Abstract
Heat stress as a yield limiting issue has become a major threat for food security as global warming progresses. Being sessile, plants cannot avoid heat stress. They respond to heat stress by activating complex molecular networks, such as signal transduction, metabolite production and expressions of heat stress-associated genes. Some plants have developed an intricate signalling network to respond and adapt it. Heat stress tolerance is a polygenic trait, which is regulated by various genes, transcriptional factors, proteins and hormones. Therefore, to improve heat stress tolerance, a sound knowledge of various mechanisms involved in the response to heat stress is required. The classical breeding methods employed to enhance heat stress tolerance has had limited success. In this era of genomics, next generation sequencing techniques, availability of genome sequences and advanced biotechnological tools open several windows of opportunities to improve heat stress tolerance in crop plants. This review discusses the potential of various functional genomic approaches, such as genome wide association studies, microarray, and suppression subtractive hybridization, in the process of discovering novel genes related to heat stress, and their functional validation using both reverse and forward genetic approaches. This review also discusses how these functionally validated genes can be used to improve heat stress tolerance through plant breeding, transgenics and genome editing approaches.
Keywords: GWAS, VIGS, T-DNA, CRISPR, Heat stress, Functional genomics
Introduction
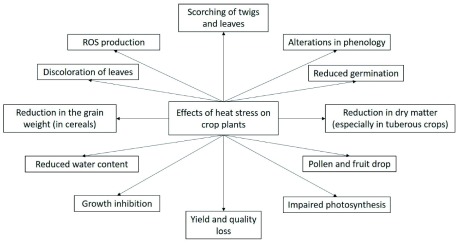
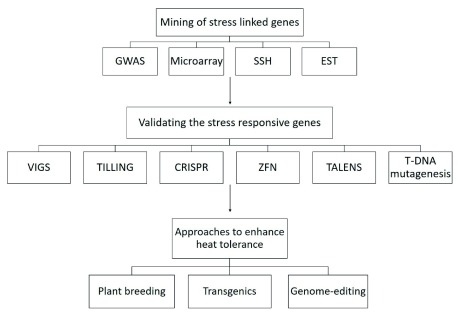
Abiotic stresses have numerous adverse effects on crop plants, which further lead to yield and quality losses ( Figure 1). To feed the whole world in the scenario of the changing climate, new and better heat tolerant varieties of various crops is needed 1. The understanding of various physiological, molecular and biochemical pathways can facilitate the development of superior heat tolerant varieties 2. However, previous efforts, aimed at improving plant heat stress tolerance, have had limited success 3, 4 because of the poor understanding of the genetics of heat tolerance. Fortunately, nowadays reference genomes of major food crops and model plant species are available publicly, which provide a solid platform for crop improvement. Moreover, wild species and various landraces of various crops have unknown heat tolerant genes that should be identified and incorporated to high yielding modern cultivars 5. The functional genomic approaches such as genome wide association studies (GWAS) and gene expression profiling using microarrays can catalyse the discovery of novel genes associated to heat stress 6– 8. In addition, suppression subtractive hybridization (SSH) is another effective and productive technique used for the screening and cloning of the genes/ESTs that express differentially under heat stress 9, 10. Reverse genetic techniques can improve the understanding of their expression patterns under heat stress. The plant breeding strategies and new biotechnological tools including genome editing techniques can use these validated genes to enhance heat stress tolerance in crop plants ( Figure 2).
Figure 1. General effects of heat stress on crop plants.
Figure 2. A systematic flow chart depicting the approaches used for the mining of genes associated with heat stress, for the functional validation of candidate genes and approaches that can take advantage of functionally validated genes to increase heat stress tolerance.
Mining of stress linked genes
Present crop varieties have limited heat tolerance because earlier domestication, green revolution and conventional breeding were focused to increase yield and qualitative traits 11. However, the knowledge of genes/markers/QTL regions associated to heat tolerance is now required to improve thermo tolerance. Previous studies suggested that vast genetic diversity still exists in the germplasms of various crops 12– 14. GWAS emerged as a powerful tool to identify the genetic basis behind complex phenotypic traits 15, 16, and it provides high mapping resolution compared with conventional genetic mapping 17, 18. So far this approach has been applied to major food crops, including wheat 7, 19, 20, rice 21, maize 22, sorghum 23and Brassica napus L. 24, to identify the natural variation associated with heat stress and to understand this genetic basis. Another way to identify and understand the key molecular mechanisms in response to heat stress is a transcriptomic study 25, 26; plants respond to heat stress by inducing various heat responsive genes, thus transcriptomic studies provide an effective screening of heat responsive candidate genes 6. For example, microarray studies allow the screening of genes on the basis of their expression patterns under stressed conditions at a particular plant developmental stage 6, 26, 27. Singh et al. (2015) 6 investigated the heat responsive genes for potato tuberization and Ginzberg et al. 28 identified the candidate heat responsive genes for potato periderm formation using microarrays. In addition, SSH is an easy and efficient approach for the identification of genes/ESTs with differential expression under heat stress. This technique is preferred when the genome sequence information is not available 9. It can identify the tissue specific differentially expressed transcripts. To identify heat responsive ESTs cDNA libraries can be generated from plants grown under heat stressed conditions 29. For example, SSH library of potato skin present 108 candidate genes for suberin and periderm formation 30. To investigate the genes/ESTs involved in heat tolerance at the stage of grain filling in wheat, SSH library was constructed by using the leaf RNA samples from heat stressed plants 9, 29. The results of these studies provided many heat responsive genes/ESTs, which can be used to develop thermo tolerant wheat varieties.
