ABSTRACT
Extracellular vesicles (EVs) have sparked tremendous interest owing to their prominent potential in diagnostics and therapeutics. Isolation of EVs from complex biological fluids with high purity is essential to the accurate analysis of EV cargo. Unfortunately, generally used isolation techniques do not offer good separation of EVs from non-EV contaminants. Hence, it is important to have a standardized method to characterise the properties of EV preparations, including size distribution, particle concentration, purity and phenotype. Employing a laboratory-built nano-flow cytometer (nFCM) that enables multiparameter analysis of single EVs as small as 40 nm, here we report a new benchmark to the quality and efficiency assessment of EVs isolated from plasma, one of the most difficult body fluids to work with. The performance of five widely used commercial isolation kits was examined and compared with the traditional differential ultracentrifugation (UC). Two to four orders of magnitude higher particle concentrations were observed for EV preparations from platelet-free plasma (PFP) by kits when compared with the EV preparation by UC, yet the purity was much lower. Meanwhile, the particle size distribution profiles of EV preparations by kits closely resembled those of PFP whereas the EV preparation by UC showed a broader size distribution at relatively large particle size. When these kits were used to isolate EVs from vesicle-depleted PFP (VD-PFP), comparable particle counts were obtained with their corresponding EV preparations from PFP, which confirmed again the isolation of a large quantity of non-vesicular contaminants. As CD9, CD63 and CD81 also exist in the plasma matrix, single-particle phenotyping of EVs offers distinct advantage in the validation of EVs compared with ensemble-averaged approaches, such as Western blot analysis. nFCM allows us to compare different isolation techniques without prejudice.
KEYWORDS: Extracellular vesicles, exosomes, nano-flow cytometry, isolation methods, platelet free plasma
Introduction
Extracellular vesicles (EVs) are nano-sized lipid bilayer vesicles (40–1000 nm in diameter) released by their cells of origin to mediate intercellular communication via delivering cargo molecules (nucleic acids, proteins, lipids, etc.) to recipient cells [1,2]. EVs are prevalent in biological fluids and recent studies have shown their promising roles in disease diagnosis and therapeutics [3,4]. Because of the abundant presence of interfering non-vesicular components such as proteins, cell debris and other particles in body fluids and cell culture media, high purity separation of EVs is a prerequisite of proteomic, genomic and lipidomic analyses for fundamental research and biomarker discovery [5–8]. Unfortunately, effective and selective separation of high purity EVs from biological fluids remains a significant challenge owning to their nanoscale size and large population heterogeneity [9,10]. The International Society for EVs has emphasized the urgent need for standardized methods in EV isolation and quality assessment [11–14].
Differential ultracentrifugation (UC) has been a classical method for EV separation, at least until recently [15,16], yet it is time-consuming, labour-intensive, and of limited accessibility. To address the obstacles in routine EV extraction, numerous separation techniques based on different principles have been applied for the purification of EVs from biological fluids, such as polymer-based precipitation [17,18], size exclusion chromatography (SEC) [19,20], ultrafiltration (UF) [21], flow field-flow fractionation [22], immunoaffinity capture [23,24], microchip-based techniques [25–27] and combinations of these techniques [28,29]. Recently, a number of commercial kits are made available and have been widely reported in the literature for EV isolation. For example, ExoQuick (System Biosciences) and Total Exosome Isolation kits (TEI, Invitrogen) rely on polymer precipitation; qEV (Izon) is an SEC column; ultrafiltration (UF, Millipore) uses centrifugal filter devices; and exoEasy (Qiagen) builds upon membrane-based affinity binding [19,29–32]. Although these kits are less time consuming, more compatible with limited volumes of biofluids, and do not require special equipment, their suitability for scientific and clinical applications is doubtful due to the uncertain quality of EV preparations [33]. This is particularly true for plasma or serum samples as there exists a considerable overlap in both the particle size and density between EVs and lipoproteins, which normally result in unintentional coisolation of these two different entities [34,35].
Several studies have attempted to compare the isolation efficiency of various techniques for EV isolation from blood samples [36–40]. This is frequently done by measuring the physical properties (size and concentration) using nanoparticle tracking analysis (NTA) or tunable resistive pulse sensing (TRPS). However, the minimum detectable EV sizes are 70–90 nm for NTA and 70–100 nm for TRPS, respectively [41]. Considering that EVs smaller than 70 nm represents a large population of EVs [42–44], NTA and TRPS can hardly reveal the full picture of all sizes of EVs. Moreover, these two methods are not suitable for measuring samples of a very broad or polydispere size distribution, which is the case for EVs isolated from biofluids. Besides physical properties, the purity and biological features of EV preparations are important factors that significantly affect the accuracy of downstream biochemical profiling of EVs [45,46]. However, these properties can hardly be characterized by ensemble-averaged methods, as they cannot establish whether nucleic acids, proteins, lipids, etc. are specifically associated with EVs or reside instead in the solution phase or other non-vesicular contaminants.
Over the past dozen years, our laboratory has been working on the development of nano-flow cytometry (nFCM) and has achieved single-particle light-scattering detection of viruses, silica nanoparticles (SiNPs) and gold nanoparticles down to 27, 24 and 7 nm size, respectively [47,48]. With the incorporation of multiparameter fluorescence detection, we recently reported sizing and protein profiling of individual EVs as small as 40 nm [43]. With comparable resolution to that of electron microscopy and the phenotyping capability of flow cytometry, here we aim to develop a standardized method to assess the quality and efficiency of EV separation by different methods. Properties such as size distribution, particle concentration, purity, recovery rate and surface proteins were measured. Human plasma was used as the model system, and the performance of several commercial isolation kits were compared with the classical differential UC method.
Materials and methods
Preparation of PFP
Peripheral blood was drawn from a healthy volunteer who had fasted for at least 12 h at the First Affiliated Hospital of Xiamen University. Informed written consent was obtained from the healthy volunteer and the collection of human blood samples was approved by the Ethics Committee of the First Affiliated Hospital of Xiamen University. Briefly, 2.7 mL blood was collected into BD Vacutainer tubes (363095) containing 0.3 mL of 0.109 M sodium citrate by using 21-gauge needles. After collection, tubes were inverted 4–5 times immediately for proper mixing with anticoagulant. Tubes were transported vertically at room temperature without agitation. And then, the blood samples were centrifuged twice at 2,500 × g for 15 min at room temperature to extract PFP within 2 h of blood collection. PFP was aliquoted and stored at −80°C until further use, and freeze-thawing was avoided as much as possible after that. Vesicle-depleted PFP (VD-PFP) was prepared by ultracentrifuging PFP without dilution at 100,000 × g for 18 h at 4°C (Beckman Coulter X-90 centrifuge; SW 41 Ti rotor), and the supernatant was recovered as VD-PFP [15].
Cell culture
Human colorectal cancer cell line (HCT15) was purchased from American Type Culture Collection (ATCC). HCT15 cells were cultured in RPMI 1640 medium (Gibco). All media were supplemented with 10% FBS and penicillin-streptomycin (Invitrogen). The FBS used above was depleted of EVs by UC at 100,000 × g for 18 h at 4°C (Beckman Coulter X-90 centrifuge, SW41 Ti rotor). For EV isolation, cells were grown in EV-depleted medium until they reached a confluency of ~90% (after approximately 24 h). The conditioned cell culture medium was collected and centrifuged at 800 × g for 5 min at 4°C to pellet the cells. The supernatant was centrifuged at 2,000 × g for 10 min at 4°C to remove cellular debris, and is called conditioned cell culture medium (CCCM) in present study.
Isolation of EVs
EVs were isolated from thawed PFP by six isolation methods including five commercially available isolation kits and the conventional differential UC (Table 1). Manufacturer’s instructions were followed for each kit. EVs were also purified from CCCM by differential UC.
Table 1.
