Abstract
The present study aimed to investigate the influence of long non-coding (lnc)RNA prostate cancer associated transcript (PCAT)19 on the progression of non-small cell lung cancer (NSCLC). It was determined that PCAT19 expression was upregulated in NSCLC tissues and also predicted poor patient survival rate. Additionally, p53 expression was downregulated in NSCLC specimens, which was negatively correlated with PCAT19 expression. Moreover, in H1993 NSCLC cells, silencing of the PCAT19 gene led to an increase in the expression of p53, whilst conversely, its overexpression led to p53 downregulation. PCAT19 silencing was associated with the decreased proliferation rate of NSCLC cells, while PCAT19 overexpression led to increased proliferation. In addition, p53 overexpression, achieved through the transfection of a p53 expression vector, attenuated the effects of PCAT19 overexpression on cell proliferation. In conclusion, the present study demonstrated that PCAT19 negatively regulates the p53 tumor-suppression pathway, promoting cancer cell proliferation in patients with NSCLC.
Keywords: non-small cell lung cancer, long non-coding RNA, prostate cancer associated transcript 19, p53, proliferation
Introduction
Lung cancer is a highly prevalent malignancy and its incidence is increasing in numerous countries (1). In the United States, lung cancer was the second most prevalent cancer type in both men and women in 2018 (2). Moreover, lung cancer is responsible for 26 and 25% of cancer-related mortalities in men and women, respectively (2). Even following active treatment (such as surgical resection, chemo- and radiation therapy), <50% patients with lung cancer at Stage I (AJCC), and <5% of patients at Stage IV live longer than five years following initial diagnosis (3). In effect, the overall survival rates of lung cancer patients have not significantly improved during the past several decades, primarily due to the fact that early diagnosis is uncommon, and that the majority of patients with lung cancer are diagnosed at advanced disease stages (4,5).
The p53 pathway is a well-characterized tumor-suppressive pathway implicated in a multitude of cancer types (6). It inhibits tumor progression via the regulation of various cancer cell properties, such as tumor cell proliferation, or the enhancement of tumor cell apoptosis (7). This pathway is frequently inactivated in cancer cells (8). The involvement of p53 in cancer development is known to be regulated by specific long non-coding (lnc)RNAs (>200 nucleotides) (9). This family of RNAs is not protein coding, but participates in numerous cellular processes by regulating gene expression (10). Prostate cancer associated transcript 19 (PCAT19) is an oncogenic lncRNA involved in the progression of prostate cancer (11). Moreover, it has been discovered that PCAT19 can also promote the development of laryngeal carcinoma (12). The present study was performed to investigate the role of PCAT19 in the progression of non-small cell lung cancer (NSCLC), a major subtype of lung cancer, using both specimens from NSCLC patients and an NSCLC cell line.
Materials and methods
Patient selection and follow-up
Jilin Province Tumor Hospital (Changchun, China) admitted a total of 155 patients with NSCLC between August 2011 and December 2013. For the present study, 66 cases (40 men and 26 women; mean age, 46- 68 years; age range, 57.7± 8.9 years) were selected according to the following inclusion criteria: i) The patients were newly diagnosed; and ii) agreed to a 5-year follow-up. The exclusion criteria were: i) Patients with recurrent NSCLC; ii) patients diagnosed with other diseases; and iii) patients that had received any other therapy <3 months before surgery. The 66 patients comprised 12, 14, 16 and 24 cases of stages I–IV cancer (AJCC), respectively. Follow-up for the 66 patients occurred 5 years after the date of admission, and patient survival conditions were monitored and recorded. Follow-up of patients that succumbed to unrelated diseases or accidents was excluded. All patients were informed of the experimental protocol and written informed consent was obtained from every participant. The present study was approved by the Ethics Committee of Jilin Province Tumor Hospital.
Tissues and NSCLC cells
NSCLC and adjacent normal tissue samples (<3 cm from the tumor) were collected from each patient during diagnosis, via a histopathological biopsy. The tissue samples were independently confirmed by ≥3 pathologists and the tissue weight ranged between 0.015 and 0.018 g. The H1993 human NSCLC cell line, obtained from the American Tissue Culture Collection, was used in the present study. Cells were cultured at 37°C and 5% CO2, in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) containing 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA).
