Abstract
Connective tissue growth factor (CTGF) is a possible key determinant of progressive fibrosis. Nanotechnology has been considered as a potential tool for developing novel drug delivery systems for various diseases, including liver fibrosis. The present study aimed to investigate the potential antifibrotic activity of CTGF small interfering RNA (siRNA) mediated by polyethyleneimine (PEI)-functionalized magnetic iron oxide (Fe3O4) nanoparticles (NPs) in LX-2 cells. PEI-Fe3O4/siRNA complexes were synthesized to facilitate siRNA delivery and were transfected into LX-2 cells. Laser confocal microscopy was employed to investigate the cell uptake of PEI-Fe3O4/siRNA complexes. Reverse transcription-quantitative PCR (RT-qPCR) and western blotting were used to verify the effect of gene silencing. The results showed that siRNA-loaded PEI-Fe3O4 exhibited low cytotoxicity. The transfection efficiency of PEI-Fe3O4/siRNA reached 73.8%, and RT-qPCR and western blotting demonstrated effective gene silencing. These results indicated that CTGF siRNA delivered by PEI-Fe3O4 NPs significantly reduces CTGF expression and collagen production in activated LX-2 cells, providing a basis for future in vivo studies.
Keywords: liver fibrosis, connective tissue growth factor, iron oxide, nanoparticles, small interfering RNA
Introduction
Liver fibrosis continues to be a major health problem worldwide and is associated with significant morbidity and mortality. If left untreated, fibrosis will develop into liver cirrhosis, leading to organ failure and ultimately death (1,2). The cytokine connective tissue growth factor (CTGF) is key in promoting liver fibrosis and plays an important role in the pathogenesis of liver fibrosis (3). Despite the advances in our understanding of the pathogenesis of fibrotic processes, the therapeutic effect of drugs used in the treatment of liver fibrosis is insufficient. Therefore, there is an urgent need for specific and effective antifibrotic therapies.
Gene therapy is a novel therapeutic tool that may provide a solution to managing chronic diseases and genetic disorders. RNA interference is a novel therapeutic strategy that may aid in the management of liver fibrosis through silencing the expression of specific genes in diseased cells (4,5). Its capacity to effectively knock down the target gene with high sequence specificity makes the use of small interfering RNA (siRNA) a promising therapeutic strategy. In previous years, there has been considerable interest in siRNA-based gene therapy for the treatment of liver fibrosis (6,7). However, gene delivery is a major challenge in gene therapy. Currently, viral vectors remain the primary gene delivery system utilized in gene therapy. However, they present important limitations, including non-specificity, immunogenicity to target cells, toxicity and enzymatic degradation (8). Therefore, the development of non-viral carriers is of considerable interest at present.
Currently, a wide variety of non-viral carriers have been developed, and mainly lipids, polymers and nanoparticles (NPs) have been utilized as siRNA delivery systems (9). NPs, which are nanostructured entities with adaptable size, shape, surface and biological properties, have been widely used in biomedical applications and diagnosis (10,11). Numerous NP core material formulations, including gold, silica, carbon, semiconductors and metal oxides, are being evaluated as potential siRNA carriers (12–16). Among them, magnetic iron oxide (Fe3O4) NPs are emerging as one of the most potent non-viral gene carriers, and have extensive applications in various biomedical fields due to their high efficiency, low toxicity and relatively simple experimental use (17–20).
In the present study, human hepatic stellate cells LX-2 were selected as target cells of fibrosis to analyze the antifibrotic efficiency of polyethyleneimine (PEI)-functionalized Fe3O4 NPs combined with CTGF siRNA, which may serve as a basis for the development of future therapeutics for hepatic fibrosis.
Materials and methods
Characterization of PEI-Fe3O4 NPs
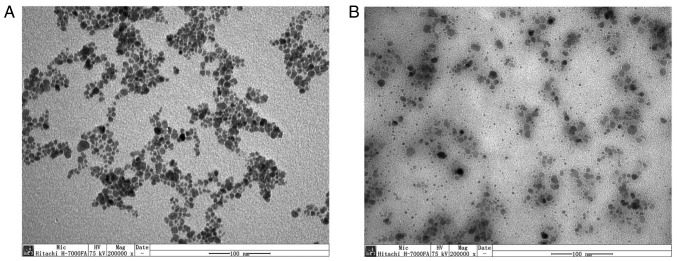
PEI-Fe3O4 NPs (Nanjing Nanoeast Biotech Co., Ltd.) were used as carriers for siRNA delivery. The NPs were dispersed in water at pH 7.4, obtaining a final concentration of 1 mg/ml, a hydrodynamic diameter of 48±5 nm and a ζ-value of +30.5±4.87 mV. The morphology of PEI-Fe3O4 NPs and PEI-Fe3O4/siRNA was characterized using a transmission electron microscope (TEM). Samples for TEM imaging were prepared by placing a drop of the NP suspension onto a copper grid and drying it at ambient temperature overnight. The samples were then visualized with a H-7000FA microscope, operating at 75 kV and 200,000× magnification (Hitachi, Ltd.).
Preparation of PEI-Fe3O4/siRNA complexes
siRNA targeting human CTGF (Genbank accession number NM-001901) was synthesized and purified by Shanghai GenePharma Co., Ltd. It is a 21-bp double-stranded RNA oligo and has the following sequence: Sense 5′-CCGACUGGAAGACACGUUUTT-3′ and antisense 5′-AAACGUGUCUUCCAGUCGGTT-3′. In addition, a cyanine (Cy)3-labeled negative control (NC) siRNA was used for evaluating the efficiency of transfection (sense 5′-Cy3-UUCUCCGAACGUGUCACGUTT-3′ and antisense 5′-ACGUGACACGUUCGGAGAATT-3′). A NC siRNA duplex was also prepared with the same sequence but without Cy3 labeling. The siRNA was complexed with PEI-Fe3O4 at different weight ratios of Fe to siRNA ranging from 0:1 to 64:1. The resulting mixture was incubated at room temperature for 30 min to allow composite formation. All complexes in the solution were freshly prepared prior to subsequent experimentation.
