Abstract
Gastric cancer (GC) ranks fifth in terms of incidence and third in terms of tumor mortality worldwide. The present study was designed to construct a Support Vector Machine (SVM) classifier and risk score system for GC. The GSE62254 (training set) and GSE26253 (validation set 2) datasets were downloaded from the Gene Expression Omnibus database. Furthermore, the gene expression profile of GC (validation set 1) was obtained from The Cancer Genome Atlas database. Differentially expressed genes (DEGs) between recurrent and non-recurrent samples were determined using the limma package. The feature genes were selected using the Caret package, and an SVM classifier was built using the e1071 package. Using the penalized package, the optimal predictive genes for constructing a risk score system were screened. Finally, stratification analysis of clinical factors and pathway enrichment analysis were performed using Gene Set Enrichment Analysis. A total of 239 DEGs were identified in GSE62254, among which 114 DEGs were significantly associated with both recurrence-free survival and overall survival. Subsequently, 21 feature genes were screened from the 114 DEGs, and an SVM classifier was built. A risk score system for survival prediction was constructed, following the selection of 10 optimal genes, including A-kinase anchoring protein 12, angiopoietin-like protein 1, cysteine-rich sequence 1, myeloid/lymphoid or mixed-lineage leukemia, translocated to chromosome 11, neuron navigator 3, neurobeachin, nephroblastoma overexpressed, pleiotrophin, tumor suppressor candidate 3 and zinc finger and SCAN domain containing 18. The stratification analysis revealed that pathological stage was an independent prognostic clinical factor in the high-risk group. Additionally, eight significant pathways were associated with the 10-gene signature. The SVM classifier and risk score system may be applied for classifying and predicting the prognosis of patients with GC, respectively.
Keywords: GC, DEGs, SVM, risk score system, stratification analysis
Introduction
Gastric cancer (GC) occurs in the inner lining of stomach, and 60% of GC cases are caused by Helicobacter pylori infection (1). Patients with GC are usually characterized by epigastric pain, heartburn, inappetence, nausea, vomiting, weight loss and dysphagia (2). In patients with advanced GC, tumor cells may migrate from the stomach to other tissues and organs, such as liver, lymph nodes, lung and bone (3). As the disease is often diagnosed late, its prognosis is usually unfavorable with a 5-year survival rate <10% worldwide in 2016 (4). Globally, stomach cancer ranks fifth in terms of incidence and third in terms of tumor mortality, affecting 950,000 new patients and resulting in 723,000 cases of mortality in 2012 (5,6). In order to improve the therapies for GC, the molecular mechanisms of GC should be further elucidated.
Astrocyte-elevated gene 1 is involved in the progression of GC and predicts the prognosis of patients with GC, and thus its targeted inhibition may be a promising strategy for treating the tumor (7). Decreased mRNA and protein expression levels of liver kinase B1 are detected in patients with GC with low survival rate, and are independent prognostic factors of GC (8,9). Nicotinamide adenine dinucleotide phosphate oxidases (NOX) family genes act as possible prognostic indicators in GC, indicating that NOX inhibitor may be useful for the treatment of patients with GC (10). Ataxia telangiectasia mutated (ATM) expression is decreased among patients with GC in Xinjiang, and thus ATM may be a potential marker of prognosis in patients with GC (11). Overexpression of fibulin-1 (FBLN1) inhibits GC cell growth and promotes apoptosis by elevating the expression of cleaved caspase-3; thus, FBLN1 is a tumor suppressor and prognostic factor in patients with GC (12). Despite these findings, the genes implicated in the pathogenesis of GC have not been thoroughly revealed.
Early diagnosis, reasonable prognostic evaluation, and timely and appropriate intervention are important for improving the outcomes of patients with GC (13). The study of prognostic markers can guide the close monitoring and further treatment of patients at high risk of recurrence and improve their survival rate (14,15). Increasing studies have identified prognostic gene signatures and developed a prognostic score model for patients with GC (16–26). However, the recurrence-associated prognostic genes in GC have not been comprehensively examined. Since recurrence is experienced in 25–40% of all patients with GC treated with surgical resection (27,28), the identification of recurrence-associated genes is significant for survival prediction in these patients. Therefore, using microarray datasets of GC samples downloaded from The National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database, differentially expressed genes (DEGs) between recurrence and non-recurrence samples were identified. Subsequently, from the selected DEGs, the present study screened the feature genes associated with the recurrence of GC. This was followed by the construction of a classifier that could accurately identify the recurrence of GC. Combined with the clinical prognostic information, the risk score system was built based on the expression level of feature genes.
Materials and methods
Data source and preliminary screening of clinical factors
Using ‘gastric cancer’ and ‘Homo sapiens’ as key words, microarray data were searched for in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/). The selected datasets met the following criteria: i) Recurrence information was available; ii) recurrence-free survival (RFS) time information was available; and iii) sample size was ≥200. Finally, GSE62254 (platform, GP570, Affymetrix Human Genome U133 Plus 2.0 Array; Thermo Fisher Scientific, Inc.) (26,29) and GSE26253 (platform, GPL8432 Illumina HumanRef-8 WG-DASL v3.0; Illumina, Inc.) (30) were selected. GSE62254 contained 300 GC tissue samples, 282 of which had recurrence information, including 125 recurrent samples and 157 non-recurrent samples. The 282 samples were used as the training set of the present study. GSE26253 (n=432) included 177 recurrent and 255 non-recurrent samples, and was used as a validation set (validation set 2).
Furthermore, in order to obtain another validation set, gene expression profiles of GC samples were downloaded from The Cancer Genome Atlas (TCGA; https://gdc-portal.nci.nih.gov/; TGCA STAD project) database based on the Illumina HiSeq 2000 RNA Sequencing platform. As a result, 421 GC tissue samples were acquired, 298 of which had corresponding recurrence information, comprising of 242 samples without recurrence and 56 samples with recurrence (validation set 1).
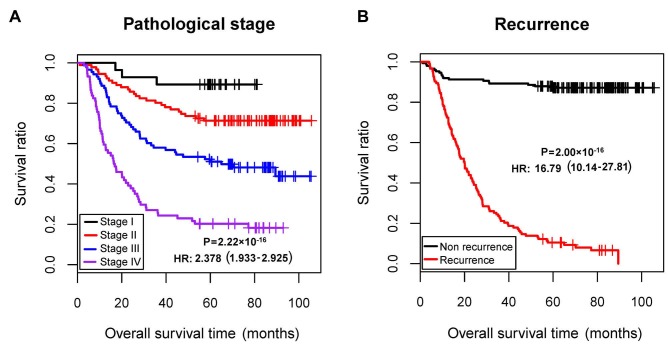
Using the training dataset (GSE62254), univariate and multivariate Cox regression analyses were conducted to evaluate the association of clinical factors with prognosis, using the survival package (version 2.41-1; http://bioconductor.org/packages/survivalr/) (31) in R (version 3.4.1; http://www.r-project.org/). P<0.05 was set as the threshold for significant association. The pathological stage and recurrence were identified to be independent prognostic clinical factors (Table I; Fig. 1). Therefore, samples of the training set were divided into recurrence and non-recurrence groups for further analysis in the present study.