Validating the stress responsive genes
The above approaches can identify potential candidate genes linked to heat stress tolerance. However, the functions of the candidate genes must be validated before incorporating them into present cultivars. Both forward and reverse genetic approaches can be employed for functional validation of genes (see examples in Table 1). Forward genetics detect variations in the nucleic acid sequence responsible for a given phenotype 31, while reverse genetics detect the gene’s functionality by observing the change in the phenotype due to alterations in known genetic sequence 32.
Table 1. Some examples of successfully validated potential heat tolerant genes in model plants and major crops.
| Plant/crop | Gene | Technique used | Reference |
|---|---|---|---|
| Arabidopsis thaliana | HSF1 and HSF3 | Transcription control (genetic
engineering using protein fusion) |
33, 34 |
| DREB2A CA | Microarray | 35 | |
| Hsp70 | antisense gene approach | 36 | |
| ATHSF1 (HSF) | Recombinant DNA technology | 33 | |
| FAD7 | T-DNA | 37 | |
| HSP101 | Transformation | 38 | |
| Rice (Oryzasativa) | spl7 | Transcription control | 39 |
| Athsp101 | Agrobacterium mediated
transformation |
40 | |
| Wheat (Triticumaestivum) | TamiR159 | MiR159 (miRNA) | 41 |
| TaGASR1 | Agrobacterium mediated
transformation |
42 | |
| Carrot (Daucuscarota) | Hsp17.7 | Hsps and molecular
Chaperones |
43 |
| Chilli pepper (Capsicum annuum) | CabZIP63 | Virus induced gene silencing | 44 |
| CaWRKY40 | Virus induced gene silencing | 45 | |
| Tomato (Solanumlycopersicum) | Hsa32 | Subtracted cDNA libraries | 46 |
| MT-sHSP | freezing transformation method | 47 | |
| ATG5, ATG7, NBR1, WRKY33 | Virus induced gene silencing | 48 | |
| 2-CP1, 2-CP2, 2-CP1/2, ATG5, ATG7 | Virus induced gene silencing | 49 | |
| RBOH1, MPK1, MPK2 | Virus induced gene silencing | 50 | |
| Hsc70.1 | Virus induced gene silencing | 51 | |
| SlLrgB | RNA interference | 51 | |
| Barley (Hordeumvulgare) | APX 1 | Agrobacterium mediated
transformation |
52 |
Virus-induced gene silencing (VIGS) is a rapid, efficient and cost effective post-transcriptional gene silencing (PTGS) technique used to study target gene(s) functionality 53. It can be used as both the forward and reverse genetic approach 48, 54. Plants can sense and then respond to heat stress by activating various transcriptional cascades 55. Being a PTGS technique, VIGS can be used to knockdown the expression of target genes after transcription. VIGS takes an advantage of a plant’s innate defence mechanism against virus infection. In this technique, a fragment of the target gene is first inserted into a suitable viral vector and then that vector is transformed into the plant, where the viral genome harbouring the fragment of target gene start replicating and produce dsRNA. Then an enzyme DICER cut this dsRNA into multiple siRNA of about 21 nucleotide long. Later these siRNA unwind into two single stranded RNAs, one out of which is degraded and the other one binds to RNA induced silencing complex (RISC), which later degrade the targeted endogenous gene and the effect on gene knockdown can be observed on by phenotypic analysis 56– 58. Many candidate genes and transcriptional factors associated with heat stress response/tolerance have been validated successfully through VIGS 58, 59. For example, TRV-VIGS based silencing of CabZIP63 gene lowered the tolerance to heat stress in pepper plants, suggesting that CabZIP63 is a positive regulator for thermos tolerance 44. The functionality of ATG5, ATG7,FAD7 and NtEDS1genes in response to heat stress have been successfully studied in tomato using tobacco rattle virus (TRV) based VIGS technique 48, 60. Recently, VIGS has also been employed to investigate the involvement of small heat shock proteins (CaHSP16.4 and CaHsp25. 9) in heat stress tolerance 61, 62.