Gel retardation assay
NC siRNA was mixed with PEI-Fe3O4 at various Fe:siRNA weight ratios. While maintaining a uniform concentration of NC siRNA, samples of PEI-Fe3O4:siRNA complexes were prepared at weight ratios of 0:1, 2:1, 4:1, 8:1, 16:1, 32:1 and 64:1 (Fe mass to NC siRNA mass). The resulting mixtures were incubated at room temperature for 30 min. Then, 20 µl of samples were loaded into 3% w/v agarose gel. The siRNA bands were visualized by 3% agarose gel electrophoresis stained with ethidium bromide (EtBr) and images were captured under UV illumination.
Heparin decomplexation assay
NC siRNA was complexed with PEI-Fe3O4 at a Fe:siRNA weight ratio of 8:1 for 30 min at room temperature. Various amounts of heparin (LEO Pharma A/S) were added (heparin:siRNA weight ratio of 0:1, 25:1, 50:1, 100:1, 200:1 and 400:1) and the mixtures were further incubated at room temperature for 30 min. After centrifugation for 15 min at 12,000 × g at 4°C, the supernatants were analyzed by 3% agarose gel electrophoresis and the released siRNA was visualized by EtBr staining and photographed under ultraviolet illumination.
Serum protection assay
NC siRNA-loaded PEI-Fe3O4 complexes prepared at a Fe:siRNA weight ratio of 8:1 were selected for a serum protection assay. Complexes containing 0.5 µg siRNA were incubated with Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences) supplemented with 50% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) for 2, 4, 8, 16 and 24 h at 37°C. The siRNA remaining in the complex was displaced with heparin at a heparin/siRNA weight ratio of 200:1 to ensure complete release of siRNA. Naked siRNA acted as a control and was treated in the same manner. The released siRNA was visualized by 3% agarose gel electrophoresis stained with EtBr.
Culture of LX-2 cells
The human hepatic stellate cell line LX-2 was obtained from Procell Life Science & Technology Co., Ltd. LX-2 cells were routinely maintained in DMEM supplemented with 10% FBS and 1% (v/v) penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere in 5% CO2.
Cytotoxicity assay
The cytotoxicity of PEI-Fe3O4 and NC siRNA complexes was evaluated by MTT assay. LX-2 cells were treated with PEI-Fe3O4/siRNA complexes and PEI-Fe3O4 NPs at concentrations 10–80 µg Fe/ml. After incubation for 6 h, the culture medium was replaced with fresh medium, and the cells were cultured for another 24 or 48 h. Then, LX-2 cells were incubated with MTT (Sigma-Aldrich; Merck KGaA) for an additional 4 h. Next, the medium in each well was removed and 100 µl/well dimethyl sulfoxide was added to dissolve the internalized purple formazan crystals. The optical density at 490 nm was measured using a microplate reader (BioTek Instruments, Inc.). All experiments were performed in triplicate and repeated ≥3 times.
Transfection efficiency
A Cy3-labeled siRNA (red fluorescence) was used to verify transfection efficiency. The transfection efficiency of PEI-Fe3O4 NPs was measured and compared with that of the naked siRNA and the standard transfection reagent Lipofectamine® 2000 (Lipo 2000; Invitrogen; Thermo Fisher Scientific, Inc.). LX-2 cells were seeded into a 6-well plate (1.5×105 cells/well) and treated with PEI-Fe3O4/Cy3-siRNA at a concentration of 15 µg Fe/ml and Cy3-siRNA at a 100 nM siRNA concentration at 37°C for 2 h. Magnetofection was performed by placing a magnetic sheet (Nanjing Nanoeast Biotech Co., Ltd.) under the plates for 4 h. The medium was replaced after 6 h, and the transfection efficiency was evaluated after 24 h. As a control experiment, the cells were also treated with Lipo 2000/Cy3-siRNA complexes. The transfection procedures were performed in accordance with the manufacturer's protocols. The transfection efficacy was evaluated by observation of the red fluorescent cells vs. total cells using fluorescence microscopy (magnification, ×200) and flow cytometry (BD Biosciences). Briefly, after transfection, the cells were rinsed with phosphate-buffered saline (PBS, pH 7.4), harvested, fixed in 4% formaldehyde solution at room temperature for 10 min, and subsequently analyzed by flow cytometer (Accuri C6 flow cytometer, BD Biosciences). Data analyses were performed using the FlowJo 10.2 software package (FlowJo LLC). The fixed cells were stained with 1 µg/ml DAPI (Sigma-Aldrich; Merck KGaA) at room temperature for 5 min. The fluorescence images were obtained with a Leica DMI 3000B fluorescence microscope (Leica Microsystems Inc.) magnification, ×200. The experiments were repeated ≥3 times.
Intracellular localization of siRNA in LX-2 cells
To determine successful siRNA internalization, LX-2 cells (1.5×105) were seeded in a 6-well tissue culture plate and cultured for 24 h (70–80% confluence). Lipo 2000 was used as a positive control for siRNA transfection, and the transfection procedure was performed in accordance with the manufacturer's protocols. The cells in each well were incubated at 37°C for 6 h in medium with 10% FBS and free Cy3-siRNA (100 nM), or with PEI-Fe3O4/Cy3-siRNA (15 µg Fe/ml). Upon incubation, the cells were washed twice with PBS (pH 7.4) and fixed for 10 min in PBS containing 4% (w/v) formaldehyde (pH 7.4). Following two further PBS washing steps, the cell nuclei were stained with 500 µl DAPI (1 µg/ml; Sigma-Aldrich; Merck KGaA) in PBS for 5 min. Then, the cells were examined under a confocal microscope magnification, ×1200 (TCS SP5; Leica Microsystems GmbH).
Transforming growth factor (TGF)-β1 treatment
LX-2 cells were seeded in a 24-well plate at a density of 1×106 cells/well and grown for 24 h until they reached 70–80% confluence. In total, three separate experimental groups were studied: Blank (no reagent) + TGF-β1 (PeproTech, Inc.); NC siRNA transfection + TGF-β1; and CTGF siRNA transfection + TGF-β1 groups. Freshly prepared PEI-Fe3O4/siRNA complexes (Fe:siRNA weight ratio of 8:1) at a concentration of 15 µg Fe/ml were incubated with the cells under standard culture conditions. After 6 h of transfection, the culture medium was removed from each well and replaced by medium with TGF-β1 (final concentration of 5 ng/ml) for 24 or 48 h.