Table I.
Preliminary screening of independent prognostic clinical factors.
| Univariate cox | Multivariate cox | ||||||
|---|---|---|---|---|---|---|---|
| Clinical characteristics | GSE62254 (n=300) | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (years, mean ± SD) | 61.94±11.36 | 1.009 | 0.993–1.025 | 2.71×10−1 | – | – | – |
| Sex (male/female) | 199/101 | 0.869 | 0.612–1.234 | 4.33×10−1 | – | – | – |
| MLH1 IHC (positive/negative/-) | 234/64/2 | 2.206 | 1.326–3.670 | 1.78×10−3 | 1.533 | 0.859–2.733 | 1.48×10−1 |
| EBV ISH (positive/negative/-) | 18/257/25 | 1.037 | 0.507–2.123 | 9.20×10−1 | – | – | – |
| Lymphovascular invasion (yes/no/-) | 205/73/22 | 2.642 | 1.602–4.357 | 7.67×10−5 | 1.659 | 0.972–2.832 | 6.34×10−2 |
| Pathologic M (M0/M1/-) | 273/27 | 3.971 | 2.517–6.266 | 1.58×10−10 | 1.609 | 0.912–2.839 | 1.01×10−1 |
| Pathologic N (N0/N1/N2/N3) | 38/131/80/51 | 2.052 | 1.698–2.480 | 2.03×10−14 | 1.206 | 0.851–1.708 | 2.92×10−1 |
| Pathologic T (T1/T2/T3/T4/-) | 2/186/91/21 | 1.847 | 1.469–2.323 | 8.37×10−8 | 1.120 | 0.809–1.550 | 4.94×10−1 |
| Pathologic stage (I/II/III/IV/-) | 30/96/95/77/2 | 2.378 | 1.933–2.925 | 2.22×10−16 | 1.660 | 1.056–2.611 | 2.81×10−2 |
| Lauren classification (diffuse/intestinal/mixed) | 135/146/17/2 | 0.828 | 0.704–0.974 | 2.19×10−2 | 0.988 | 0.829–1.177 | 8.92×10−1 |
| Recurrence (yes/no) | 125/157/18 | 16.790 | 10.14–27.81 | 2.00×10−16 | 13.61 | 7.704–24.041 | 2.00×10−16 |
| Mortality (dead/alive/-) | 135/148//17 | – | – | – | – | – | – |
| Overall survival time (months, mean ± SD) | 50.59±31.42 | – | – | – | – | – | – |
Cox regression analysis was not performed for mortality and overall survival time, as they are dependent variables and not independent variables. HR, hazard ratio; MLH1 IHC, MutL homolog 1 immunohistochemistry; EBV ISH, Epstein-Barr virus in situ hybridization.
Figure 1.
KM survival curves based on pathological stage and recurrence. (A) KM curves according to pathological stage. (B) KM curves based on recurrence. KM, Kaplan-Meier; HR, hazard ratio.
Data normalization
The expression matrices of the three datasets were stacked, and each matrix was scaled based on expression levels. The unit specification was scaled and a sample vector was given as follows:
In the formula, ||v||22 stands for the 2-norm of vector (norm).
Combined with the sqrt [sum(data2)] function (32) in R, the square root of the eigenvalue of matrix B=A*AT was extracted to acquire the samples scaled to 1. Based on the median and median absolute deviation (MAD) of each gene, the gene expression level was centralized and normalized using median scaling. The details were shown as follows: Giving an eigenvector x=(x1, …, xn); and defining median scale normalization as:
Identification of DEGs between recurrence and non-recurrence samples
As aforementioned, the GSE62254 dataset was classified into recurrent and non-recurrent groups. The DEGs between the two groups were analyzed using the limma package (version 3.34.7; http://bioconductor.org/packages/release/bioc/html/limma.html) (33) in R. The strict cut-off was a false discovery rate (FDR) <0.05 and |log2 fold change (FC)|>0.263. Subsequently, bidirectional hierarchical clustering based on centered Pearson correlation algorithm was performed on the DEGs using the pheatmap package (version 1.0.8; http://cran.r-project.org/web/packages/pheatmap/index.html) (34) in R.
Construction of the Support Vector Machine (SVM) classifier
Using Cox regression analysis in the survival package (31), the DEGs that were significantly associated with RFS time and overall survival (OS) time were selected from the GSE62254 dataset. P<0.05 was set as the threshold. The DEGs significantly associated with both RFS time and OS time were used for subsequent analysis.
The recursive feature elimination algorithm in the Caret package (version 6.0–76; http://cran.r-project.org/web/packages/caret) (35) in R was used to identify the optimal combination of feature genes. During the 100-fold cross validation, the gene combination corresponding to the highest accuracy and the smallest Root Mean Square Error (RMSE) was considered as the optimal combination of feature genes.
Combined with the eigenvalues in each sample, the supervised classification algorithm SVM evaluates the probability of a sample belonging to one type (36). Using the SVM algorithm (Cross, 100-fold cross validation; Core, Sigmoid Kernel) in the e1071 package (version 1.6–8; http://cran.r-project.org/web/packages/e1071) (37) in R, an SVM classifier was built on account of the feature gene combination.
In GSE62254, GSE26253 and the TCGA dataset, the classification efficiency of the SVM classifier was assessed based on Concordance index (C-index), Brier score, log-rank P-value of Cox-proportional hazard (Cox-PH) regression and area under the receiver operating characteristic (AUROC) curve. Using the survcomp package (version 1.30.0; http://www.bioconductor.org/packages/release/bioc/html/survcomp.html) (38) in R, the C-index (the score of all individual pairs that predicted the correct order of survival time) (39) and the Brier score (a scoring function for measuring the accuracy of probability prediction) (40) were calculated.
Using the Kaplan-Meier (KM) curve analysis of the survival package (31), KM curves were drawn for the two groups predicted using the SVM classifier, and the log-rank P-value was calculated. Combined with the pROC package (version 1.12.1; http://cran.r-project.org/web/packages/pROC/index.html) (41) in R, the indexes including sensitivity, specificity, positive prediction value and negative prediction value were calculated for ROC curves.
Construction of risk score system
Using the Cox-PH model of the penalized package (version 0.9–50; http://bioconductor.org/packages/penalized/) (42) in R, the optimal combination of prognosis-associated genes was further screened from the selected combination of feature genes. The optimized parameter ‘lambda’ in the screening model was calculated through 1,000 cross-validation likelihood (cvl).
Combined with prognostic coefficients of the prognosis-associated DEGs in the optimal combination, a risk score system was constructed based on gene expression level. Furthermore, the risk score was calculated for each sample using the following formula:
CoefDEG and ExpDEG represent regression coefficient and the corresponding gene expression level, respectively.
