Abstract
Thoracic ossification of the posterior longitudinal ligament (T-OPLL) is one of the most common factors that causes thoracic spinal stenosis, resulting in intractable myelopathy and radiculopathy. Our previous study reported that the rs201153092 polymorphism present in the collagen 6A1 (COL6A1) gene was a potentially pathogenic locus for the development of T-OPLL. The present study aimed to determine whether the rs201153092 mutation causes abnormal expression of COL6A1 in Han Chinese patients with T-OPLL, and to examine the effects of this mutation on osteogenesis by establishing a model of osteogenic differentiation. COL6A1 gene mutant and wild-type mouse 3T3-E1 embryonic osteoblast models were constructed to induce the differentiation of these cells into osteoblasts. The potential of the mutation site to induce abnormal expression of the COL6A1 gene and osteogenic markers was assessed via reverse transcription-quantitative PCR and western blot analyses. The results demonstrated that the rs201153092A mutation site resulted in significantly increased COL6A1 gene expression levels in the OPLL tissues obtained following clinical surgery. This mutation was shown to play an important role in the development of T-OPLL by regulating the overexpression of the COL6A1 gene and significantly increasing the expression levels of osteogenic markers. The findings of the present study suggested that the rs201153092A mutant variant could increase the expression levels of COL6A1 and consequently play a role in the pathogenesis of T-OPLL.
Keywords: thoracic ossification of the posterior longitudinal ligament, collagen 6A1, genetics
Introduction
Thoracic ossification of the posterior longitudinal ligament (T-OPLL) is a rare disease of the spine with a high disability rate (1,2). The disease is concealed and slowly progressive, and the majority of the patients develop myelopathy only when the spinal cord is severely compressed (3). Direct removal of the massive PLL is the only effective option for the treatment; however, surgery for T-OPLL is complicated and the risk is extremely high (4). It is considered one of the most challenging spinal surgeries, mainly owing to its unique anatomy and pathophysiological factors. The main disadvantages of this operation can be summarized as follows: i) The incidence of thoracic natural kyphosis may limit the positive effect of spinal cord retreat following posterior decompression; ii) the thoracic spinal cord is characterized by relatively low blood supply, leading to spinal cord ischemia and consequently spinal cord injury; and iii) OPLL with tight adhesion to the dural sac increases the difficulty of removing the ossified ligament and increases the risk of intraoperative spinal cord injury (3–8). Therefore, patients with T-OPLL that proceed to surgical treatment often result in unsatisfactory prognosis (9,10).
The pathogenesis of OPLL remains unclear, and genetic factors are considered to play a predominant role in the development of cervical OPLL (C-OPLL) (11,12). To date, more than 10 susceptibility genes/loci have been shown to be associated with susceptibility to C-OPLL (10,13–25). However, few studies have assessed the contribution of T-OPLL susceptibility genes. Previous studies have shown that genetic factors may also play an important role in the development of T-OPLL, the rs201153092A mutation site in the collagen 6A1 (COL6A1) gene was identified as a susceptibility locus for T-OPLL (26,27).
COL6A1 is a crucial component of the extracellular matrix and is involved in membranous or endochondral ossification (28). Although COL6A1 has been identified as a potentially pathogenic gene for C-OPLL, the mutations reported in previous studies were located in the promoter regions or intronic regions of the COL6A1 gene, and the data were not supported by relevant functional validation (11). The rs201153092A mutation site is located in the exonic region of the COL6A1 gene, and may affect the expression levels of the protein by altering the amino acid sequence composition.
The present study aimed to verify the functional role of the COL6A1 rs201153092A mutant variant and to determine whether this mutation causes abnormal expression of the COL6A1 gene in patients with T-OPLL. The patients were selected from a Han Chinese population and the experiments aimed to determine whether the rs201153092A mutation site could promote osteogenesis. This was achieved by establishing a cell model of osteogenic differentiation. This study will provide a theoretical basis for the early detection and diagnosis of T-OPLL diseases, and the investigation of treatments other than surgery.
Materials and methods
Inclusion criteria and patient selection
The study protocol was approved by The Ethics Committee for Human Subjects of the Peking University Third Hospital (permit no. 2014036). Informed consent was provided by all participants. Unrelated Northern Chinese Han patients with T-OPLL carrying the rs201153092A site mutation in COL6A1 and unrelated Northern Chinese Han patients with T-OPLL carrying the wild-type rs201153092G site were enrolled at Peking University Third Hospital. The subjects were recruited between May 2015 and December 2018. The diagnosis of T-OPLL was performed by orthopedic spine specialists based on clinical symptoms and by computed tomography scans of the thoracic spine. The neurological status was evaluated by the Japanese Orthopedic Association (JOA) score for thoracic myelopathy (maximum 11 points). The posterior longitudinal ligament specimens of the thoracic spine in patients with T-OPLL were collected during circumferential decompression surgery (Fig. 1).
Figure 1.
Specimens of the thoracic spine. T-OPLL specimens removed by circumferential decompression surgery in patients with T-OPLL. T-OPLL, thoracic ossified posterior longitudinal ligament.