The role of candidate genes in heat stress tolerance can also be verified through the generation of transfer (T)-DNA mutants 63. Like VIGS, insertion of T-DNA in the target gene’s sequence disrupt its functionality, which results in the change of phenotype. This approach is widely accepted because of the genome wide distribution of transposable elements with superior insertions in the gene sequences, resulting in the direct gene knockout 64. In addition, T-DNA can also be used as a gain of function approach to study the target gene’s functionality, called as activation tagging 65. For example, T-DNA having a tetramer of cauliflower mosaic virus 35S promoter can cause gene activation mutations 65. Since plant responses to environmental stresses are polygenic and complex traits, model plants, such as Arabidopsis, are used to first study adaptive responses 66. T-DNA mutant lines of many heat tolerant ecotypes of Arabidopsis have been discovered and are available at Nottingham Arabidopsis Stock Centre (NASC) or The Arabidopsis Information Resource (TAIR).
Targeting induced local lesions in genome (TILLING) is another non-transgenic approach that allows the PCR based identification of directed mutations in the target gene sequence and the function of target gene can be analysed from the modified phenotype due to that mutation 67, 68. It is a fast and cost efficient technique for the screening of point mutations and for the functional validation gene of interest 69. It take advantage of conventional insertional mutagenesis and availability of genomic sequences 70. These point mutations can be generated with the help of chemical mutagens such as ethyl methane sulfonate (EMS) 71. Nowadays, the genome sequences of many crop plant species are available, which make this technique more effective. The plants under heat stress exhibit different phenotypes associated with allelic variations in their genomic sequence. The TILLING approach used to study these natural variations or SNP mutations in individuals is called as EcoTILLING. The next generation sequencing techniques allow inexpensive TILLING by sequencing method to screen SNP variations 72. Recently, the functionality of heat shock binding protein 1 (HSBP1) was examined with TILLING. The chemically induced mutations disrupted the functionality of HSBP1 partially and the mutant plants exhibited increased heat stress tolerance. These findings confirmed that HSBP1 is a negative regulator of heat stress response in tomato 73.
In addition, the genome-editing techniques such as transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs) and clustered regularly interspace short palindromic repeat (CRISPR) can also be used as reverse genetic approaches to study the target gene function. TALENs using sequence specific nucleases (SSNs) became a powerful genome editing technique, which can also be applied as a reverse genetic approach to understand the function of a target gene. It consists of one customizable DNA-binding domain and a nuclease, which generate double stranded DNA breaks (DSBs) at the target gene sequence 74. Similar to CRISPR, these DSB are repaired either via NHEJ pathway or via homologous recombination. Both these recovery pathways allow insertion, deletion and intentional replacements in the target gene sequence. These modifications in the target gene’s sequence may cause a variation in the phenotype, which suggests the function of that gene 74, 75. The ZFNs are the synthetic proteins having a DNA binding domain that consists two finger modules and a DNA cleaving domain. ZFNs causes DSBs in the targeted DNA sequence and facilitate site-specific mutagenesis, or base substation, which alter or may knockout the gene expression 76. The ZFNs have revealed the function of various genes in model plants as well as in crop species 77.
Among all genome-editing techniques, CRISPR-Cas9 has emerged as a powerful tool for precise genome editing to study the molecular pathways linked to heat stress and to enhance thermo tolerance in crop plants 78, 79. It is comparatively simpler, more accurate and faster than other genome editing techniques. In brief, CRISPR involves designing of a guide RNA of ~20 nucleotides complementary to the gene of interest and a Cas9 nuclease enzyme that cut 3–4 bases next to the protospacer adjacent motif, which is later repaired either by homology directed repair pathway or via error prone non-homologous end joining 80, 81. Therefore, this technique can be used to generate gene knockout mutant lines to study the function of targeted gene(s). For example, annexin gene OsAnn3 knockout mutant lines developed via CRISPR-Cas9 technique revealed the role of OsAnn3 gene in cold stress tolerance in rice 82.
Approaches to enhance heat tolerance
Plants have inherent mechanisms to survive under heat stressed conditions but the heat tolerance capacity of plants varies species-to-species and even within the species. If heat tolerant genes are present in sexually compatible species, then marker-assisted selection (MAS), new generation molecular breeding, precision breeding and genome editing techniques can be used.