Comparison of EV isolation methods.
| Principle of EV isolation | Method | Abbr. | Particle conc. from PFP (particles/mL) | Median size (nm) | Purity (%) | Particle conc. from VD-PFP (particles/mL) | (%) |
|---|---|---|---|---|---|---|---|
| PFP | (1.4 ± 0.1) × 1013 | 56 | |||||
| VD-PFP | (1.3 ± 0.1) × 1013 | ||||||
| Sedimentation | Differential ultracentrifugation | UC | (4.9 ± 0.2) × 108 | 81 | 78.2 ± 0.6 | (1.0 ± 0.2) × 107 | 2.0 |
| Polymer precipitation | ExoQuick (System Biosciences) | ExoQuick | (8.6 ± 0.2) × 1012 | 56 | 5.3 ± 2.6 | (8.5 ± 0.1) × 1012 | 98.8 |
| Polymer precipitation | Total Exosome Isolation (Invitrogen) | TEI | (5.5 ± 0.2) × 1012 | 58 | 18.5 ± 1.5 | (4.6 ± 0.1) × 1012 | 83.6 |
| Size exclusion chromatography | qEV Columns (Izon Science) | qEV | (1.4 ± 0.1) × 1011 | 64 | 28.1 ± 0.8 | (9.2 ± 0.3) × 1010 | 65.7 |
| Membrane filtration | Ultrafiltration (Millipore) | UF | (3.2 ± 0.1) × 1012 | 56 | 11.4 ± 0.7 | (2.9 ± 0.2) × 1012 | 90.6 |
| Membrane affinity | exoEasy (Qiagen) | exoEasy | (8.9 ± 0.2) × 1010 | 64 | N.A. | (8.2 ± 0.1) × 1010 | 92.1 |
Differential UC
Freshly prepared CCCM of HCT15 cells (12.5 mL) or thawed PFP (2.0 mL) diluted to 12.5 mL with PBS were centrifuged at 100,000 × g for 2 h at 4°C in a Beckman Coulter XE-90K Ultracentrifuge using an SW 41 Ti rotor. The pellet was washed with 12.5 mL of PBS and followed by a second UC at 100,000 × g for 2 h at 4°C. Afterwards, the supernatant was discarded and EVs were resuspended in 50–100 μL PBS.
ExoQuick isolation kit (ExoQuick)
ExoQuick PLUS isolation kit (System Biosciences, EXOQ5TM-1) was used in present study. Briefly, 4 μL of thrombin (611 U/mL) was added into 500 μL PFP to a final concentration of 5 U/mL. Then the sample was incubated at room temperature for 5 min while gently flicking the tube. After thrombin treatment, the sample was centrifuged at 10,000 × g for 5 min at room temperature and the supernatant was transferred to a clean tube. It was stated by the manufacturer that ‘When isolating exosomes from plasma, fibrinogen and fibrin can impede efficient recovery. By pre-treating plasma with thrombin, the fibrinogen can be converted to fibrin and easily pre-cleared by a simple centrifugation step’. Then 126 μL of ExoQuick exosome precipitation solution was added into thrombin-treated PFP and the mixture was incubated for 30 min at 4°C. Afterwards, the sample was centrifuged at 1,500 × g for 30 min room temperature. The supernatant was removed carefully and the pellet was resuspended in PBS.
Total exosome isolation kit (TEI)
Total Exosome Isolation (from plasma) reagent (Invitrogen, 4484450) was used. Briefly, thawed PFP was centrifuged at 10,000 × g for 20 min at room temperature to remove debris. Then 100 μL supernatant was transferred into a clean tube and 50 μL PBS was added into the plasma. The sample was mixed thoroughly by vortexing. 5 μL proteinase K was added into the sample. The sample was vortexed and incubated at 37°C for 10 min. Then 30 μL Exosome Precipitation Reagent was added to the proteinase K-treated PFP. After vortexing, the mixture was incubated at 4°C for 30 min, then centrifuged at 10,000 × g for 5 min at room temperature. The supernatant was carefully discarded and the pellet was resuspended in PBS.
qEV column (qEV)
The qEV columns (Izon, 1000871) were equilibrated with at least 10 mL PBS before using. Then 500 μL plasma was pipetted onto the column, and fractions were immediately collected with a volume of 500 μL into each tube. PBS was used to elute EVs during the purification process. The first six fractions were discarded because they do not contain EVs. The 7th–9th fractions were combined as the EV preparation for downstream analysis. Prior to the Western blot and transmission electron microscopy analyses, a fraction of this EV preparation was concentrated by UF as described below.
Ultrafiltration (UF)
Amicon® ultra-0.5 gentrifugal filter devices (Millipore, Amicon® Ultra 100 K device) were used here to purify and concentrate EVs from PFP. Briefly, 25 μL PFP was diluted to 500 μL with PBS. The diluted plasma was added into the Amicon® ultra-0.5 devices rinsed with PBS before using. The sample was concentrated from 500 μL to 50 μL by centrifugation at 14,000 × g for 10 min at 4°C. Then 450 μL PBS was added into the 50 μL retentate and followed by a second UC at 14,000 × g for 10 min at 4°C. To recover the concentrate, the Amicon® ultra-0.5 device was turned upside down in a clean microcentrifuge tube and centrifuged at 1,000 × g for 5 min to transfer the concentrated sample from the device to the tube.
exoEasy Maxi kit (exoEasy)
The exoEasy Maxi kit (Qiagen, 76064) was used according to the quick-start protocol. Briefly, the PFP was filtered using a 0.8-μm pore filter (Millipore Millex-AA, SLAA033SB) to exclude large particles. Equivalent volume XBP buffer was added to 500 μL PFP and the sample was mixed well by gently inverting the tube 5 times. The mixture was added into the exoEasy spin column to centrifuge at 500 × g for 1 min at room temperature, and the flow-through was discarded. Then 10 mL buffer XWP was added and the column was centrifuged at 5,000 × g for 5 min to remove residual buffer from the column. After that, the column was transferred to a fresh collection tube. Then, 400 μL XE buffer was added to the column membrane with an incubation for 1 min. The column was centrifuged at 500 × g for 5 min to collect the eluate.
nFCM analysis
The laboratory-built nFCM reported before was used in the present study, and Figure 1(a) shows the schematic diagram of the instrument design [43,47]. Briefly, two single-photon counting avalanche photodiodes (APDs) were used for the simultaneous detection of the side scatter (SSC) (bandpass filter: FF01 − 524/24) and orange fluorescence (bandpass filter: FF01 − 579/34) of individual EVs, respectively. Unless stated otherwise, each distribution histogram or dot-plot was derived from data collected 1 min unless stated otherwise. Ultrapure water supplied by an ultrapure water system (PURELAB Ultra FLC00006307, ELGA) served as the sheath fluid via gravity feed.
Figure 1.