Immunocytochemical staining for α-smooth muscle actin (SMA)
Immunocytochemical staining for α-SMA in LX-2 cells was carried out using a horseradish peroxidase-conjugated streptavidin system (Dako; Agilent Technologies, Inc.). In brief, the cells were fixed in 4% formaldehyde for 30 min at 37°C and then washed three times with PBS. Subsequently, cells were blocked with 3% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. The cells were treated with 1:100 dilution of the primary antibody, α-SMA monoclonal antibody (cat. no. 412021; Nichirei Corporation) at 4°C overnight. The cells were washed in PBS and then incubated with a biotinylated secondary antibody (diluted 1:100, cat. no. 5220-0336, Seracare Life Sciences) and streptavidin peroxidase for 60 min at room temperature. Subsequently, the cells were developed with DAB and H2O2, followed by counterstaining with hematoxylin for 3 min at room temperature (Sigma-Aldrich; Merck KGaA). Then, images were captured of the cells via an Olympus IX71 inverted microscopy (Olympus Corporation, magnification ×400).
Reverse transcription-quantitative PCR (RT-qPCR)
After treatment for 48 h, total RNA was extracted from LX-2 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. RNA samples were converted into first-strand cDNA with PrimeScript™ RT Reagent kit with gDNA Eraser (Takara Bio, Inc.). RT-qPCR analysis was performed with the StepOne™ Real-Time PCR Detection System (Thermo Fisher Scientific, Inc.) using SYBR® Premix Ex Taq™ (Takara Bio, Inc.). The primer sequences used to amplify the desired cDNA were as follows: CTGF forward 5′-GGAAAAGATTCCCACCCAAT-3′, and reverse 5′-TGCTCCTAAAGCCACACCTT-3′; type collagen I forward, 5′-CCCGGGTTTCAGAGACAACTTC-3′, and reverse 5′-TCCACATGCTTTATTCCAGCAAT-3′; tissue inhibitor of matrix metalloproteinase-1 (TIMP-1) forward, 5′-GCTTCTGGCATCCTGTTGTTG-3′, and reverse, 5′-CTTCTGGTGTCCCCACGAACT-3; and β-actin forward, 5′-AGAGCTACGAGCTGCCTGAC-3′, and reverse 5′-AGCACTGTGTTGGCGTACAG-3′. The thermal cycling conditions were as follows: cDNA synthesis, 15 min at 50°C and 5 sec at 85°C; reverse transcriptase inactivation, 10 min at 95°C; thermal cycling and detection (up to 40 cycles), 15 sec at 95°C and 60 sec at 60°C (data collection). The mRNA expression of the target gene was evaluated against β-actin mRNA. The quantification cycle (Cq) value was calculated using the 2−ΔΔCq method (21) (IQ5 software version 2.0; Bio-Rad Laboratories, Inc.). The experiments were repeated three times.
Western blot analysis
Western blotting was performed on the total protein extracts of the LX-2 cells 48 h after treatment. For the total protein fraction, the harvested cells were washed three times with ice-cold PBS and lysed in RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA), and then centrifuged at 12,000 × g at 4°C for 30 min. The total protein concentration of the supernatant was determined using the Bradford method with a Bio-Rad protein assay kit II (Bio-Rad Laboratories, Inc.). Briefly, equal amounts (30 µg) of protein samples were separated by 10% SDS-PAGE and subsequently electrotransferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% fat-free milk in PBS plus 0.05% (v/v) Tween-20 at 37°C for 2 h. After blocking, the membranes were incubated with 1:1,000 dilution of anti-CTGF (cat. no. ab6992; Abcam), anti-TIMP-1 (cat. no. ab61224; Abcam), anti-collagen I (cat. no. ab34710; Abcam) and anti-β-actin (cat. no. ab8226; Abcam) antibodies overnight at 4°C with gentle rocking. After 3 further washing steps with TBS, the membranes were incubated with 1:5,000-diluted horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no. SA00001-2; ProteinTech Group, Inc.). The antibody-antigen complexes were visualized using the enhanced chemiluminescence detection system (Pierce™ ECL Plus Western Blotting Substrate; Thermo Fisher Scientific, Inc.). All western blotting experiments were performed ≥3 times. Western blot data were quantitatively analyzed using Image Lab 2.0 software (Bio-Rad Laboratories, Inc.). The relative protein abundance in each sample was normalized to that of β-actin.
Statistical analysis
The results were expressed as the mean ± SD. One-way ANOVA followed by Tukey's post hoc test was carried out for the statistical analyses. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS 16.0 software (SPSS, Inc.).
Results
TEM images of PEI-Fe3O4 NPs and PEI-Fe3O4/siRNA complexes
TEM observations were performed to assess the morphology of PEI-Fe3O4 NPs and their association with siRNA. The images indicated that PEI-Fe3O4 NPs and PEI-Fe3O4/NC siRNA complexes (Fe:siRNA weight ratio of 8:1) were of spherical morphology and well-dispersed (Fig. 1).
Figure 1.
Visualization of PEI-Fe3O4 NPs in complex with siRNA. Transmission electron microscopy images of (A) PEI-Fe3O4 NPs and (B) PEI-Fe3O4/siRNA complexes (Fe:siRNA weight ratio of 8:1). PEI-Fe3O4 NPs, polyethyleneimine-functionalized magnetic iron oxide nanoparticles; siRNA, small interfering RNA.