Thermo tolerance is a complex multigenic trait, which is influenced by genotype X environment interactions 83. Development of heat tolerant crop varieties through traditional breeding is very labours and time consuming. However, precision breeding with the help of MAS can accelerate the plant breeding programs with high efficiency 84. SNPs and simple sequence repeats (SSR) are being used widely in plant breeding experiments aimed to enhance abiotic stress tolerance. Presently the use of SNPs become more common in plant breeding than SSR markers 85, 86. Garg et al. 87 found one SNP in the sequence of heat shock protein (HSP16.9) between a heat tolerant and heat susceptible wheat genotypes. This SNP contribute 29.89% phenotypic variation for grain weight per spike. Recently, many SNPs associated to heat stress tolerance have been identified in major food crops 7, 88– 90. However, heat stress tolerance is a polygenic trait and a single molecular marker contribute little to improve it. Therefore, it is important to incorporate several SNPs associated to various QTLs that are controlling the heat stress tolerance mechanisms 89, 91. However, the accessibility of genome editing techniques opened various new windows to introduce targeted editing of plant genomes to understand the molecular aspects involved in heat stress tolerance 92, 93. For example, ethylene response factors (ERFs) are the stress induced transcriptional factors that take part in abiotic stress tolerance. The CRISPR-Cas9 based genome editing of one such ethylene response factor from AP2/ERF superfamily enhanced abiotic stress tolerance in crop plants 94.
In cases when heat tolerant genes do not exist in sexually compatible species, these methods cannot be applied. Advanced biotechnological tools can increase the limited heat stress tolerance in crop plants. The transfer of heat tolerant genes through recombinant DNA technology can generate heat tolerant transgenic lines in a short amount of time. This method also allows utilisation of potential genes from other species to enhance thermo tolerance in target crops, e.g. AmDREB2C, from Ammopiptanthus mongolicus has been used to increase heat stress tolerance in transgenic Arabidopsis plants 95. In addition, many genes responsible for heat stress tolerance have been identified and validated in model plants and also in major food crops that can be introduced to heat susceptible cultivars or their expression levels can be increased by generating their stable overexpression lines. For instance, the overexpression TaPEPKR2 gene enhanced heat stress tolerance in wheat and Arabidopsis plants 96.
Conclusion
Heat stress affects crop production significantly. Plants respond to heat stress by activating complex molecular networks, such as signal transduction, metabolite production and expressions of heat stress-associated genes. With the developments in plant functional genomics techniques, many novel genes related to heat stress tolerance have been identified and are being used to improve stress tolerance with the help of advanced biotechnological approaches. Next generation sequencing and genome-editing techniques will play crucial roles in crop improvement. In the near future, the scientists will have a better understanding of plant heat tolerant mechanisms and farmers will be able to grow better high yielding heat tolerant crop varieties in the fields.
Data availability
No data is associated with this article.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; peer review: 2 approved
References
- 1. Lesk C, Rowhani P, Ramankutty N: Influence of extreme weather disasters on global crop production. Nature. 2016;529(7584):84–7. 10.1038/nature16467 [DOI] [PubMed] [Google Scholar]
- 2. Fahad S, Bajwa AA, Nazir U, et al. : Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front Plant Sci. 2017;8:1147. 10.3389/fpls.2017.01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lamaoui M, Jemo M, Datla R, et al. : Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front Chem. 2018;6:26. 10.3389/fchem.2018.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grover A, Mittal D, Negi M, et al. : Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013;205–206:38–47. 10.1016/j.plantsci.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 5. Mammadov J, Buyyarapu R, Guttikonda SK, et al. : Wild Relatives of Maize, Rice, Cotton, and Soybean: Treasure Troves for Tolerance to Biotic and Abiotic Stresses. Front Plant Sci. 2018;9:886. 10.3389/fpls.2018.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh A, Siddappa S, Bhardwaj V, et al. : Expression profiling of potato cultivars with contrasting tuberization at elevated temperature using microarray analysis. Plant Physiol Biochem. 2015;97:108–16. 10.1016/j.plaphy.