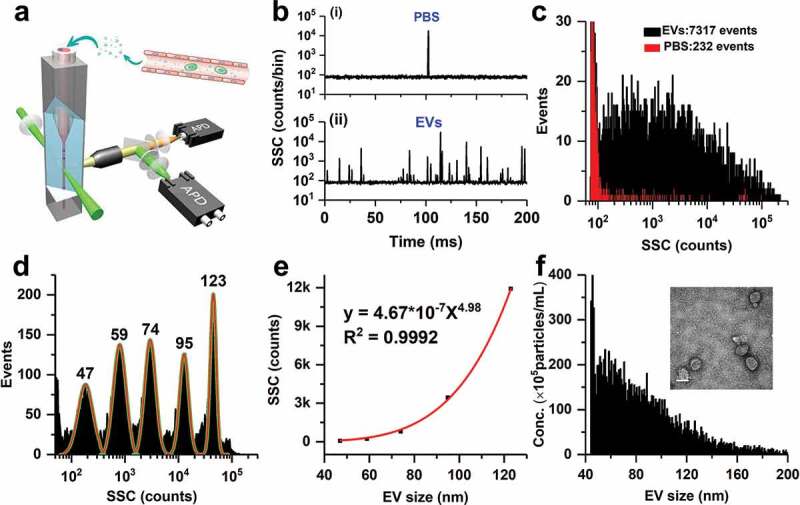
Principle of particle size (diameter) distribution and concentration measurement for EV preparations by nFCM. (a) Schematic diagram of the Laboratory-built nFCM; (b) Representative SSC burst traces of PBS (i) and EV preparation from PFP by UC (ii); (c) SSC distribution histograms of PBS (red) and EVs (black) derived from data collected over 2 min each; (d) SSC distribution histogram of a mixture of monodisperse SiNPs of five different diameters ranging between 47 and 123 nm, and fit to a sum of Gaussian peaks; (e) Plot of the Gaussian-fitted SSC intensity (after refractive index correction) as a function of EV particle size; (f) Histogram of particle size with a bin width of 1 nm for the EV sample (n = 7317) along with a representative TEM micrograph (inset, scale bar: 100 nm).
For nFCM analysis, the sample stream is completely illuminated within the central region of the focused laser beam, and the detection efficiency is approximately 100%, which leads to accurate particle concentration measurement via single particle enumeration [49,50]. The concentration of each EV sample was determined by employing 100 nm orange FluoSpheres of known particle concentration to calibrate the sample flow rate. Several dilutions were made to the orange FluoSpheres solution, and a linear correlation between particle concentration and detected event rate was obtained with R2 of 0.999 (data not shown). Regarding immunofluorescent staining, the following antibodies were purchased from BD Biosciences: PE-conjugated mouse anti-human CD9 antibody (clone M-L13), PE-conjugated mouse anti-human CD63 antibody (clone H5C6), PE-conjugated mouse anti-human CD81 antibody (clone JS-81), PE-conjugated mouse anti-human CD235a (clone GA-R2), PE-conjugated mouse anti-human CD45 (clone HI30), PE-conjugated mouse anti-human CD41a (clone HIP8), PE-conjugated mouse anti-human CD144 (clone 55-7H1), PE-conjugated mouse IgG1, κ (clone MOCP-21), and PE-conjugated mouse IgG2b, κ (clone 27–35). For phenotyping of EVs derived by HCT15 cells, EVs were isolated from 1 mL CCCM diluted to 12.5 mL with PBS by centrifugation once at 100,000 × g for 2 h at 4°C (Beckman Coulter X-90 centrifuge, SW 41 Ti rotor), and the pellet was resuspended in 50 μL PBS. EV preparation from PFP by UC or commercial kit was divided into 50 μL with a particle concentration of 6 × 108 particles/mL. Into each 50 μL EV sample, 20 μL of PE-conjugated antibody against CD9, CD63, CD81, CD235a, CD45, CD41a, CD144, IgG1 or IgG2b was added. The mixture was incubated at 37°C for 30 min and then washed twice with 1 mL PBS by UC at 100,000 × g for 17 min at 4°C (Beckman Coulter MAX-XP centrifuge, TLA-120.2 rotor). The pellet was resuspended in 50 μL PBS for nFCM analysis. For fluorescent labelling of phosphatidylserine (PS), all the other experimental procedures were the same as immunofluorescent staining except for replacing 20 μL PE-conjugated antibody with 5 μL PE-conjugated Annexin V (BD Bioscience). Meanwhile, the PBS buffer was replaced with 1 × annexin-binding buffer (Invitrogen, V13241) starting from EV preparation.
Light scattering correction for the refractive index difference between SiNPs and EVs
Monodisperse SiNPs of five different sizes were used as the size reference standard to calibrate the size of EVs. Because the refractive index of SiNPs (1.461) is slightly different from that of EVs (1.40), the intensity ratio between light scattered by a SiNP to that of an EV of the same particle size was calculated based on the Mie theory for every size of the SiNP standard. These ratios were used as the correction factors to derive the calibration curve between the scatted light intensity and particle size of EVs from the correlation between the SSC intensity and particle size of SiNPs [43]. Note that in reality there is refractive index heterogeneity in the EV population based on size and composition differences, and 1.40 used here is an approximation. Simulation of side-scattered light detected by the nFCM system was performed with MiePlot, a computer program for scattering of light from a sphere using Mie theory and the Debye series [51]. For more detailed calculation, one can refer to the article by Zhang et al. [52].
Transmission electron microscopy (TEM)
A 3-μL aliquot of the EV preparation with a concentration around 1011 particles/mL was placed on a formvar-/carbon-coated grid and allowed to settle for 2 min. The sample was negative-stained with 2% phosphotungstic acid for 1 min. Grids were imaged with a Tecnai G2 Spirit BioTwin transmission electron microscope operating at 120 kV.
Western blot analysis
The protein concentration of PFP, VD-PFP, or EV preparation was measured using Qubit 4.0 protein assay kit (Thermo Scientific, Q33211, which is designed specifically for use with the Qubit Fluorometer to quantitate samples ranging from 12.5 µg⁄mL to 5 mg⁄mL with 1 and 20 µL of sample volume). For each sample, the protein concentration was adjusted to 10 μg/20 μL and 10 μg protein was loaded onto a 12% polyacrylamide gel. Following electrophoresis, the proteins were transferred from gel onto a polyvinylidene fluoride membrane (PVDF, Millipore) by using a Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked with 5% non-fat dry milk in TBST for 30 min at room temperature and incubated with primary antibodies overnight at 4°C. Following incubation with horseradish peroxidase-conjugated secondary antibody, the blot was developed with chemiluminescent reagents from Advansta. Images were captured on an Amersham Imager 600 (GE Healthcare Life Sciences). The following antibodies used for immunoblotting were purchased from Abcam: rabbit monoclonal anti-human CD9 antibody (clone EPR2949, dilution 1:2000), rabbit monoclonal anti-human CD63 antibody (clone EPR5702, dilution 1:1000), rabbit monoclonal anti-human CD81 antibody (clone EPR4244, dilution 1:500), rabbit monoclonal anti-human ApoB (clone EPR2914, dilution 1:10,000), and the secondary antibodies: horseradish peroxidase-labelled goat anti-rabbit.
Results
Particle size and concentration analysis of EV preparations from plasma
The nFCM measures the particle size and concentration via light scattering detection of single particles at a throughput up to 10,000 particles per minute. Figure 1(b) (i) and (ii) show the representative SSC burst trace of PBS filtered through a 220-nm filter and an EV preparation from PFP by UC, respectively. Figure 1(c) depicts the SSC burst area distribution histograms for PBS (232 events) and EVs (7317 events). A mixture of five different sizes (47, 59, 74, 95 and 123 nm) of monodisperse SiNPs was analysed on the nFCM, and the SSC burst area distribution histogram is shown in Figure 1(d). When the centroids of the SSC intensity obtained from the fitted Gaussian curves were corrected for each size population of the SiNPs, a calibration curve of the scattered light intensity and the particle size of EVs was obtained, with a size dependence of 4.98th order (Figure 1(e)). Then, the SSC distribution histogram of the EV sample (Figure 1(c)) was converted to particle size distribution, and the bin width was set at 1 nm (Figure 1(f)). Meanwhile, the event rate of EVs counted within 1 nm for every size was converted to particle concentration by using the 100 nm orange FluoSpheres of known particle concentration to calibrate the sample flow rate. A typical TEM image of EVs isolated from PFP by UC is shown as the inset of Figure 1(f). By using EVs isolated from cell culture supernatant as an example, we demonstrated before that the size distribution profile measured by nFCM closely resembled those obtained by cryo-TEM [43]. Although large size EVs up to 1000 nm have been observed by cryo-TEM [53], Figure 1(f) indicates that most EVs in preparation from PFP by UC fall in the size range of 40–200 nm. Note that if there exists particles of large size that saturate the APD detector, these particles can also be accurately counted by nFCM (Figure S1).