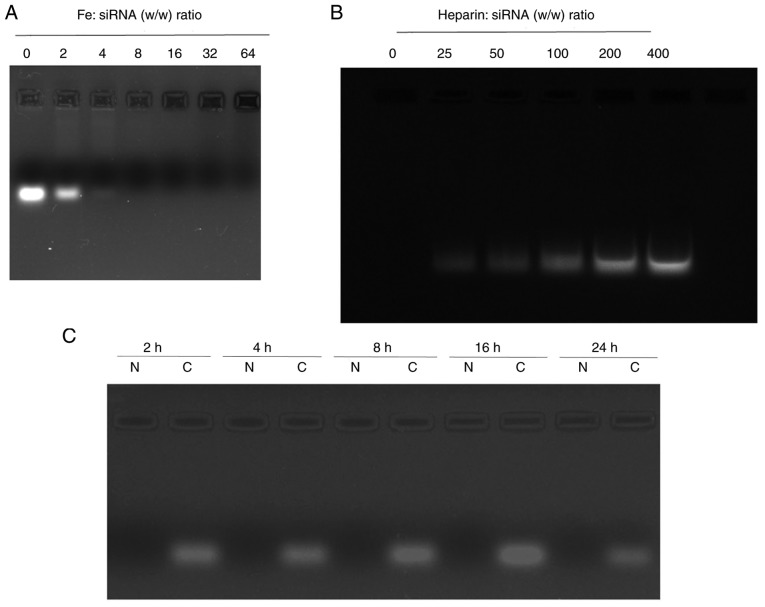
siRNA-loading capacity of PEI-Fe3O4 NPs
The migration of naked NC siRNA and PEI-Fe3O4/siRNA complexes at weight ratios ranging from 0:1 to 64:1 are shown in Fig. 2A. The siRNA band tended to disappear in the gel as the weight ratio of PEI-Fe3O4 to siRNA increased, indicating that binding occurred. When the ratio of Fe mass to siRNA mass was >4, the migration of siRNA was completely blocked, indicating that the siRNA was largely complexed with the PEI-Fe3O4 NPs. At a weight ratio of 8:1, no free siRNA was detected in the gel retardation assay. Therefore, the PEI-Fe3O4 NPs exhibited stable binding capability. In the following experiments, 8:1 was selected as the optimized ratio of PEI-Fe3O4/siRNA complexes.
Figure 2.
Agarose gel electrophoresis. (A) siRNA were complexed with PEI-Fe3O4 at different Fe:siRNA (w/w) ratios. (B) siRNA release assayed by heparin decomplexation. Various w/w ratios of heparin to siRNA were added, and the mixtures were further incubated for 30 min. (C) Effect of the different incubation times on serum stability of siRNA. siRNA (0.5 µg/sample), either in naked form or complexed with PEI-Fe3O4, was mixed with fresh fetal bovine serum (1:1, v/v) to achieve a 50% serum concentration, and was incubated at 37°C for various periods of time. N represents naked siRNA, while C represents PEI-Fe3O4/siRNA complexes. PEI-Fe3O4, polyethyleneimine-functionalized magnetic iron oxide; siRNA, small interfering RNA.
Heparin induces complex dissociation
To investigate the effect of PEI-Fe3O4 NPs on complex dissociation, PEI-Fe3O4/siRNA complexes were challenged by exposure to different quantities of heparin. As expected, the complex was dissociated in the presence of negatively charged heparin molecules, and the amount of siRNA released increased with increasing levels of heparin (Fig. 2B).
Serum protection assay
As shown in Fig. 2C, naked NC siRNA degradation was completely degraded within 2 h of incubation. By contrast, the PEI-Fe3O4/siRNA complexes displayed significant stability under the same conditions, with no notable degradation detected within 24 h. Therefore, these data suggested that PEI-Fe3O4 had strong siRNA affinity and effectively protected siRNA from the enzymatic activity of serum components.
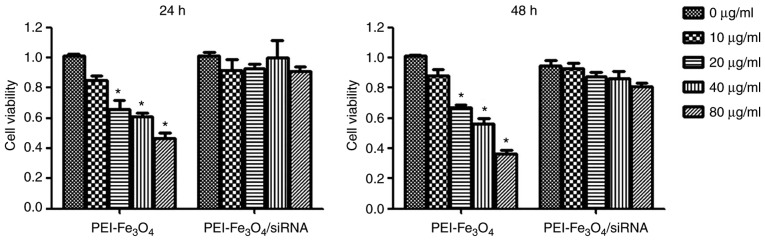
Cytotoxicity assay
The MTT results indicated that the PEI-Fe3O4/siRNA complexes had no obvious cytotoxic effect on LX-2 cells when added in the range of 0–80 µg Fe/ml for 24 or 48 h, while empty PEI-Fe3O4 exhibited a significant cytotoxicity on LX-2 cells in a concentration-dependent manner (Fig. 3).
Figure 3.
Cytotoxicity assay. LX-2 cells were incubated with either PEI-Fe3O4 or PEI-Fe3O4/siRNA at different concentrations ranging from 10 to 80 µg/ml (Fe concentration) for 24 and 48 h, and cell viability was then measured by MTT assay. The data are displayed as the mean ± SD (n=3). *P<0.05 vs. 0 µg/ml. PEI-Fe3O4, polyethyleneimine-functionalized magnetic iron oxide; siRNA, small interfering RNA.
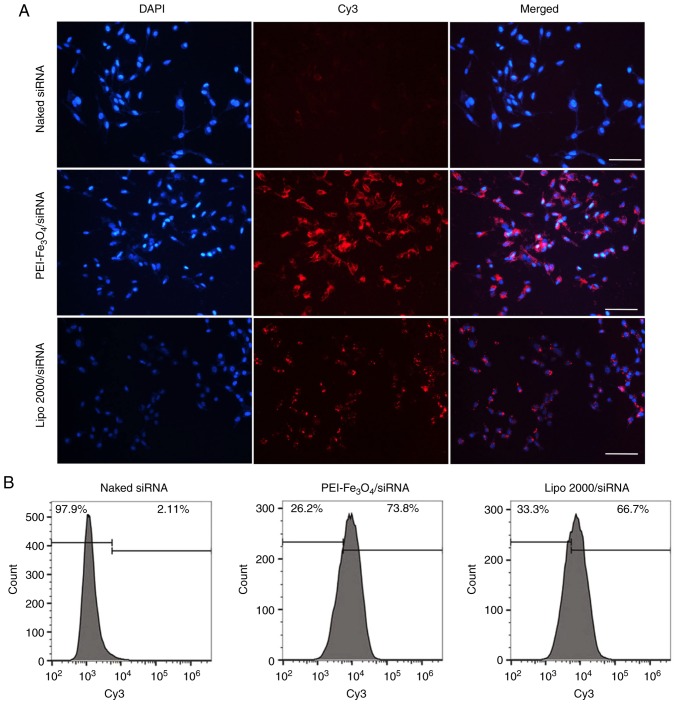
Transfection efficiency
Fluorescence microscopy was performed to examine cellular uptake of siRNA when complexed with PEI-Fe3O4 NPs. When the cells were transfected with PEI-Fe3O4/Cy3-siRNA complexes for 6 h, red fluorescence was observed inside the cells exposed to the complexes (Fig. 4A), demonstrating that Cy3-siRNA has been delivered into the cells. To exclude nonspecific uptake of siRNA, the cells were incubated with Cy3-siRNA alone. However, no siRNA-derived fluorescence was observed inside the cells.