With the median of risk scores as the demarcation point, the samples in GSE62254 were classified into high- and low-risk groups. Using the KM curve analysis of the survival package (31), correlation analysis for the risk score system and prognosis was carried out. Additionally, the risk score system was further validated in the GSE26253 and TCGA datasets.
Stratification analysis of clinical factors
Combined with the univariate and multivariate Cox regression analysis of the survival package (31), the independent prognostic clinical factors in GSE62254 were selected. Combined with the high- and low-risk samples determined by the risk score system, stratification analysis was further carried out.
Pathway enrichment analysis
According to the risk scores of the samples in GSE62254, the samples were divided into high-risk and low-risk groups. Under FDR<0.05 and |log2 FC|>0.263, the DEGs between the two groups were identified using the limma package (33). Using Gene Set Enrichment Analysis (http://software.broadinstitute.org/gsea/index.jsp) (43), pathway enrichment analysis was conducted for the DEGs, with the screening criterion of nominal P<0.05.
Results
Identification of DEGs
Following data normalization, 239 DEGs between recurrent and non-recurrent samples in the GSE62254 dataset were identified (Fig. 2A). The Kernel density curve of the DEGs revealed that 79.08% (189/239) of the DEGs were upregulated and 20.92% (50/239) of the DEGs were downregulated in recurrent samples (Fig. 2B). A bidirectional hierarchical clustering heatmap, based on the expression levels of the identified DEGs, indicated that the samples clustered into two groups (Fig. 2C).
Figure 2.
Screening results of DEGs. (A) Scatter diagram of the DEGs (red dots represent DEGs; green horizontal dashed line represents FDR <0.05, and the two green vertical dashed lines represent log2 (FC)>0.263. (B) Kernel density curve of DEGs. (C) Bidirectional hierarchical clustering heatmap of the DEGs (pink and green sample bars represent recurrent samples and non-recurrent samples, respectively; red and blue represent upregulation and downregulation, respectively). DEGs, differentially expressed genes; FC, fold change; FDR, false discovery rate.
Construction of SVM classifier
A total of 124 recurrence-associated DEGs and 127 overall survival-associated DEGs were screened in GSE62254. Following comparison of the two sets of DEGs, 114 DEGs were found to be significantly associated with both RFS time and OS time.
The 114 DEGs were further screened for feature genes. When min RMSE=0.148 and max Accuracy=0.842, the gene combination involving 21 genes was considered as the optimal one. Based on the 21 feature genes, an SVM classifier was built in GSE62254.
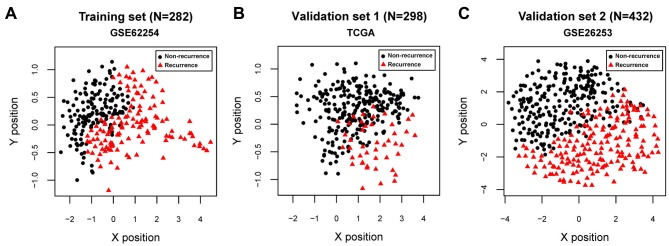
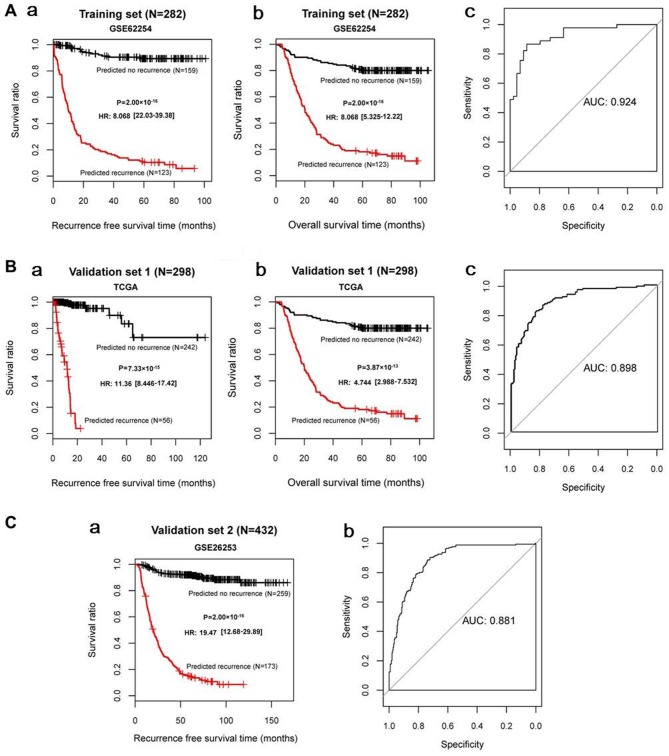
For GSE62254, GSE26253 and the TCGA datasets, all C-index values were >0.80 and all Brier scores were <0.30 for RFS time and OS time. The classification results of the samples, based on the SVM classifier, are presented in scatter diagrams (Fig. 3). KM survival curves demonstrated that the log-rank P-values for RFS time and OS time in the training and validation sets were all <0.05 (Fig. 4), suggesting significantly different RFS time and OS time between predicted recurrence and non-recurrence samples in the GSE62254 and TCGA datasets, and significantly different RFS time in GSE26253 (the samples in GSE26253 had no OS information). The predicted results of the SVM classifier were consistent with the actual outcomes of patients with GC in these datasets. The AUROC curves revealed that all AUROC values for the training and validation sets were >0.8 (Table II; Fig. 4). These results suggested that the SVM classifier based on the 21 feature genes could accurately determine the recurrence type of GC samples.
Figure 3.
Scatter diagrams showing classification results of Support Vector Machine classifier. Scatter diagram of (A) GSE62254, (B) TCGA and (C) GSE26253 datasets. Red triangles and black dots represent recurrent and non-recurrent samples, respectively. TCGA, The Cancer Genome Atlas.
Figure 4.
KM survival curves and AUROC curves based on the Support Vector Machine classifier. (A-a and A-b) KM curves and (A-c) AUROC curve of the GSE62254 dataset. (B-a and B-b) KM curves and (B-c) AUROC curve of the TCGA dataset. (C-a) KM curve and (C-b) AUROC curve of the GSE26253 dataset. For KM curves, red and black curves represent recurrent samples and non-recurrent samples, respectively. KM, Kaplan-Meier; AUROC, area under the receiver operating characteristic; AUC, area under the curve; TCGA, The Cancer Genome Atlas.
