Phylogenetic analysis of this ORF and all of the HGT-KAP genes identified to date, revealed that it was separated from all known HGT-KAP families and the distance of separation suggested it should be designated as a new KAP family (Figure 2). Despite only 28 KAP families (KAP1 to KAP28) have been confirmed across mammalian species [2,7,14], the names KAP29-KAP35 have been used for some sequences reported in public databases. In this context and to avoid confusion this ORF was named SHEEP-KRTAP36-1, according to the updated KRTAP/KAP nomenclature [15].
Figure 2.
Phylogenetic tree of the HGT-KAPs identified in sheep and human. The tree was constructed using the predicted amino acid sequences. The numbers at the forks indicate the bootstrap confidence values and only those equal to or higher than 50% are shown. The sheep KAPs are indicated with a prefix “s”, whereas the human sequences are indicated with “h”. The newly identified ORF is designated as sheep KAP36-1 gene and is indicated in box. The GenBank accession numbers for other sheep HGT-KAPs are NM_001193399 (sKAP6-1), KT725832 (sKAP6-2), KT725837 (sKAP6-3), KT725840 (sKAP6-4), KT725845 (sKAP6-5), X05638 (sKAP7-1), X05639 (sKAP8-1), KF220646 (sKAP8-2), MH243552 (sKAP20-1), MH071391 (sKAP20-2) and KX377616 (sKAP22-1). The GenBank accession numbers for human HGT-KAPs are: NM_181602 (hKAP6-1), NM_181604 (hKAP6-2), NM_181605 (hKAP6-3), AJ457063 (hKAP7-1), AJ457064 (hKAP8-1), AJ457067 (hKAP19-1), NM_181608 (hKAP19-2), NM_181609 (hKAP19-3), NM_181610 (hKAP19-4), NM_181611 (hKAP19-5), NM_181612 (hKAP19-6), NM_181614 (hKAP19-7), NM_181615 (hKAP20-1), NM_181616 (hKAP20-2), NM_181619 (hKAP21-1), NM_181617 (hKAP21-2) and NM_181620 (hKAP22-1).
A BLAST search of the human Genome Assembly GRCh38.p13 using this ORF did not reveal any homologue in the human genome, and the closest similarity (84%) was to KRTAP19-3.
3.2. Variation in Ovine KRTAP36-1
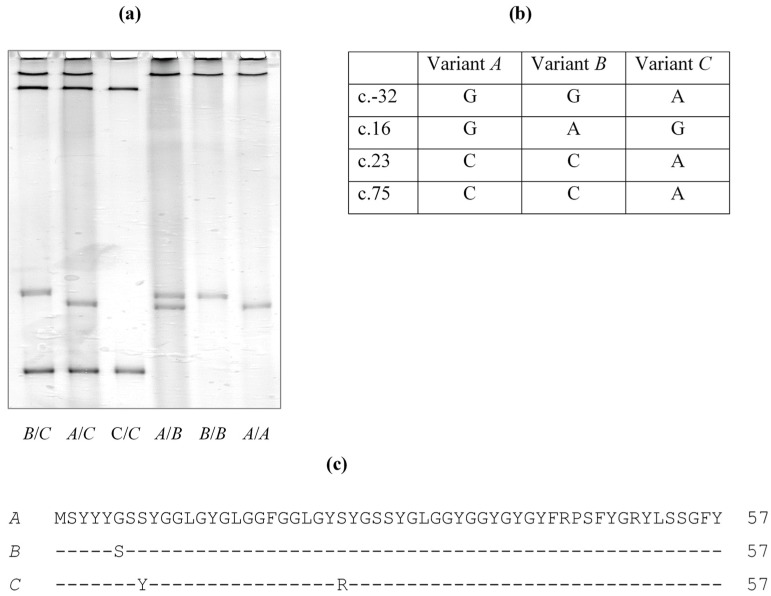
None of the ovine EST sequences were identical to the ORF sequence reported in the sheep assembly sequence, with each having one or two nucleotides different when compared to the ORF sequence, and with a total of four nucleotide differences being observed at positions c.16, c.23, c.75 and c.-32.