Figure 4.
Transfection efficiency of PEI-Fe3O4/siRNA complexes. (A) Fluorescence microscopy image of LX-2 cells transfected with PEI-Fe3O4/siRNA complexes (original magnification, ×200). Red fluorescence indicates Cy3-labeled siRNA, while blue fluorescence represents DAPI-stained nuclei. The scale bar represents 100 µm. (B) Transfection efficiency (percentage of cells transfected) of LX-2 cells transfected with PEI-Fe3O4/siRNA complexes and Lipo 2000/siRNA. PEI-Fe3O4, polyethyleneimine-functionalized magnetic iron oxide; siRNA, small interfering RNA; cy3, Cyanine 3; Lipo 2000, Lipofectamine 2000.
Transfection efficiency was assessed by quantifying the number of red fluorescence-expressing cells with flow cytometry. As shown in Fig. 4B, siRNA alone translated into almost no uptake in LX-2 cells, which also confirmed the limitation of siRNA alone in cellular uptake. PEI-Fe3O4 NPs achieved 73.8% uptake of siRNA, which is comparable with that of Lipo 2000 (liposome transfection, 66.7%), which is one of the most efficient commercially available transfection reagents.
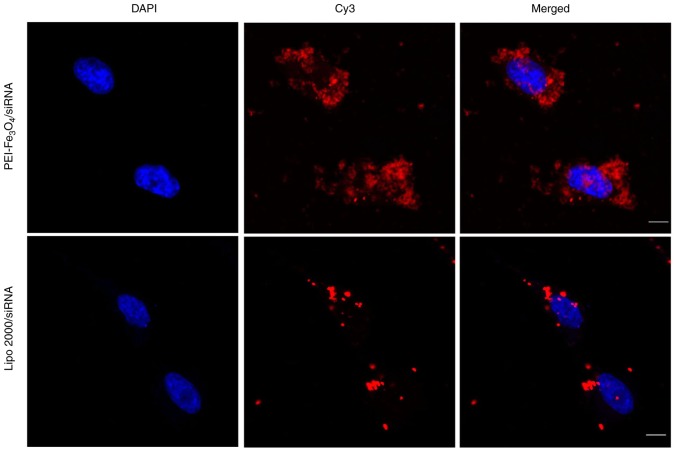
Intracellular localization of Cy3-siRNA in LX-2 cells
The cellular localization of siRNA is shown in Fig. 5. The overlay images revealed that the red fluorescence was predominantly localized in the perinuclear region of LX-2 cells treated with PEI-Fe3O4/Cy3-siRNA complexes, which indicates disruption of endosomes and release of Cy3-siRNA into the cytoplasm of the cells. The siRNAs that were delivered by Lipo 2000 were also localized to regions near the nuclear membrane and were distributed in a non-homogeneous pattern at the periphery of the nucleus.
Figure 5.
Confocal fluorescence images of cells for evaluation of Cy3-labeled siRNA internalization. LX-2 cells were transfected with PEI-Fe3O4/siRNA or Lipo 2000/siRNA. The cell nuclei are stained in blue, and siRNA in red (original magnification, ×1,200). The scale bar represents 10 µm. siRNA, small interfering RNA; PEI-Fe3O4, polyethyleneimine-functionalized magnetic iron oxide; cy3, cyanine 3; Lipo 2000, Lipofectamine 2000.
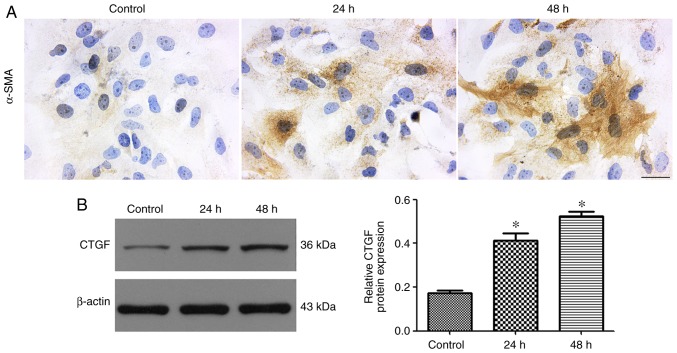
Activation of LX-2 cells mediated by TGF-β1
The expression of α-SMA is a characteristic feature of activated liver stellate cells and considered as a marker for the triggering of hepatic fibrosis (22). Immunocytochemical staining for α-SMA demonstrated strong positive staining in LX-2 cells stimulated with TGF-β1 (Fig. 6A). CTGF acts both as a profibrotic marker and a downstream effector of TGF-β. The western blot results showed that LX-2 cells treated with TGF-β1 exhibited a markedly upregulated expression of CTGF (Fig. 6B).
Figure 6.
Effect of TGF-β1 on the activation of LX-2 cells. (A) Representative immunocytochemistry of α-SMA in LX-2 cells before or after treatment. The brown color indicates α-SMA staining (original magnification, ×400). The scale bar represents 50 µm. (B) Western blotting showed that the expression of CTGF protein was significantly increased in activated cells, compared with that in quiescent LX-2 cells. *P<0.05 vs. TGF-β1-non-treated cells. CTGF; connective tissue growth factor; TGF-β1, transforming growth factor-β1; α-SMA, α-smooth muscle actin.
