Abstract
Background
MicroRNAs (miRNAs) in milk-derived exosomes may reflect pathophysiological changes caused by mastitis. This study profiled miRNAs in exosomes from both normal milk and mastitic milk infected by Staphylococcus aureus (S. aureus). The potential targets for differentially expressed (DE) miRNAs were predicted and the target genes for bta-miR-378 and bta-miR-185 were also validated.
Results
Total RNA from milk exosomes was collected from healthy cows (n = 3, the control group) and S. aureus infected cows (n = 6, the SA group). Two hundred ninety miRNAs (221 known and 69 novel ones) were identified. Among them, 22 known and 15 novel miRNAs were differentially expressed. Target genes of DE miRNAs were significantly enriched in intracellular protein transport, endoplasmic reticulum and identical protein binding. The expression of two miRNAs (bta-miR-378 and bta-miR-185) with high read counts and log2 fold changes (> 3.5) was significantly higher in mastitic milk infected with S. aureus. One target gene (VAT1L) of bta-miR-378 and five target genes (DYRK1B, MLLT3, HP1BP3, NPR2 and PGM1) of bta-miR-185 were validated.
Conclusion
DE miRNAs in exosomes from normal and S. aureus infected milk were identified. The predicted targets for two DE miRNAs (bta-miR-378 and bta-miR-185) were further validated. The linkage between the validated target genes and diseases suggested that we should pay particular attention to exosome miRNAs from mastitic milk in terms of milk safety.
Keywords: Milk, Exosome, Bta-miR-378, Bta-miR-185, Staphylococcus aureus
Background
MicroRNAs (miRNAs) are short noncoding (~ 22 nucleotide in length), regulatory RNAs that modulate gene expression at the post-transcriptional level, mostly via binding to perfectly/partially complementary sites at the 3′-UTR of target mRNAs [1]. Among different body fluids, milk contains the highest amount of miRNAs [2]. Milk is an essential source of nutrients to all mammalian offspring. Bovine milk and dairy products have long traditions in human nutrition. In addition to providing nutrition, milk has long been known to protect the infant from infections and to play developmental functions integral to the infant, in which miRNAs are likely to be highly involved [3].
The majority of milk’s miRNAs are transported and protected by the lipid bilayer of extracellular vesicles, predominantly exosomes of about 100 nm in diameter secreted by mammary epithelial cells [4]. Exosomes are cell-derived vesicles that are present in all biological fluids including blood, saliva, urine, amniotic fluid, bronchoalveolar lavage fluid and milk [5, 6]. Milk exosomes have been reported in cows [7], buffalos [8], goats [9], pigs [10], marsupial tammar wallabies [11] and humans [12]. Exosomes protect miRNA molecules from effects of RNase digestion and low pH [13]. Thus, miRNAs in milk exosomes may be transferred into the gastrointestinal tract of infants and likely contribute to infant development and protection against infections [14].
Cells can take up exosomes through a variety of endocytic pathways, including clathrin-dependent endocytosis, clathrin-independent pathways such as caveolin-mediated uptake, macropinocytosis and phagocytosis [15]. Uptake of milk exosomes including their miRNAs has been demonstrated in human colon carcinoma Caco-2 cells and rat intestinal epithelial cell (IEC) IEC-6 cells [16]. Further, orally administered exosomes escaped re-packaging in the intestinal mucosa, and accumulated in liver and spleen. The same group later reported that labelled RNA derived from the milk exosomes was detected in mouse brain, kidney and lung [17]. Porcine milk exosomes promoted IEC proliferation in mice and increased mouse villus height, crypt depth and ratio of villus length to crypt depth of intestinal tissues were associated with miRNA-mediated gene regulatory changes in IECs [18]. In another study, oral delivery of bovine milk exosomes ameliorated experimentally induced arthritis [19]. Correctively, these data suggest that miRNA in milk exosomes can get into the body.
Accumulating evidence suggests that exosomal miRNAs play crucial roles in numerous diseases such as hepatocellular carcinoma [20], breast cancer [21] and Alzheimer’s disease [22]. Secretion of milk exosomes is affected by bacterial infections in mammary glands. Staphylococcus aureus (S. aureus) is one of the most important etiologic agents for chronic bovine mastitis. Our previous in vitro study showed that 5 miRNAs (miR-2339, miR-21-3p, miR-92a, miR-23a and miR-365-3p) were up-regulated in bovine mammary epithelial cells when challenged with S. aureus [23]. In bovine mammary gland infected with S. aureus, a total of 77 miRNAs showed significant differences compared to the control group [24]. Previous studies have also investigated milk exosomal miRNAs following S. aureus induced bovine mastitis [25, 26]. However, no study has focused on miRNAs in exosomes derived from milk naturally infected with S. aureus. More importantly, previous studies focused on the profiling of miRNAs in milk exosomes, without experimental confirmation of the predicted target genes by bioinformatics. In addition, how miRNAs in exosomes affect milk safety has not been considered.
The objective of this study, therefore, was to characterize the miRNA expression profiles comprehensively in exosomes from normal and uninfected milk (the control group) and S. aureus infected milk (the SA group) and to predict potential targets for DE miRNAs and explore their possible functions.
Results
Identification of S. aureus in bovine milk
Based on colony counting and PCR results for nuc and bacterial 16S rRNA genes, 13.95% (42/301) milk samples were infected with S. aureus. At the cow level, the infection rate was 31.58% (24/76) (Additional file 8: Table S1).
Isolation and detection of exosome from bovine milk
Bovine milk exosomes with approximately 100 nm in diameter were observed (Additional file 1: Figure S1a). Exosomes with a particle diameter in the range of 20 nm to 200 nm amounted to 84.1% of the total (Additional file 1: Figure S1b). The expression of CD63 and CD81 on the surface of exosomes was at a positive rate of 72.0 and 77.9%, respectively (Additional file 2: Figure S2).
Characterization of bovine milk exosomal miRNAs
Average RNA contents of the exosomes from 40 mL of the control or S. aureus infected milk samples were 1301 ± 38.7 ng (n = 3) and 1223 ± 56.6 ng (n = 6), respectively. Bovine milk exosomal RNA contained little or no 28S and 18S ribosomal RNA (data not shown).
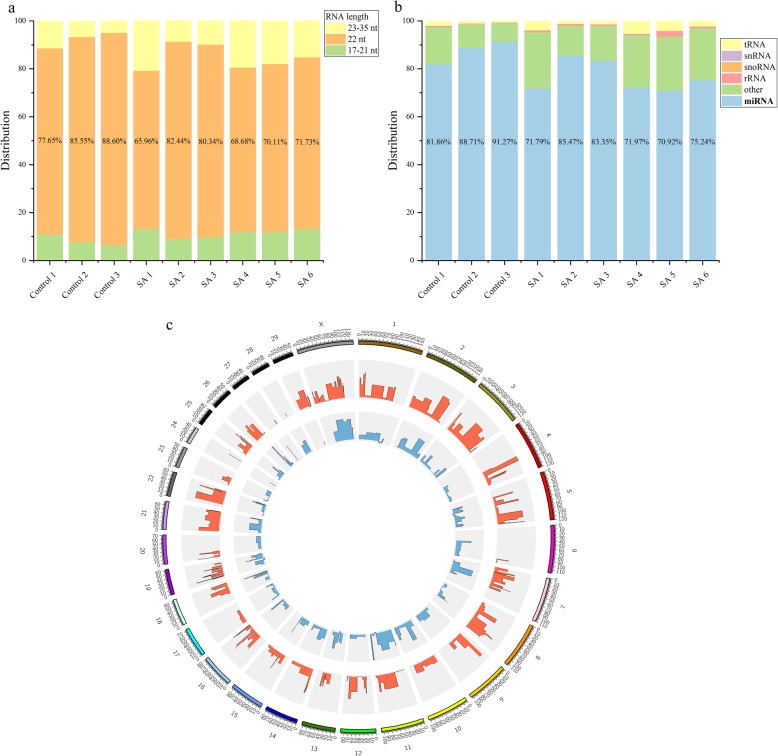
The total raw read count from the sequencing of 9 libraries was 101,392,712 with an average of 11,265,857 reads per sample. After removing linker reads, reads containing N and poly A/T structure, length-anomalous reads, low-quality reads and reads greater than 35 nt or less than 17 nt, the resulting high quality clean data accounted for 83 to 96% of the original raw read counts. The majority of retained reads were 22 nt in length (Fig. 1a).