Transient transfection
pcDNA3 vectors expresssing PCAT19 and p53, and empty pcDNA3 vectors were purchased from Sangon Biotech Co., Ltd. PCAT19 small interfering (si)RNA (5′-GAUCCUUAGGUUCUCAGAAAC-3′) and negative control (NC) siRNA (5′-CGCUUACCCAAAUGCAUUGGC-3′) were purchased from Shanghai GenePharma Co., Ltd. H1993 cells were harvested at a confluence of 70–80%, and 1×105 cells were transfected with 10 nM PCAT19- and p53-expressing pcDNA3 vectors, (10 nM empty pcDNA3 vector was used as the NC) 40 nM PCAT19 siRNA and 40 nM NC siRNA, using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Untransfected cells were used as the control (C). Subsequent experiments were performed using cells collected at 24 h post-transfection.
Reverse transcription-quantitative (RT-q)PCR
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was used to extract the total RNA from patient tissue samples (1 ml TRIzol®/0.015 g tissue) and H1993 cells (1 ml TRIzol®/1×105 cells). All RNA samples were quantified and DNase I digestion was performed (1 h at 37°C). The SensiFAST™ cDNA Synthesis Kit (Bioline, Meridian Bioscience Inc.) was used to reverse-transcribe RNA into cDNA according to the manufacturer's instructions, and the SYBR® Green Master Mix (Bio-Rad Laboratories, Inc.) was used to prepare the PCR reaction mixtures according to the manufacturer's instructions. 18S rRNA was selected as the endogenous control and the expression of PCAT19 and p53 mRNA were detected. The primer sequences were as follows: PCAT19 forward, 5′-TCAGAACAGGGAACCATTGG-3′ and reverse, 5′-CAAGAAGATTCTTATCAGCT-3′; p53 forward, 5′-GCCCAACAACACCAGCTCCT-3′ and reverse, 5′-CCTGGGCATCCTTGAGTTCCT-3′; and human 18S rRNA forward, 5′-CTACCACATCCAAGGAAGCA-3′ and reverse, 5′-TTTTTCGTCACTACCTCCCCG-3′. The thermocycling conditions were: 95°C for 30 sec, 40 cycles of 95°C for 10 sec followed by 55°C for 40 sec. The mRNA levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene (13).
Cell proliferation assay
H1993 cells were harvested at 24 h post-transfection, and 3×104 cells were resuspended in 1 ml Eagle's Minimum Essential Medium (Sigma-Aldrich; Merck KGaA) containing 10% FBS (Sigma-Aldrich; Merck KGaA)) to produce a single-cell suspension. Cell suspensions were added into a 96-well cell culture plate at 0.1 ml per well. Triplicate wells were set for each cell transfection group, and the cells were incubated at 37°C and 5% CO2 for 24, 48, 72 or 96 h. Cell Counting Kit-8 solution (10 µl; Sigma-Aldrich; Merck KGaA) was added into the wells 4 h before the end of each time point according to the manufacturer's instructions. Following the addition of 10 µl DMSO, the optical density values were measured at 450 nm using a microplate reader.
Western blotting
PCAT19 cells were harvested at 24 h post-transfection and 2×105 cells per sample were treated with 1 ml RIPA solution (Thermo Fisher Scientific, Inc.) to extract the total protein. The samples were quantified using a bicinchoninic acid assay. After denaturation, the protein samples were separated via SDS-PAGE on a 10% gel and transferred onto PVDF membranes, which were subsequently blocked with 5% non-fat milk for 20 min at room temperature. The membranes were then incubated with rabbit polyclonal primary antibodies against p53 (cat. no. ab131442; 1:800) and the endogenous control GAPDH (cat. no. ab9485; 1:800; both Abcam) overnight at 4°C. Incubation with an IgG-horse radish peroxidase secondary antibody (cat. no. MBS435036; 1:1,000; MyBioSource, Inc.) was used to further probe the membranes at 25°C for 2 h. Bands were visualized using ECL reagents (Sigma-Aldrich; Merck KGaA) and processed using ImageJ software (version 1.46; National Institutes of Health).
Statistical analysis
All experiments were performed in triplicate and mean values were calculated to perform data comparisons. Data were expressed as the means ± standard deviation. Differences in gene expression between normal and NSCLC tissues were analyzed using a paired Student's t-test. Differences in cell proliferation or gene expression among different cell groups were analyzed using ANOVA (one-way) followed by Tukey's post-hoc test. Linear regression was used to conduct correlation analysis. Based on Youden's index, the 66 cases were grouped into low- and high-PCAT19 expression level groups, with 30 and 36 patients in each group, respectively. Kaplan-Meier analysis and the log-rank test were used to construct and compare survival curves. P<0.05 was considered to indicate a statistically significant difference.