Table II.
| RFS/OS | ROC | |||||||
|---|---|---|---|---|---|---|---|---|
| Datasets | C-index | Brier score | Log rank P-value | AUROC | Sensitivity | Specificity | PPV | NPV |
| Training set (GSE62254; n=282) | 0.966/0.871 | 0.0108/0.0255 | 2.00×10−16/2.00×10−16 | 0.924 | 0.896 | 0.929 | 0.911 | 0.918 |
| Validation set 1 (TCGA; n=295) | 0.929/0.807 | 0.0272/0.0283 | 7.33×10−15/3.87×10−13 | 0.898 | 0.844 | 0.929 | 0.779 | 0.871 |
| Validation set 2 (GSE26253; n=432) | 0.950 | 0.0115 | 2.00×10−16 | 0.881 | 0.853 | 0.914 | 0.873 | 0.899 |
SVM, Support Vector Machine; TCGA, The Cancer Genome Atlas; RFS, recurrence-free survival; OS, overall survival; C-index, Concordance index; ROC, receiver operating characteristic; AUROC, area under the receiver operating characteristic curve; PPV, positive prediction value; NPV, negative prediction value.
Construction of risk score system
Using the Cox-PH model, the optimal combination of prognostic genes was further screened from the 21 feature genes. When the optimized parameter ‘lambda’ was 2.2604, the cvl value was largest (−757.1749; Fig. 5A). When ‘lambda’=2.2604, 10 optimal genes were obtained [A-kinase anchoring protein 12 (AKAP12), angiopoietin-like protein (ANGPTL) 1, cysteine-rich sequence 1 (CYS1), myeloid/lymphoid or mixed-lineage leukemia, translocated to chromosome 11 (MLLT11), neuron navigator 3 (NAV3), neurobeachin (NBEA), nephroblastoma overexpressed (NOV), pleiotrophin (PTN), tumor suppressor candidate 3 (TUSC3), zinc finger and SCAN domain containing 18 (ZSCAN18); Fig. 5B; Table III].
Figure 5.
Selection of the optimal gene combination. (A) Curve for selecting the optimized parameter ‘lambda’. The horizontal and vertical axes represent values of ‘lambda’ and cvl, respectively. The crossing of red dashed lines represents the value of ‘lambda’ parameter (2.2604), where cvl takes the maximum value (−757.1749). (B) Coefficient distribution diagram of the 10 optimal genes. AKAP12, A-kinase anchoring protein 12; ANGPTL1, angiopoietin-like protein 1; CYS1, cysteine-rich sequence 1; MLLT11, myeloid/lymphoid or mixed-lineage leukemia; translocated to chromosome 11; NAV3, neuron navigator 3; NBEA, neurobeachin; NOV, nephroblastoma overexpressed; PTN, pleiotrophin; TUSC3, tumor suppressor candidate 3; ZSCAN18, zinc finger and SCAN domain containing 18; cvl, cross-validation likelihood.
Table III.
Top 10 optimal genes selected for building the risk score system.
| Gene | Coef | HR (95% CI) | P-value |
|---|---|---|---|
| AKAP12 | 0.3340 | 1.559 (1.278–3.112) | 2.07×10−2 |
| ANGPTL1 | −0.5826 | 0.256 (0.121–0.541) | 3.53×10−4 |
| CYS1 | 0.1153 | 1.466 (1.149–3.311) | 3.58×10−2 |
| MLLT11 | 0.4899 | 1.623 (1.537–3.498) | 2.16×10−2 |
| NAV3 | 0.4681 | 2.243 (1.007–4.996) | 4.79×10−2 |
| NBEA | 0.3292 | 1.706 (1.361–3.379) | 1.26×10−2 |
| NOV | 0.2839 | 1.317 (1.187–2.525) | 4.07×10−2 |
| PTN | 0.1638 | 1.563 (1.215–3.418) | 2.63×10−2 |
| TUSC3 | 0.0332 | 1.188 (1.053–1.711) | 3.76×10−2 |
| ZSCAN18 | 0.6275 | 2.308 (1.107–4.812) | 2.56×10−2 |
HR, hazard ratio; AKAP12, A-kinase anchoring protein 12; ANGPTL1, angiopoietin-like protein 1; CYS1, cysteine-rich sequence 1; MLLT11, myeloid/lymphoid or mixed-lineage leukemia; translocated to chromosome 11; NAV3, neuron navigator 3; NBEA, neurobeachin; NOV, nephroblastoma overexpressed; PTN, pleiotrophin; TUSC3, tumor suppressor candidate 3; ZSCAN18, zinc finger and SCAN domain containing 18.
Based on prognostic coefficients of the 10 optimal genes, a risk score system was built and risk scores were calculated using the following formula:
Risk score=(0.3340) × ExpAKAP12 + (−0.5826) × ExpANGPTL1 + (0.1153) × ExpCYS1 + (0.4899) × ExpMLLT11 + (0.4681) × ExpNAV3 + (0.3292) × ExpNBEA + (0.2839) × ExpNOV + (0.1638) × ExpPTN + (0.0332) × ExpTUSC3 + (0.6275) × ExpZSCAN18
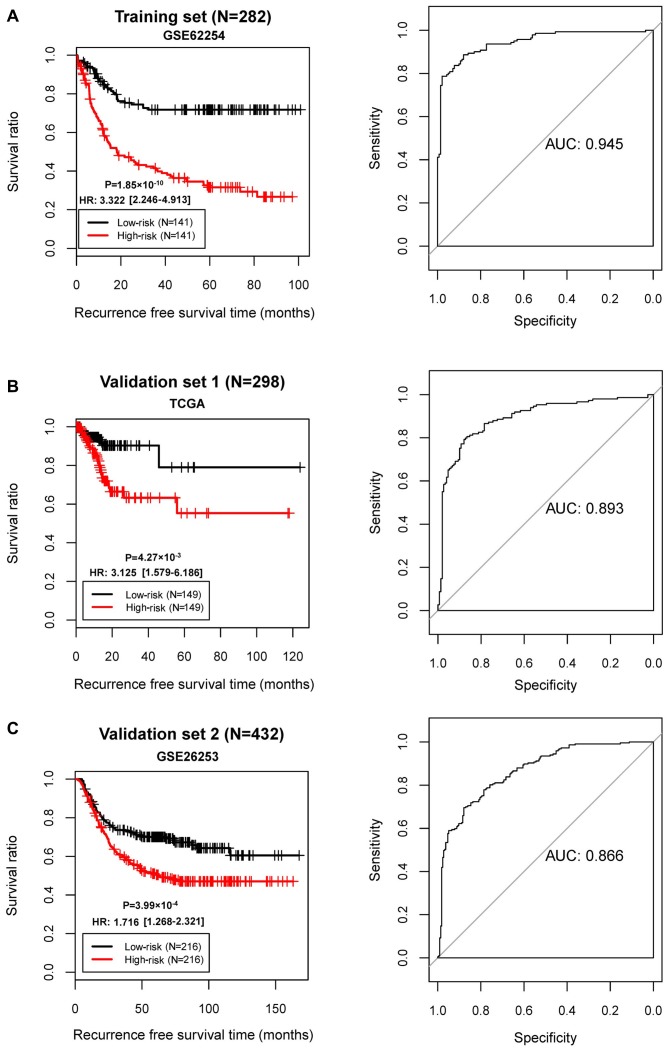
The samples in GSE62254 were divided into high- and low-risk groups. KM survival curves revealed that the high- and low-risk groups determined by the risk score system had significantly different RFS time in all three datasets (GSE62254, P=1.85×10−10; AUC=0.945; TCGA set, P=4.27×10−3, AUC=0.893; GSE26253, P=3.99×10−4, AUC=0.866; Fig. 6). These results revealed robust prognostic power of the 10-gene risk score.