To determine whether these EST nucleotide differences result from sequence variation in the gene, potential variation in ovine KRTAP36-1 was screened for using a PCR-SSCP approach. Three banding patterns representing three variants (A to C) were detected (Figure 3) and four single nucleotide polymorphism (SNPs) were detected, including one SNP (c.-32G/A) upstream of the coding region, and three SNPs (c.16G/A, c.23C/A and c.75C/A) in the coding region. All of the coding SNPs were nonsynonymous and would result in the amino acid substitutions p.Gly6Ser, p.Ser8Tyr and p.Ser25Arg. These coding region SNPs match well with three (c.16, c.23 and c.75) of the four nucleotide differences between the ORF sequence and the sheep genome assembly, and with the EST sequences. These variant sequences were deposited into GenBank with accession numbers MK770620-MK770622.
Figure 3.
Polymorphism of ovine KRTAP36-1. (a) Three variants (A to C) in either homozygous or heterozygous forms were detected by PCR-SSCP. (b) Four single nucleotide polymorphisms (SNPs) were found in these variants. (c) Three of the SNPs would lead to amino acid changes and result in three different amino acid sequences.
All of the KRTAP36-1 variants were found in the Merino and Romney breeds, but at different frequencies. Of the 48 Merino sheep, three were AA, five were AB, ten were AC, four were BB, 14 were BC and 12 were CC, with variants A, B and C being present at frequencies of 21.9%, 28.1% and 50.0%, respectively. Of the 46 Romney sheep, one was AA, eight were AB, three were AC, four were BB, 15 were BC and 15 were CC, with frequencies of 14.1%, 33.7% and 52.2% being detected for variants A, B and C, respectively.
Of the four SNPs identified, three (c.-32G/A, c.23C/A, and c.75C/A) were found to be in linkage. Near to these SNPs, a Chi-like sequence (5’-GCTGGTGA-3’) was found at positions c.-66 to c.-59.
3.3. Effect of KRTAP36-1 Variation on Wool Traits
When only KRTAP36-1 was considered in GLMMs, the presence of variant A was found to be associated with increased GFW. Variant B was found to be associated with decreased PF in the single-variant GLMMs, but the association disappeared in the multivariant GLMMs. Variant C was associated with an increase in MFD and PF, and the association with PF persisted in the multivariant GLMMs (Table 1).
Table 1.
Association between the absence or presence of KRTAP36-1 variants and various wool traits.
| Trait 1 | Variant Assessed 2 | Other Variants Fitted | Other KRTAP Fitted | (Mean ± SE) 3 | P | |
|---|---|---|---|---|---|---|
| Absent | Present | |||||
| GFW | A | None | None | 2.39 ± 0.04 | 2.49 ± 0.04 | 0.035 |
| (kg) | B | None | None | 2.46 ± 0.04 | 2.43 ± 0.04 | 0.616 |
| C | None | None | 2.46 ± 0.04 | 2.44 ± 0.04 | 0.680 | |
| A | None | KRTAP20-1 | 2.44 ± 0.05 | 2.49 ± 0.04 | 0.345 | |
| CFW (kg) |
A | None | None | 1.76 ± 0.03 | 1.81 ± 0.03 | 0.172 |
| B | None | None | 1.79 ± 0.03 | 1.79 ± 0.03 | 0.968 | |
| C | None | None | 1.81 ± 0.04 | 1.78 ± 0.03 | 0.359 | |
| Yield | A | None | None | 73.2 ± 0.62 | 72.2 ± 0.56 | 0.133 |
| (%) | B | None | None | 72.4 ± 0.56 | 72.8 ± 0.62 | 0.572 |
| C | None | None | 72.7 ± 0.65 | 72.5 ± 0.54 | 0.742 | |
| MSL (mM) |
A | None | None | 83.2 ± 1.22 | 83.6 ± 1.11 | 0.723 |
| B | None | None | 83.3 ± 1.11 | 83.7 ± 1.23 | 0.765 | |
| C | None | None | 83.0 ± 1.28 | 83.7 ± 1.08 | 0.634 | |
| MFD | A | None | None | 19.5 ± 0.18 | 19.5 ± 0.16 | 0.913 |
| (µM) | B | None | None | 19.6 ± 0.16 | 19.4 ± 0.18 | 0.208 |