Results
PCAT19 is upregulated in NSCLC tissues and correlates with poor survival time in patients with NSCLC
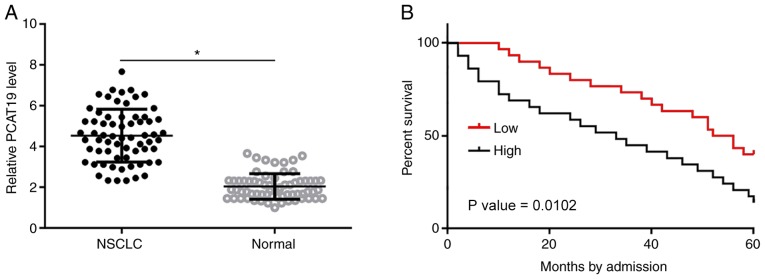
PCAT19 expression levels in NSCLC and paratumor tissues collected from patients with NSCLC (n=66) were determined using RT-qPCR. Differences in expression levels between these two groups were compared using the paired Student's t-test. The results indicated that the expression level of PCAT19 was significantly higher in 59/66 NSCLC tissues compared with the paratumor tissues (P<0.05; Fig. 1A). Survival curves were plotted for high- and low-PCAT19 expression groups. It was found that the overall survival rate of the high-expression level group was significantly lower than that of the low-expression level group (P<0.05; Fig. 1B). Expression levels of PCAT19 increased slightly in patients at more advanced clinical stages, but the changes were not statistically significant (data not shown).
Figure 1.
PCAT19 is upregulated in NSCLC tissues, which correlates with poor overall survival time. (A) PCAT19 levels determined using reverse transcription-quantitative PCR and compared using the Paired Student's t-test indicated that the expression levels of PCAT19 were significantly higher in NSCLC, compared with normal tissues. (B) Kaplan-Meier survival curve analysis showed that the overall survival rate of the high-PCAT19 group was significantly lower than that of low-PCAT19 group. *P<0.05. PCAT19, prostate cancer associated transcript 19; NSCLC, non-small cell lung cancer.
p53 expression is negatively correlated with that of PCAT19
Expression levels of p53 in the paratumor and tumor tissues of NSCLC patients (n=66) were also quantified using RT-qPCR. The data between the two tissue types were compared using the paired Student's t-test. Expression levels of p53 mRNA were significantly lower in NSCLC tissues compared with normal tissues (P<0.05; Fig. 2A). The correlation between p53 and PCAT19 expression levels was analyzed using linear regression. It was found that the expression levels of p53 and PCAT19 exhibited a significant negative correlation in normal (Fig. 2B) and NSCLC tissues (Fig. 2C).
Figure 2.
p53 expression is negatively correlated with PCAT19 expression. (A) p53 expression levels determined using reverse transcription-quantitative PCR and compared using the Paired Student's t-test, indicated that expression levels of PCAT19 were significantly lower in NSCLC, compared with normal tissues. Linear regression revealed that p53 and PCAT19 were inversely and significantly correlated in (B) normal and (C) NSCLC tissues. *P<0.05. PCAT19, prostate cancer associated transcript 19; NSCLC, non-small cell lung cancer.