Hematoxylin-eosin (H&E) staining and immunohistochemistry (IHC) analysis
T-OPLL tissue was fixed with 10% paraformaldehyde (cat. no. P1110; Beijing Solarbio Science & Technology Co., Ltd.) at room temperature for 24 h. Serial 5-µm sections were prepared from paraffin-embedded thoracic spine specimens for staining. H&E staining was performed at 35°C for 80 min in an autostainer machine (Leica Microsystems GmbH) using standard procedures. The sections for IHC staining were deparaffinized using xylene and dehydrated in serially graded ethanol solutions. The sections were washed in distilled water, treated with a 0.3% H2O2 solution, dissolved in absolute methanol at 20°C for 15 min and finally rinsed with PBS (pH 7.4). Antigen retrieval was performed using a high temperature and high-pressure method. Blocking was conducted with 10% FBS (cat. no. 16210064; Thermo Fisher Scientific, Inc.) at room temperature for 2 h. The sections were incubated with primary polyclonal rabbitanti-human COL6A1 antibody (1:200; cat. no. ab182744; Abcam) at 4°C overnight in a humidified chamber. The sections were washed with PBS three times for 5 min each time and subsequently incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2,000; cat. no. ab205718; Abcam) in a humidified chamber for 30 min at room temperature. The sections were rinsed with PBS (pH 7.4) and antibody binding was visualized by incubation with a diaminobenzidine (DAB) solution (cat. no. ZLI-9017; OriGene Technologies, Inc.). The sections were washed in water to remove excess DAB and counterstained with hematoxylin at room temperature for 4 min to visualize the nuclei. The sections were examined using an inverted light microscope (cat. no. DMIL; Leica Microsystems GmbH; magnification, ×20). Negative control sections were incubated with PBS instead of the primary antibody under the same conditions.
Cell lines and cell culture
The 3T3-E1 mouse embryonic osteoblast cell line (American Type Culture Collection) was cultured in MEM-α (cat. no. 51985-034; Thermo Fisher Scientific, Inc.) with ribonucleosides, deoxyribonucleosides, 2 mM L-glutamine and 1 mM sodium pyruvate in the absence of ascorbic acid (cat no. A1049001; Thermo Fisher Scientific, Inc). The complete growth medium was prepared by the addition of FBS (cat. no. 10099141; Thermo Fisher Scientific, Inc.) to the base medium to reach a final concentration of 10% under 37°C and a humidified atmosphere containing 5% CO2, and the culture medium was replaced every 2 days.
Construction of the COL6A1 gene vector
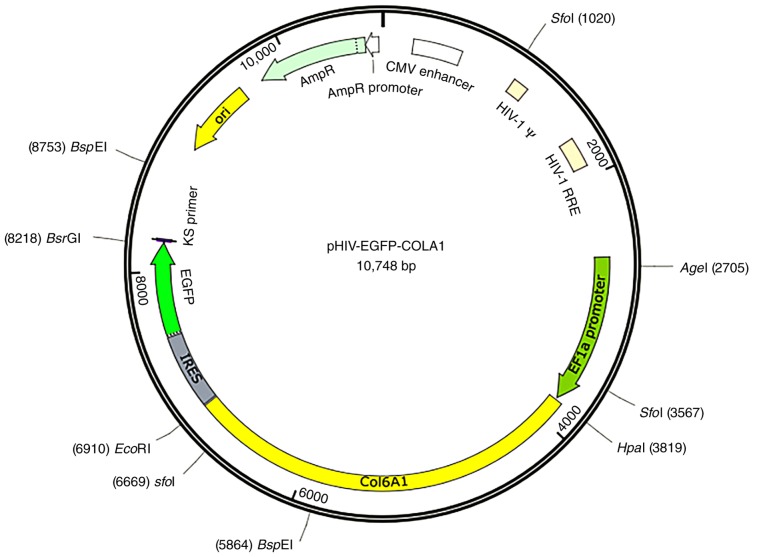
3T3-E1 cells were collected for total DNA extraction using QIAamp DNA Mini kit (Qiagen GmbH) according to the maunfacturer's protocol. The Q5 high-fidelity DNA polymerase (New England BioLabs, Inc.) was utilized to amplify the sequence. The PCR reaction was performed using the following amplification profile: One cycle at 98°C for 5 min, 33 cycles of 98°C for 15 sec, 60°C for 15 sec and 72°C for 1 min followed by one cycle of 72°C for 5 min. Primers were designed and synthesized based on the human COL6A1 sequence. Upstream HpaI and downstream EcoRI restriction sites were added. The primer sequences are presented in Table I. A pHIV-EGFP vector contained the EF1a-MCS-IRES-EGFP component sequence (Addgene). Moreover, HpaI and EcoRI enzyme cleavage sites were produced and the following control insert sequence was used: 5′-TATTCTAGGGATCCAACCCTCGAGTGACCCGTCTAGAGGGCTAG-3′. The vector map is presented in Fig. 2. GFP is located downstream of the target gene separated by the IRES sequence. The positive bacterial Sanger sequencing primer sequences are shown in Table II. The LA taq DNA polymerase (Takara Bio, Inc.) was utilized to identify the positive clones. The PCR reaction was performed using the following amplification profile: One cycle at 95°C for 10 min, 33 cycles of 98°C for 30 sec, 60°C for 30 sec and 72°C for 2 min followed by one cycle of 72°C for 10 min. Ethidium bromide was added to 1% agarose gels for PCR product visualization, and visualized using Image Lab software (version no. 1709690; Bio-Rad Laboratories, Inc.). Mutant primers were designed based on the human COL6A1 gene sequence information and wild-type plasmids were used as templates to design mutant primers according to the primer design principles of the Site-Directed Mutagenesis kits (cat. no. 210514; Agilent Technologies, Inc.). The mutant and wild-type COL6A1 primer sequences are presented in Table III. The COL6A1 wild-type and mutant plasmids were subjected to endotoxin plasmid extraction and the procedure was completed using Plasmid DNA Purification kits (cat. no. K210006; Thermo Fisher Scientific, Inc.).
Table I.
COL6A1 gene primer sequences.
| Gene | Primer sequence | PCR product (bp) |
|---|---|---|
| COL6A1 forward | 5′-ATCGGTTAACATGAGGGCGGCCCGTGCTCTGCT-3′ | 3,087 |
| COL6A1 reverse | 5′-CGGAATTCTTAGCCCAGCGCCACCTTCCTGGA-3′ |
COL6A1, collagen 6A1.
Figure 2.
pHIV-EGFP lentiviral vector information. The component sequence is EF1a-MCS-IRES-EGFP and the enzyme restriction sites are HpaI and EcoRI.