Figure 6.
KM and AUROC curves based on the risk score system. KM curve (left) and AUROC curve (right) of (A) GSE62254, (B) the TCGA dataset and (C) GSE26253. KM, Kaplan-Meier; AUROC, area under the receiver operating characteristic; AUC, area under the curve; TCGA, The Cancer Genome Atlas.
Stratification analysis
Cox regression analysis demonstrated that pathological stage and risk status were independent prognostic clinical factors in GSE62254 (Table IV). Consequently, all samples were stratified into high- and low-risk groups. Furthermore, stratification analysis revealed that pathological stage was an independent prognostic clinical factor in the high-risk group (Table V). In addition, patients at different pathological stages in the high-risk group had significantly different RFS time (P=4.40×10−9; hazard ratio, 2.455; 95% confidence interval, 1.807–3.335; Fig. 7).
Table IV.
Results of Cox regression analysis for the GSE62254 dataset.
| Univariate Cox | Multivariate Cox | |||||
|---|---|---|---|---|---|---|
| Clinical characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (years, mean ± SD) | 1.003 | 0.987–1.02 | 6.76×10−1 | – | – | – |
| Sex (male/female) | 0.967 | 0.669–1.401 | 8.61×10−1 | – | – | – |
| MLH1 IHC (positive/negative/-) | 2.096 | 1.241–3.544 | 4.72×10−3 | 1.023 | 0.564–1.855 | 9.39×10−1 |
| EBV ISH (positive/negative/-) | 1.044 | 0.509–2.141 | 9.07×10−1 | – | – | – |
| Lymphovascular invasion (yes/no/-) | 2.409 | 1.456–3.987 | 4.15×10−4 | 1.552 | 0.899–2.680 | 1.15×10−1 |
| Pathologic M (M0/M1/-) | 3.839 | 2.364–6.236 | 5.01×10−9 | 1.293 | 0.719–2.324 | 3.91×10−1 |
| Pathologic N (N0/N1/N2/N3) | 2.024 | 1.661–2.465 | 5.82×10−13 | 1.049 | 0.733–1.503 | 7.93×10−1 |
| Pathologic T (T1/T2/T3/T4/-) | 1.816 | 1.435–2.298 | 4.06×10−7 | 0.867 | 0.599–1.252 | 4.46×10−1 |
| Pathologic stage (I/II/III/IV/-) | 2.414 | 1.939–3.005 | 2.22×10−16 | 2.082 | 1.270–3.415 | 3.65×10−3 |
| Lauren classification (diffuse/intestinal/mixed) | 0.874 | 0.739–1.033 | 1.14×10−1 | – | – | – |
| Risk status (high/low) | 3.322 | 2.246–4.913 | 1.85×10−10 | 2.535 | 1.656–3.882 | 1.86×10−5 |
HR, hazard ratio; MLH1 IHC, MutL homolog 1 immunohistochemistry; EBV ISH, Epstein-Barr virus in situ hybridization.
Table V.
Results of stratification analysis of clinical factors.
| A, Low risk | ||||||
|---|---|---|---|---|---|---|
| Univariate cox | Multivariate cox | |||||
| Clinical characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (years, mean ± SD) | 1.029 | 0.992–1.067 | 1.21×10−1 | – | – | – |
| Sex (male/female) | 1.374 | 0.644–2.933 | 4.09×10−1 | – | – | – |
| MLH1 IHC (positive/negative/-) | 2.59 | 1.075–6.241 | 2.77×10−2 | 2.297 | 0.779–6.775 | 1.32×10−1 |
| EBV ISH (positive/negative/-) | 2.399 | 0.926–6.218 | 6.29×10−2 | – | – | – |
| Lymphovascular invasion (yes/no/-) | 3.796 | 1.333–10.81 | 7.19×10−3 | 2.782 | 0.965–8.022 | 5.82×10−2 |
| Pathologic M (M0/M1/-) | 5.649 | 2.167–14.73 | 6.48×10−5 | 2.256 | 0.787–6.471 | 1.30×10−1 |
| Pathologic N (N0/N1/N2/N3) | 2.39 | 1.675–3.41 | 4.15×10−7 | 1.977 | 0.845–4.623 | 1.16×10−1 |
| Pathologic T (T1/T2/T3/T4/-) | 1.34 | 0.816–2.2 | 2.45×10−1 | – | – | – |
| Pathologic stage (I/II/III/IV/-) | 2.203 | 1.537–3.158 | 6.08×10−6 | 0.961 | 0.397–2.326 | 9.30×10−1 |
| Lauren classification (diffuse/intestinal/mixed) | 0.869 | 0.628–1.205 | 4.01×10−1 | – | – | – |
| B, High risk | ||||||
| Uni-variate cox | Multi-variate cox | |||||
| Clinical characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (years, mean ± SD) | 1.009 | 0.991–1.028 | 3.31×10−1 | – | – | – |
| Sex (male/female) | 0.868 | 0.566–1.33 | 5.15×10−1 | – | – | – |
| MLH1 IHC (positive/negative/-) | 0.727 | 0.376–1.406 | 3.42×10−1 | – | – | – |
| EBV ISH (positive/negative/-) | 0.539 | 0.170–1.711 | 2.87×10−1 | – | – | – |
| Lymphovascular invasion (yes/no/-) | 1.787 | 1.003–3.183 | 4.58×10−2 | 1.297 | 0.676–2.487 | 4.34×10−1 |
| Pathologic M (M0/M1/-) | 2.847 | 1.612–5.027 | 1.63×10−4 | 1.115 | 0.555–2.239 | 7.59×10−1 |
| Pathologic N (N0/N1/N2/N3) | 1.706 | 1.332–2.186 | 1.85×10−5 | 0.987 | 0.666–1.463 | 9.48×10−1 |
| Pathologic T (T1/T2/T3/T4/-) | 1.722 | 1.262–2.348 | 5.21×10−4 | 0.977 | 0.630–1.513 | 9.15×10−1 |
| Pathologic stage (I/II/III/IV/-) | 2.455 | 1.807–3.335 | 4.40×10−9 | 2.245 | 1.241–4.062 | 7.48×10−3 |
| Lauren classification (diffuse/intestinal/mixed) | 1.018 | 0.841–1.232 | 8.59×10−1 | – | – | – |
HR, hazard ratio; MLH1 IHC, MutL homolog 1 immunohistochemistry; EBV ISH, Epstein-Barr virus in situ hybridization.
Figure 7.
KM survival curves for pathological stage. (A) KM curve revealing the association between pathological stage and survival in the in low-risk group. (B) KM curve demonstrating the association between pathological stage and recurrence-free survival in the high-risk group. KM, Kaplan-Meier.