PCAT19 negatively regulates p53 expression in H1993 cells
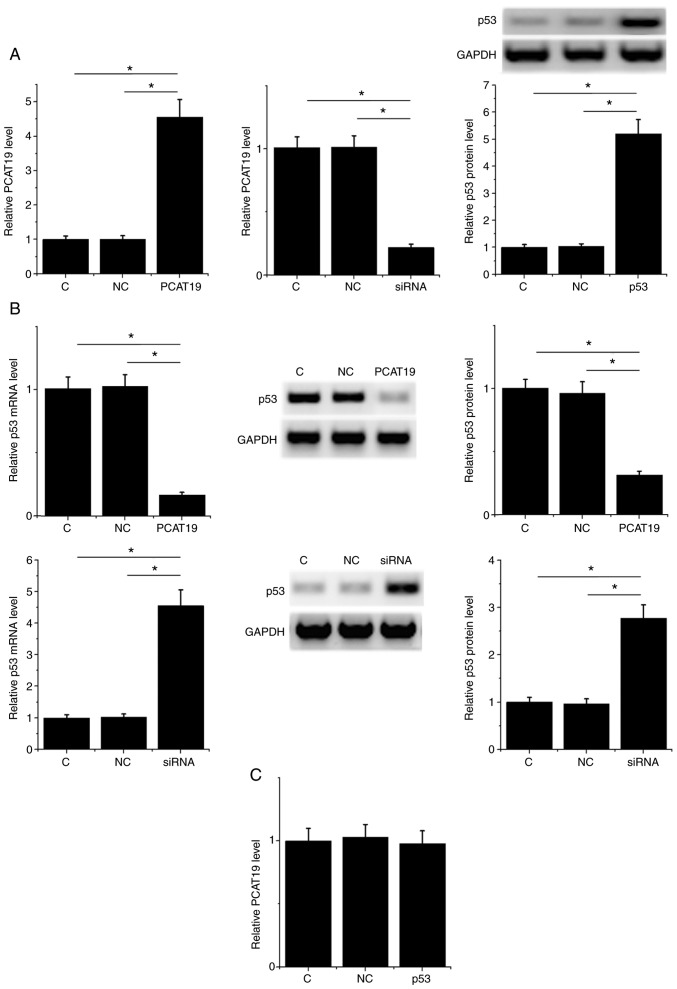
PCAT19 and p53 expression vectors, and PCAT19-siRNA, were used to transfect H1993 cells. PCAT19 and p53 expression levels were significantly altered at 24 h post-transfection compared with the control (non-transfected cells) and negative control groups (cells transfected with an empty vector or NC siRNA) (P<0.05; Fig. 3A). Moreover, in the two control groups, PCAT19-silencing resulted in the upregulation, whilst PCAT19 overexpression led to the downregulation of p53 at both the mRNA and protein levels (P<0.05; Fig. 3B). However, expression of PCAT19 in H1993 cells was not significantly affected by the transfection of the p53 expression vector (Fig. 3C).
Figure 3.
PCAT19 negatively regulates p53 expression in H1993 cells. (A) PCAT19 and p53 expression levels were significantly altered 24 h after the transfection of PCAT19 and p53 expression vectors, and PCAT19 siRNA, compared with the control and negative control groups. (B) PCAT19 silencing led to upregulated, while PCAT19 overexpression led to downregulated p53 expression at both the mRNA and protein levels. (C) Expression of PCAT19 in H1993 cells was not significantly affected by transfection with the p53 expression vector. *P<0.05. PCAT19, prostate cancer associated transcript 19; NSCLC, non-small cell lung cancer; siRNA, short interfering RNA.
PCAT19 regulates H1993 cell proliferation through p53
Cell proliferation data were compared among groups by performing one-way ANOVA followed by Tukey's post-hoc test. It was observed that compared to the two controls, PCAT19-silencing led to a decrease, whilst PCAT19 overexpression led to an increase in the proliferation rate of NSCLC cells. Furthermore, p53 overexpression attenuated the effects of PCAT19 overexpression on cellular proliferation (P<0.05;Fig. 4).
Figure 4.
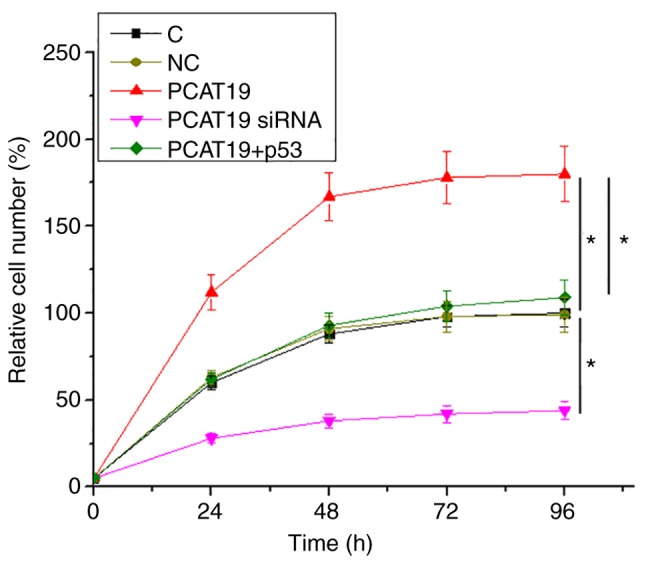
PCAT19 regulates H1993 cell proliferation via the p53 pathway. Cell proliferation data were analyzed using one-way ANOVA and Tukey's post-hoc test, and indicated that PCAT19-silencing led to decreased NSCLC-cell proliferation, compared with the two control groups. Additionally, PCAT19 overexpression led to an increased proliferation rate. p53 overexpression attenuated the effects of PCAT19 overexpression. *P<0.05. PCAT19, prostate cancer associated transcript 19; NSCLC, non-small cell lung cancer; siRNA, short interfering RNA.