Table II.
Sanger sequencing primer sequences.
| Gene | Primer sequence | Length (bp) |
|---|---|---|
| COL6A1 forward | 5′-AAGCCTCAGACAGTGGTTCAAAG-3′ | 24 |
| COL6A1 reverse | 5′-CAAGCGGCTTCGGCCAGTAACGT-3′ | 25 |
COL6A1, collagen 6A1.
Table III.
Mutant and wild-type gene primer sequences.
| Gene | Primer sequence | PCR product (bp) |
|---|---|---|
| COL6A1-rs201153092G-F | 5′-GGGGAGAAGACGGCCCCGCTGGAAATGGCA-3′ | 3,087 |
| COL6A1-rs201153092G-R | 5′-TCCAGCGGGGCCGTCTTCTCCCCTTTCACC-3′ | |
| COL6A1-rs201153092A-F | 5′-GGGGAGAAGACAGCCCCGCTGGAAATGGCA-3′ | 3,087 |
| COL6A1-rs201153092A-R | 5′-TCCAGCGGGGCTGTCTTCTCCCCTTTCACC-3′ |
COL6A1, collagen 6A1; F, forward; R, reverse.
Transfection of the pHIV-COL6A1-rs201153092A, pHIV-COL6A1-rs201153092G and pHIV-GFP expression vectors into 3T3-E1 cells
A total of 5 ml poly-D-lysine (PDL; cat. no. P7280; Sigma-Aldrich; Merck KGaA) was pre-added to a 10-cm dish, which was incubated at 37°C for 30 min. PDL was aspirated and the dish was washed twice with PBS (cat. no. AM9625; Thermo Fisher Scientific, Inc.). The samples were stored in a 37°C incubator for 7 days. 293 cells (American Type Culture Collection; ATCC no. CRL-1573) that were passaged up to 20 times were used for lentivirus production. that were passaged up to 20 times were used. The cells were seeded into 10-cm dishes (70% confluent) following trypsin digestion. For these experiments only cells at the logarithmic growth phase were used. The cells were cultured in DMEM (cat. no. 11885-084; Thermo Fisher Scientific, Inc.) at 37°C overnight and the medium was discarded. A small quantity of Opti-MEM (cat. no. 51985034; Thermo Fisher Scientific, Inc.) was added to rinse the dish. Subsequently, 6 ml Opti-MEM were added to the dish, which was placed in an incubator for further use. The helper plasmid pMD2G (Addgene), pSPAX2 (Addgene), and the wild-type or mutant expression plasmids pHIV-EGFP-COL6A1 (Addgene) were added to 500 µl Opti-MEM medium at a ratio of 3:6:9 µg, respectively. A total of 15 µl PLUS™ reagent (cat. no. 11514015; Invitrogen; Thermo Fisher Scientific, Inc.) was added to the mixture. An additional sample was prepared using 500 µl Opti-MEM medium and 30 µl Lipofectamine® Transfection Reagent (cat. no. L3000008 Thermo Fisher Scientific, Inc.). The reagents were mixed thoroughly and the resulting sample was allowed to stand for 5 min at room temperature. The final volume of the solution was 1 ml. The liposomes were mixed with the plasmids and subsequently coated at room temperature for 30 min. The liposome and 18 µg plasmid mixtures were added to the 5×104/ml 293 cells in the presence of Opti-MEM and complete DMEM containing 10% FBS (cat. no. 10099141; Thermo Fisher Scientific, Inc.). The FBS was replaced following 6–8 h of cell culture. The cells were cultured for 48 h at 37°C in a 5% CO2 incubator, then the solution containing the virus was concentrated and purified. The cells were infected with the high quality viral solutions, namely pHIV-COL6A1-rs201153092G (titer, 2.6×108 TU/ml) and pHIV-COL6A1-rs201153092A (titer, 2.9×108 TU/ml) for 24 h. The viral supernatant was aspirated and fresh DMEM was added. 3T3-E1 cells were plated in 6-well plates (30% confluent) and following 12 h of incubation, the concentrated viral solution was added to infect the cells, at a multiplicity of infection value of 50. This cutoff was used for all the groups. GFP expression was monitored using a fluorescent microscope (magnification, ×20) at 48 h following exposure. The cells with GFP expression were sorted using a CytoFLEX flow cytometer (Beckman Coulter, Inc.) 72 h following exposure and data were analyzed using CytExpert (version no. 1.1.10.0; Beckman Coulter, Inc.). An aliquot of the cells were collected for protein expression analysis performed via western blotting (WB). Once the cells had grown to a confluence of 80–90%, they were subcultured.
Differentiation of 3T3-E1 cells into osteoblasts
3T3-E1 cells were divided into various groups according to the type of plasmid as follows: pHIV-COL6A1-rs201153092G (wild-type group), pHIV-COL6A1-rs201153092A (mutation group), pHIV-GFP (empty lentivirus control group) and no vector (mock group). 3T3-E1 cells were cultured as in our previous study (29). The cells were harvested at 21 days following osteogenic induction.
Alkaline phosphatase (ALP) activity assay and Alizarin red staining
The cells were seeded in 24-well plates at a density of 1×105 cells/well and cultured in osteogenic medium for 21 days. Osteogenic identification was assessed using an ALP activity staining kit (cat. no. GMS80033.1; Genmed Scientifics, Inc.) as in our previous study (29). Cell staining was analyzed via visual inspection. Mineralization was assessed using an Alizarin Red S kit (cat. no. GMS80046.3; Genmed Scientific) as in our previous study (29), and cell staining was analyzed by visual inspection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and RT was performed using a M-MLV Reverse Transcriptase kit (Promega Corporation) according to the manufacturer's instructions. qPCR was applied to quantify the mRNA levels of COL6A1, ALP and GAPDH using SYBR Green Real-Time PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) under a 2-step cycling program (95°C for 10 min hold, 40 cycles of 95°C for 15 sec and 60°C for 60 sec). The reaction was performed as in our previous study (29). All experiments were normalized to GAPDH, and the relative gene expression was calculated based on the 2−ΔΔCq method (30). The primers are listed in Table IV.