Pathway enrichment analysis
Based on the risk score system, the samples in GSE62254 were divided into high- and low-risk groups. A total of 671 DEGs were identified between the two groups, including 656 upregulated genes and 15 downregulated genes. Pathway enrichment analysis revealed that eight significant pathways were enriched for the DEGs (Table VI). According to the nominal P-value, the top three significant pathways were ‘vascular smooth muscle contraction’, ‘regulation of actin cytoskeleton’ and ‘tyrosine metabolism’.
Table VI.
Significant pathways enriched for the differentially expressed genes between high- and low-risk groups.
| Pathway | Gene count, n | ES | NES | Nominal P-value |
|---|---|---|---|---|
| Vascular smooth muscle contraction | 9 | 0.5535 | 1.7480 | 8.30×10−3 |
| Regulation of actin cytoskeleton | 8 | 0.4851 | 1.6408 | 1.45×10−2 |
| Tyrosine metabolism | 3 | 0.6451 | 1.6072 | 1.72×10−2 |
| Metabolism of xenobiotics by cytochrome p450 | 2 | 0.6845 | 1.5654 | 1.80×10−2 |
| Leukocyte transendothelial migration | 5 | 0.5237 | 1.5113 | 3.04×10−2 |
| Tight junction | 9 | 0.4560 | 1.4869 | 3.37×10−2 |
| Adherens junction | 2 | 0.6969 | 1.4027 | 3.78×10−2 |
| Cytokine-cytokine receptor interaction | 10 | 0.3808 | 1.4334 | 4.29×10−2 |
ES, enrichment score; NES, normalized enrichment score.
Discussion
In the present study, 239 DEGs (189 upregulated and 50 downregulated) were identified between the recurrent and non-recurrent samples in the GSE62254 dataset. From the 114 DEGs that were significantly associated with both RFS and OS, 21 feature genes were further screened. Subsequently, an SVM classifier was built in GSE62254, which could accurately determine the recurrence type of GC samples. Additionally, the optimal set of 10 prognostic genes (AKAP12, ANGPTL1, CYS1, MLLT11, NAV3, NBEA, NOV, PTN, TUSC3 and ZSCAN18) was obtained, followed by the construction of a risk score system. The stratification analysis demonstrated that pathological stage was an independent prognostic clinical factor in the high-risk group.
AKAP12A expression decreases colony formation and causes apoptotic cell death; thus, AKAP12A may be a critical mediator of survival in patients with GC (44). AKAP12 is usually inactivated in patients with GC and several other types of cancer, serves a role in regulating cytokinesis progression and functions as a tumor suppressor (45). The expression of ANGPTL2 is associated with GC progression, and the overexpression of ANGPTL2 at both the invasive margin and tumor center is an independent marker of prognosis in patients with GC (46,47). Elevated expression of cytoplasmic ANGPTL2 has been associated with invasion, metastasis and unfavorable survival in patients with GC, and thus ANGPTL2 may be used as a promising indicator for predicting postoperative recurrence of GC (48). Therefore, AKAP12 and ANGPTL1 may be associated with the outcomes of patients with GC.
The oncogenic factor MLLT11 is associated with tumor progression and adverse survival, exhibiting pro-tumorigenic activity in patients with ovarian cancer (49). Signal transducer and activator of transcription 3 (STAT3) is involved in tumor formation, development, migration and motility, and MLLT11 overexpression promotes pYSTAT3 expression in invasive carcinoma cells through activating the Src kinase (50). Copy number changes of NAV3 are often detected in adenomas and colorectal cancer (CRC), and NAV3 acts in connecting colon inflammation with CRC development (51). NOV and cysteine-rich protein 61 (CYR61) are upregulated in GC, and elevated CYR61 levels are responsible for unfavorable outcome (52). Additionally, increased NOV contributes to cell proliferation and invasion in GC (52). These findings indicate that MLLT11, NAV3 and NOV may also act in the development and progression of GC.
Increased PTN is significantly associated with poor OS time and RFS time of patients with GC, and may serve as an independent prognostic indicator (53). TUSC3 serves an oncogenic role in CRC, and may affect proliferation, aggression, invasion and metastasis of CRC via mediating PI3K/Akt, p38 mitogen-activated protein kinase and Wnt/β-catenin signaling pathways (54). Decreased levels of TUSC3 contribute to cell proliferation, invasion and metastasis in pancreatic cancer (PC), which predicts unfavorable outcomes in patients with PC (55,56). Gene expression and promoter methylation of ZSCAN18, cysteine dioxygenase 1 and zinc-finger protein 331 are negatively associated, and these genes have epigenetic similarity and may be potential biomarkers of gastrointestinal cancer (57). Therefore, PTN, TUSC3 and ZSCAN18 may be implicated in the pathogenesis of GC.
In order to unveil possible biological functions of the 10 prognostic genes in GC, the present study screened the DEGs between the two risk groups, classified by the 10-gene risk score. Pathway enrichment analysis revealed that the resulting DEGs were significantly enriched with several pathways, including ‘vascular smooth muscle contraction’, ‘regulation of actin cytoskeleton’ and ‘tyrosine metabolism’. The ‘vascular smooth muscle contraction’ and ‘regulation of actin cytoskeleton’ pathways serve critical roles in cancer cell migration and invasion (58,59). Tyrosine phosphorylation enhances the Warburg effect and promotes tumor growth (60). Therefore, it can be inferred that the 10 prognostic genes may affect GC prognosis by modulating cancer migration and growth.
The present study was a secondary analysis based on 282 samples with recurrence information in the GSE62254 dataset. A study by Cristescu et al (29) used GSE62254 to investigate the molecular alterations in four subtypes of GC by using targeted sequencing and genome-wide copy number microarrays. Wang et al (26) determined a six-gene signature (RNA binding protein, MRNA processing factor 2, Hes related family BHLH transcription factor with YRPW motif like, nestin, thiopurine S-Methyltransferase, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily D, member 3 and family with sequence similarity 127, member A), based on GSE62254, as a prognostic biomarker in patients with GC. The six survival-associated genes were selected using a robust likelihood-based survival model from the prognosis-associated genes identified by univariate survival analysis (26). By contrast, the present study focused on recurrence-associated DEGs to identify prognostic genes and acquired a prognostic 10-gene signature. The application of different analysis methods, analysis processes and screening thresholds is another underlying factor of the different results obtained by the two studies.
Although complex bioinformatics analyses were conducted for the gene expression profile of GC, the limitations of the present study should not be neglected. The primary limitation of the present study was the lack of experiments. In subsequent studies, experiments such as quantitative PCR and western blotting should be performed to validate the findings of the present study.