| C | None | None | 19.3 ± 0.19 | 19.7 ± 0.16 | 0.025 | |
| C | None | KRTAP20-1 | 19.7 ± 0.20 | 20.0 ± 0.17 | 0.081 | |
| FDSD | A | None | None | 4.10 ± 0.06 | 4.10 ± 0.06 | 0.922 |
| (µM) | B | None | None | 4.13 ± 0.06 | 4.04 ± 0.06 | 0.164 |
| C | None | None | 4.01 ± 0.07 | 4.14 ± 0.06 | 0.053 | |
| C | B | None | 4.01 ± 0.07 | 4.13 ± 0.06 | 0.099 | |
| C | B | KRTAP20-1 | 4.12 ± 0.07 | 4.22 ± 0.06 | 0.179 | |
| CVFD | A | None | None | 20.9 ± 0.23 | 20.9 ± 0.21 | 0.930 |
| (%) | B | None | None | 21.0 ± 0.21 | 20.8 ± 0.23 | 0.403 |
| C | None | None | 20.8 ± 0.24 | 21.0 ± 0.20 | 0.395 | |
| MSS | A | None | None | 23.7± 0.79 | 23.8 ± 0.72 | 0.951 |
| (N/ktex) | B | None | None | 24.2 ± 0.72 | 23.1 ± 0.80 | 0.190 |
| C | None | None | 23.7 ± 0.83 | 23.8 ± 0.70 | 0.876 | |
| PF | A | None | None | 2.72 ± 0.32 | 2.57 ± 0.29 | 0.681 |
| (%) | B | None | None | 2.91 ± 0.29 | 2.25 ± 0.32 | 0.061 |
| C | None | None | 2.04 ± 0.33 | 2.99 ± 0.28 | 0.007 | |
| B | C | None | 2.69 ± 0.31 | 2.31 ± 0.32 | 0.765 | |
| C | B | None | 2.10 ± 0.33 | 2.91 ± 0.29 | 0.031 | |
| C | B | KRTAP20-1 | 2.71 ± 0.37 | 3.49 ± 0.32 | 0.038 | |
| CURV | A | None | None | 87.9 ± 1.61 | 88.3 ± 1.47 | 0.817 |
| (°/mM) | B | None | None | 88.3 ± 1.46 | 88.0 ± 1.62 | 0.891 |
| C | None | None | 86.9 ± 1.68 | 88.9 ± 1.42 | 0.269 | |
1 GFW: Greasy fleece weight; CFW: Clean fleece weight; Yield: wool yield; MFD: Mean fibre diameter; FDSD: Fibre diameter standard deviation; CVFD: Coefficient of variation of fibre diameter; MSL: Mean staple length; MSS: Mean staple strength; CURV: Curvature; PF: Prickle factor (percentage of fibres over 30 microns). 2 Of the 321 Southdown × Merino-cross lambs, KRTAP36-1 variant A was present in 195 lambs and absent in 126 lambs, variant B was present in 139 lambs and absent in 182 lambs, and variant C was present in 194 lambs and absent in 127 lambs. 3 Predicted means and standard error of these means derived from GLMMs with various factors being included into the models for different wool traits as described in Materials and Methods.
Given that variation in KRTAP20-1 (a gene located near to KRTAP36-1) has been reported to affect wool weight and mean fibre diameter-associated traits [8], and to test whether the associations detected above were because of the effect of KRTAP20-1, the GLMMs were then corrected for variation in KRTAP20-1. All of the associations disappeared or became a trend, except for the association between KRTAP36-1 and PF, where the association persisted (Table 1).
4. Discussion
This study has identified a new KAP gene on sheep chromosome 1. This gene is comprised of one exon, appears to be expressed, and the protein encoded for is rich in glycine and tyrosine. The gene is clustered with all of the other known HGT-KAP genes on sheep chromosome 1, but it does not share high sequence similarity to any known HGT-KAP gene. It would however appear to be phylogenetically related to the HGT-KAP genes (Figure 2).
These characteristics led us to identify this gene as a HGT-KAP gene, and to assign it into a new KAP family. While two GenBank sequences (KF543056.1 and AY502950.1) that are similar to this sequence are designated as KRTAP16-1, the assignment of these sequences into the KAP16 family is inappropriate, as KAP16 is a HS-KAP family, and in sheep the name has already been used for other HS-KAP genes [16] that are not related to this newly identified HGT-KAP gene. This gene was therefore named SHEEP-KRTAP36-1, this being a KAP family name that has not been used previously.