Table IV.
Primer sequences for RT-qPCR.
| Gene | Primer sequence | PCR product (bp) |
|---|---|---|
| COL6A1-RT-F | 5′-ACAGTGACGAGGTGGAGATCA-3′ | 122 |
| COL6A1-RT-R | 5′-GATAGCGCAGTCGGTGTAGG-3′ | |
| ALP-RT-F | 5′-GTGAACCGCAACTGGTACTC-3′ | 81 |
| ALP-RT-R | 5′-GAGCTGCGTAGCGATGTCC-3′ | |
| GAPDH-RT-F | 5′-TGGGTGTGAACCATGAGAAGT-3′ | 126 |
| GAPDH-RT-R | 5′-TGAGTCCTTCCACGATACCAA-3′ |
COL6A1, collagen 6A1; ALP, alkaline phosphatase; F, forward; R, reverse; RT-q, reverse transcription-quantitative.
WB analysis
Tissue and cell lysates were obtained using ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology) containing 10 mM PMSF as a protease inhibitor. WB was performed as described in our previous study (29) using the following primary antibodies: Anti-COL6A1 (1:2,000; cat. no. ab182744; Abcam); anti-ALP (1:1,000; cat. no. ab67228; Abcam); anti-GFP (1:10,000; cat. no. ab183734; Abcam); and anti-GAPDH (1:2,500; cat. no. ab9485; Abcam). In addition, the following secondary antibodies were used: Goat anti-mouse antibody (1:2,500; cat. no. CW0102M; Beijing Kangwei Biotech Co. Ltd.) and goat anti-rabbit antibody (1:2,500; cat. no. CW0103M; Beijing Kangwei Biotech Co. Ltd.). The blots were detected using a Kodak film developer (Fujifilm). The protein levels were quantified by densitometric analysis using Image-Pro Plus software version 6.0 (Media Cybernetics, Inc.). GAPDH was used as the endogenous control.
Statistical analysis
All statistical analyses were performed using SPSS software version 17.0 (SPSS, Inc.). Descriptive data for continuous variables are presented as the mean ± standard deviation. Student's t-test was used to compare mean values between two groups. The differences in T-OPLL subtypes between patients with or without COL6A1 gene mutation were determined using one-way ANOVA with Fisher's post hoc test. Statistical significance was determined by using one-way ANOVA and post hoc Tukey tests for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed three times.
Results
Genotype-phenotype analysis
A total of 20 unrelated Northern Chinese Han patients with T-OPLL carrying the rs201153092A mutation in the COL6A1 gene (8 men, mean age 54.38±10.77 years; 12 women, mean age 57.92±11.19 years) and 20 unrelated Northern Chinese Han patients with T-OPLL carrying the wild-type rs201153092G variant (10 men, mean age 62.30±7.27 years; 10 women, mean age 52.00±6.55 years) were enrolled in the present study. Phenotype-genotype associations were analyzed among the patients with T-OPLL with or without the rs201153092A mutation (n=20/group). No significant differences were noted between these two groups with regard to sex, age and JOA score at diagnosis (Table V).
Table V.
Clinical features of patients with thoracic ossification of the posterior longitudinal ligament with or without rs201153092A mutations.
| Indices | rs201153092A, n=20 | rs201153092G, n=20 | P-value |
|---|---|---|---|
| Age, years | 56.50±10.88 | 57.15±8.56 | NS |
| Male/female | 8/12 | 10/10 | NS |
| Continuous | 6 (30.0%) | 5 (25.0%) | NS |
| Local | 1 (5.0%) | 1 (5.0%) | NS |
| Segmental | 3 (15.0%) | 4 (20.0%) | NS |
| Mixed | 10 (50.0%) | 10 (50.0%) | NS |
| JOA score | 3.79±0.74 | 3.96±0.86 | NS |
Data are presented as the mean ± SDs or n (%). NS, not significant; JOA, Japanese Orthopedic Association.
Functional assays of COL6A1 variants in patients with T-OPLL
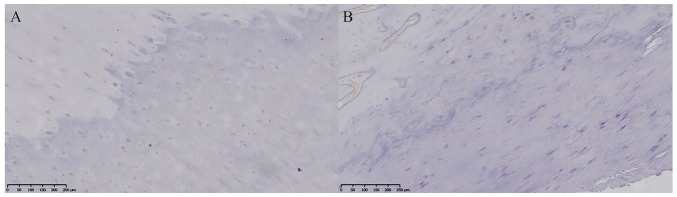
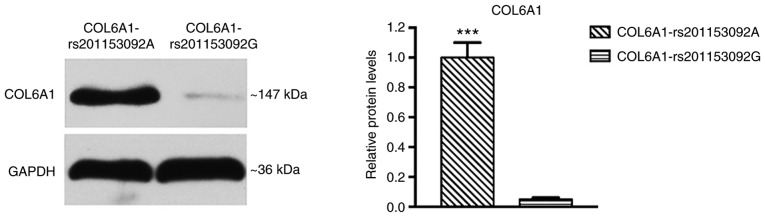
H&E staining revealed a number of chondrocytes in the ossification zone (open arrow) with zigzag tidal traces that were formed by calcification near the chondrocytes (solid arrow) in the patients with T-OPLL (Fig. 3). IHC revealed that the COL6A1 protein was positively expressed in the ossified areas of the patients with T-OPLL possessing the rs201153092A mutation, although no nuclear reactivity was noted in the normal fibers (Fig. 4A). However, weak positive expression of COL6A1 was noted in the patients with T-OPLL carrying the wild-type rs201153092G variant (Fig. 4B). WB analysis revealed that the expression of the COL6A1 protein was significantly higher in thoracic posterior longitudinal ligament specimens from patients with T-OPLL possessing the rs201153092A mutation compared with that of patients carrying the wild-type rs201153092G variant (Fig. 5).