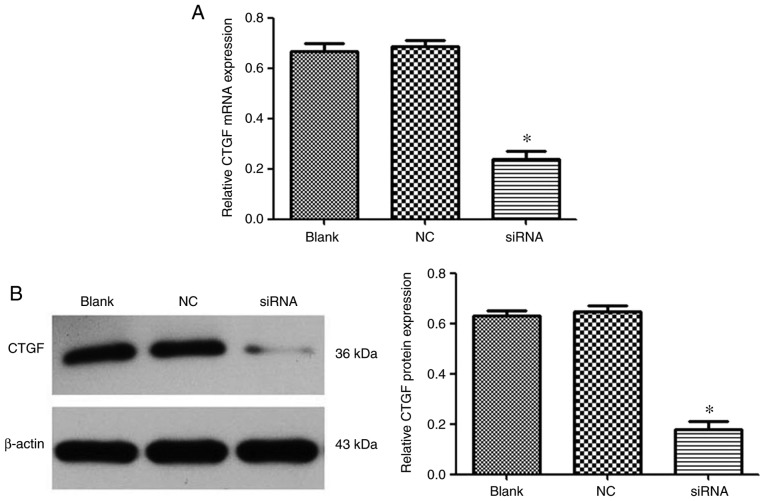
Evaluation of gene silencing efficiency mediated by PEI-Fe3O4/CTGF-siRNA complexes in activated LX-2 cells
The mRNA and protein expression levels of CTGF were measured by RT-qPCR and western blotting, respectively. As shown in Fig. 7, PEI-Fe3O4/CTGF-siRNA complexes achieved 62 and 71% gene silencing efficiency at the mRNA and protein level, respectively. This result indicated that PEI-Fe3O4 could efficiently deliver siRNA to cells and induce specific gene silencing.
Figure 7.
Efficacy of the polyethyleneimine-functionalized magnetic iron oxide/CTGF-siRNA complexes (w/w=8) on suppressing CTGF expression in activated LX-2 cells. (A) Reduced CTGF mRNA levels, as quantified by reverse transcription-quantitative PCR analysis (n=3). (B) Reduced protein expression of CTGF, as evaluated by western blot analysis (n=3). *P<0.05 vs. the blank and NC siRNA groups. NC, negative control; siRNA, small interfering RNA; CTGF, connective tissue growth factor.
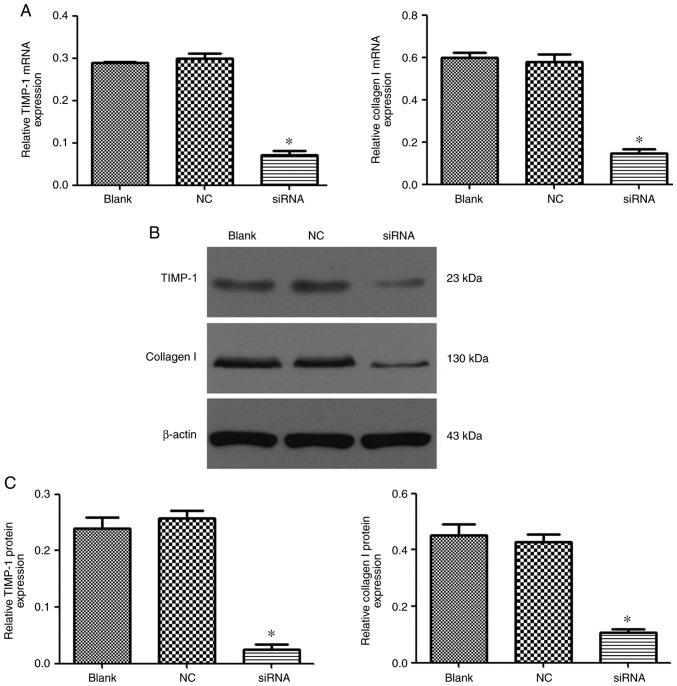
Effect of PEI-Fe3O4/CTGF-siRNA complexes on type I collagen and TIMP-1 expression in activated LX-2 cells
The ability of PEI-Fe3O4/CTGF-siRNA complexes to reduce mRNA and protein expression levels of collagen I and TIMP-1 was analyzed via RT-qPCR and western blot analysis. As shown in Fig. 8, the collagen I mRNA and protein expression levels in CTGF-knockdown LX-2 cells were markedly reduced, while no interference effects were observed in cells treated with siRNA-NC only. Similarly, TIMP-1 gene transcription and protein expression were also significantly knocked down by PEI-Fe3O4/CTGF-siRNA.
Figure 8.
Efficacy of the polyethyleneimine-functionalized magnetic iron oxide/connective tissue growth factor-siRNA complexes on suppressing collagen I and TIMP-1 expression in activated LX-2 cells. (A) Suppression of collagen I and TIMP-1 mRNA levels was quantified by reverse transcription-quantitative PCR analysis (n=3). *P<0.05 vs. the blank and negative control siRNA groups. (B) Suppression of the protein expression of collagen I and TIMP-1 was evaluated by western blot analysis. (C) Relative protein expression levels of collagen I and TIMP-1 were significantly downregulated when compared with the blank and negative control siRNA groups. *P<0.05 vs. the blank and negative control siRNA groups. siRNA, small interfering RNA; TIMP-1, tissue inhibitor of matrix metalloproteinase-1; NC, negative control.
Discussion
Liver fibrosis is the final stage of all chronic hepatic diseases, and may develop into cirrhosis and hepatic carcinoma, eventually inducing liver failure (23,24). Previous studies have shown that hepatic fibrosis is a reversible wound-healing process, and effective drugs against fibrosis should be used as early as possible (25,26). However, there is an imperative requirement to develop a radical therapy for hepatic fibrosis, and new effective drugs for fibrosis should be applied in the clinic.
siRNAs are expected to serve as a powerful molecular therapy for the treatment of hepatic fibrosis (4,7,27). Previously, several lines of evidence have indicated that CTGF is a central mediator and promising therapeutic target in hepatic fibrosis (28–30). However, an effective and safe delivery vector is considered as the key to successful delivery of siRNA into target cells. Gene delivery system-based NPs could be an efficient treatment strategy for the management of liver fibrosis (31). In the present study, PEI-Fe3O4 NPs were used as the CTGF siRNA carrier, and their anti-fibrosis effect in vitro was investigated.
Agarose gel electrophoresis is one of the simplest methods available to visualize a change in electromobility of DNA and RNA strands before and after complexation with cationic polymers (32). In the present study, the migration of siRNA was completely retarded in the PEI-Fe3O4/siRNA complexes, at a mass ratio of 8:1. This indicated that positively charged PEI-Fe3O4 NPs effectively interacted with negatively charged siRNA molecules via electrostatic interaction. In addition, PEI-Fe3O4/siRNA complexes were notably more stable than naked siRNA in the presence of serum. The ability of a nanocarrier to protect its cargo from nuclease degradation is an important property for efficient gene delivery. siRNA must be protected from nuclease digestion for maximum activity in cells.