The gene is located in a chromosome region near to KRTAP20-1, a gene for which the location and transcription direction differ between sheep and humans [8]. This suggests this region of the chromosome may have evolved via different pathways in sheep and humans, and thus it is perhaps not surprising that KRTAP36-1 is present in sheep and goats, but absent in humans.
The protein (KAP36-1) putatively produce by this gene appears to possess a high content (63.2 mol%) of glycine and tyrosine, but it does not contain cysteine. Cysteines are commonly found in keratins and KAPs, and they are thought to form disulphide bonds that cross-link the KIFs and KAPs. The absence of cysteine in KAP36-1, suggests that the other forms of cross-linking may occur. In this respect, tyrosine is an aromatic amino acid containing a benzene ring. The possession of this stable ring structure may allow tyrosine to interact with other tyrosine residues and other aromatic amino acids via a ring-stacking mechanism. This has been reported for other aromatic amino acid-containing proteins [17]. In the HGT-KAPs, the tyrosine residues are usually surrounded by glycine residues. Having the smallest residue (glycine) in proximity to the tyrosine, will allow the tyrosine residues greater conformational freedom to move their benzene rings into a preferred orientation, and thus enable the formation of stronger amino acid to amino acid interactions. Tyrosine also possesses a hydroxyl group, which can act as a hydrogen donor and form hydrogen bond interactions with the centre of the benzene ring from another tyrosine, or other aromatic amino acids [18]. This would make the ring-stacking interaction even stronger. This kind of interaction is expected to result in the wool fibre being strengthened, while simultaneously giving some degree of pliability [19].
Unlike the covalent disulphide bonding, ring-stacking and hydrogen bonding do not require any additional covalent bond formation and they could readily form soon after the proteins are synthesised. Therefore, we hypothesise that these types of interaction may serve as the primary interactions that cross-link the KIFs and stabilise the wool fibre structure, and that this occurs prior to the formation of disulphide bonds. This is in agreement with the observation that the HGT-KAPs are expressed first among the KAPs in the wool follicle [20], and immediately following intermediate filament synthesis. In that capacity, they may therefore play a key role in the assembly of KIFs, and hence act as a key determinant of fibre structure.
When the effect from a nearby KRTAP was corrected for, there was a loss or weakening of the associations with GFW, MFD and FDSD. This suggests that these associations are not due to the effect of KRTAP36-1, but instead result from the linkage with other KRTAPs nearby. This highlights the importance of correcting for the effect of other KRTAPs that are in proximity on the same chromosome, and such that a more precise indication of how any given gene may be affecting wool traits can be obtained. The persistence of the association with PF after the correction for the effect of KRTAP20-1 variation (which has also been shown to affect PF), strengthens the finding.
Though the potential functional effect of the SNP in the 5’-UTR should not be ignored, as 5’-UTR SNPs may affect gene expression [21], it is interesting to note that all of the coding region SNPs were nonsynonymous. Of these, two were in linkage with the 5’-UTR SNP. The only coding-region SNP that was not in the linkage with other SNPs was c.16G/A, which was the only sequence difference between variants A and B.
Variant B was found to be associated with a high PF, but variant A was not. The SNP c.16G/A would result in the substitution of glycine by serine at the 6th amino acid residue in the protein encoded by variant B, with a string of three tyrosine residues at positions 3 to 5. The substitution of glycine by serine at position 6 may have an impact on the conformational freedom of this string of tyrosine residues, with this potentially impacting ring-stacking and/or hydrogen bonding, and with a consequent reduction in fibre compactness or density, and consequently a higher PF.
The percentage of fibres over a given diameter threshold (typically 30 microns), is an indicator of the relative comfort of wool fibres worn next to the skin. Fibres over 30 microns in diameter tend to bend less and produce a “prickle” sensation on the skin’s surface, and with more than 5% of the total number of fibres, the effect tends to be quite noticeable (SGS Wool Testing Services 2011) [22]. The finding of association between KRTAP36-1 and PF suggest that KRTAP36-1 has potential to be used as a gene marker for breeding sheep to produce wool with reduced PF, and that this could add value to fine wool production.
Sheep have primary and secondary wool follicles. The fibres produced by secondary follicles are finer while the fibres produced by primary follicles are usually much larger. The observation that KRTAP36-1 variation is only associated with variation in PF, and no other fibre diameter-associated traits such as MFD, FDSD and CVFD, suggests that KRTAP36-1 may affect or reflect the secondary to primary wool follicle ratio (S/P ratio), and that this consequently affects PF. This would require confirmation with an analysis of the S/P ratios in sheep carrying the different variants of KRTAP36-1. A putative function for KRTAP36-1 in regulating or determining S/P ratios, or as a consequence of variation in S/P ratio that has come about for another reason, may explain the absence of this gene in humans, as humans only have one type of follicle.