Figure 3.
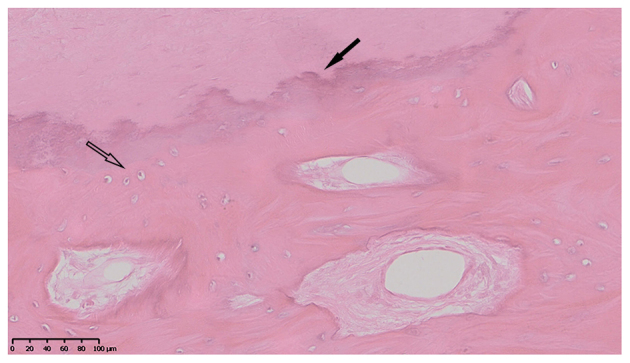
Hematoxylin-eosin staining. Representative hematoxylin-eosin staining of thoracic ossified posterior longitudinal ligament tissue. Scale bar, 100 µm.
Figure 4.
IHC staining for COL6A1. (A) Patients with T-OPLL carrying the COL6A1 gene rs201153092A mutation. (B) Patients with T-OPLL possessing the rs201153092G variant. Scale bar, 250 µm. COL6A1, collagen 6A1; IHC, immunohistochemistry; T-OPLL, thoracic ossified posterior longitudinal ligament.
Figure 5.
Protein expression of COL6A1 in T-OPLL. Expression of COL6A1 protein in thoracic posterior longitudinal ligament specimens from patients with T-OPLL carrying the COL6A1 gene rs201153092A mutation was significantly higher than in those carrying the rs201153092G variant; ***P<0.001 vs. COL6A1-rs201153092G. COL6A1, collagen 6A1; T-OPLL, thoracic ossified posterior longitudinal ligament.
Wild-type and mutant bacterial COL6A1 variant analysis via Sanger sequencing
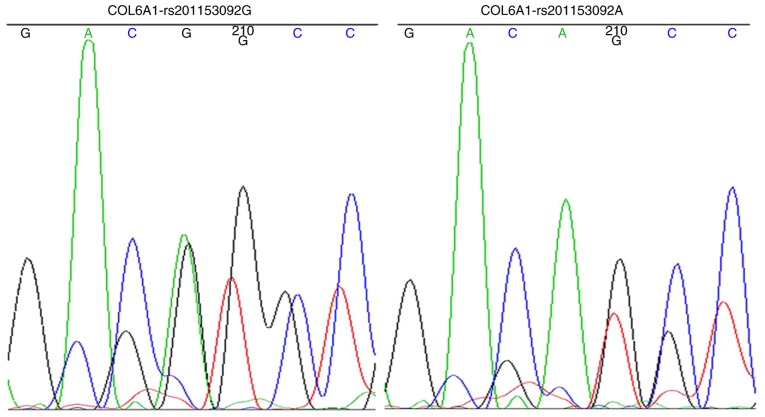
The strains that were identified as positive using the electrophoresis of the PCR products were sequenced to obtain the wild-type and mutant COL6A1 gene clones (Fig. 6). The data indicated that the wild-type and mutant COL6A1 gene lentiviral packaging vectors were successfully constructed.
Figure 6.
Wild-type and mutant COL6A1 gene bacteria Sanger sequencing. Sanger sequencing of the strains that were identified as positive using electrophoresis of the PCR products. A allele, green; T allele, red; C allele, blue; G allele, black. COL6A1, collagen 6A1.
Lentiviral infection of the 3T3-E1 cells
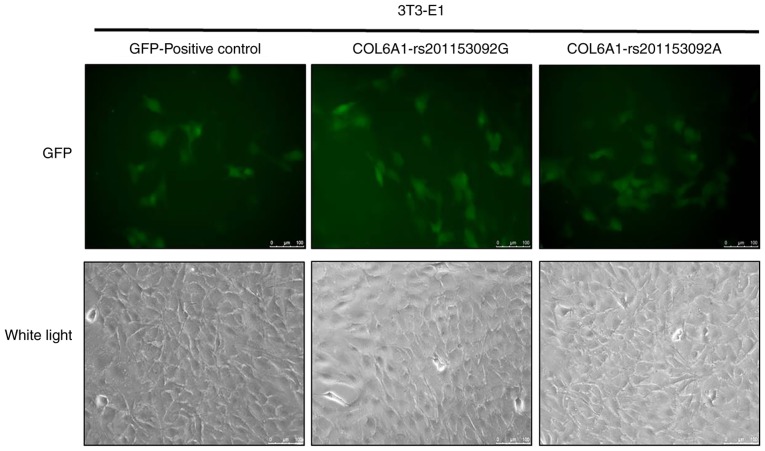
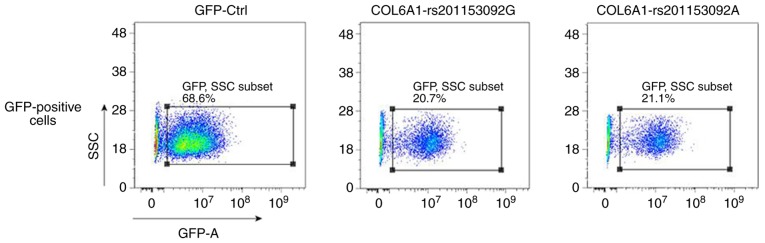
Notable levels of GFP expression were observed using fluorescence microscopy at 48 h following cell exposure (Fig. 7), which indicated that 3T3-E1 cells were successfully infected. 3T3-E1 cells were trypsinized at 72 h following infection and the GFP-positive cells were sorted by flow cytometry, as shown in Fig. 8. The positive cells were plated, cultured and subjected to the subsequent osteogenesis induction experiments. Cell lines expressing the wild-type and mutant COL6A1 gene variants were thus obtained.