Fig. 1.
Sequencing data for small RNAs in milk-derived exosomes. (a) Length (nt) distribution of the read counts. (b) Different categories of small RNAs in 9 studied samples. (c) Intuitive map of miRNA distribution across bovine chromosomes (the outermost circle, one unit of the scale stands for one million base-pairs). The middle circle (red lines) represents known miRNAs and the innermost circle (blue lines) represents novel miRNAs. The height of the column is proportional to the expression level and the position of the column corresponds to the miRNA location on chromosome
Approximately 95% (range from 92.22 to 96.97%) of clean reads were successfully aligned to the bovine Reference Genome (UMD 3.1) (Additional file 9: Table S2). miRNAs are the dominant small RNAs (Fig. 1b, Additional file 10: Table S3).
A total of 221 known and 69 novel miRNAs satisfied the conditions of having at least 1 transcript per million clean tags and were present in a minimum of four libraries. These 290 miRNAs were used for differently expressed (DE) analysis (Additional file 11: Table S4).
Twenty-five miRNAs having > 0.1% of the total read counts in both control and SA groups were regarded as abundantly expressed miRNA (Table 1). Seven most abundantly expressed miRNAs (bta-miR-148a, bta-miR-30a-5p, bta-let-7f, bta-miR-21-5p, bta-miR-26a, bta-let-7a-5p and bta-let-7 g) accounted for 93.80 and 90.91% of the total read counts in the control and SA groups, respectively. Bta-miR-148a had the highest miRNA read counts in both groups. Bta-miR-11_2406 was the most highly expressed novel miRNA, which accounted for 0.139‰ of total read counts (Additional file 11: Table S4b).
Table 1.
Twenty-five most abundantly expressed miRNAs in bovine milk exosomes
| miRNA ID | Control group (n = 3) | SA group (n = 6) | Overall | |||
|---|---|---|---|---|---|---|
| Mean reads | % | Mean reads | % | Mean reads | % | |
| bta-miR-148a | 5,704,941.00 | 83.444 | 4,487,889.67 | 77.659 | 4,893,573.44 | 79.809 |
| bta-miR-30a-5p | 234,243.67 | 3.426 | 260,044.17 | 4.500 | 251,444.00 | 4.101 |
| bta-let-7f | 131,014.00 | 1.926 | 126,834.67 | 2.195 | 128,227.78 | 2.091 |
| bta-miR-21-5p | 93,483.33 | 1.367 | 132,236.83 | 2.288 | 119,319.00 | 1.946 |
| bta-miR-26a | 102,636.33 | 1.501 | 94,022.50 | 1.627 | 96,893.78 | 1.580 |
| bta-let-7a-5p | 78,246.67 | 1.144 | 83,324.33 | 1.442 | 81,631.78 | 1.331 |
| bta-let-7 g | 68,033.00 | 0.995 | 69,446.83 | 1.202 | 68,975.56 | 1.125 |
| bta-miR-200a | 47,714.67 | 0.698 | 59,328.50 | 1.027 | 55,457.22 | 0.904 |
| bta-let-7b | 47,204.67 | 0.690 | 59,017.83 | 1.021 | 55,080.11 | 0.898 |
| bta-miR-200c | 32,256.33 | 0.472 | 38,897.00 | 0.673 | 36,683.44 | 0.598 |
| bta-miR-30d | 24,246.67 | 0.355 | 28,719.33 | 0.497 | 27,228.44 | 0.444 |
| bta-miR-99a-5p | 19,972.33 | 0.292 | 26,966.17 | 0.467 | 24,634.89 | 0.402 |
| bta-miR-26b | 24,645.67 | 0.360 | 21,275.50 | 0.368 | 22,398.89 | 0.365 |
| bta-let-7i | 18,153.00 | 0.266 | 22,036.17 | 0.381 | 20,741.78 | 0.338 |
| bta-miR-27b | 19,007.00 | 0.278 | 20,082.17 | 0.348 | 19,723.78 | 0.322 |
| bta-miR-151-3p | 15,890.00 | 0.232 | 18,085.17 | 0.313 | 17,353.44 | 0.283 |
| bta-miR-186 | 9732.33 | 0.142 | 20,533.17 | 0.355 | 16,932.89 | 0.276 |
| bta-miR-200b | 11,909.67 | 0.174 | 12,660.67 | 0.219 | 12,410.33 | 0.202 |
| bta-let-7c | 9782.67 | 0.143 | 12,613.83 | 0.218 | 11,670.11 | 0.190 |
| bta-miR-103 | 6039.00 | 0.088 | 10,320.67 | 0.179 | 8893.44 | 0.145 |
| bta-miR-375 | 7205.33 | 0.105 | 9041.00 | 0.156 | 8429.11 | 0.137 |
| bta-miR-182 | 6910.00 | 0.101 | 8220.67 | 0.142 | 7783.78 | 0.127 |
| bta-miR-92a | 8571.33 | 0.125 | 6102.33 | 0.106 | 6925.33 | 0.113 |
| bta-miR-532 | 5888.67 | 0.086 | 7368.33 | 0.128 | 6875.11 | 0.112 |
| bta-miR-423-5p | 4426.67 | 0.064 | 7251.67 | 0.125 | 6310.00 | 0.103 |
A higher number of known miRNAs were located on Chr X (36 miRNAs), Chr 19 (29 miRNAs), and Chr 21 (27 miRNAs), while the highest number of novel miRNAs were located on Chr 5 (15 miRNAs) (Fig. 1c).
DE miRNAs in exosomes between the control and S. aureus infected milk
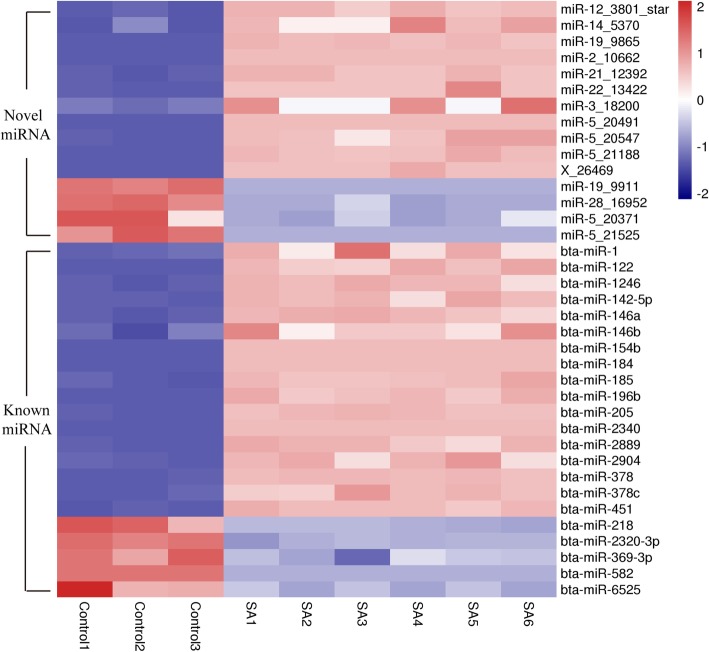
Thirty seven miRNAs (twenty two known and fifteen novel) were significantly differentially expressed (p < 0.05) between the control and the SA groups. Out of these, twenty-eight miRNAs were significantly (p < 0.05) up-regulated, whereas nine miRNAs were significantly (p < 0.05) down-regulated (Fig. 2). Notably, four miRNAs (bta-miR-2_10662, bta-miR-5_20491, bta-miR-184 and bta-miR-2340) were only expressed in the SA group, while one miRNA (bta-miR-5_21525) was only expressed in the control group (Table 2). Furthermore, three known (bta-miR-185, bta-miR-2904 and bta-miR-378) and eight novel (bta-miR-12_3801, bta-miR-14_5370, bta-miR-21_12392, bta-miR-22_13422, bta-miR-3_18200, bta-miR-5_20547, bta-miR-5_21188 and bta-miR-X_26469) miRNAs were highly expressed (log2foldchange > 3) in the SA group as compared to the control group.