EVs were separated from PFP of a single healthy donor using five different commercial kits in parallel with UC (Table 1). The original PFP sample and EV preparations by different kits were analysed by nFCM and TEM. Of note, appropriate dilutions were made to the PFP and EV preparations prior to the nFCM analysis to make the final particle concentration fall in the range of 108–109 particles/mL to avoid coincidence of two particles inside the probe volume. For example, 10,000-fold dilution was made to the PFP sample due to its high particle concentration. The original particle concentrations of PFP and EV preparations were calculated based on the measured concentrations of sample diluents and the dilution factors. Figure 2(a–g) show the particle size distribution histograms along with the representative TEM micrographs (insert and Figure S2) for PFP and EV preparations by different isolation methods, respectively. Figure 2(h) depicts the particle concentration of PFP and EV preparations. The particle concentration of PFP was measured to be 1.4 × 1013 particles/mL whereas the particle concentration of EVs isolated by UC was only 4.8 × 108 particles/mL, suggesting that EVs were less than 0.0035% of the total particles with diameter larger than 40 nm in plasma. Nonetheless, it is worth noting that if there exists a substantial loss of EVs during UC isolation, the particle number of the EV proportion in PFP could be severely underestimated. Compared with the narrow size distribution of PFP particles, with a median size of 56 nm, the particle size distribution of EVs isolated by UC was much broader, with a continuous profile ranging from 40 to 200 nm in diameter, and the median size was measured to be 81 nm. Meanwhile, the ‘cup-shaped EVs’ are clearly seen in the TEM micrograph for EVs isolated by UC. The measured particle concentrations for EV preparations by commercial kits were 2–4 orders of magnitude higher than that of UC, ranging from 8.5 × 1012 particles/mL for ExoQuick to 8.9 × 1010 particles/mL for exoEasy. Moreover, their size distribution profiles closely resembled those of the source PFP: very narrow, with the majority of particles smaller than 80 nm in diameter. Meanwhile, dense protein particulates are evident in their TEM micrographs. For EV preparation by exoEasy, a population with increased diameter, centred around 185 nm, was observed, suggesting aggregation or fusion occurred during the isolation process. It is worth noting that no matter how we tried, it was very difficult to obtained good TEM images for EVs isolated by exoEasy, which could be ascribed to the proprietary reagents used in the kit.
Figure 2.
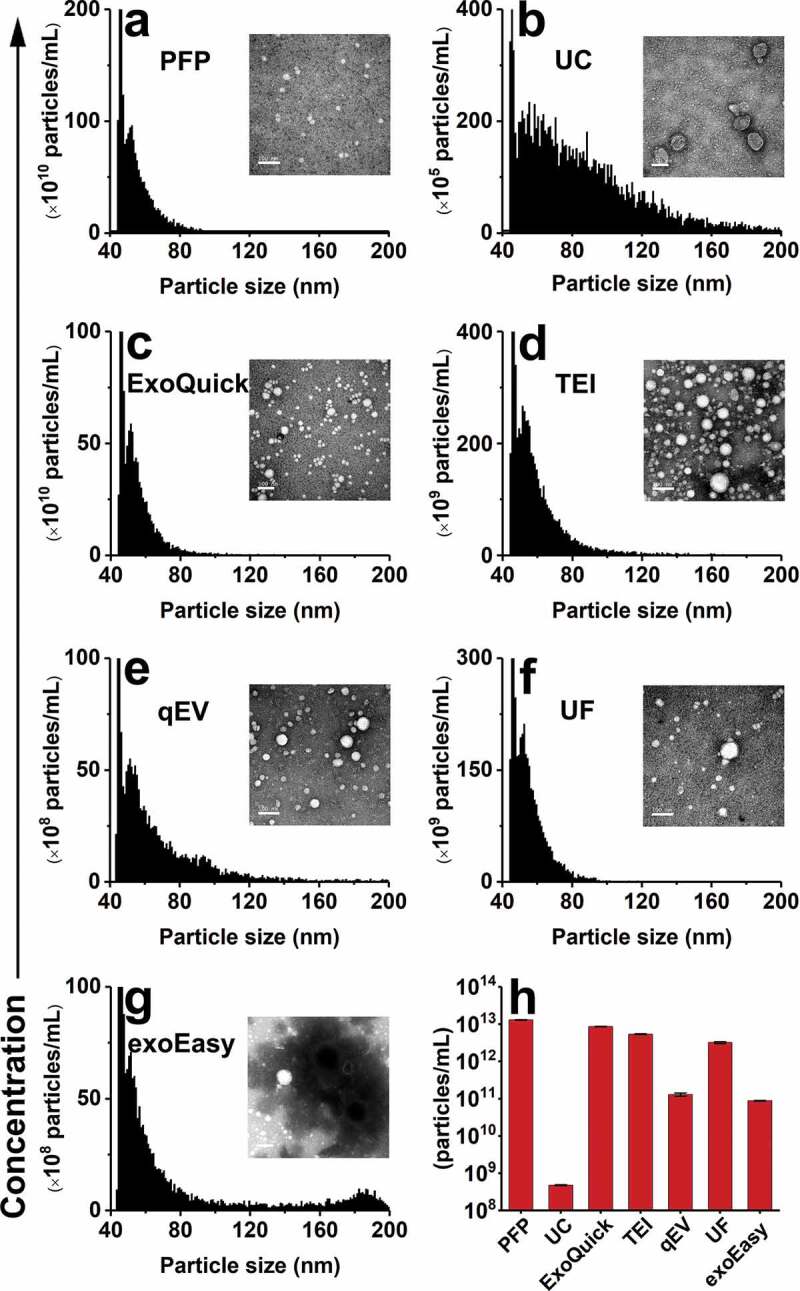
nFCM measurement of the particle size (diameter) distribution and concentration of EV preparations from PFP by different isolation methods. (a) Particle size distribution histogram of PFP and the typical TEM micrograph (scale bar: 100 nm). (b-g) Particle size distribution histograms of EV preparations from PFP by UC, ExoQuick, TEI, qEV, UF and exoEasy, respectively. (h) Particle concentrations measured for PFP and its EV preparations. The error bars represent the standard deviation (s. d.) of three repetitive experiments for each isolation methods. Note: Because the particle concentration of EV preparation by different isolation methods varies largely, the Y-axes of panels a-g are plotted in different scale.
Purity assessment of EV preparations from plasma
Blood plasma EVs are unavoidably co-isolated with a complex assortment of non-vesicular materials such as protein aggregates and lipoproteins. EVs are membrane vesicles that are susceptible to be lysed by detergents, while non-vesicular contaminants may remain largely unchanged [54,55]. To assess the purity of EV preparations, we proposed to use non-ionic surfactant Triton X-100 to lyse the phospholipid bilayer of EVs, and then measure the particle counts before and after Triton X-100 treatment [43]. The EV preparations by UC and five commercial kits were diluted to particle concentration around 2 × 1010 particles/mL. To 45 μL of the EV diluent, 5 μL of 10% Triton X-100 was added to make the final Triton X-100 concentration of 1%. After 1 hr incubation on ice, the treated EV sample was diluted 20-fold prior to the nFCM analysis. Figure 3(a) (i) and (ii) depict the representative SSC burst traces of EVs isolated from PFP by UC prior to and after 1% Triton X-100 treatment. A remarkable reduction in event rate was observed upon treatment. The SSC burst area distribution histograms before (5062 events) and after (1124 events) Triton X-100 treatment are plotted in Figure 3(b,c). Note that the event rate of PBS measured at the same experimental condition has been deducted for each EV sample. The purity of EVs is defined as P = 1 – C2/C1, of which C1 and C2 denote the particle number measured in 1 min before and after Triton X-100 treatment, respectively. Using this approach, the purity of EVs isolated from PFP by UC was measured to be 77.8%, which is easy to understand due to the abundant presence of lipoprotein particles in plasma. To verify the accuracy of the as-proposed purity assessment approach, the effect of Triton X-100 treatment on lipoproteins needs to be studied as they can be co-isolated with EVs.