Figure 7.
Lentivirus infection of 3T3-E1 cells. 3T3-E1 cells were infected with pHIV-COL6A1-rs201153092G and pHIV-COL6A1-rs201153092A lentiviruses and GFP expression was observed. COL6A1, collagen 6A1.
Figure 8.
Flow cytometric sorting of the GFP-positive cells. After the 3T3-E1 cells were infected with the lentivirus, flow cytometry was used to detect the GFP-positive cells. COL6A1, collagen 6A1; SSC, side scatter.
COL6A1 rs201153092A mutation site induces the osteogenic differentiation of 3T3-E1 cells
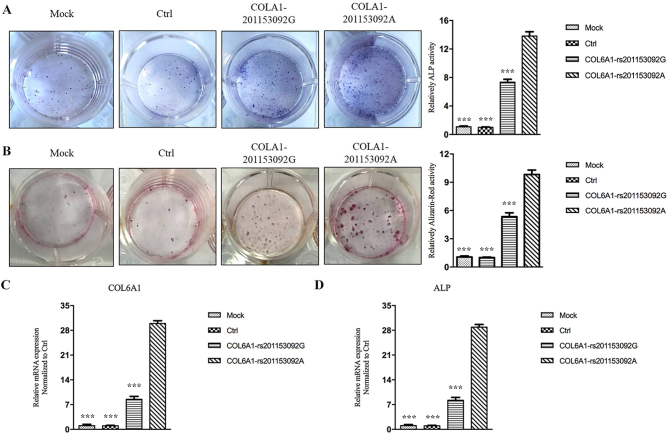
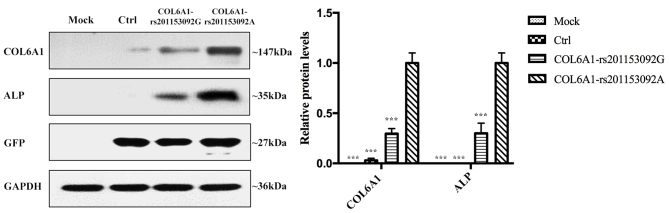
Following 21 days of osteogenic induction, the ALP expression levels of the COL6A1 mutant group were significantly higher than those of the mock, empty lentivirus control and the wild-type groups (Fig. 9A). The Alizarin red content in the COL6A1 mutant group was significantly higher than that of the mock, the empty lentivirus control and the wild-type groups (Fig. 9B). RT-qPCR analysis revealed that the mRNA expression levels of COL6A1 and ALP in the COL6A1 mutant group were significantly higher than those of the mock, empty lentivirus control and the wild-type groups (Fig. 9C and D). The expression levels of the exogenous COL6A1 protein in the cells was confirmed via WB analysis of GFP tags. The expression levels of COL6A1 and ALP in the rs201153092A transgenic COL6A1 3T3-E1 cell group were significantly higher than those noted in the mock, empty lentivirus control and rs201153092G transgenic COL6A1 gene groups (Fig. 10).
Figure 9.
COL6A1 gene rs201153092A site mutation induces osteogenic differentiation of 3T3-E1 cells. After 21 days of osteogenic induction, (A) ALP staining and (B) Alizarin red staining were performed in the COL6A1 gene mutation, non-mutation, control and mock groups. ***P<0.001 vs. COL6A1-rs201153092A. mRNA expression levels of (C) COL6A1 and (D) ALP in the COL6A1 gene mutation and non-mutation groups were normalized to the control group. ***P<0.001 vs. COL6A1-rs201153092A. ALP, alkaline phosphatase; COL6A1, collagen 6A1; Ctrl, control.
Figure 10.
Expression levels of GFP-tagged COL6A1 and ALP proteins detected by western blot. The relative protein levels of COL6A1 and ALP in the COL6A1 gene mutation, non-mutation, control and mock groups. ***P<0.001 vs. COL6A1-rs201153092A. ALP, alkaline phosphatase; COL6A1, collagen 6A1; Ctrl, control.
Discussion
Despite advances in surgical techniques, the treatment of T-OPLL remains challenging. Previous studies conducted by our group identified the rs201153092A mutation site in the COL6A1 gene as a potentially pathogenic genetic locus for T-OPLL (26,27). The present study revealed the functional impact of the rs201153092A mutation of the COL6A1 gene.
In the present study, H&E staining demonstrated a number of chondrocytes in the ossification area, suggesting that intraosseous ossification may be involved in the pathological process of OPLL. IHC and WB assays were performed to determine COL6A1 protein expression in patients with T-OPLL. The results demonstrated that the expression levels of the COL6A1 protein were higher in the patients with T-OPLL possessing the mutation compared to those with the wild-type gene, suggesting that this is a potential pathogenic genetic locus that alters COL6A1 gene expression in cells.
To assess the pathogenic potential of the mutation site, COL6A1-induced mouse embryonic osteoblast 3T3-E1 cells were established via infection with lentiviral vectors encoding the COL6A1 gene possessing either the rs201153092A and rs201153092G sites. The data indicated that the presence of the rs201153092A mutation led to increased expression of the COL6A1 protein. Based on the comparison of osteogenic markers, the data further demonstrated that the rs201153092A mutant promoted the osteogenic differentiation of 3T3-E1 cells, suggesting that this mutation may enhance this process via the overexpression of the corresponding COL6A1 gene.
The rs201153092 site of the COL6A1 gene is located in an exonic region. The sequence of COL6A1 mRNA exhibited no difference between the wild-type and mutant variants. Therefore, RT-qPCR and WB analyses could not distinguish between the wild-type and mutant COL6A1. The mutation in the exonic region enhanced the expression levels of the COL6A1 mRNA, which was further translated into the COL6A1 protein.