The cytotoxicity of gene carriers is one of their main disadvantages, and needs to be considered. Therefore, it is important to evaluate the cytotoxicity of the PEI-Fe3O4/siRNA complexes. An MTT assay revealed that the PEI-Fe3O4/siRNA complexes had no obvious toxicity in LX-2 cells, while empty PEI-Fe3O4 NPs showed dose-dependent cytotoxicity. It is well known that PEI is a type of typical cationic polymer. The toxicity of PEI-Fe3O4 NPs could be associated with the positively charged nature of the PEI. The reduced toxicity of PEI-Fe3O4/siRNA may be partially attributed to mild cationic charge upon the loading of siRNA. These data illustrated that PEI-Fe3O4 NPs could be a genetic drug carrier for further studies.
In order to investigate whether PEI-Fe3O4 NPs can successfully deliver Cy3-labeled siRNA into cells, siRNA transfection efficiency was evaluated via flow cytometry and confocal microscopy. Flow cytometric analyses showed that the transfection efficiency of PEI-Fe3O4/Cy3-siRNA was strongly improved compared with that of naked Cy3-siRNA. Fluorescence from Cy3 was obviously detected in the cytoplasm, suggesting that PEI-Fe3O4 NPs could effectively carry Cy3-siRNA and lead to a subsequent disruption of the endosome. These results suggested that PEI-Fe3O4 NPs could be effective carriers for further studies.
Activated hepatic stellate cells (HSCs) are considered to be the major effector cells in the development of hepatic fibrosis (33). TGF-β1 participates in the initiation and maintenance of fibrogenesis in the liver (34). Stimulation of activated HSCs by TGF-β1 is considered to be the key fibrogenic response in liver fibrosis (35). In the present study, the immunocytochemical staining result suggested that TGF-β1 treatment increased α-SMA protein expression, which is an important surrogate marker for activated myofibroblasts during liver fibrogenesis compared with the non-treated groups (36). CTGF expression was also remarkably upregulated in LX-2 cells stimulated with TGF-β1. Moreover, the data demonstrated that the vector efficiently conveyed PEI-Fe3O4/siRNA into HSCs and inhibited the gene expression of CTGF in activated LX-2 cells.
Activated HSCs not only secrete excess type I collagen, but also exhibit markedly increased TIMP expression, leading to a shift towards excessive extracellular matrix (ECM) synthesis and fibrogenesis (37). Previous studies have suggested that the downregulation of CTGF expression may inhibit CTGF- and TGF-β1-mediated ECM production both in vitro and in vivo (31,38). The present study observed that the siRNA knockdown of CTGF delivered by PEI-Fe3O4 NPs could significantly attenuate TIMP-1 and type I collagen expression in activated LX-2 cells. The data demonstrated that disruption of CTGF expression mediated by PEI-Fe3O4 NPs inhibits the production of ECM. Therefore, PEI-Fe3O4/CTGF-siRNA showed a remarkable antifibrotic effect in vitro.
Overall, the data of the present study indicated that PEI-Fe3O4/CTGF-siRNA complexes can be used as a safe and effective method to deliver siRNA to target cells in vitro, providing a basis for gene delivery in vivo. However, additional studies using animal models are required to gain further insight into the biological effects of hepatic fibrosis.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81402640 and 81502816), the Natural Science Foundation of Hubei province (grant no. 2014CFB406) and the Health and Family Planning commission of Wuhan city (grant no. WX15B23).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QY and QW participated in the study design, wrote the paper and submitted the manuscript, and QW revised the manuscript, QY, XX, LZ, TX and QW performed the experiments. QY analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Atta HM. Reversibility and heritability of liver fibrosis: Implications for research and therapy. World J Gastroenterol. 2015;21:5138–5148. doi: 10.3748/wjg.v21.i17.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol. 2006;21(Suppl 3):S84–S87. doi: 10.1111/j.1440-1746.2006.04584.x. [DOI] [PubMed] [Google Scholar]
- 3.Gressner OA, Gressner AM. Connective tissue growth factor: A fibrogenic master switch in fibrotic liver diseases. Liver Int. 2008;28:1065–1079. doi: 10.1111/j.1478-3231.2008.01826.x. [DOI] [PubMed] [Google Scholar]
- 4.Salazar-Montes AM, Hernández-Ortega LD, Lucano-Landeros MS, Armendariz-Borunda J. New gene therapy strategies for hepatic fibrosis. World J Gastroenterol. 2015;21:3813–3825. doi: 10.3748/wjg.v21.i13.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Rodriguez A, Valverde AM. RNA interference as a therapeutic strategy for the treatment of liver diseases. Curr Pharm Des. 2015;21:4574–4586. doi: 10.2174/138161282131151013190740. [DOI] [PubMed] [Google Scholar]
- 6.Cheng K, Yang N, Mahato RI. TGF-beta1 gene silencing for treating liver fibrosis. Mol Pharm. 2009;6:772–779. doi: 10.1021/mp9000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omar R, Yang J, Liu H, Davies NM, Gong Y. Hepatic stellate cells in liver fibrosis and siRNA-based therapy. Rev Physiol Biochem Pharmacol. 2016;172:1–37. doi: 10.1007/112_2016_6. [DOI] [PubMed] [Google Scholar]