In conclusion, 239 DEGs were identified between the recurrent and non-recurrent samples of GSE62254. Furthermore, the SVM classifier may be applied for distinguishing recurrent from non-recurrent patients with GC. Additionally, the risk score system involving 10 optimal genes may be used for predicting the prognosis of patients with GC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- GC
gastric cancer
- SVM
Support Vector Machine
- DEGs
differentially expressed genes
- NOX
nicotinamide adenine dinucleotide phosphate oxidases
- ATM
ataxia telangiectasia mutated
- FBLN1
fibulin-1
- KM
Kaplan-Meier
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HJ performed data analyses and wrote the manuscript. JG, JD, XQ and CQ contributed significantly in data analyses and manuscript revision. BF conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forman D. Helicobacter pylori and gastric cancer. Scand J Gastroenterol Suppl. 1996;215:48–51. doi: 10.3109/00365529609094533. [DOI] [PubMed] [Google Scholar]
- 2.Nunobe S, Sasako M, Saka M, Fukagawa T, Katai H, Sano T. Symptom evaluation of long-term postoperative outcomes after pylorus-preserving gastrectomy for early gastric cancer. Gastric Cancer. 2007;10:167–172. doi: 10.1007/s10120-007-0434-7. [DOI] [PubMed] [Google Scholar]
- 3.Araya M, Terashima M, Takagane A, Abe K, Nishizuka S, Yonezawa H, Irinoda T, Nakaya T, Saito K. Microvessel count predicts metastasis and prognosis in patients with gastric cancer. J Surg Oncol. 1997;65:232–236. doi: 10.1002/(SICI)1096-9098(199708)65:4<232::AID-JSO2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Liu S, Feng F, Xu G, Liu Z, Tian Y, Guo M, Lian X, Cai L, Fan D, Zhang H. Clinicopathological features and prognosis of gastric cancer in young patients. BMC Cancer. 2016;16:478. doi: 10.1186/s12885-016-2489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Martel C, Forman D, Plummer M. Gastric cancer: Epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42:219–240. doi: 10.1016/j.gtc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Mcguire S. World Cancer Report 2014. Geneva, Switzerland: World health organization, international agency for research on cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jian-bo X, Hui W, Yu-long H, Chang-hua Z, Long-juan Z, Shi-rong C, Wen-hua Z. Astrocyte-elevated gene-1 overexpression is associated with poor prognosis in gastric cancer. Med Oncol. 2011;28:455–462. doi: 10.1007/s12032-010-9475-6. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Ling B, Xu X, Ma R, Li G, Cao X, Ling W, Yang Z, Hoffman RM, Lu J. Decreased expression of tumor-suppressor gene LKB1 correlates with poor prognosis in human gastric cancer. Anticancer Res. 2016;36:869–875. [PubMed] [Google Scholar]
- 9.Ma LG, Bian SB, Cui JX, Xi HQ, Zhang KC, Qin HZ, Zhu XM, Chen L. LKB1 inhibits the proliferation of gastric cancer cells by suppressing the nuclear translocation of Yap and β-catenin. Int J Mol Med. 2016;37:1039–1048. doi: 10.3892/ijmm.2016.2494. [DOI] [PubMed] [Google Scholar]
- 10.You X, Ma M, Hou G, Hu Y, Shi X. Gene expression and prognosis of NOX family members in gastric cancer. Onco Targets Ther. 2018;11:3065–3074. doi: 10.2147/OTT.S161287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han M, Ma L, Qu Y, Tang Y. Decreased expression of the ATM gene linked to poor prognosis for gastric cancer of different nationalities in Xinjiang. Pathol Res Pract. 2017;213:908–914. doi: 10.1016/j.prp.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Feng L, Yao C, Li P, Feng Y, Wang F, Liu YF, Guo YB, Mao QS, Xue WJ. Low expression of fibulin-1 correlates with unfavorable prognosis in gastric cancer. Tumor Biol. 2016;37:9399–9410. doi: 10.1007/s13277-015-4537-1. [DOI] [PubMed] [Google Scholar]
- 13.Baĭramov RB, Abdullaeva RT. The impact of early gastric cancer diagnosis on indices of survival in patients after radical surgical intervention. Klin Khir. 2013:18–21. (In Russian) [PubMed] [Google Scholar]
- 14.Kim JW, Hwang I, Kim MJ, Jang SJ. Clinicopathological characteristics and predictive markers of early gastric cancer with recurrence. J Korean Med Sci. 2009;24:1158–1164. doi: 10.3346/jkms.2009.24.6.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Cai H, Wang Y. Prognostic significance of tumor markers in T4a gastric cancer. World J Surg Oncol. 2012;10:68. doi: 10.1186/1477-7819-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Yan Z, Zhang B, Rao Z, Zhang Y, Liu J, Yu L, Zhao Y, Yang B, Wu T, Gao J. Identification of a 5-gene signature for clinical and prognostic prediction in gastric cancer patients upon microarray data. Med Oncol. 2013;30:678. doi: 10.1007/s12032-013-0678-5. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Wei J, Wang H, Yue G, Yu L, Yang Y, Xie L, Zou Z, Qian X, Ding Y, et al. A three-gene signature as potential predictive biomarker for irinotecan sensitivity in gastric cancer. J Transl Med. 2013;11:73. doi: 10.1186/1479-5876-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Cai H, Wang X, Ma L. Discovery of signature genes in gastric cancer associated with prognosis. Neoplasma. 2016;63:239–245. doi: 10.4149/209_150531N303. [DOI] [PubMed] [Google Scholar]
- 21.Hou JY, Wang YG, Ma SJ, Yang BY, Li QP. Identification of a prognostic 5-Gene expression signature for gastric cancer. J Cancer Res Clin Oncol. 2017;143:619–629. doi: 10.1007/s00432-016-2324-z. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Chen G, Wang Q, Lu W, Xu M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget. 2017;8:73826–73836. doi: 10.18632/oncotarget.17764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Yu Q, Zhu R, Wang Y, Li J, Wang Q, Guo W, Fu S, Zhu L. A six-gene signature predicts clinical outcome of gastric adenocarcinoma. Comb Chem High Throughput Screen. 2018;21:444–452. doi: 10.2174/1871524918666180531085713. [DOI] [PubMed] [Google Scholar]
- 24.Deng X, Xiao Q, Liu F, Zheng C. A gene expression-based risk model reveals prognosis of gastric cancer. PeerJ. 2018;6:e4204. doi: 10.7717/peerj.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min L, Zhao Y, Zhu S, Qiu X, Cheng R, Xing J, Shao L, Guo S, Zhang S. Integrated analysis identifies molecular signatures and specific prognostic factors for different gastric cancer subtypes. Transl Oncol. 2017;10:99–107. doi: 10.1016/j.tranon.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Gao P, Sun J, Shi J. A six-gene prognostic predictor for patients with gastric cancer. Oncotarget. 2018;5 [Google Scholar]