Figure 3.
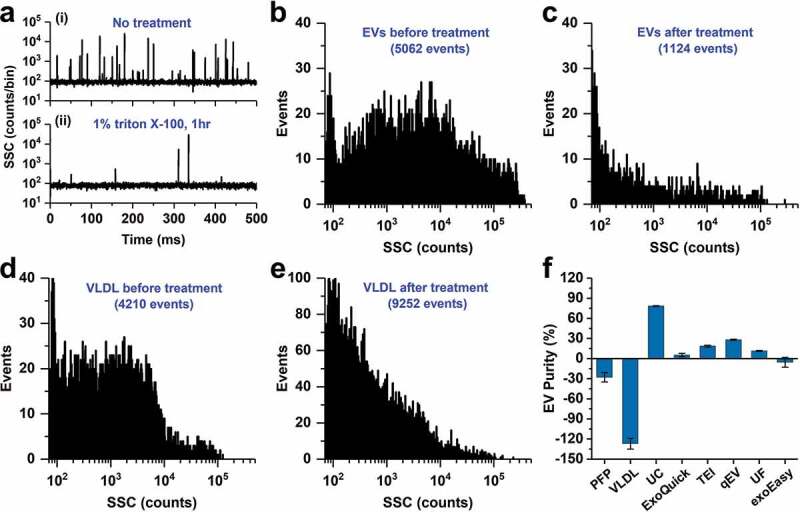
Purity assessment of EV preparations from PFP by UC and five commercial isolated kits and the effect of Triton X-100 treatment on PFP and VLDL. (a) Representative SSC burst traces of EV preparation from PFP by UC before (i) and after (ii) 1% Triton X-100 treatment for 1 h on ice; (b and c) SSC distribution histograms of EV preparation from PFP by UC before (b) and after (c) Triton X-100 treatment. (d and e) SSC distribution histograms of VLDL before (d) and after (e) Triton X-100 treatment. (f) Purity measurement for EVs in PFP, VLDL and preparations from PFP by UC and five commercial isolation kits (n = 3, mean ± s.d.).
Lipoprotein particles in the peripheral blood plasma include high-density lipoprotein (HDL, 5–12 nm, 1.063–1.210 g/cm3), low-density lipoprotein (LDL, 18–25 nm, 1.019–1.063 g/cm3), intermediate-density lipoprotein (IDL, 25–35 nm, 1.006–1.019 g/cm3), very low-density lipoproteins (VLDL, 30–80 nm, 0.930–1.006 g/cm3) and chylomicrons (75–1200 nm, <0.930 g/cm3) [56]. The density of EVs has been reported to be 1.110–1.190 g/cm3 [56] or 1.08–1.21 g/cm3 [35], and we can see from Figure 1(f) that the measured particle size of EVs mainly fall in the range of 40–200 nm. Therefore, EVs significantly overlap with VLDL and chylomicrons in particle size and with HDL in particle density. For blood drawn after overnight-fasting, the quantity of chylomicrons can be significantly minimized. As the particle size of HDL is too small to be detected by nFCM, we think the measured impurity particles for EV preparations from PFP by UC were mainly contributed by VLDL. So, the same Triton X-100 treatment was conducted for VLDL as for the EV preparation. Purified VLDL (American Research Products, 12–4020, purity > 95%, 1.48 mg/mL) was diluted 100-fold with PBS and filtered through a 0.22-μm filter. The particle concentration of VLDL was adjusted to 2 × 1010 particles/mL before the Triton X-100 treatment. The VLDL samples before and after treatment were diluted 20-fold and analysed on the nFCM. We can see from the SSC burst area distribution histograms (Figure 3(d,e)) that upon Triton X-100 treatment, the numbers of medium- to large-size particles decreased along with an increase of small-size particles. As one medium- to large-size VLDL could break up into several small-size particles upon treatment, the total particle numbers measured in 1 min increased from 4210 to 9252. Therefore, the real purity of EV preparation from PFP by UC is actually higher than what measured by the as-proposed method, i.e. 77.8%.
The effect of Triton X-100 treatment on PFP was also investigated and a slight increase in particle number was observed. Using the same approach, the purities of EVs isolated from PFP by five commercial isolations kits were measured and the results are plotted in Figure 3(f). Upon three replicate measurements for each isolation method, the average purities of EV preparations by UC, ExoQuick, TEI, qEV and UF were 78.2, 5.3, 18.5, 28.1 and 11.4%, respectively. It is interesting to note that for EV preparation by exoEasy, the particle concentration after Triton X-100 treatment increased slightly after Triton X-100 treatment, which implicates that some of the relatively large-sized aggregates formed during the isolation process (Figure 2(g)) were broken apart upon Triton X-100 treatment. However, it is important to note that caution needs to be taken when using number-only measurements to assess the purity of an EV preparation. If there exist lipoprotein particles or other impurity particles that can break up into small-size particles rather than being fully lysed upon detergent treatment, especially for the case when EVs are isolated from PFP by commercial kits, the measured purity is lower than they actually are. Moreover, because PFP samples can differ both in the number of EVs and the number and types of lipoprotein particles, it would be tricky to compare the purity of EV preparations generated from different PFP samples.
Efficacy and recovery of various isolation methods for pure EVs
The very high particle concentrations of EV preparations from plasma sample by commercial kits raised our curiosity about the recovery rate of these isolation methods applied on an EV preparation isolated by UC from cell culture supernatant. EVs isolated from CCCM by UC (purity of 88.5%) was used as the source material and particle concentration was adjusted to 1011/mL. For ExoQuick and TEI kits, as recommended by the manufactures, thrombin or proteinase K treatment was only conducted for EV isolation from PFP and not for EV isolation from pure EVs. Figure 4 (a–g) shows the particle size distribution histograms of purified EVs (a) and their EV preparations by different isolation methods (b–g) along with the representative TEM micrographs. After another UC step at 100,000 × g for 2 h at 4°C, no obvious change was observed for the particle size distribution profile of isolated EVs (Figure 4(b)) when compared to the source material (EVs purified from CCCM by UC via carrying out twice UC at 100,000 × g for 2 h at 4°C) (Figure 4(a)). Note that we did not observe noticeable aggregation or fusion of EVs induced by high-speed UC, of which cryo-EM reveals that EV aggregates have an overall size ranging from about 500 nm to several micrometers [57]. Compared to EM analysis, directly measuring EVs in liquid suspension by nFCM can more likely reflect the real situation of EVs dispersed in solution. Meanwhile, the data is more quantitative and statistically representative. For EVs isolated by different methods, the recovery rate of EVs was defined as R = Ni/N, of which N denotes the particle number of pure EVs used as the source material and Ni denotes the particle number recovered by each isolation method. Ni and N were measured by nFCM via single-particle enumeration. Meanwhile, protein assay was conducted in parallel to measure the protein concentration of each EV preparation. Figure 4(h) depicts the recovery rates of different isolation methods obtained by both nFCM and Qubit protein assay. We can see that for methods that do not bring in other particles during the isolation process, such as UC, qEV and UF, the particle size distribution profiles of isolated EVs closely resembled those of the source material. Meanwhile, the recovery rates measured by nFCM and Qubit protein assay agreed well with each other. By nFCM measurement, qEV yielded the highest recovery rate of 64.7 ± 13.1% whereas UC and UF yielded recovery rates of 39.6 ± 4.6% and 36.9 ± 2.4%, respectively. For ExoQuick and TEI methods, polymer precipitation resulted in the formation of impurity particles, the recovery rates measured by nFCM were slightly higher than those of the Qubit protein assay. For exoEasy kit, as fusion or aggregation occurred during the isolation process (see the TEM micrograph), a large peak centred around 185 nm was observed. The aggregation resulted in a decreased particle number and a low recovery rate measured by nFCM. Yet, the Qubit protein assay indicates that the recovery rate of protein was as high as 93.2 ± 8.7%. It has been speculated above that the extremely low proportion (less than 0.0035%) of EVs detected among all the particles with diameter larger than 40 nm in PFP could be ascribed to the substantial loss of EVs during UC isolation. Figure 4(a,b,h) indicate that the recovery rate of a single UC step at 100,000 × g for 2 h at 4°C was ~40%. Thus the recovery rate of EV preparation by UC (twice UC at 100,000 × g for 2 h at 4°C) would be ~16%. So, the actual proportion of EVs among the total particles with diameter larger than 40 nm in plasma could be around 0.02%, which could be largely attributed to the natural low abundance of EVs in plasma.