Fig. 2.
Heat map of DE miRNAs expression profile. The intensity of each colour indicates the log2 (the miRNA expression in each sample/ the average of miRNA expression across all 9 samples). Red colour pixels indicate an increased abundance of miRNA in the indicated samples whereas blue pixels indicate decreased miRNA levels in log2 scale
Table 2.
DE miRNAs between the control group and the SA group
| miRNA ID | Log2fold changea | p-value |
|---|---|---|
| Novel miRNAs | ||
| bta-miR-12_3801 | 3.002 | 2.48 × 10−4 |
| bta-miR-14_5370 | 3.807 | 1.14 × 10−2 |
| bta-miR-19_9865 | 2.663 | 2.94 × 10−2 |
| bta-miR-19_9911 | −1.585 | 3.83 × 10−2 |
| bta-miR-2_10662 | $ | 5.15 × 10−5 |
| bta-miR-21_12392 | 4.644 | 7.64 × 10−4 |
| bta-miR-22_13422 | 4.170 | 1.22 × 10−3 |
| bta-miR-28_16952 | −1.280 | 3.15 × 10−2 |
| bta-miR-3_18200 | 3.807 | 1.22 × 10−2 |
| bta-miR-5_20371 | −1.078 | 2.87 × 10−2 |
| bta-miR-5_20491 | $ | 1.20 × 10−2 |
| bta-miR-5_20547 | 4.000 | 2.27 × 10− 3 |
| bta-miR-5_21188 | 4.000 | 2.10 × 10− 3 |
| bta-miR-5_21525 | $$ | 1.75 × 10−3 |
| bta-miR-X_26469 | 5.170 | 2.93 × 10−6 |
| Known miRNAs | ||
| bta-miR-1 | 1.926 | 2.40 × 10−2 |
| bta-miR-122 | 1.838 | 3.17 × 10−5 |
| bta-miR-1246 | 2.140 | 5.35 × 10−3 |
| bta-miR-142-5p | 1.864 | 1.49 × 10−2 |
| bta-miR-146a | 1.978 | 3.01 × 10−2 |
| bta-miR-146b | 1.482 | 4.46 × 10−2 |
| bta-miR-154b | 2.848 | 1.90 × 10−2 |
| bta-miR-184 | $ | 2.31 × 10−8 |
| bta-miR-185 | 3.585 | 3.74 × 10−2 |
| bta-miR-196b | 2.841 | 4.92 × 10−2 |
| bta-miR-205 | 2.883 | 2.05 × 10−2 |
| bta-miR-218 | −1.078 | 3.40 × 10−2 |
| bta-miR-2320-3p | −2.322 | 4.02 × 10−2 |
| bta-miR-2340 | $ | 1.21 × 10−2 |
| bta-miR-2889 | 2.816 | 1.56 × 10−2 |
| bta-miR-2904 | 3.484 | 1.66 × 10−3 |
| bta-miR-369-3p | −1.625 | 1.01 × 10−2 |
| bta-miR-378 | 4.044 | 1.46 × 10−3 |
| bta-miR-378c | 2.193 | 9.65 × 10−3 |
| bta-miR-451 | 1.554 | 1.85 × 10−3 |
| bta-miR-582 | −2.000 | 7.50 × 10−3 |
| bta-miR-6525 | −2.585 | 2.37 × 10−2 |
a Values represent the amount of expression of the miRNA in the SA group divided by the amount in the control group, followed by a base-2 logarithmic value
$ miRNA only expressed in the SA group
$$ miRNA only expressed in the control group
Predicted target genes of known DE miRNAs and GO and KEGG pathway annotations
Twenty-two known DE miRNAs were predicted to target 2678 genes (Additional file 12: Table S5). Bta-miR-185 had the highest number of target genes (515 genes) (Additional file 3: Figure S3), while the MTMR3 gene was the most popular target for DE miRNAs (targeted by 8 DE miRNAs). Other common target genes for DE miRNAs were USP12, SYT13, PDHA1, FRMD8, KLHL29, MCAT, ABAT, CHFT8 and CELF3 (each targeted by 6 DE miRNAs).
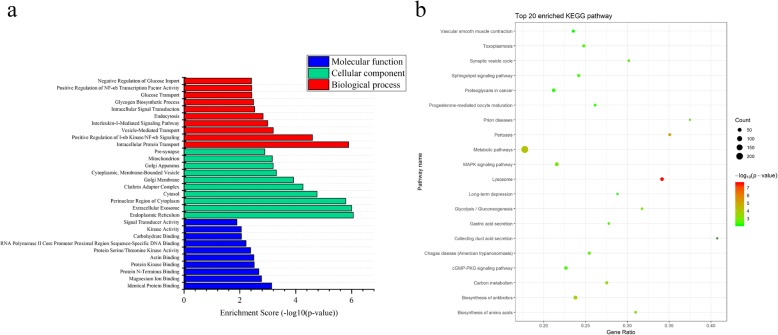
Target genes of DE miRNAs were significantly (p < 0.05) enriched in 121 GO terms (63 biological process GO terms, 34 cellular component GO terms and 24 molecular function GO terms) (Additional file 13: Table S6). The most enriched biological process, cellular component and molecular function GO terms were intracellular protein transport (p = 1.29 × 10− 6), endoplasmic reticulum (p = 8.79 × 10− 7) and identical protein binding (p = 7.28 × 10− 4), respectively (Fig. 3a). Moreover, 49 KEGG pathways were significantly enriched for the target genes of DE miRNAs (Additional file 14: Table S7). The lysosome pathway (p = 2.73 × 10− 8) was the most significantly enriched KEGG pathway (Fig. 3b).
Fig. 3.
GO and KEGG analyses of 22 known DE miRNA. a Top 10 GO terms of 22 known DE miRNAs target genes in each three GO categories (biological process, cellular component, and molecular function). Enrichment score is presented as -log10(p-value). b Top 20 enriched KEGG pathways for target genes of 22 known DE miRNAs. The sizes of the dots represent the counts of genes. The gene ratio indicates the ratio between the number of target genes associated with a KEGG term and the total number of genes in the KEGG term
Target genes for bta-miR-378 and bta-miR-185 were validated
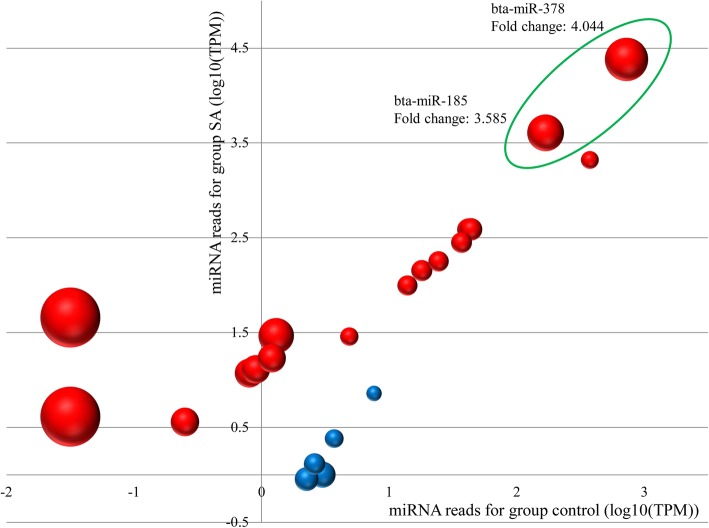
Read counts of DE miRNAs showed that bta-miR-378, bta-miR-185 and bta-miR-146b were 3 top DE miRNAs in the SA group as compared to the control group. Considering the potential importance based on read counts and log2foldchange values, the potential targets for bta-miR-378 and bta-miR-185 were further validated (Fig. 4).