COL6 is an important component of the extracellular matrix of several tissues, including muscle, tendons and cartilage (29). Disorders caused by mutations in the COL6A1 gene mainly affect the ligament and muscle tissues, whereas mutations in the COL6A1 polypeptide chain are causative of a broad spectrum of diseases in humans, including Ullrich congenital muscular dystrophy and C-OPLL (31). Tanaka et al (28) identified that certain molecular variants of extracellular proteins may be implicated in the ectopic ossification observed in OPLL. The COL6A1 gene may increase bone mass and promote bone formation via the regulation of various pathways. Izu et al (32) demonstrated that cell-cell interactions played an essential role in bone formation, COL6A1 regulates bone formation by establishing the communication of several cell networks that contribute to this process. Cheng et al (33) indicated that the COL6A1 gene exerted its biological functions via the Akt/PI3K pathway. However, the mechanism by which the COL6A1 gene facilitates the development of T-OPLL remains unclear and future studies are required to fully clarify this interaction.
In conclusion, the present study demonstrated that the expression levels of the COL6A1 protein were higher in the patients with T-OPLL carrying a mutant genotype of the corresponding COL6A1 gene compared to those with the wild-type variant. In addition, the rs201153092A mutation was able to induce osteogenic differentiation of mouse embryonic osteoblasts via overexpression of the COL6A1 gene. The present findings provided potential insight into the pathogenesis of T-OPLL and the pathogenic role of COL6A1 in the development of this disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Natural Science Foundation of China (grant no. 81672201) and China Postdoctoral Science Foundation (grant no. 2019M650771).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PW, XGL and SL conceived and designed the study. PW and ZT performed the experiments. PW, XL and CK analyzed the data. PW drafted the manuscript. SL revised the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to participate
The study was approved by The Ethics Committee for Human Subjects of the Peking University Third Hospital (permit no. 2014036). Informed consent was provided by all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Koda M, Furuya T, Okawa A, Inada T, Kamiya K, Ota M, Maki S, Takahashi K, Yamazaki M, Aramomi M, et al. Mid- to long-term outcomes of posterior decompression with instrumented fusion for thoracic ossification of the posterior longitudinal ligament. J Clin Neurosci. 2016;27:87–90. doi: 10.1016/j.jocn.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Hu P, Yu M, Liu X, Liu Z, Jiang L. A circumferential decompression-based surgical strategy for multilevel ossification of thoracic posterior longitudinal ligament. Spine J. 2015;15:2484–2492. doi: 10.1016/j.spinee.2015.08.060. [DOI] [PubMed] [Google Scholar]
- 3.Kong W, Ao J, Cao G, Xia T, Liu L, Liao W. Local spinal cord decompression through a full endoscopic percutaneous transcorporeal approach for cervicothoracic ossification of the posterior longitudinal ligament at the T1-T2 level. World Neurosurg. 2018;112:287–293. doi: 10.1016/j.wneu.2018.01.099. [DOI] [PubMed] [Google Scholar]
- 4.Xu N, Yu M, Liu X, Sun C, Chen Z, Liu Z. A systematic review of complications in thoracic spine surgery for ossification of the posterior longitudinal ligament. Eur Spine J. 2017;26:1803–1809. doi: 10.1007/s00586-015-4097-5. [DOI] [PubMed] [Google Scholar]
- 5.Xu ZW, Hu YC, Sun CG, Shang XP, Lun DX, Li F, Ji XB, Liu DY, Chen NW, Zhuang QS. Treatment for thoracic ossification of posterior longitudinal ligament with posterior circumferential decompression. Orthop Surg. 2017;9:206–214. doi: 10.1111/os.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu JW, Yun SO, Hsieh CS, Lee SH. Minimally invasive anterior decompression technique without instrumented fusion for huge ossification of the posterior longitudinal ligament in the thoracic spine: Technical note and literature review. J Korean Neurosurg Soc. 2017;60:597–603. doi: 10.3340/jkns.2017.0404.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Yang B, Wang Y, He X, Li H. Treatment for thoracic ossification of posterior longitudinal ligament with posterior circumferential decompression: Complications and managements. J Orthop Surg Res. 2016;11:153. doi: 10.1186/s13018-016-0489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onishi E, Yasuda T, Yamamoto H, Iwaki K, Ota S. Outcomes of surgical treatment for thoracic myelopathy: A Single-institutional study of 73 patients. Spine (Phila Pa 1976) 2016;41:E1356–E1363. doi: 10.1097/BRS.0000000000001622. [DOI] [PubMed] [Google Scholar]
- 9.Imagama S, Ando K, Takeuchi K, Kato S, Murakami H, Aizawa T, Ozawa H, Hasegawa T, Matsuyama Y, Koda M, et al. Perioperative complications after surgery for thoracic ossification of posterior longitudinal ligament-nationwide multicenter prospective study. Spine (Phila Pa 1976) 2018;43:E1389–E1397. doi: 10.1097/BRS.0000000000002703. [DOI] [PubMed] [Google Scholar]