- 27.Kim S, Lim DH, Lee J, Kang WK, MacDonald JS, Park CH, Park SH, Lee SH, Kim K, Park JO, et al. An observational study suggesting clinical benefit for adjuvant postoperative chemoradiation in a population of over 500 cases after gastric resection with D2 nodal dissection for adenocarcinoma of the stomach. Int J Radiat Oncol Biol Phys. 2005;63:1279–1285. doi: 10.1016/j.ijrobp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Yoo CH, Noh SH, Shin DW, Choi SH, Min JS. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236–242. doi: 10.1046/j.1365-2168.2000.01360.x. [DOI] [PubMed] [Google Scholar]
- 29.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, Park YS, Lim HY, Sohn TS, Bae JM, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One. 2014;9:e90133. doi: 10.1371/journal.pone.0090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Therneau TM. A Package for Survival Analysis in S. R package version 2.41–3. http://cran.nexr.com/web/packages/survival/index.html
- 32.Kouznetsov D, Trappmann H. Superfunctions and sqrt of factorial. Moscow Univ Phys Bull. 2010;65:6–12. doi: 10.3103/S0027134910010029. [DOI] [Google Scholar]
- 33.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn M. Building predictive models in R using the caret package. J Stat Software. 2008;28:1–26. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 36.Mavroforakis ME, Theodoridis S. Proceedings of the 2005 13th European Signal Processing Conference. IEEE; Antalya: 2013. Support Vector Machine (SVM) classification through geometry; pp. 1–4. [Google Scholar]
- 37.Meyer D. Support vector machines the interface to libsvm in package e1071. R News. 2013;1:1–3. [Google Scholar]
- 38.Schröder MS, Culhane AC, Quackenbush J, Haibe-Kains B. Survcomp: An R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalderstam J, Edén P, Bendahl PO, Strand C, Fernö M, Ohlsson M. Training artificial neural networks directly on the concordance index for censored data using genetic algorithms. Artif Intell Med. 2013;58:125–132. doi: 10.1016/j.artmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Ferro CAT, Fricker TE. A bias-corrected decomposition of the Brier score. Quarterly J Royal Meteorological Soc. 2012;138:1954–1960. doi: 10.1002/qj.1924. [DOI] [Google Scholar]
- 41.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripatti S, Palmgren J. Estimation of multivariate frailty models using penalized partial likelihood. Biometrics. 2000;56:1016–1022. doi: 10.1111/j.0006-341X.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- 43.Alexeyenko A, Lee W, Pernemalm M, Guegan J, Dessen P, Lazar V, Lehtiö J, Pawitan Y. Network enrichment analysis: Extension of gene-set enrichment analysis to gene networks. BMC Bioinformatics. 2012;13:226. doi: 10.1186/1471-2105-13-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim TY, Kim NK, Bang YJ. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095–7103. doi: 10.1038/sj.onc.1207932. [DOI] [PubMed] [Google Scholar]
- 45.Choi MC, Lee YU, Kim SH, Park JH, Kim HA, Oh DY, Im SA, Kim TY, Jong HS, Bang YJ. A-kinase anchoring protein 12 regulates the completion of cytokinesis. Biochem Biophys Res Commun. 2008;373:85–89. doi: 10.1016/j.bbrc.2008.05.184. [DOI] [PubMed] [Google Scholar]
- 46.Shimura T, Tanaka K, Saigusa S, Kondo S, Kitajima T, Toiyama Y, Okugawa Y, Inoue Y, Araki TH, Uchida K, et al. Abstract 1139: Prognostic value of angiopoietin-like protein 2 (ANGPTL2) at the tumor margins in patients with gastric cancer. Cancer Res. 2013;73:1139. [Google Scholar]
- 47.Sheng WZ, Chen YS, Tu CT, He J, Zhang B, Gao WD. ANGPTL2 expression in gastric cancer tissues and cells and its biological behavior. World J Gastroenterol. 2016;22:10364–10370. doi: 10.3748/wjg.v22.i47.10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimura T, Toiyama Y, Tanaka K, Saigusa S, Kitajima T, Kondo S, Okigami M, Yasuda H, Ohi M, Araki T, et al. Angiopoietin-like protein 2 as a predictor of early recurrence in patients after curative surgery for gastric cancer. Anticancer Res. 2015;35:4633–4639. [PubMed] [Google Scholar]
- 49.Tiberio P, Lozneanu L, Angeloni V, Cavadini E, Pinciroli P, Callari M, Carcangiu ML, Lorusso D, Raspagliesi F, Pala V, et al. Involvement of AF1q/MLLT11 in the progression of ovarian cancer. Oncotarget. 2017;8:23246–23264. doi: 10.18632/oncotarget.15574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J, Kim S, Joh J, Remick SC, Miller DM, Yan J, Kanaan Z, Chao JH, Krem MM, Basu SK, et al. MLLT11/AF1q boosts oncogenic STAT3 activity throughSrc-PDGFR tyrosine kinase signaling. Oncotarget. 2016;7:43960–43973. doi: 10.18632/oncotarget.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlsson E, Ranki A, Sipilä L, Karenko L, Abdel-Rahman WM, Ovaska K, Siggberg L, Aapola U, Ässämäki R, Häyry V, et al. Potential role of a navigator gene NAV3 in colorectal cancer. Br J Cancer. 2012;106:517–524. doi: 10.1038/bjc.2011.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Gao X, Ji K, Sanders AJ, Zhang Z, Jiang WG, Ji J, Ye L. Differential expression of CCN family members CYR611, CTGF and NOV in gastric cancer and their association with disease progression. Oncol Rep. 2016;36:2517–2525. doi: 10.3892/or.2016.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu H, Li C, Cai S, Zhu C, Tian Y, Zheng J, Hu J, Chen C, Liu W. Increased expression of pleiotrophin is a prognostic marker for patients with gastric cancer. Hepatogastroenterology. 2014;61:1478–1482. [PubMed] [Google Scholar]
- 54.Gu Y, Wang Q, Guo K, Qin W, Liao W, Wang S, Ding Y, Lin J. TUSC3 promotes colorectal cancer progression and epithelial-mesenchymal transition (EMT) through WNT/β-catenin and MAPK signalling. J Pathol. 2016;239:60–71. doi: 10.1002/path.4697. [DOI] [PubMed] [Google Scholar]
- 55.Fan X, Zhang X, Shen J, Zhao H, Yu X, Chen Y, Zhuang Z, Deng X, Feng H, Wang Y, Peng L. Decreased TUSC3 promotes pancreatic cancer proliferation, invasion and metastasis. PLoS One. 2016;11:e0149028. doi: 10.1371/journal.pone.0149028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vašíčková K, Horak P, Vaňhara P. TUSC3: Functional duality of a cancer gene. Cell Mol Life Sci. 2017;75:849–857. doi: 10.1007/s00018-017-2660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vedeld HM, Andresen K, Eilertsen IA, Nesbakken A, Seruca R, Gladhaug IP, Thiis-Evensen E, Rognum TO, Boberg KM, Lind GE. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Louis SF, Zahradka P. Vascular smooth muscle cell motility: From migration to invasion. Exp Clin Cardiol. 2010;15:e75–e85. [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.