Fig. 4.
The fold change and read counts (log10(TPM)) of DE miRNAs in the control and SA groups. The size of bubble stands for the DE miRNA fold change. The up- and down-regulated miRNAs are colored in red and blue, respectively. Bta-miR-378 and bta-miR-185 are shown inside of the green ellipse
A total of 441 and 814 target genes were predicted for bta-miR-378 and bta-miR-185 by TargetScan and miRanda programs, respectively (Additional file 15: Table S8). Among them, 8 and 23 genes were predicted by both programs, and considered as more plausible targets of bta-miR-378 and bta-miR-185, respectively (Table 3, Additional file 4: Figure S4).
Table 3.
Plausible target genes of bta-miR-378 and bta-miR-185 predicted by both TargetScan and miRanda programs
| miRNA | Predicted target gene | Gene name |
|---|---|---|
| bta-miR-378 | UBE2W | ubiquitin-conjugating enzyme E2W (putative) |
| TCF12 | transcription factor 12 | |
| TLK2 | tousled-like kinase 2 | |
| TBX6 | T-box 6 | |
| VAT1L | vesicle amine transport 1-like | |
| AQP3 | aquaporin 3 (Gill blood group) | |
| RNF144B | ring finger protein 144B | |
| PDIA4 | protein disulfide isomerase family A, member 4 | |
| bta-miR-185 | PAK7 | p21 protein (Cdc42/Rac)-activated kinase 7 |
| CAPZB | capping protein (actin filament) muscle Z-line, beta | |
| KDM2A | lysine (K)-specific demethylase 2A | |
| PHYHIP | phytanoyl-CoA 2-hydroxylase interacting protein | |
| NFATC3 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | |
| E2F6 | E2F transcription factor 6 | |
| CHMP7 | charged multivesicular body protein 7 | |
| DGKI | diacylglycerol kinase, iota | |
| CYP4V2 | cytochrome P450, family 4, subfamily V, polypeptide 2 | |
| SF1 | splicing factor 1 | |
| FOSL2 | FOS-like antigen 2 | |
| CABP4 | calcium binding protein 4 | |
| NR1D1 | nuclear receptor subfamily 1, group D, member 1 | |
| HP1BP3 | heterochromatin protein 1, binding protein 3 | |
| DYRK1B | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | |
| MLLT3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | |
| STX4 | syntaxin 4 | |
| SGMS1 | sphingomyelin synthase 1 | |
| NPR2 | natriuretic peptide receptor B | |
| SPATA2 | spermatogenesis associated 2 | |
| AGFG1 | ArfGAP with FG repeats 1 | |
| PGM1 | phosphoglucomutase 1 | |
| SDHC | succinate dehydrogenase complex |
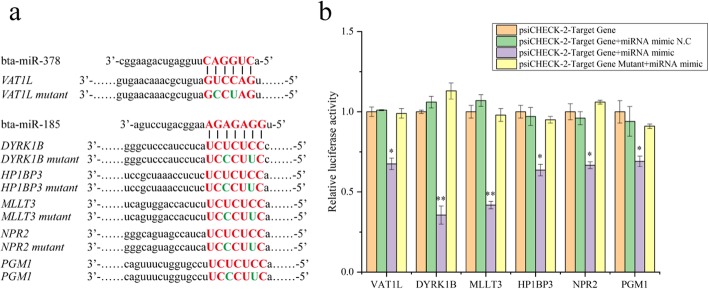
The binding sites for bta-miR-378 and bta-miR-185 in the 3′-UTR of commonly predicted target genes were analyzed by bioinformatics methods (microrna.org and TargetScan) (Additional file 16: Table S9). To biochemically confirm the in silico predicted targets, the 3′-UTRs of predicted candidate mRNAs were cloned into a dual luciferase vector. The luciferase activity of the psiCHECK-2 vector with 3′-UTR of VAT1L was strongly inhibited by bta-miR-378 (p < 0.05) (Additional file 5: Figure S5). Similarly, luciferase activities of psiCHECK-2 vectors with 3′-UTR of DYRK1B, MLLT3, HP1BP3, NPR2 or PGM1 were significantly down-regulated by bta-miR-185 (p < 0.05) (Additional file 6: Figure S6). To validate these results, the miRNA target sites in the 3′-UTRs of VAT1L, DYRK1B, MLLT3, HP1BP3, NPR2 and PGM1 were mutated (Fig. 5a). After the mutation, transfection of the miRNA mimics (bta-miR-378 or bta-miR-185) did not change the luciferase activities (Fig. 5b). These results suggested that VAT1L was the target of bta-miR-378, while DYRK1B, MLLT3, HP1BP3, NPR2 and PGM1 were targets of bta-miR-185.
Fig. 5.
Detection of bta-miR-378 and bta-miR-185 target genes. a Binding sites of bta-miR-378 and bta-miR-185 and their target gene vectors. The seed region is indicated by red bases and the green bases are the inserted mutations. b Relative luciferase activities of target genes for bta-miR-378 and bta-miR-185. The values represent the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, N. C, negative control
Discussion
Milk provides important nutrients that are of benefit to most people throughout life. Due to the direct effects of the protein, fat, lipid, vitamin, and mineral fractions, milk has a specific growth promoting effect in children in both developing and developed countries [27].
Pasteurization is widely used in commercial milk production and it destroys all known pathogens and most of the spoilage bacteria in raw milk. Nowadays, there is compelling evidence that milk exosomes are retained in pasteurized commercial milk [17] and reach the systemic circulation and tissues of the human milk consumer [28]. Furthermore, pasteurization did not affect the profile expression of miRNA in bovine milk [29]. Bovine milk exosomes miRNAs which resist the harsh conditions in the gastrointestinal tract [30] are taken up via receptor mediated endocytosis by intestinal epithelial cells [16] and vascular endothelial cells [31]. More importantly, in vivo studies confirmed that milk exosomes miRNAs could reach distant tissues [19] and human plasma [32].
In our study, two miRNAs (bta-miR-378 and bta-miR-185) with high read counts were up-regulated significantly in exosomes from bovine milk infected by S. aureus. These two miRNAs have been reported to be associated with health. MiRNA-378 facilitates the development of hepatic inflammation and fibrosis [33]. In addition, expression of the miR-378 was reported to promote tumor growth [34]. MiR-185-5p may inhibit ameloblast and osteoblast differentiations and result in cleidocranial dysplasia [35], and promotes lung epithelial cell apoptosis [36]. How these miRNAs affect health parameters is not clear and it is plausible that their target genes are involved.
Consistent with two previous studies [25, 26], the expression level of bta-miR-148a was the highest among all milk-derived exosomal miRNAs in our study. In these two previous studies, the miRNAs with the greatest differences in expression in milk-derived exosomes after S. aureus infection were bta-miR-142-5p [25] and bta-miR-223 [26], respectively. While the expression level of bta-miR-142-5p was also up-regulated significantly in our study, it was not the most differentially expressed one. In addition, expression of bta-miR-223 was not significantly changed in our study. These discrepancies between our study and the other two studies could be due to the fact that exosomes were isolated from mastitic milk naturally infected with S. aureus in this study, while the other two studies used milk samples from the mammary gland challenged with S. aureus.