- 10.Xu C, Chen Y, Zhang H, Chen Y, Shen X, Shi C, Liu Y, Yuan W. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci Rep. 2016;6:21580. doi: 10.1038/srep21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegawa S. Genetics of ossification of the posterior longitudinal ligament of the spine: A Mini Review. J Bone Meta. 2014;21:127–132. doi: 10.11005/jbm.2014.21.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikegawa S. Genomic study of ossification of the posterior longitudinal ligament of the spine. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:405–412. doi: 10.2183/pjab.90.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Shi L, Shi G, Guo Y, Chen D, Chen D, Shi J. Connexin 43 affects osteogenic differentiation of the posterior longitudinal ligament cells via regulation of ERK activity by stabilizing Runx2 in ossification. Cell Physiol Biochem. 2016;38:237–247. doi: 10.1159/000438625. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima M, Kou I, Ohashi H, Genetic Study Group of the Investigation Committee on the Ossification of Spinal Ligaments. Ikegawa S. Identification and functional characterization of RSPO2 as a susceptibility gene for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2016;99:202–207. doi: 10.1016/j.ajhg.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, He HL, Chen CY, Zhao Y, Jiang HL, Liu WT, Du ZF, Chen XL, Shi SY, Zhang XN. Whole exome sequencing implicates PTCH1 and COL17A1 genes in ossification of the posterior longitudinal ligament of the cervical spine in Chinese patients. Genet Mol Res. 2014;13:1794–1804. doi: 10.4238/2014.March.17.7. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima M, Takahashi A, Tsuji T, Karasugi T, Baba H, Uchida K, Kawabata S, Okawa A, Shindo S, Takeuchi K, Taniguchi Y, et al. A genome-wide association study identifies susceptibility loci for ossification of the posterior longitudinal ligament of the spine. Nat Genet. 2014;46:1012–1016. doi: 10.1038/ng.3045. [DOI] [PubMed] [Google Scholar]
- 17.Guo Q, Lv SZ, Wu SW, Tian X, Li ZY. Association between single nucleotide polymorphism of IL15RA gene with susceptibility to ossification of the posterior longitudinal ligament of the spine. J Orthop Surg Res. 2014;9:103. doi: 10.1186/s13018-014-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z, Zhu H, Ding L, Xiao H, Chen D, Xue F. Association of NPP1 polymorphism with postoperative progression of ossification of the posterior longitudinal ligament in Chinese patients. Genet Mol Res. 2013;12:4648–4655. doi: 10.4238/2013.October.18.3. [DOI] [PubMed] [Google Scholar]
- 19.Ren Y, Feng J, Liu ZZ, Wan H, Li JH, Lin X. A new haplotype in BMP4 implicated in ossification of the posterior longitudinal ligament (OPLL) in a Chinese population. J Orthop Res. 2012;30:748–756. doi: 10.1002/jor.21586. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Liu ZZ, Feng J, Wan H, Li JH, Wang H, Lin X. Association of a BMP9 haplotype with ossification of the posterior longitudinal ligament (OPLL) in a Chinese population. PLoS One. 2012;7:e40587. doi: 10.1371/journal.pone.0040587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung WS, Nam DH, Jo DJ, Lee JH. Association of toll-like receptor 5 gene polymorphism with susceptibility to ossification of the posterior longitudinal ligament of the spine in Korean population. J Korean Neurosurg Soc. 2011;49:8–12. doi: 10.3340/jkns.2011.49.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Zhao Y, Chen Y, Shi G, Yuan W. RUNX2 polymorphisms associated with OPLL and OLF in the Han population. Clin Orthop Relat Res. 2010;468:3333–3341. doi: 10.1007/s11999-010-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao M, Mao F, Zhao Z, Ma G, Xu G, Xu W, Chen H, Zhu M. COL6A1 mutation leading to Bethlem myopathy with recurrent hematuria: A case report. BMC Neurol. 2019;19:32. doi: 10.1186/s12883-019-1263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Guo J, Cai T, Zhang F, Pan S, Zhang L, Wang S, Zhou F, Diao Y, Zhao Y, et al. Targeted next-generation sequencing reveals multiple deleterious variants in OPLL-associated genes. Sci Rep. 2016;6:26962. doi: 10.1038/srep26962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong Q, Ma X, Li F, Guo Z, Qi Q, Li W, Yuan H, Wang Z, Chen Z. COL6A1 polymorphisms associated with ossification of the ligamentum flavum and ossification of the posterior longitudinal ligament. Spine (Phila Pa 1976) 2007;32:2834–2838. doi: 10.1097/BRS.0b013e31815b761c. [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Liu X, Zhu B, Ma Y, Yong L, Teng Z, Wang Y, Liang C, He G, Liu X. Identification of susceptibility loci for thoracic ossification of the posterior longitudinal ligament by whole-genome sequencing. Mol Med Rep. 2018;17:2557–2564. doi: 10.3892/mmr.2017.8171. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Liu X, Zhu B, Ma Y, Yong L, Teng Z, Liang C, He G, Liu X. Association of IL17RC and COL6A1 genetic polymorphisms with susceptibility to ossification of the thoracic posterior longitudinal ligament in Chinese patients. J Orthop Surg Res. 2018;13:109. doi: 10.1186/s13018-018-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T, Ikari K, Furushima K, Okada A, Tanaka H, Furukawa K, Yoshida K, Ikeda T, Ikegawa S, Hunt SC, et al. Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2003;73:812–822. doi: 10.1086/378593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Liu X, Kong C, Liu X, Teng Z, Ma Y, Yong L, Liang C, He G, Lu S. Potential role of the IL17RC gene in the thoracic ossification of the posterior longitudinal ligament. Int J Mol Med. 2019;43:2005–2014. doi: 10.3892/ijmm.2019.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Capitanio D, Moriggi M, De Palma S, Bizzotto D, Molon S, Torretta E, Fania C, Bonaldo P, Gelfi C, Braghetta P. Collagen VI Null mice as a model for early onset muscle decline in aging. Front Mol Neurosci. 2017;10:337. doi: 10.3389/fnmol.2017.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izu Y, Ezura Y, Koch M, Birk DE, Noda M. Collagens VI and XII form complexes mediating osteoblast interactions during osteogenesis. Cell Tissue Res. 2016;364:623–635. doi: 10.1007/s00441-016-2385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng IH, Lin YC, Hwang E, Huang HT, Chang WH, Liu YL, Chao CY. Collagen VI protects against neuronal apoptosis elicited by ultraviolet irradiation via an Akt/Phosphatidylinositol 3-kinase signaling pathway. Neuroscience. 2011;183:178–188. doi: 10.1016/j.neuroscience.2011.03.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.