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 7. Maulana F, Ayalew H, Anderson JD, et al. : Genome-Wide Association Mapping of Seedling Heat Tolerance in Winter Wheat. Front Plant Sci. 2018;9:1272. 10.3389/fpls.2018.01272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan S, Liu B, Zhang Y, et al. : Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genomics. 2019;20(1):257. 10.1186/s12864-019-5617-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goswami S, Kumar RR, Dubey K, et al. : SSH Analysis of Endosperm Transcripts and Characterization of Heat Stress Regulated Expressed Sequence Tags in Bread Wheat. Front Plant Sci. 2016;7:1230. 10.3389/fpls.2016.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rampuria S, Joshi U, Palit P, et al. : Construction and analysis of an SSH cDNA library of early heat-induced genes of Vigna aconitifolia variety RMO-40. Genome. 2012;55(11):783–96. 10.1139/g2012-064 [DOI] [PubMed] [Google Scholar]
- 11. Hussain B: Modernization in plant breeding approaches for improving biotic stress resistance in crop plants. Turk J Agric For. 2015;39(4):515–530. 10.3906/tar-1406-176 [DOI] [Google Scholar]
- 12. Menguer PK, Sperotto RA, Ricachenevsky FK: A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet Mol Biol. 2017;40(1 suppl 1):238–252. 10.1590/1678-4685-GMB-2016-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopes MS, El-Basyoni I, Baenziger PS, et al. : Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J Exp Bot. 2015;66(12):3477–86. 10.1093/jxb/erv122 [DOI] [PubMed] [Google Scholar]
- 14. Machida-Hirano R: Diversity of potato genetic resources. Breed Sci. 2015;65(1):26–40. 10.1270/jsbbs.65.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Visscher PM, Wray NR, Zhang Q, et al. : 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101(1):5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Challa S, Neelapu NRR: Genome-Wide Association Studies (GWAS) for Abiotic Stress Tolerance in Plants. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants2018;135–50. 10.1016/B978-0-12-813066-7.00009-7 [DOI] [Google Scholar]
- 17. Zargar SM, Raatz B, Sonah H, et al. : Recent advances in molecular marker techniques: Insight into QTL mapping, GWAS and genomic selection in plants. Journal of Crop Science and Biotechnology. 2015;18(5):293–308. 10.1007/s12892-015-0037-5 [DOI] [Google Scholar]
- 18. Lu F, Romay MC, Glaubitz JC, et al. : High-resolution genetic mapping of maize pan-genome sequence anchors. Nat Commun. 2015;6:6914. 10.1038/ncomms7914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valluru R, Reynolds MP, Davies WJ, et al. : Phenotypic and genome-wide association analysis of spike ethylene in diverse wheat genotypes under heat stress. New Phytol. 2017;214(1):271–283. 10.1111/nph.14367 [DOI] [PubMed] [Google Scholar]
- 20. Li L, Mao X, Wang J, et al. : Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell Environ. 2019;42(9):2540–2553. 10.1111/pce.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patishtan J, Hartley TN, Fonseca de Carvalho R, et al. : Genome-wide association studies to identify rice salt-tolerance markers. Plant Cell Environ. 2018;41(5):970–982. 10.1111/pce.12975 [DOI] [PubMed] [Google Scholar]
- 22. Gao J, Wang S, Zhou Z, et al. : Linkage mapping and GWAS reveal candidate genes conferring thermotolerance of seed-set in maize. J Exp Bot. 2019; pii: erz171. 10.1093/jxb/erz171 [DOI] [PubMed] [Google Scholar]
- 23. Chen J, Chopra R, Hayes C, et al. : Genome-Wide Association Study of Developing Leaves’ Heat Tolerance during Vegetative Growth Stages in a Sorghum Association Panel. Plant Genome. 2017;10(2). 10.3835/plantgenome2016.09.0091 [DOI] [PubMed] [Google Scholar]
- 24. Rahaman M, Mamidi S, Rahman M: Genome-wide association study of heat stress-tolerance traits in spring-type Brassica napus L. under controlled conditions. Crop J. 2018;6(2):115–125. 10.1016/j.cj.2017.08.003 [DOI] [Google Scholar]
- 25. Zhang SS, Yang H, Ding L, et al. : Tissue-Specific Transcriptomics Reveals an Important Role of the Unfolded Protein Response in Maintaining Fertility upon Heat Stress in Arabidopsis. Plant Cell. 2017;29(5):1007–1023. 10.1105/tpc.16.00916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin D, Wu H, Peng H, et al. : Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat ( Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics. 2008;9:432. 10.1186/1471-2164-9-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sham A, Moustafa K, Al-Ameri S, et al. : Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS One. 2015;10(5):e0125666. 10.1371/journal.pone.0125666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ginzberg I, Barel G, Ophir R, et al. : Transcriptomic profiling of heat-stress response in potato periderm. J Exp Bot. 2009;60(15):4411–21. 10.1093/jxb/erp281 [DOI] [PubMed] [Google Scholar]