Figure 4.
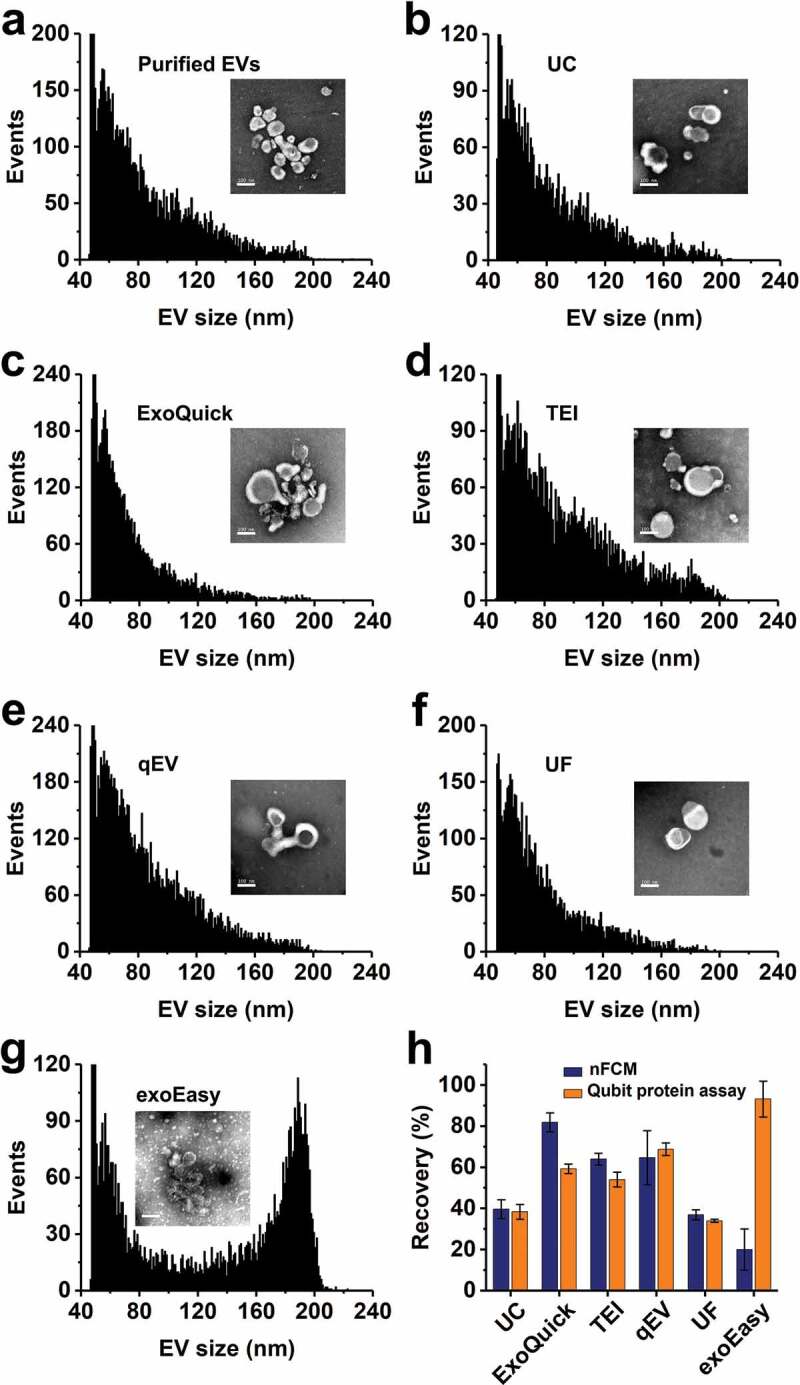
Measurement of the recovery rates of different isolation methods for pure EVs. (a) Particle size (diameter) distribution histogram of EVs purified from the conditioned medium of human colorectal cancer HCT15 cell culture by UC, the typical TEM micrograph (scale bar: 100 nm) is plotted as an inset. (b-g) Particle size distribution histograms of EV preparations from pure EVs by different isolation methods. (h) Recovery assessment of different isolation methods for pure EVs by both nFCM and Qubit protein assay. The error bars represent the standard deviation of three repetitive experiments for each isolation methods (mean ± s.d.). Note: Because the particle concentration of EV preparation by different isolation methods varies, the Y-axes of panels a-g are plotted in different scale.
What particles are found in plasma EV preparations?
Figures 2 and 3 indicate that, for PFP sample, the particle concentrations of EVs separated by commercial isolation kits were 2–4 orders of magnitude higher than that of UC, whereas the purities of these isolates were much lower (see Table 1 for details). To investigate the nature of the non-EV particles present in such large numbers in EV preparations by commercial isolation kits, we depleted vesicles from PFP by ultracentrifuging PFP at 100,000 × g for 18 h at 4°C, and used this VD-PFP as the starting material for EV isolation. Representative TEM micrograph of VD-PFP is shown in Figure S3. Figure 5(a) shows the comparison of particle concentrations between the EV preparations from PFP and from VD-PFP by these six different isolation methods. We can see that the particle concentration of VD-PFP was 1.3 × 1013 particles/mL, which was comparable to the original particle concentration of PFP (1.4 × 1013 particles/mL). These data suggest that EVs only take a very small fraction of particle number in the PFP. The particle concentration of EV preparations from VD-PFP by UC was 1.0 × 107 particles/mL. Comparing with the 4.9 × 108 particles/mL concentration of EVs separated from PFP by UC, we know that 98% of EVs were depleted from PFP during the 18 h UC process of VD-PFP preparation. The ratios between the particle concentrations of EV preparations from VD-PFP and those from PFP are plotted in Figure 5(b) for these six isolation methods along with the concentration ratio between VD-PFP and PFP (marked as plasma). Clearly, there was only a negligible amount of EVs left in VD-PFP. The high particle concentration of EV preparations from VD-PFP by commercial kits arises from the abundant non-vesicular particles residing in VD-PFP. For example, though qEV outperformed other commercial kits, the particle concentration for preparation from VD-PFP was as high as 66% of that from PFP. These results indicate that the majority of particles isolated by kits are not EVs but are contaminants.
Figure 5.