Merino sheep usually have a higher S/P ratio and the wool produced has a lower PF than Romney sheep. In the sheep populations investigated in this study, B was found at a higher frequency in Romney sheep, than Merino sheep. This supports the contention that variant B of KRTAP36-1 was associated with increased PF.
The presence of four SNPs in a 327-bp PCR fragment (excluding the primer binding regions) corresponds to a density of 12.2 SNPs per kb. This is much higher than the average density of 4.9 SNPs per kb across the sheep genome suggested by Kijas et al. [23]. This suggests that KRTAP36-1 has far less functional constraint than other parts of the genome. This is consistent with the trend reported for many other KRTAPs [24,25,26,27,28]. Little is known of how the variation in KRTAPs has come about, but the lineage of SNPs and the presence of a Chi-like sequence suggests that gene conversion or nonreciprocal genetic exchange may have occurred in KRTAP36-1. Gene conversion or nonreciprocal genetic exchange has been suggested previously to be a mechanism for generating variation in other KRTAPs [24,29,30].
The HGT-KRTAPs are clustered on one chromosome region surrounding by HS-KRTAPs. With the identification of KRTAP36-1, the number of HGT-KRTAPs identified in sheep has risen from eleven to twelve. Of these, the effect on wool traits has been investigated for eight HGT-KRTAPs: KRTAP6-1 [31,32], KRTAP6-3 [33], KRTAP8-1 [34], KRTAP8-2 [35], KRTAP20-2 [6], KRTAP20-1 [8], KRTAP22-1 [9] and now KRTAP36-1. In Merino-cross sheep, KRTAP6-1 is reported to affect wool yield, MFD, FDSD, CVFD and PF [31]; KRTAP6-3 is reported to affect MFD, FDSD and PF [32]; KRTAP8-1 is reported to affect wool fibre staple strength and curvature [34]; KRTAP22-1 is reported to affect wool yield [9], KRTAP20-2 is reported to affect MFC [6], KRTAP20-1 is reported to affect GFW, wool yield, MFD, FDSD and PF [8]; and, in this study, KRTAP36-1 is found to affect PF. In the early life of Chinese Tan sheep, KRTAP6-1 is reported to affect wool growth, crimp number and the degree of crimping [32], while KRTAP8-2 is found to affect wool growth and degree of crimping [35].
The finding that the HGT-KRTAPs have similar but unique effects on wool traits, suggests that all the HGT-KRTAPs may contribute to cross-linking roles in the wool fibre, but with different HGT-KRTAPs contributing differently to the extent, timing and/or mechanisms of cross-linking.
Acknowledgments
The authors thank Freeman Fang for technical support and Andrea Hogan for collecting wool trait data.
Author Contributions
H.G., H.Z., Y.L. and J.G.H.H. conceived and designed the project. H.G., H.Z. and S.L. performed the experiments. H.G., H.Z. and J.W. analysed the data. H.G., H.Z. and J.G.H.H. wrote the manuscript. All authors reviewed and commented on the manuscript.