- 29. Vishwakarma H, Junaid A, Manjhi J, et al. : Heat stress transcripts, differential expression, and profiling of heat stress tolerant gene TaHsp90 in Indian wheat ( Triticum aestivum L.) cv C306. PLoS One. 2018;13(6):e0198293. 10.1371/journal.pone.0198293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soler M, Serra O, Fluch S, et al. : A potato skin SSH library yields new candidate genes for suberin biosynthesis and periderm formation. Planta. 2011;233(5):933–45. 10.1007/s00425-011-1350-y [DOI] [PubMed] [Google Scholar]
- 31. Ji Q: Gene Identification: Forward Genetics.In: Diagnostics in Plant Breeding Dordrecht: Springer Netherlands;2013;41–60. 10.1007/978-94-007-5687-8_3 [DOI] [Google Scholar]
- 32. Bahuguna RN, Gupta P, Bagri J, et al. : Forward and reverse genetics approaches for combined stress tolerance in rice. Indian Journal of Plant Physiology. 2018;23(4):630–646. 10.1007/s40502-018-0418-0 [DOI] [Google Scholar]
- 33. Lee JH, Hübel A, Schöffl F: Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 1995;8(4):603–12. 10.1046/j.1365-313X.1995.8040603.x [DOI] [PubMed] [Google Scholar]
- 34. Prändl R, Hinderhofer K, Eggers-Schumacher G, et al. : HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet. 1998;258(3):269–78. 10.1007/s004380050731 [DOI] [PubMed] [Google Scholar]
- 35. Sakuma Y, Maruyama K, Qin F, et al. : Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci. 2006;103(49):18822–7. 10.1073/pnas.0605639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee JH, Schöffl F: An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Mol Gen Genet. 1996;252(1–2):11–9. 10.1007/s004389670002 [DOI] [PubMed] [Google Scholar]
- 37. Murakami Y, Tsuyama M, Kobayashi Y, et al. : Trienoic fatty acids and plant tolerance of high temperature. Science. 2000;287(5452):476–9. 10.1126/science.287.5452.476 [DOI] [PubMed] [Google Scholar]
- 38. Queitsch C, Hong SW, Vierling E, et al. : Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2007;12(4):479–92. 10.1105/tpc.12.4.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamanouchi U, Yano M, Lin H, et al. : A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci. 2002;99(11):7530–5. 10.1073/pnas.112209199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katiyar-Agarwal S, Agarwal M, Grover A: Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol. 2003;51(5):677–86. 10.1023/a:1022561926676 [DOI] [PubMed] [Google Scholar]
- 41. Wang Y, Sun F, Cao H, et al. : TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One. 2012;7(11):e48445. 10.1371/journal.pone.0048445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang L, Geng X, Zhang H, et al. : Isolation and characterization of heat-responsive gene TaGASR1 from wheat ( Triticum aestivum L.). J Plant Biol. 2017;60(1):57–65. 10.1007/s12374-016-0484-7 [DOI] [Google Scholar]
- 43. Malik MK, Slovin JP, Hwang CH, et al. : Modified expression of a carrot small heat shock protein gene, hsp17. 7, results in increased or decreased thermotolerancedouble dagger. Plant J. 1999;20(1):89–99. 10.1046/j.1365-313X.1999.00581.x [DOI] [PubMed] [Google Scholar]
- 44. Shen L, Liu Z, Yang S, et al. : Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J Exp Bot. 2016;67(8):2439–51. 10.1093/jxb/erw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dang FF, Wang YN, Yu L, et al. : CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013;36(4):757–74. 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- 46. Liu NY, Ko SS, Yeh KC, et al. : Isolation and characterization of tomato Hsa32 encoding a novel heat-shock protein. Plant Sci. 2006;170(5):976–985. 10.1016/j.plantsci.2006.01.008 [DOI] [Google Scholar]
- 47. Sanmiya K, Suzuki K, Egawa Y, et al. : Mitochondrial small heat-shock protein enhances thermotolerance in tobacco plants. FEBS Lett. 2004;557(1–3):265–8. 10.1016/s0014-5793(03)01494-7 [DOI] [PubMed] [Google Scholar]
- 48. Zhou J, Wang J, Yu JQ, et al. : Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 2014;5:174. 10.3389/fpls.2014.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao J, Liu Q, Hu P, et al. : An efficient Potato virus X -based microRNA silencing in Nicotiana benthamiana. Sci Rep. 2016;6(1):20573. 10.1038/srep20573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nie WF, Wang MM, Xia XJ, et al. : Silencing of tomato RBOH1 and MPK2 abolishes brassinosteroid-induced H 2O 2 generation and stress tolerance. Plant, Cell Environ. 2013;36(4):789–803. 10.1111/pce.12014 [DOI] [PubMed] [Google Scholar]
- 51. Zhang S, Ai G, Li M, et al. : Tomato LrgB regulates heat tolerance and the assimilation and partitioning of carbon. Plant Sci. 2018;274:309–319. 10.1016/j.plantsci.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 52. Shi WM, Muramoto Y, Ueda A, et al. : Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene. 2001;273(1):23–7. 10.1016/s0378-1119(01)00566-2 [DOI] [PubMed] [Google Scholar]