We have functionally validated VAT1L as a target gene of bat-miR-378. Through a network-based analysis of three independent schizophrenia genome-wide association studies, Chang et al. reported that VAT1L may be one of the genes associated with schizophrenia [37]. In addition, DYRK1B, HP1BP3, MLLT3, NPR2 and PGM1 were identified as the target genes of bta-miR-185 in our study. Surprisingly, deficiency of these target genes also leads to a variety of diseases. DYRK1B belongs to the Dyrk family of proteins, a group of evolutionarily conserved protein kinases that are involved in cell differentiation, survival, and proliferation [38]. Mutations in DYRK1B were associated with a clinical phenotype that is characterized by central obesity, hypertension, type II diabetes and early-onset coronary artery disease [38]. HP1BP3 was identified as a novel modulator of cognitive aging and HP1BP3 protein levels were significantly reduced in the hippocampi of cognitively impaired elderly humans relative to cognitively intact controls [39]. Targeted knockdown of HP1BP3 in the hippocampus induced cognitive deficits [40]. MLLT3 gene is required for normal embryogenesis in mice, and an MLLT3 null mutation caused perinatal lethality [41]. A loss-of-function mutation of the AF9/MLLT3 gene was hypothesized to relate to neuromotor development delay, cerebellar ataxia and epilepsy [42]. Homozygous inactivating mutations of NPR2 caused a severe skeletal dysplasia, acromesomelic dysplasia and Maroteaux type [43]. PGM1 deficiency has been described in a patient with myopathy and exercise induced hypoglycemia [44, 45]. PGM1 deficiency causes a non-neurological disorder of glycosylation as well as a rare muscular glycolytic defect [46].
In addition to bta-miR-378 and bta-miR-185, several other miRNAs were also differentially expressed, including miR-1, miR-122, miR-1246, miR-142-5p, miR-146a, miR-154, miR-184, miR-196 and miR-205. They were also associated with a variety of human diseases. For example, circulating miR-122 is strongly associated with the risk of developing metabolic syndrome and type II diabetes [47]. MiR-196 was overexpressed in the inflammatory intestinal epithelia of individuals with Crohn’s disease [48]. While it is beyond the scope of this study to confirm the linkage between discussed miRNAs and health parameters, the above discussion about increased expression of certain miRNAs in exosomes from S. aureus infected milk argues strongly to be vigilant about the safety of mastitis milk, even after pasteurization.
Conclusions
In conclusion, we characterized the miRNA profiles in exosomes derived from control and S. aureus infected bovine milk, and 37 miRNAs (22 known and 15 novel) were significantly differentially expressed between the control group and SA group. This is the first report of functional validation of VAT1L, and DYRK1B, MLLT3, HP1BP3, NPR2 and PGM1 as target genes for bta-miR-378 and bta-miR-185, respectively. Finally, the potential safety hazards of mastitic milk were discussed, in the context of miRNAs within milk exosomes.
Methods
Milk sample collection and bacteria identification
Milk samples from seventy-six 3- to 4-year-old Holstein cows in the middle stage of lactation from 4 dairy farms (the Shaanxi Academy of Agricultural Sciences Farm, the farm of Delikang Dairy Co., Ltd., the Duzhai dairy farm and the Cuidonggou dairy farm) in Shaanxi Province were collected for this study with the approval of the Animal Use and Care Committee of Northwest A&F University (NWAFAC3751). Milk samples from all four quarters of each cow were aseptically collected and stored at − 80 °C.
To select the samples for the control and the SA groups, 100 μL of each milk sample were plated on the Plate Count Agar (BD Diagnostics, Sparks, MD, USA) and incubated at 32 °C for 48 h. The control group samples (n = 3) were randomly selected from the samples among which the colony count was zero. The milk samples with colony counts more than 1000 were marked as samples with bacterial infection for further detection. To rule out the interference caused by Escherichia coli (E. coli) infection for subsequent experiments, samples were cultured on the BactiCard™ E. coli (Thermo Oxoid Remel, Lenexa, USA). The milk samples without E. coli infection were selected for the identification of S. aureus by the Baird-Parker agar (Oxoid, Basingstoke, Hampshire, UK) as described previously [49]. Briefly, aliquots of individual milk samples were added to an equal volume of a double-strength enrichment broth (a trypticase soy broth supplemented with 10% NaCl and 1% sodium pyruvate) (Oxoid, Basingstoke, Hampshire, UK). After 24 h incubation at 35 °C, the enrichment broth was streaked onto the Baird-Parker (Oxoid) agar containing 30% egg yolk with 1% tellurite (Oxoid) and onto the phenol red mannitol salt agar plates. Following 48 h incubation at 35 °C, the colonies on the plates were counted, and one to three presumptive staphylococcal colonies from each plate were transferred to trypticase soy agar plates. Yellow colored colonies from the phenol red mannitol salt agar plates were assumed to be S. aureus. Further identification of these presumptive staphylococcal colonies was first based on conventional methods including Gram stain staining, colony morphology, a catalase test and a coagulase test with rabbit plasma. The culture result was further confirmed by using a polymerase chain reaction (PCR) assay targeting S. aureus-specific region of the thermonuclease gene (nuc) [49] and bacterial 16S rRNA genes. The samples with nuc positive S. aureus, which was also confirmed by sequencing the PCR products of 16S rRNA genes, were selected for the SA group (n = 6).
Preparation and purification of milk exosomes
Milk exosomes from the SA group (n = 6) and the control group (n = 3) were isolated by differential centrifugation as described previously [50]. Briefly, milk samples were centrifuged at 5000×g for 60 min at 4 °C to remove milk fat and milk somatic cell. For the removal of casein and other cell debris, skimmed milk samples were subjected to three successive centrifugations at 4 °C for 1 h each at 12,000×g, 35,000×g and 70,000×g (Beckman Coulter, USA). The whey was collected and centrifuged at 135,000×g at 4 °C for 90 min (Beckman Coulter) to remove large particles and micro-vesicles. The supernatant was carefully collected and filtered through a 0.22 μm syringe-driven filter unit (Merck KGaA, Darmstadt, Germany). The percolate was collected and centrifuged at 150,000×g for 90 min at 4 °C (Beckman Coulter). The exosome pellet was re-suspended in 1 mL sterile PBS and filtered through a 0.22 μm syringe-driven filter unit (Merck KGaA). Finally, exosomes were stored in aliquots of 200 μL at − 80 °C until being used.
Identification of bovine milk exosomes
Dynamic light scattering analysis was used for analyzing nanoparticle sizes. Aliquot of 200 μL stored exosomes was diluted to 1 mL volume with sterile PBS stored on ice. The exosome solution was slowly injected into the sample cell of the Malvern Zetasizer Nano ZS90 (Malvern Panalytical Ltd., United Kingdom) system and measurements were taken according to manufacturer’s instructions.
For transmission electron microscopy (TEM), milk exosomes were fixed in 3% (w/v) glutaraldehyde and 2% paraformaldehyde in a cacodylate buffer, pH 7.3. The fixed exosomes were then applied to a continuous carbon grid and negatively stained with 2% uranyl acetate. The samples were examined with a HT7700 transmission electron microscope (HITACHI, Japan).
Exosome marker proteins (CD63 and CD81) were detected via flow cytometry (Accuri™ C6, BD Biosciences, USA) using anti-CD63 (BD Biosciences, USA) and anti-CD81 (BD Biosciences, USA) antibodies, according to manufacturer’s protocols.
Extraction of total RNA from bovine milk exosomes
Total RNA was extracted from bovine milk exosomes using the Trizol reagent (TAKARA, Japan) according to manufacturer’s protocol and dissolved in RNase free water. Quality and quantity of RNA was examined using a NanoDrop 2000/2000C (Thermo Fisher Scientific, Waltham, MA, USA) and integrity was detected using agarose gel electrophoresis.