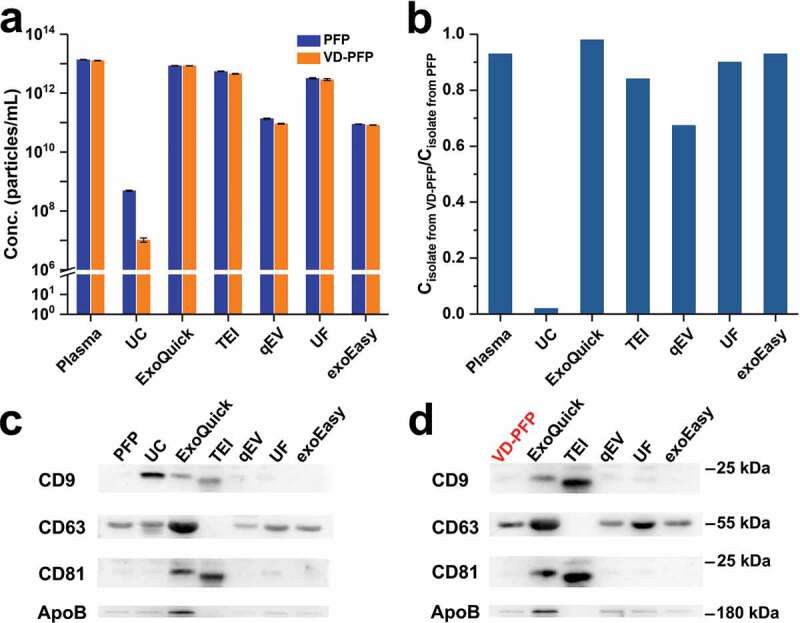
Comparison between particle concentration and protein markers for EV preparations from PFP and VD-PFP by UC and five commercial isolation kits. (a) Particle concentration of EV preparations from PFP and VD-PFP (n = 3, mean ± s.d.). (b) Ratio between the particle concentrations of EV preparations from PFP and from VD-PFP for different isolation methods. (c) Western blot analysis of EV preparations from PFP. (d) Western blot analysis of EV preparations from VD-PFP.
For commercial isolation kits, high concentration of the classical protein markers of EVs such as CD9, CD63 and CD81 in EV preparations from plasma or other sources has been widely used to demonstrate the high recovery and high purity of EVs [58]. Figure 5(c,d), and S4 show the Western blot analysis of CD9, CD63, CD81 and apolipoprotein B (ApoB) proteins for EV preparations from PFP and VD-PFP, respectively. Here ApoB, the primary apolipoprotein of chylomicrons, VLDL, IDL and LDL particles, served as a contaminating marker of lipoproteins. Note that the protein concentration of EV preparation from VD-PFP by UC was too low to enable Western blot analysis. Figure 5(c,d) indicate that CD63 and ApoB were clearly positive for PFP and VD-PFP. The abundant presence of CD63 could be ascribed to the residual platelets in PFP preparation and the abundant quantities of CD63 in platelets [59]. For EV preparation from PFP by UC, CD9, CD63 and ApoB were detected positive and the intensity for CD81 was very weak. The presence of ApoB confirms the co-isolation of lipoproteins with EVs by UC. Interestingly, for all five commercial isolation kits, the Western blot patterns of CD9, CD63 and CD81 for EV preparations from VD-PFP resembled well with those of EV preparations from PFP. These results suggest that CD9, CD63 and CD81 not only reside on the surface of EVs but are also present in presumably non-EV materials. As proteinase K was used in TEI isolation, proteins with large molecular weight such as CD63 and ApoB were digested and could not be detected by Western blot analysis.
Single-particle phenotyping of EV preparations
EVs in complex biological fluids are highly heterogeneous in terms of cell of origin, release pathway and physiological status. Because nFCM can detect a single antibody conjugated with PE [43], phenotyping of single EVs by nFCM was used to assess the quality of EV preparations from PFP by measuring the expression of CD9, CD63, CD81, CD235a, CD45, CD41a and CD144 via immunofluorescent labelling. EV preparation from CCCM of HCT15 cells by UC was also included for comparison. Among which, CD235a, CD45, CD41a and CD144 are protein markers of EVs derived from erythrocyte, leukocytes, platelets and vascular endothelial cells. Meanwhile, surface expression of PS was examined by PE-conjugated annexin V. Concentration optimization of PE-conjugated antibodies or annexin V is given in Figure S5. IgG isotype control and reagent control (PE conjugated antibody or annexin V) are shown in Figures S6 and S7, respectively. Figure 6(a,b) show the bivariate dot-plots of PE fluorescence versus SSC for EV preparations by UC from CCCM of HCT15 cells and PFP, respectively. As expected, compared to the classical EV protein markers such as CD9, CD63, CD81 and PS, the expression of blood cell markers (CD235a, CD45, CD41a and CD144) in EVs secreted by HCT15 was very low. For EVs isolated from PFP, the ratio of EVs derived by erythrocytes (CD235a+) exhibited the highest percentage up to 57.0%, which could be ascribed to the fact that red blood cells are the most abundant type of blood cells in the human body. Meanwhile, the ratios for CD9+, CD63+, CD81+, PS+, CD45+, CD41a+ and CD144+ EVs ranged between 10 and 25%. The bivariate dot-plots of PE fluorescence versus SSC for EV preparations from PFP by five commercial kits are displayed in Figures S8-1 and S8-2. Figure 6(c) displays the total event rates measured in 1 min for all six isolation methods along with their normalized event rates, i.e. the ratios between the event rate measured in 1 min for each isolation method and that of UC (the total event rate of reagent control was deducted from both the numerator and the denominator). We can see that the events were much fewer for EV preparations by kits when compared to that of UC. Note that upon fluorescent labelling, these EV preparations by kits were all washed twice by UC to remove unbound antibodies or annexin V as for EVs isolated by UC. The reduced particle numbers of commercial kits can be attributed to the removal of impurity particles upon UC. For example, the particle numbers measured for qEV, UF and exoEasy were only 14, 8 and 5% of that of UC, respectively. However, for ExoQuick and TEI, the ratios increased to 22 and 57%, respectively. We suspect that the polymer precipitates introduced during the isolation process could also be pelleted upon UC. The reason why TEI exhibits a much higher ratio could be because the relatively large particle size of EV preparation from PFP by TEI (Figure 2(d)) when compared to that of ExoQuick (Figure 2(c)). It is worth noting that besides the removal of unbound antibodies by two washes of UC, the large amounts of impurity particles presented in EV preparations by commercial isolation kits were also efficiently removed. Thus, for EV preparations from PFP by commercial kits, the proportions of EVs expressing a specific protein marker were comparable to those of EV preparation by UC (Figure 6(d)).
Figure 6.
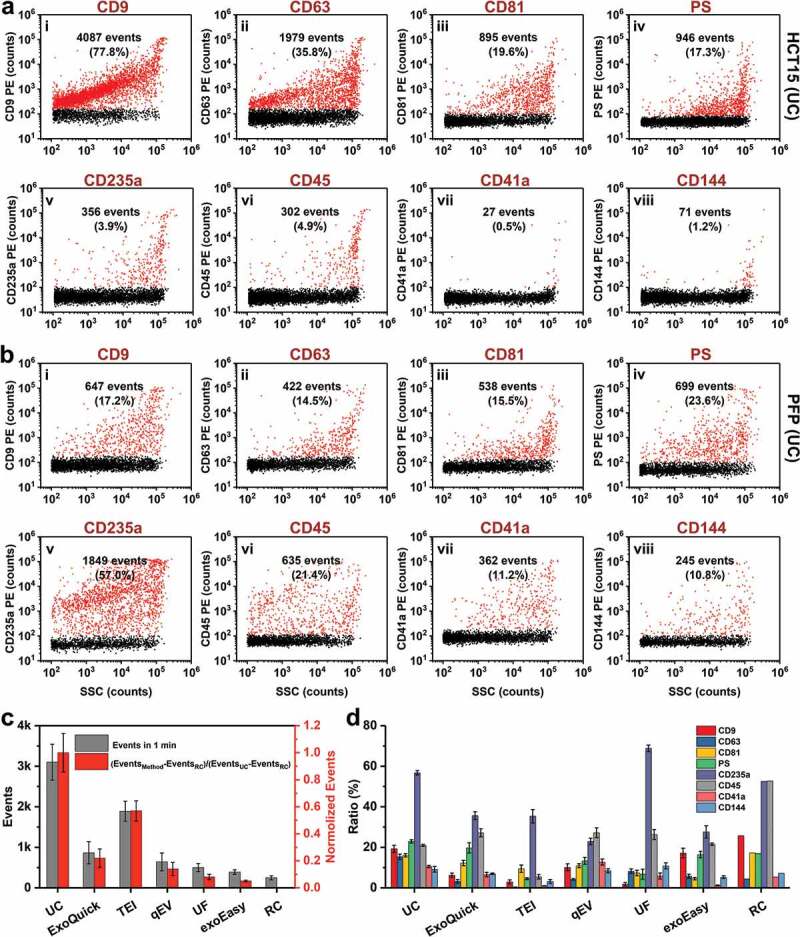
Single-particle phenotyping of EVs isolated by UC from CCCM of HCT15 cells and PFP. (a and b) Bivariate dot-plots of PE fluorescence versus SSC for EV preparations from CCCM of HCT15 cells (a) and PFP (b), respectively. The EVs were fluorescently labelled with PE-conjugated mAbs specific to CD9 (i), CD63 (ii), CD81 (iii), CD235a (v), CD45 (vi), CD41a (vii), or CD144 (vii), or PE-conjugated annexin V specific to PS (iv). The particle numbers of phenotype-positive EVs along with their percentages are provided in each plot. (c) Total particle numbers detected in 1 min for all six isolation methods and reagent control (RC), and their normalized event rates (mean ± s.d.). (d). Measured percentages of a specific phenotype-positive EVs (n = 3) for eight different markers and for six different isolation methods and RC (mean ± s.d.).