- 53. Senthil-Kumar M, Mysore KS: New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16(12):656–65. 10.1016/j.tplants.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 54. Senthil-Kumar M, Lee HK, Mysore KS: VIGS-Mediated Forward Genetics Screening for Identification of Genes Involved in Nonhost Resistance. J Vis Exp. 2013; (78):e51033. 10.3791/51033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohama N, Sato H, Shinozaki K, et al. : Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017;22(1):53–65. 10.1016/j.tplants.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 56. Movahedi A, Zhang J, Sun W, et al. : Plant small RNAs: definition, classification and response against stresses. Biologia (Poland). 2018;73(3):285–94. 10.2478/s11756-018-0034-5 [DOI] [Google Scholar]
- 57. Senthil-Kumar M, Mysore KS: Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc. 2014;9(7):1549–62. 10.1038/nprot.2014.092 [DOI] [PubMed] [Google Scholar]
- 58. Singh B, Kukreja S, Salaria N, et al. : VIGS: a flexible tool for the study of functional genomics of plants under abiotic stresses. J Crop Improv. 2019;1–38. 10.1080/15427528.2019.1640821 [DOI] [Google Scholar]
- 59. Ramegowda V, Mysore KS, Senthil-Kumar M: Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front Plant Sci. 2014;5:323. 10.3389/fpls.2014.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hiremath SS, Sajeevan RS, Nataraja KN, et al. : Silencing of fatty acid desaturase ( FAD7) gene enhances membrane stability and photosynthetic efficiency under heat stress in tobacco ( Nicotiana benthamiana). Indian J Exp Biol. 2017;55(8):532–41. Reference Source [Google Scholar]
- 61. Feng XH, Zhang HX, Ali M, et al. : A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper ( Capsicum annuum L.). Plant Physiol Biochem. 2019;142:151–62. 10.1016/j.plaphy.2019.07.001 [DOI] [PubMed] [Google Scholar]
- 62. Huang LJ, Cheng GX, Khan A, et al. : CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance. Protoplasma. 2019;256(1):39–51. 10.1007/s00709-018-1280-7 [DOI] [PubMed] [Google Scholar]
- 63. Radhamony RN, Prasad AM, Srinivasan R: T-DNA insertional mutagenesis in Arabidopsis: A tool for functional genomics. Electron J Biotechnol. 2005;8(1). Reference Source [Google Scholar]
- 64. An G, Lee S, Kim SH, et al. : Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005;46(1):14–22. 10.1093/pcp/pci502 [DOI] [PubMed] [Google Scholar]
- 65. Lo SF, Fan MJ, Hsing YI, et al. : Genetic resources offer efficient tools for rice functional genomics research. Plant Cell Environ. 2016;39(5):998–1013. 10.1111/pce.12632 [DOI] [PubMed] [Google Scholar]
- 66. Shindo C, Bernasconi G, Hardtke CS: Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot. 2007;99(6):1043–54. 10.1093/aob/mcl281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suprasanna P, Mirajkar SJ, Patade VY, et al. : 17. Induced mutagenesis for improving plant abiotic stress tolerance. In: Mutagenesis: exploring genetic diversity of crops2014;345–376. 10.3920/978-90-8686-796-7_17 [DOI] [Google Scholar]
- 68. Gilliham M, Able JA, Roy SJ: Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 2017;90(5):898–917. 10.1111/tpj.13456 [DOI] [PubMed] [Google Scholar]
- 69. Akpinar BA, Lucas SJ, Budak H: Genomics approaches for crop improvement against abiotic stress. ScientificWorldJournal. 2013;2013:361921. 10.1155/2013/361921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kurowska M, Daszkowska-Golec A, Gruszka D, et al. : TILLING: a shortcut in functional genomics. J Appl Genet. 2011;52(4):371–90. 10.1007/s13353-011-0061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barkley NA, Wang ML: Application of TILLING and EcoTILLING as Reverse Genetic Approaches to Elucidate the Function of Genes in Plants and Animals. Curr Genomics. 2008;9(4):212–26. 10.2174/138920208784533656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guo Y, Abernathy B, Zeng Y, et al. : TILLING by sequencing to identify induced mutations in stress resistance genes of peanut ( Arachis hypogaea). BMC Genomics. 2015;16:157. 10.1186/s12864-015-1348-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marko D, El-shershaby A, Carriero F, et al. : Identification and Characterization of a Thermotolerant TILLING Allele of Heat Shock Binding Protein 1 in Tomato. Genes (Basel). 2019;10(7): pii: E516. 10.3390/genes10070516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sprink T, Metje J, Hartung F: Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr Opin Biotechnol. 2015;32:47–53. 10.1016/j.copbio.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 75. Mahfouz MM, Piatek A, Stewart CN, Jr: Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol J. 2014;12(8):1006–14. 10.1111/pbi.12256 [DOI] [PubMed] [Google Scholar]