MiRNA library preparation and sequencing
Deep sequencing was performed on all 9 individual samples. For each library, 1 μg of high-quality RNA per sample was used as the input material for a small RNA library construction using the NEXTflex™ Small RNA Sequencing Kit V3 (Illumina, San Diego, CA) according to the manufacturer’s instruction. Small RNA libraries were gel purified and pooled together in equimolar concentrations and subjected to 50 bp single read sequencing on an Illumina HiSeq 2500 system (Illumina, San Diego, CA). Read quality (adaptor removal and size selection) was assessed using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and the cutadapt [51].
Known miRNAs identification and novel miRNAs discovery
Identification of known miRNAs was performed with miRBase v21 (http://www.mirbase.org/) [52], while novel miRNA discovery was achieved with miRDeep2 v2.0.0.8 (https://github.com/rajewsky-lab/mirdeep2) [53]. The core and quantifier modules of miRDeep2 were applied to discover novel miRNAs in the pooled dataset of all the libraries while the quantifier module was used to profile the detected miRNAs in each library. The amount of miRNA expression was calculated by the transcript per million (TPM) metric, which is calculated as number of reads per miRNA alignment/number of reads of the total sample alignment× 106. MiRDeep2 score > 1 was used as a cuff point for identification of novel miRNAs. Subsequently, a threshold of ≥1 TPM of total reads and present in ≥4 libraries was applied to remove lowly expressed miRNAs. MiRNAs meeting these criteria were further used in downstream analyses including differential expression analyses.
Differential miRNA expression and predicted target genes
DE miRNAs were detected with DeSeq2 (v1.14.1) (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) [54]. Following normalization, miRNAs read counts in the SA group were compared with corresponding values in the control group. Significant DE miRNAs between the control and the SA groups were defined as having a Benjamini and Hochberg [55] corrected p-value< 0.05.
In order to investigate the potential functions of DE miRNAs, their target genes were predicted using the miRanda algorithm [56]. Predicted target genes with tot scores above 150 and tot energy below − 15 were further used for pathway analyses. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations of their target genes [57].
Bta-miR-378 and bta-miR-185 target genes validation and function analysis
Among all DE miRNAs, two miRNAs (bta-miR-378 and bta-miR-185) with high expression levels and the highest log2fold change values between the two groups were further investigated. Target genes of bta-miR-378 and bta-miR-185 were predicted by Target Scan 7.0 (http://www.targetscan.org), miRDB (http://www.mirdb.org/miRDB) and miRanda (http://www.microrna.org/microrna/home.do). Then, 3′-UTRs of target gene transcripts for bta-miR-378 and bta-miR-185 were amplified with specific primers (Additional file 17: Table S10). Furthermore, the seed regions in the 3′-UTRs of the genes were mutated with mutagenic primers by using overlapping extensions (Additional file 17: Table S10). The wild-type and mutated 3′-UTRs were sub-cloned into the restriction endonuclease NotI and XhoI site of the psiCHECK-2 vector (Promega, Madison, WI, USA).
The HEK293-T cell line (ATCC, Manassas, VA, USA) was used for transfection. The bta-miR-378 mimic (5’ACUGGACUUGGAGUCAGAAGGC3’), bta-miR-185 mimic (5’UGGAGAGAAAGGCAGUUCCUGA3’) and a miRNA mimic negative control (N.C, 5’UUGUACUACACAAAAGUACUG3’) were synthesized by GenePharma (Shanghai, China). Transfection of the miRNA mimic and psiCHECK-2 was achieved using the Lipofectamine® 3000 reagent (Invitrogen, USA) according to the manufacturer’s instructions (Additional file 7: Figure S7). Twenty-four hours after transfection, the medium was changed and cells were grown for an additional 24 h before the luciferase assay.
Firefly and Renilla luminescent signals arising from transfected cells were quantified according to the manufacturer’s instructions using a Dual Luciferase assay system (Promega) with a Multilabel Counter luminometer (Varioskan Flash, Thermo Fisher Scientific). Renilla luciferase activities to firefly luciferase activities in cells transfected with an empty psiCHECK-2 vector without a 3′-UTR fragment was set to 100%. The experiment was repeated 3 times.
Statistical analysis
Data were analyzed with the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The statistical significance between experimental groups was analyzed using One-Way ANOVA. p < 0.05 and p < 0.01 were defined to be statistically significant and extremely significant, respectively.
Supplementary information
Additional file 1: Figure S1. Bovine milk exosomes. (a) Electron microscopy images of exosomes isolated from bovine milk. (b) Particle size analysis of exosomes isolated from bovine milk by ultracentrifugation.
Additional file 2: Figure S2. Expression of CD63 and CD81 on exosome surfaces by flow cytometry.
Additional file 3: Figure S3. The number of target genes for 22 known DE miRNAs predicted by using miRanda.
Additional file 4: Figure S4. Number of predicted target genes of bta-miR-378 (A) and bta-miR-185 (B) by TargetScan (blue) and miRanda (red) programs.
Additional file 5: Figure S5. Relative luciferase activities of target genes for bta-miR-378. The values represent the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, N.C. stands for negative control.
Additional file 6: Figure S6. Relative luciferase activities of target genes for bta-miR-185. The values represent the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, N.C. stands for negative control.
Additional file 7: Figure S7. Verification of in silico predicted bta-miR-378 and bta-miR-185 targets with a dual luciferase assay. HEK293 cells were transfected with bta-miR-378/bta-miR-185 mimics together with a vector carrying the firefly luciferase gene (hluc+) as well as the Renilla luciferase gene (hRluc) fused to the 3′-UTR fragment containing either the predicted bta-miR-378/bta-miR-185 binding site or the mutated seed sequence. The assays were performed 48 h later, and the ratio of Renilla luciferase activities to firefly luciferase activities in cells transfected with an empty expression vector without 3′-UTR fragment of the target gene was set to 100%.
Additional file 8: Table S1. Results for bacterial culture and nuc detection.
Additional file 9: Table S2. Alignment rates of clean reads with the bovine Reference Genome (UMD 3.1).
Additional file 10: Table S3. Classification statistics for small RNA sequences.
Additional file 11: Table S4. a. Read counts and read counts expressed as transcript per million (TPM) for known miRNAs. b. Read counts and read counts expressed as transcript per million (TPM) for novel miRNAs.
Additional file 12: Table S5. Predicted target genes of DE miRNAs.
Additional file 13: Table S6. a. Gene ontologies (biological process) enriched for target genes of 22 known DE miRNAs. b. Gene ontologies (cellular component) enriched for target genes of 22 known DE miRNAs. c. Gene ontologies (molecular function) enriched for target genes of 22 known DE miRNAs.
Additional file 14: Table S7. KEGG pathways enriched for the target genes of DE miRNAs.
Additional file 15: Table S8. a. Predicted target genes of bta-miR-378 by TargetScan and miRanda. b. Predicted target genes of bta-miR-185 by TargetScan and miRanda.
Additional file 16: Table S9. a. The binding sites between bta-miR-378 and 3′-UTR of target genes. b. The binding sites between bta-miR-185 and 3′-UTR of target genes.
Additional file 17: Table S10. PCR primers.
Acknowledgements
We would like to thank Genergy Bio Co., Ltd. (Shanghai, China) for assistance with bioinformatics analysis.
Abbreviations
- bta
Bos taurus
- Chr
Chromosome
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DE
Differentially expressed
- DMEM
Dulbecco’s modified eagle medium
- FBS
Fetal bovine serum
- GO
Gene Ontology
- IEC
Intestinal epithelial cell
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miRNA
microRNA
- PBS
Phosphate buffer saline
- S. aureus
Staphylococcus aureus
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- TPM
Transcript per million
- UTR
Untranslated region
Authors’ contributions
SYM and XZ conceived and designed the experiments and wrote the manuscript. SYM and CT collected the data and performed the analysis. EMI participated in data collection and manuscript preparation. All authors have read and approved the final manuscript.