Funding
This research was funded by the Lincoln University Gene marker Laboratory, the AGMARDT Postdoctoral Fellowship to HG and the New Zealand Guardian Trust for the Vernon Willey Trust Fellowship to HZ.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Strnad P., Usachov V., Debes C., Gräter F., Parry D.A., Omary M.B. Unique amino acid signatures that are evolutionarily conserved distinguish simple-type, epidermal and hair keratins. J. Cell Sci. 2011;124:4221–4232. doi: 10.1242/jcs.089516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong H., Zhou H., Forrest R.H., Li S., Wang J., Dyer J.M., Luo Y., Hickford J.G.H. Wool keratin-associated protein genes in Sheep—A review. Genes. 2016;7:24. doi: 10.3390/genes7060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell B.C., Rogers G.E. The role of keratin proteins and their genes in the growth, structure and properties of hair. In: Jollès P., Zahn H., Höcker H., editors. Formation and Structure of Human Hair. Birkhäuser Verlag; Basel, Switzerland: 1997. pp. 59–148. [DOI] [PubMed] [Google Scholar]
- 4.Fraser R., MacRae T.P., Sparrow L.G., Parry D. Disulphide bonding in α-keratin. Int. J. Biol. Macromol. 1988;10:106–112. doi: 10.1016/0141-8130(88)90017-7. [DOI] [Google Scholar]
- 5.Gong H., Zhou H., Dyer J.M., Hickford J.G.H. The sheep KAP8-2 gene, a new KAP8 family member that is absent in humans. SpringerPlus. 2014;3:1–5. doi: 10.1186/2193-1801-3-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai L., Gong H., Zhou H., Tao J., Hickford J.G.H. A nucleotide substitution in the ovine KAP20-2 gene leads to a premature stop codon that affects wool fibre curvature. Anim. Genet. 2018;49:357–358. doi: 10.1111/age.12668. [DOI] [PubMed] [Google Scholar]
- 7.Rogers M.A., Schweizer J. Human KAP genes, only the half of it? Extensive size polymorphisms in hair keratin-associated protein genes. J. Investig. Dermatol. 2005;124:vii–ix. doi: 10.1111/j.0022-202X.2005.23728.x. [DOI] [PubMed] [Google Scholar]
- 8.Gong H., Zhou H., Bai L., Li W., Li S., Wang J., Luo Y., Hickford J.G.H. Associations between variation in the ovine high glycine-tyrosine keratin-associated protein gene KRTAP20-1 and wool traits. J. Anim. Sci. 2019;97:587–595. doi: 10.1093/jas/sky465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Luo Y., Hickford J.G.H. Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits. Genes. 2017;8:27. doi: 10.3390/genes8010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H., Gong H., Wang J., Dyer J.M., Luo Y., Hickford J.G.H. Identification of four new gene members of the KAP6 gene family in sheep. Sci. Rep. 2016;6:24074. doi: 10.1038/srep24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H., Hickford J.G.H., Fang Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006;354:159–161. doi: 10.1016/j.ab.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 12.Byun S.O., Fang Q., Zhou H., Hickford J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009;385:174–175. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Gong H., Zhou H., Hickford J.G.H. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011;38:31–35. doi: 10.1007/s11033-010-0074-6. [DOI] [PubMed] [Google Scholar]
- 14.Bai L., Wang J., Zhou H., Gong H., Tao J., Hickford J.G.H. Identification of ovine KRTAP28-1 and its association with wool weight and mean fibre diameter-associated traits. Animals. 2019;9:142. doi: 10.3390/ani9040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong H., Zhou H., McKenzie G.W., Yu Z., Clerens S., Dyer J.M., Plowman J.E., Wright M.W., Arora R., Bawden C.S., et al. An updated nomenclature for Keratin-Associated Proteins (KAPs) Int. J. Biol. Sci. 2012;8:258–264. doi: 10.7150/ijbs.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers M.A., Langbein L., Winter H., Ehmann C., Praetzel S., Korn B., Schweizer J. Characterization of a cluster of human high/ultrahigh sulfur keratin-associated protein genes embedded in the type I keratin gene domain on chromosome 17q12-21. J. Biol. Chem. 2001;276:19440–19451. doi: 10.1074/jbc.M100657200. [DOI] [PubMed] [Google Scholar]
- 17.McGaughey G.B., Gagné M., Rappé A.K. π-Stacking interactions alive and well in proteins. J. Biol. Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 18.Levitt M., Perutz M.F. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 19.Fraser R.B., Parry D.A. Filamentous structure of hard β-keratins in the epidermal appendages of birds and reptiles. In: Parry D., Squire J., editors. Fibrous Proteins: Structures and Mechanisms. Volume 82. Springer; Cham, Switzerland: 2017. pp. 231–252. [DOI] [PubMed] [Google Scholar]
- 20.Rogers G.E. Biology of the wool follicle: An excursion into a unique tissue interaction system waiting to be re-discovered. Exp. Dermatol. 2006;15:931–949. doi: 10.1111/j.1600-0625.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 21.Sugimoto M., Watanabe T., Sugimoto Y. The molecular effects of a polymorphism in the 5′ UTR of solute carrier family 44, member 5 that is associated with birth weight in Holsteins. PLoS ONE. 2012;7:e41267. doi: 10.1371/journal.pone.0041267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SGS Wool Testing Services Laboratory Fleece Measurements. [(accessed on 12 July 2019)];2011 Info Bulletin 5.2b. Available online: https://www.sgs.co.nz/~/media/Local/New%20Zealand/Documents/Technical%20Documents/Technical%20Bulletins/Wool%20Testing%20Info%20Bulletins/SGS-AGRI-5-2b-Lab-Fleece-Measurements-A4-EN-11-V1.