- 76. Li WT, He M, Wang J, et al. : Zinc finger protein (ZFP) in plants-A review. Plant OMICS. 2013;6(6):474–480. Reference Source [Google Scholar]
- 77. Kim S, Kim JS: Targeted genome engineering via zinc finger nucleases. Plant Biotechnol Rep. 2011;5(1):9–17. 10.1007/s11816-010-0161-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Biswal AK, Mangrauthia SK, Reddy MR, et al. : CRISPR mediated genome engineering to develop climate smart rice: Challenges and opportunities. Semin Cell Dev Biol. 2019; pii: S1084-9521(18)30114-9. 10.1016/j.semcdb.2019.04.005 [DOI] [PubMed] [Google Scholar]
- 79. Nguyen HC, Lin KH, Ho SL, et al. : Enhancing the abiotic stress tolerance of plants: from chemical treatment to biotechnological approaches. Physiol Plant. 2018;164(4):452–466. 10.1111/ppl.12812 [DOI] [PubMed] [Google Scholar]
- 80. Jaganathan D, Ramasamy K, Sellamuthu G, et al. : CRISPR for Crop Improvement: An Update Review. Front Plant Sci. 2018;9:985. 10.3389/fpls.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jiang F, Doudna JA: CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–529. 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- 82. Shen C, Que Z, Xia Y, et al. : Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J Plant Biol. 2017;60(6):539–547. 10.1007/s12374-016-0400-1 [DOI] [Google Scholar]
- 83. Tayade R, Nguyen T, Oh SA, et al. : Effective Strategies for Enhancing Tolerance to High-Temperature Stress in Rice during the Reproductive and Ripening Stages. Plant Breed Biotechnol. 2018;6(1):1–18. 10.9787/PBB.2018.6.1.1 [DOI] [Google Scholar]
- 84. Zafar SA, Hameed A, Nawaz MA, et al. : Mechanisms and molecular approaches for heat tolerance in rice ( Oryza sativa L.) under climate change scenario. J Integr Agr. 2018;17(4):726–38. 10.1016/S2095-3119(17)61718-0 [DOI] [Google Scholar]
- 85. Mammadov J, Aggarwal R, Buyyarapu R, et al. : SNP markers and their impact on plant breeding. In: The Role of Bioinformatics in Agriculture2014;387–413. Reference Source [Google Scholar]
- 86. Singh N, Choudhury DR, Singh AK, et al. : Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS One. 2013;8(12):e84136. 10.1371/journal.pone.0084136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Garg D, Sareen S, Dalal S, et al. : Heat shock protein based snp marker for terminal heat stress in wheat ( Triticum aestivum L.). Aust J Crop Sci. 2012;6(11):1516–1521. Reference Source [Google Scholar]
- 88. Ruggieri V, Calafiore R, Schettini C, et al. : Exploiting Genetic and Genomic Resources to Enhance Heat-Tolerance in Tomatoes. Agronomy. 2019;9(1):22 10.3390/agronomy9010022 [DOI] [Google Scholar]
- 89. Kilasi NL, Singh J, Vallejos CE, et al. : Heat Stress Tolerance in Rice ( Oryza sativa L.): Identification of Quantitative Trait Loci and Candidate Genes for Seedling Growth Under Heat Stress. Front Plant Sci. 2018;9:1578. 10.3389/fpls.2018.01578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yuan Y, Cairns JE, Babu R, et al. : Genome-Wide Association Mapping and Genomic Prediction Analyses Reveal the Genetic Architecture of Grain Yield and Flowering Time Under Drought and Heat Stress Conditions in Maize. Front Plant Sci. 2019;9:1919. 10.3389/fpls.2018.01919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Paul PJ, Samineni S, Thudi M, et al. : Molecular Mapping of QTLs for Heat Tolerance in Chickpea. Int J Mol Sci. 2018;19(8): pii: E2166. 10.3390/ijms19082166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Parmar N, Singh KH, Sharma D, et al. : Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech. 2017;7(4):239. 10.1007/s13205-017-0870-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Haque E, Taniguchi H, Hassan MM, et al. : Application of CRISPR/Cas9 Genome Editing Technology for the Improvement of Crops Cultivated in Tropical Climates: Recent Progress, Prospects, and Challenges. Front Plant Sci. 2018;9:617. 10.3389/fpls.2018.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Debbarma J, Sarki YN, Saikia B, et al. : Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR-Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol Biotechnol. 2019;61(2):153–172. 10.1007/s12033-018-0144-x [DOI] [PubMed] [Google Scholar]
- 95. Yin Y, Jiang X, Ren M, et al. : AmDREB2C, from Ammopiptanthus mongolicus, enhances abiotic stress tolerance and regulates fatty acid composition in transgenic Arabidopsis. Plant Physiol Biochem. 2018;130:517–528. 10.1016/j.plaphy.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 96. Zang X, Geng X, He K, et al. : Overexpression of the Wheat ( Triticum aestivum L.) TaPEPKR2 Gene Enhances Heat and Dehydration Tolerance in Both Wheat and Arabidopsis. Front Plant Sci. 2018;9:1710. 10.3389/fpls.2018.01710 [DOI] [PMC free article] [PubMed] [Google Scholar]