- 8.Motoyama H, Ogawa S, Kubo A, Miwa S, Nakayama J, Tagawa Y, Miyagawa S. In vitro reprogramming of adult hepatocytes into insulin-producing cells without viral vectors. Biochem Biophys Res Commun. 2009;385:123–128. doi: 10.1016/j.bbrc.2009.04.146. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Shum KT, Burnett JC, Rossi JJ. Nanoparticle-based delivery of RNAi therapeutics: Progress and challenges. Pharmaceuticals (Basel) 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poilil Surendran SP, Thomas RG, Moon MJ, Jeong YY. Nanoparticles for the treatment of liver fibrosis. Int J Nanomedicine. 2017;12:6997–7006. doi: 10.2147/IJN.S145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li TT, Shen X, Chen Y, Zhang CC, Yan J, Yang H, Wu CH, Zeng HJ, Liu YY. Polyetherimide-grafted Fe3O4@SiO2 nanoparticles as theranostic agents for simultaneous VEGF siRNA delivery and magnetic resonance cell imaging. Int J Nanomedicine. 2015;10:4279–4291. doi: 10.2147/IJN.S85095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirazi AN, Paquin KL, Howlett NG, Mandal D, Parang K. Cyclic peptide-capped gold nanoparticles for enhanced siRNA delivery. Molecules. 2014;19:13319–13331. doi: 10.3390/molecules190913319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Zhao Q, Han N, Bai L, Li J, Liu J, Che E, Hu L, Zhang Q, Jiang T, Wang S. Mesoporous silica nanoparticles in drug delivery and biomedical applications. Nanomedicine. 2015;11:313–327. doi: 10.1016/j.nano.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Reina G, Gismondi A, Carcione R, Nanni V, Peruzzi C, Angjellari M, Chau NDQ, Canini A, Terranova ML, Tamburri E. Oxidized and amino-functionalized nanodiamonds as shuttle for delivery of plant secondary metabolites: Interplay between chemical affinity and bioactivity. Appl Surface Sci. 2019;470:744–754. doi: 10.1016/j.apsusc.2018.11.161. [DOI] [Google Scholar]
- 15.Getz T, Qin J, Medintz IL, Delehanty JB, Susumu K, Dawson PE, Dawson G. Quantum dot-mediated delivery of siRNA to inhibit sphingomyelinase activities in brain-derived cells. J Neurochem. 2016;139:872–885. doi: 10.1111/jnc.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta AK, Gupta M. Synthesis and surface engineering of iron oxide nanoparticles for biomedical applications. Biomaterials. 2005;26:3995–4021. doi: 10.1016/j.biomaterials.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Wu W, He QG, Jiang CZ. Magnetic iron oxide nanoparticles: Synthesis and surface functionalization strategies. Nanoscale Res Lett. 2008;3:397–415. doi: 10.1007/s11671-008-9174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao S, Castro R, Rodrigues J, Shi X, Tomás H. PAMAM dendrimer/pDNA functionalized-magnetic iron oxide nanoparticles for gene delivery. J Biomed Nanotechnol. 2015;11:1370–1384. doi: 10.1166/jbn.2015.2101. [DOI] [PubMed] [Google Scholar]
- 19.Saeed M, Ren W, Wu A. Therapeutic applications of iron oxide based nanoparticles in cancer: Basic concepts and recent advances. Biomater Sci. 2018;6:708–725. doi: 10.1039/C7BM00999B. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Zou S, Gao J, Liang J, Zhou H, Liang L, Wu W. Block copolymer conjugated Au-coated Fe3O4 nanoparticles as vectors for enhancing colloidal stability and cellular uptake. J Nanobiotechnology. 2017;15:56. doi: 10.1186/s12951-017-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Friedman SL. Cytokines and fibrogenesis. Semin Liver Dis. 1999;19:129–140. doi: 10.1055/s-2007-1007105. [DOI] [PubMed] [Google Scholar]
- 23.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI200524282C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elpek GÖ. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J Gastroenterol. 2014;20:7260–7276. doi: 10.3748/wjg.v20.i23.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trautwein C, Friedman SL, Schuppan D, Pinzani M. Hepatic fibrosis: Concept to treatment. J Hepatol. 2015;62(1 Suppl):S15–S24. doi: 10.1016/j.jhep.2015.02.039. [DOI] [PubMed] [Google Scholar]
- 26.Zhang CY, Yuan WG, He P, Lei JH, Wang CX. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuppan D, Surabattula R, Wang XY. Determinants of fibrosis progression and regression in NASH. J Hepatol. 2018;68:238–250. doi: 10.1016/j.jhep.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Lipson KE, Wong C, Teng Y, Spong S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair. 2012;5(Suppl 1):S24. doi: 10.1186/1755-1536-5-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao C, Xie Y, Peng M, Ma L, Zhou Y, Zhang Y, Kang W, Wang J, Bai X, Wang P, Jia Z. Inhibition of connective tissue growth factor suppresses hepatic stellate cell activation in vitro and prevents liver fibrosis in vivo. Clin Exp Med. 2014;14:141–150. doi: 10.1007/s10238-013-0229-6. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Xie Q, Shi Y, Li D, Zhang M, Jiang S, Zhou H, Lu H, Jin Y. Inhibition of connective tissue growth factor by siRNA prevents liver fibrosis in rats. J Gene Med. 2006;8:889–900. doi: 10.1002/jgm.894. [DOI] [PubMed] [Google Scholar]
- 31.Taymouri S, Taheri A. Use of nanotechnology in diagnosis and treatment of hepatic fibrosis: A review. Curr Drug Deliv. 2016;13:662–672. doi: 10.2174/1567201812666150907115404. [DOI] [PubMed] [Google Scholar]
- 32.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Zern MA. Hepatic stellate cells: A target for the treatment of liver fibrosis. J Gastroenterol. 2000;35:665–672. doi: 10.1007/s005350070045. [DOI] [PubMed] [Google Scholar]
- 34.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–d807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 35.Yang N, Dang S, Shi J, Wu F, Li M, Zhang X, Li Y, Jia X, Zhai S. Caffeic acid phenethyl ester attenuates liver fibrosis via inhibition of TGF-β1/Smad3 pathway and induction of autophagy pathway. Biochem Biophys Res Commun. 2017;486:22–28. doi: 10.1016/j.bbrc.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 36.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang G, Brigstock DR. Regulation of hepatic stellate cells by connective tissue growth factor. Front Biosci (Landmark Ed) 2012;17:2495–2507. doi: 10.2741/4067. [DOI] [PubMed] [Google Scholar]
- 38.Yuhua Z, Wanhua R, Chenggang S, Jun S, Yanjun W, Chunqing Z. Disruption of connective tissue growth factor by short hairpin RNA inhibits collagen synthesis and extracellular matrix secretion in hepatic stellate cells. Liver Int. 2008;28:632–639. doi: 10.1111/j.1478-3231.2008.01730.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.