Discussion
In the present study, the functionality and clinical value of PCAT19 for use in the treatment of NSCLC was evaluated. It was discovered that PCAT19 expression was upregulated in NSCLC tissues, and may promote cancer cell proliferation by negatively regulating the expression level of p53. In addition, high expression levels of PCAT19 predicted poorer overall survival times in NSCLC patients compared with patients with low expression levels.
The same lncRNAs are likely to play similar roles in different types of cancer. For example, HOX transcript antisense RNA (HOTAIR) is upregulated in most, if not all types of cancer, and the overexpression of HOTAIR promotes cancer cell invasion and migration, and suppresses apoptosis (14). However, the pathogenesis of different types of cancer is different. Therefore, it is not reasonable to use the function of a specific lncRNA in one type of cancer to speculate or predict its role in another. For example, lncRNA taurine-upregulated gene 1 (TUG1) is upregulated in a number of malignancies, and promotes these cancers in similar ways, such as initiating cell invasion and proliferation (15). However, in a recent study, Li et al (16) reported that TUG1 promoted cancer cell apoptosis in glioma, indicating its tumor-suppressive function in this disease. The oncogenic role of PCAT19 has been published in prostate cancer and laryngeal carcinoma (11,12). To the best of our knowledge, the present study is the first to report the upregulated expression of PCAT19 in NSCLC. The positive regulation of cancer cell proliferation by PCAT19 was also revealed. Therefore, PCAT19 is likely to be an oncogenic lncRNA in NSCLC.
Early diagnosis of NSCLC is uncommon and problematic, primarily because of non-specific and late-developing symptoms, but also due to a lack of reliable biomarkers (17). To improve survival rates in patients with NSCLC, it is critical to improve diagnosis, and more accurately understand disease progression and the risk of mortality. The present study indicates that patients with high expression levels of PCAT19 were significantly more likely to exhibit low survival rates. Therefore, measurement of pre-treatment levels of PCAT19 may help to improve the prognosis of patients with NSCLC.
p53 signaling in cancer biology can be regulated by certain specific lncRNAs (18). In the current study, it was determined that PCAT19 is a negative regulator of p53 and influences the proliferation of NSCLC cells. Moreover, PCAT19 has previously been found to regulate micro RNA (miR)-182, which exhibits crosstalk with p53 (19). Therefore, miR-182 may be a mediator of the interaction between PCAT19 and p53.
In conclusion, the present study demonstrates that PCAT19 is upregulated in NSCLC tissues and may promote the proliferation of NSCLC cells via the downregulation of p53.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and LZ designed the study. XZ, QW, YX, BW, CJ, LW and HS performed the experiments. HZ, ZW, QZ and SS analyzed the data. LZ drafted the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
All patients were informed of the experimental protocol and written informed consent was obtained from every participant. The present study was approved by the Jilin Province Tumor Hospital Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 5.Postmus PE, Kerr KM, Oudkerk M, Senan S, Waller DA, Vansteenkiste J, Escriu C, Peters S, ESMO Guidelines Committee Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(Suppl_4):iv1–iv21. doi: 10.1093/annonc/mdx222. [DOI] [PubMed] [Google Scholar]
- 6.Labuschagne CF, Zani F, Vousden KH. Control of metabolism by p53-cancer and beyond. Biochim Biophys Acta Rev Cancer. 2018;1870:32–42. doi: 10.1016/j.bbcan.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 8.Joerger AC, Fersht AR. The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 9.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 10.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F, Lu J, Zhou S, Wang M, Li H, et al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. 2018;174:564–575.e18. doi: 10.1016/j.cell.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Guo J, Zhang W. lncRNA PCAT19 promotes the proliferation of laryngocarcinoma cells via modulation of the miR-182/PDK4 axis. J Cell Biochem. 2019;120:12810–12821. doi: 10.1002/jcb.28552. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Shen J, Chan MT, Wu WK. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–475. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Zhang M, An G, Ma Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp Biol Med (Maywood) 2016;241:644–649. doi: 10.1177/1535370215622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canadian Task Force on Preventive Health Care, corp-author. Recommendations on screening for lung cancer. CMAJ. 2016;188:425–432. doi: 10.1503/cmaj.151421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 19.Handa H, Hashimoto A, Hashimoto S, Sugino H, Oikawa T, Sabe H. Epithelial-specific histone modification of the miR-96/182 locus targeting AMAP1 mRNA predisposes p53 to suppress cell invasion in epithelial cells. Cell Commun Signal. 2018;16:94. doi: 10.1186/s12964-018-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.