Funding
This work was supported by a grant from National Science Foundation of China (31372282). The funder had no role in study design, data collection, analysis, interpretation of data, or manuscript preparation.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the NCBI Sequence Read Archive (BioProject No.: PRJNA589206, SRA Accession: SRR10439412, SRR10439411, SRR10439410, SRR10439409, SRR10439408, SRR10439407, SRR10439406, SRR10439405 and SRR10439404).
Ethics approval and consent to participate
Animal management and use procedures were performed in accordance with the guidelines and regulations of the animal ethics committee of Northwest Agriculture and Forestry (A&F) University, Yangling, Shaanxi, China. The whole study, including the animal use protocol, was approved by the Animal Use and Care Committee of Northwest A&F University (NWAFAC3751). The animal use in this study was consented by each of the 4 dairy farms in writing.
Consent for publication
Not applicable.
Competing interests
Eveline M. Ibeagha-Awemu is a member of the editorial board for BMC Genomics. All other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shaoyang Ma, Email: conanhorse@126.com.
Chao Tong, Email: xyq150957@163.com.
Eveline M. Ibeagha-Awemu, Email: Eveline.ibeagha-awemu@canada.ca
Xin Zhao, Email: xin.zhao@mcgill.ca.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6338-1.
References
- 1.Steinkraus BR, Toegel M, Fulga TA. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdiscip Rev Dev Biol. 2016;5:311–362. doi: 10.1002/wdev.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsaweed M, Hartmann PE, Geddes DT, Kakulas F. MicroRNAs in Breastmilk and the lactating breast: potential Immunoprotectors and developmental regulators for the infant and the mother. Int J Environ Res Public Health. 2015;12:13981–14020. doi: 10.3390/ijerph121113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci Rep. 2016;6:20680–20692. doi: 10.1038/srep20680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 7.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 8.Baddela VS, Nayan V, Rani P, Onteru SK, Singh D. Physicochemical biomolecular insights into Buffalo Milk-derived Nanovesicles. Appl Biochem Biotechnol. 2016;178:544–557. doi: 10.1007/s12010-015-1893-7. [DOI] [PubMed] [Google Scholar]
- 9.Na RS, GX E, Sun W, Sun XW, Qiu XY, Chen LP, Huang YF. Expressional analysis of immune-related miRNAs in breast milk. Genet Mol Res. 2015;14:11371–11376. doi: 10.4238/2015.September.25.4. [DOI] [PubMed] [Google Scholar]
- 10.Chen T, Xi QY, Ye RS, Cheng X, Qi QE, Wang SB, Shu G, Wang LN, Zhu XT, Jiang QY, Zhang YL. Exploration of microRNAs in porcine milk exosomes. BMC Genomics. 2014;15:100. doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modepalli V, Kumar A, Hinds LA, Sharp JA, Nicholas KR, Lefevre C. Differential temporal expression of milk miRNA during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii) BMC Genomics. 2014;15:1012. doi: 10.1186/1471-2164-15-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 13.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine Milk Exosomes is mediated by endocytosis in human Colon carcinoma Caco-2 cells and rat small intestinal IEC-6 cells. J Nutr. 2015;145:2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, Zempleni J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep. 2018;8:11321. doi: 10.1038/s41598-018-29780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen T, Xie MY, Sun JJ, Ye RS, Cheng X, Sun RP, Wei LM, Li M, Lin DL, Jiang QY, Xi QY, Zhang YL. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci Rep. 2016;20:33862. doi: 10.1038/srep33862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arntz OJ, Pieters BC, Oliveira MC, Broeren MG, Bennink MB, De Vries M, Van Lent PL, Koenders MI, Van den Berg WB, Van der Kraan PM, Van de Loo FA. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol Nutr Food Res. 2015;59:1701–1712. doi: 10.1002/mnfr.201500222. [DOI] [PubMed] [Google Scholar]
- 20.Pan JH, Zhou H, Zhao XX, Ding H, Li W, Qin L, Pan YL. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: potential in diagnosis and antitumour treatments (review) Int J Mol Med. 2018;41:1809–1816. doi: 10.3892/ijmm.2018.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Q, Stevic I, Pan C, Müller V, Oliviera-Ferrer L, Pantel K, Schwarzenbach H. Different signatures of miR-16, miR-30b and miR-93 in exosomes from breast cancer and DCIS patients. Sci Rep. 2018;28:12974. doi: 10.1038/s41598-018-31108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC. The serum exosome derived microRNA-135a, −193b, and −384 were potential Alzheimer's disease biomarkers. Biomed Environ Sci. 2018;31:87–96. doi: 10.3967/bes2018.011. [DOI] [PubMed] [Google Scholar]
- 23.Jin W, Ibeagha-Awemu EM, Liang G, Beaudoin F, Zhao X, Guan LL. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genomics. 2014;15:181. doi: 10.1186/1471-2164-15-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Zhang CL, Liao XX, Chen D, Wang WQ, Zhu YH, Geng XH, Ji DJ, Mao YJ, Gong YC, Yang ZP. Transcriptome microRNA profiling of bovine mammary glands infected with Staphylococcus aureus. Int J Mol Sci. 2015;16:4997–5013. doi: 10.3390/ijms16034997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J, Aswath K, Schroeder SG, Lippolis JD, Reinhardt TA, Sonstegard TS. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics. 2015;16:806. doi: 10.1186/s12864-015-2044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai M, He H, Jia X, Chen S, Wang J, Shi Y, Liu B, Xiao W, Lai S. Genome-wide microRNA profiling of bovine milk-derived exosomes infected with Staphylococcus aureus. Cell Stress Chaperones. 2018;23:663–672. doi: 10.1007/s12192-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaelsen KF. Cow's milk in the prevention and treatment of stunting and wasting. Food Nutr Bull. 2013;34:249–251. doi: 10.1177/156482651303400219. [DOI] [PubMed] [Google Scholar]
- 28.Melnik BC, Schmitz G. Exosomes of pasteurized milk: potential pathogens of Western diseases. J Transl Med. 2019;17:3. doi: 10.1186/s12967-018-1760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golan-Gerstl R, Elbaum SY, Moshayoff V, Schecter D, Leshkowitz D, Reif S. Characterization and biological function of milk-derived miRNAs. Mol Nutr Food Res. 2017;61:10. doi: 10.1002/mnfr.201700009. [DOI] [PubMed] [Google Scholar]
- 30.Benmoussa A, Lee CH, Laffont B, Savard P, Laugier J, Boilard E, Gilbert C, Fliss I, Provost P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J Nutr. 2016;146:2206–2215. doi: 10.3945/jn.116.237651. [DOI] [PubMed] [Google Scholar]