- 23.Kijas J.W., Townley D., Dalrymple B.P., Heaton M.P., Maddox J.F., McGrath A., Wilson P., Ingersoll R.G., McCulloch R., McWilliam S. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE. 2009;4:e4668. doi: 10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong H., Zhou H., Hickford J.G.H. Polymorphism of the ovine keratin-associated protein 1-4 gene (KRTAP1-4) Mol. Biol. Rep. 2010;37:3377–3380. doi: 10.1007/s11033-009-9925-4. [DOI] [PubMed] [Google Scholar]
- 25.Gong H., Zhou H., McKenzie G.W., Hickford J.G., Yu Z., Clerens S., Dyer J.M., Plowman J.E. Emerging issues with the current keratin-associated protein nomenclature. Int. J. Trichology. 2010;2:104–105. doi: 10.4103/0974-7753.77519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong H., Zhou H., Dyer J.M., Hickford J.G.H. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011;38:5429–5433. doi: 10.1007/s11033-011-0697-2. [DOI] [PubMed] [Google Scholar]
- 27.Gong H., Zhou H., Plowman J.E., Dyer J.M., Hickford J.G.H. Search for variation in the ovine KAP7-1 and KAP8-1 genes using polymerase chain reaction–single-stranded conformational polymorphism screening. DNA Cell Biol. 2011;31:367–370. doi: 10.1089/dna.2011.1346. [DOI] [PubMed] [Google Scholar]
- 28.Gong H., Zhou H., Yu Z., Dyer J., Plowman J.E., Hickford J.G.H. Identification of the ovine keratin-associated protein KAP1-2 gene (KRTAP1-2) Exp. Dermatol. 2011;20:815–819. doi: 10.1111/j.1600-0625.2011.01333.x. [DOI] [PubMed] [Google Scholar]
- 29.Rogers G.R., Hickford J.G.H., Bickerstaffe R. Polymorphism in two genes for B2 high sulfur proteins of wool. Anim. Genet. 1994;25:407–415. doi: 10.1111/j.1365-2052.1994.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H., Visnovska T., Gong H., Schmeier S., Hickford J.G.H., Ganley A.R.D. Contrasting patterns of coding and flanking region evolution in mammalian keratin associated protein-1 genes. Mol. Phylogenet. Evol. 2019;133:352–361. doi: 10.1016/j.ympev.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H., Gong H., Li S., Luo Y., Hickford J.G.H. A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter. J. Anim. Breed. Genet. 2015;132:301–307. doi: 10.1111/jbg.12138. [DOI] [PubMed] [Google Scholar]
- 32.Tao J., Zhou H., Gong H., Yang Z., Ma Q., Cheng L., Ding W., Li Y., Hickford J.G.H. Variation in the KAP6-1 gene in Chinese Tan sheep and associations with variation in wool traits. Small Rumin. Res. 2017;154:129–132. doi: 10.1016/j.smallrumres.2017.08.001. [DOI] [Google Scholar]
- 33.Li S., Zhou H., Gong H., Zhao F., Wang J., Luo Y., Hickford J.G.H. Variation in the ovine KAP6-3 gene (KRTAP6-3) is associated with variation in mean fibre diameter-associated wool traits. Genes. 2017;8:204. doi: 10.3390/genes8080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong H., Zhou H., Li W., Wang J., Li S., Luo Y., Hickford J.G.H. Variation in ovine KRTAP8-1 affects wool fibre staple strength and curvature. J. Agric. Sci. 2019 doi: 10.1017/S0021859619000741. in press. [DOI] [Google Scholar]
- 35.Tao J., Zhou H., Yang Z., Gong H., Ma Q., Ding W., Li Y., Hickford J.G.H. Variation in the KAP8-2 gene affects wool crimp and growth in Chinese Tan sheep. Small Rumin. Res. 2017;149:77–80. doi: 10.1016/j.smallrumres.2017.01.001. [DOI] [Google Scholar]