- 31.Kusuma RJ, Manca S, Friemel T, Sukreet S, Nguyen C, Zempleni J. Human vascular endothelial cells transport foreign exosomes from cow's milk by endocytosis. Am J Physiol Cell Physiol. 2016;310:C800–C807. doi: 10.1152/ajpcell.00169.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang T, Hu J, Wang X, Zhao X, Li Z, Niu J, Steer CJ, Zheng G, Song G. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. J Hepatol. 2019;70:87–96. doi: 10.1016/j.jhep.2018.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang H, Wang Y, Liu H, Nan X, Wong S, Peng S, Gu Y, Zhao H, Feng H. Mutant Runx2 regulates amelogenesis and osteogenesis through a miR-185-5p-Dlx2 axis. Cell Death Dis. 2017;8:3221. doi: 10.1038/s41419-017-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Lee H, Cao Y, Dela Cruz CS, Jin Y. miR-185 mediates lung epithelial cell death after oxidative stress. Am. J. Physiol. Lung cell. Mol. Physiol. 2016;310:L700–L710. doi: 10.1152/ajplung.00392.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang S, Fang K, Zhang K, Wang J. Network-based analysis of schizophrenia genome-wide association data to detect the joint functional association signals. PLoS One. 2015;10:e0133404. doi: 10.1371/journal.pone.0133404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keramati AR, Fathzadeh M, Go GW, Singh R, Choi M, Faramarzi S, Mane S, Kasaei M, Sarajzadeh-Fard K, Hwa J, Kidd KK, Babaee Bigi MA, Malekzadeh R, Hosseinian A, Babaei M, Lifton RP, Mani A. A form of the metabolic syndrome associated with mutations in DYRK1B. N Engl J Med. 2014;370:1909–1919. doi: 10.1056/NEJMoa1301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuner SM, Garfinkel BP, Wilmott LA, Ignatowska-Jankowska BM, Citri A, Orly J, Lu L, Overall RW, Mulligan MK, Kempermann G, Williams RW, O’Connell KM, Kaczorowski CC. Systems genetics identifies Hp1bp3 as a novel modulator of cognitive aging. Neurobiol Aging. 2016;46:58–67. doi: 10.1016/j.neurobiolaging.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuner SM, Ding S, Kaczorowski CC. Knockdown of heterochromatin protein 1 binding protein 3 recapitulates phenotypic, cellular, and molecular features of aging. Aging Cell. 2019;18(1):e12886. doi: 10.1111/acel.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pramparo T, Grosso S, Messa J, Zatterale A, Bonaglia MC, Chessa L, Balestri P, Rocchi M, Zuffardi O, Giorda R. Loss-of-function mutation of the AF9/MLLT3 gene in a girl with neuromotor development delay, cerebellar ataxia, and epilepsy. Hum Genet. 2005;118:76–81. doi: 10.1007/s00439-005-0004-1. [DOI] [PubMed] [Google Scholar]
- 43.Bartels CF, Bükülmez H, Padayatti P, Rhee DK, Van Ravenswaaij-Arts C, Pauli RM, Mundlos S, Chitayat D, Shih LY, Al-Gazali LI, Kant S, Cole T, Morton J, Cormier-Daire V, Faivre L, Lees M, Kirk J, Mortier GR, Leroy J, Zabel B, Kim CA, Crow Y, Braverman NE, Van den Akker F, Warman ML. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojkovic T, Vissing J, Petit F, Piraud M, Orngreen MC, Andersen G, Claeys KG, Wary C, Hogrel JY, Laforêt P. Muscle glycogenosis due to phosphoglucomutase 1 deficiency. N Engl J Med. 2009;361:425–427. doi: 10.1056/NEJMc0901158. [DOI] [PubMed] [Google Scholar]
- 45.Tegtmeyer LC, Rust S, Van Scherpenzeel M, Ng BG, Losfeld ME, Timal S, Raymond K, He P, Ichikawa M, Veltman J, Huijben K, Shin YS, Sharma V, Adamowicz M, Lammens M, Reunert J, Witten A, Schrapers E, Matthijs G, Jaeken J, Rymen D, Stojkovic T, Laforêt P, Petit F, Aumaître O, Czarnowska E, Piraud M, Podskarbi T, Stanley CA, Matalon R, Burda P, Seyyedi S, Debus V, Socha P, Sykut-Cegielska J, Van Spronsen F, de Meirleir L, Vajro P, DeClue T, Ficicioglu C, Wada Y, Wevers RA, Vanderschaeghe D, Callewaert N, Fingerhut R, Van Schaftingen E, Freeze HH, Morava E, Lefeber DJ, Marquardt T. Multiple phenotypes in phosphoglucomutase 1 deficiency. N Engl J Med. 2014;370:533–542. doi: 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez B, Medrano C, Ecay MJ, Ruiz-Sala P, Martínez-Pardo M, Ugarte M, Pérez-Cerdá C. A novel congenital disorder of glycosylation type without central nervous system involvement caused by mutations in the phosphoglucomutase 1 gene. J Inherit Metab Dis. 2013;36:535–542. doi: 10.1007/s10545-012-9525-7. [DOI] [PubMed] [Google Scholar]
- 47.Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM, Goedeke L, Rotllan N, Bonora E, Hughes AD, Santer P, Fernández-Hernando C, Tilg H, Willeit J, Kiechl S, Mayr M. Circulating MicroRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes. 2017;66:347–357. doi: 10.2337/db16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuternel X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in irgm alters a binding site for mir-196 and causes deregulation of irgm-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Zhou L, Wang L, Xue H, Zhao X. Characterization of methicillin-resistant and -susceptible staphylococcal isolates from bovine milk in northwestern China. PLoS One. 2015;10:e0116699. doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371(1):48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 52.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic. Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 56.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2013;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Bovine milk exosomes. (a) Electron microscopy images of exosomes isolated from bovine milk. (b) Particle size analysis of exosomes isolated from bovine milk by ultracentrifugation.
Additional file 2: Figure S2. Expression of CD63 and CD81 on exosome surfaces by flow cytometry.
Additional file 3: Figure S3. The number of target genes for 22 known DE miRNAs predicted by using miRanda.
Additional file 4: Figure S4. Number of predicted target genes of bta-miR-378 (A) and bta-miR-185 (B) by TargetScan (blue) and miRanda (red) programs.
Additional file 5: Figure S5. Relative luciferase activities of target genes for bta-miR-378. The values represent the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, N.C. stands for negative control.
Additional file 6: Figure S6. Relative luciferase activities of target genes for bta-miR-185. The values represent the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, N.C. stands for negative control.
Additional file 7: Figure S7. Verification of in silico predicted bta-miR-378 and bta-miR-185 targets with a dual luciferase assay. HEK293 cells were transfected with bta-miR-378/bta-miR-185 mimics together with a vector carrying the firefly luciferase gene (hluc+) as well as the Renilla luciferase gene (hRluc) fused to the 3′-UTR fragment containing either the predicted bta-miR-378/bta-miR-185 binding site or the mutated seed sequence. The assays were performed 48 h later, and the ratio of Renilla luciferase activities to firefly luciferase activities in cells transfected with an empty expression vector without 3′-UTR fragment of the target gene was set to 100%.
Additional file 8: Table S1. Results for bacterial culture and nuc detection.
Additional file 9: Table S2. Alignment rates of clean reads with the bovine Reference Genome (UMD 3.1).
Additional file 10: Table S3. Classification statistics for small RNA sequences.
Additional file 11: Table S4. a. Read counts and read counts expressed as transcript per million (TPM) for known miRNAs. b. Read counts and read counts expressed as transcript per million (TPM) for novel miRNAs.
Additional file 12: Table S5. Predicted target genes of DE miRNAs.
Additional file 13: Table S6. a. Gene ontologies (biological process) enriched for target genes of 22 known DE miRNAs. b. Gene ontologies (cellular component) enriched for target genes of 22 known DE miRNAs. c. Gene ontologies (molecular function) enriched for target genes of 22 known DE miRNAs.
Additional file 14: Table S7. KEGG pathways enriched for the target genes of DE miRNAs.
Additional file 15: Table S8. a. Predicted target genes of bta-miR-378 by TargetScan and miRanda. b. Predicted target genes of bta-miR-185 by TargetScan and miRanda.
Additional file 16: Table S9. a. The binding sites between bta-miR-378 and 3′-UTR of target genes. b. The binding sites between bta-miR-185 and 3′-UTR of target genes.
Additional file 17: Table S10. PCR primers.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI Sequence Read Archive (BioProject No.: PRJNA589206, SRA Accession: SRR10439412, SRR10439411, SRR10439410, SRR10439409, SRR10439408, SRR10439407, SRR10439406, SRR10439405 and SRR10439404).