Discussion
Isolation of pure EVs from biological fluids is of critical importance as coisolation of soluble proteins and lipoproteins may impede the interpretation of experimental findings. For example, genomic, proteomic and lipidomic studies require the EVs to be as pure as possible to ensure that what are measured are compositions associated with EVs rather than interfering impurities. No matter what type biological fluids to isolate from and what kind of separation methods to use, knowing the properties of EV preparations, such as size distribution, particle concentration, purity and phenotype is of critical importance in ensuring the sensitivity and accuracy of downstream analysis. Among which, the purity of EVs isolated from blood sample is of major concern, as the concentration of lipoprotein particles is several orders of magnitude higher than that of vesicles [35,56]. Of note, compared with serum, plasma is usually the preferred source of EVs because additional EVs are released from platelet during the clot formation when preparing serum [11,60].
As ensemble-averaged approaches cannot differentiate whether the presence of nucleic acids, proteins or lipids is associated with EVs or reside in the other non-EV materials, and indeed no biomarkers have been established for EVs, there has not been an efficient approach to analyse the purity of EV preparations. Recently, the ratio of particle counts (measured by NTA) to protein content (acquired by BCA-assay) has been proposed and used as an adequate marker to estimate the purity of EV preparations [45,61]. However, NTA can neither detect vesicles smaller than 70 nm nor differentiate between vesicles and similarly sized lipoproteins or protein aggregates, which could easily lead to an inaccurate enumeration of EV counts. In the meantime, BCA-assay measures total protein concentration of EV preparation, which could include great amounts of lipoproteins and other non-vesicular proteins. Upon a systematic study of the yield, purity and functional potential of EVs isolated from plasma by UC and SEC, it was found that particle/protein ratio is not an accurate measure of purity of plasma EV preparations, and the importance of measuring the protein levels of specific EV markers and contaminating factors (e.g. ApoB lipoproteins) was emphasized [62]. Since the double-layered lipid membrane is the most prominent feature of EVs, we proposed a straightforward and simple approach to quantify the purity of EVs by using nFCM to enumerate particle counts before and after EV membrane lysis with Triton X-100 [43]. This single-particle approach is based on light-scattering detection and thus is label-free.
The purity of EVs isolated from PFP by UC was quantified to be 78.2% (Table 1) in the present study, which is only slightly lower than the normally 90% purity for EVs isolated from conditioned cell culture medium. Due to the partial dissociation of VLDL upon Triton X-100 treatment, the actual purity of EV preparations from PFP should be higher than what measured by nFCM. Considering the abundant presence of lipoproteins in plasma (≈1016/mL) [35], UC demonstrated its great efficiency in separating large quantities of non-vesicular contaminants. Although it has been anticipated that the particle concentrations of EV preparations from PFP by commercial kits could be higher than that by UC, the two to four orders of magnitude greater concentrations were still striking. In contrast to the much higher yields of commercial isolation kits, the measured purity (ranging from 28.1 to 5.3%) was markedly lower in comparison to UC, though an underestimation of their purity could occur. It is interesting to note that when these kits were used to isolate EVs from VD-PFP, comparable particle counts were obtained with their corresponding EV preparations from PFP. Meanwhile, the particle size distribution profiles of EVs isolated by kits closely resembled those of PFP, whereas EVs isolated by UC showed broader size distribution at relatively larger particle size. This may be due to the presence of more, smaller lipoprotein particles in EV preparations by kits. Compared with polymer precipitation, membrane filtration and membrane affinity, SEC is gaining increasing popularity owning to its simple procedure and relatively good separation of EVs from contaminants. However, although the qEV kit provided the best performance among all five tested commercial kits, its purity of 28.1% is compromised when compared to the 78.2% purity of UC. In studies using UC and SEC for isolation of EVs from rat blood plasma, Takov et al. found that plasma EVs obtained by UC appear to be of superior purity to those isolated by SEC. Meanwhile, the particle modal size (96.6 ± 3.1 nm) of plasma EVs isolated by UC was significantly higher than that of the EVs isolated by SEC (81.5 ± 3.3 nm) according to the NTA measurement [62]. Accordingly, nFCM measurements indicate that the 81 nm median size of human plasma EVs isolated by UC was much larger than the 64 nm median size of the EV preparations by SEC (qEV kit) and the 56 nm median size of PFP. Collectively, these data indicate that a high degree of non-vesicular contaminants exists in plasma EV isolation by commercial kits. It is worth noting that a complete separation of EVs from plasma is an arduous task due to the extensive overlap in particle size and density between EVs and lipoproteins. In order to isolate EVs from blood with minimal contamination by plasma proteins and lipoprotein particles, a combination of several separation methods can be applied such as UC followed by density cushion and SEC [56], or UC followed by gradient density UC [63].
Regarding phenotyping of EVs, ensemble-averaged analysis of tetraspanin (CD9, CD63 and CD81) have been widely used to assess the purity and recovery rate of EVs. However, we demonstrate that these tetraspannin membrane proteins also present in VD-PFP and can be co-isolated by all five commercial EV isolation kits. Thus the mere presence of these proteins in EV preparations via Western blot analysis cannot be used to claim a high recovery rate of EVs. Compared with ensemble-averaged analysis of protein markers, single-particle phenotyping exhibits distinct advantages, as the result is straightforward and conclusive.
In summary, nFCM analysis highlighted that ExoQuick, TEI, qEV, UF and exoEasy failed to isolate high-purity EVs from plasma, and UC is the most appropriate isolation method among the ones tested. We hope that this work will raise awareness about the quality of EV preparations obtained with commercial isolation kits and stress the need for caution when making conclusive statements on the molecular composition and biological functions of EVs. By analysing single particles at a rate of up to 10,000 particles/min, in a resolution comparable to that of TEM, and with concurrent light scattering and fluorescence measurement, nFCM provides quantitative analysis of EV size distribution, concentration, purity and phenotype without prejudice. Thus, nFCM could serve as a new benchmark for the quality and efficiency assessment of EV isolation methods and will be a great asset to the further development of EV isolation protocols.
Supplementary Material
Conflict of interest
X.Y. and S.Z. declare competing financial interests as cofounders of NanoFCM Inc., a company committed to commercializing the nano-flow cytometry (nFCM) technology. D. A. declares competing financial interests as an employee of NanoFCM Inc.
References
- [1].Thery C, Zitvogel L, Amigorena S.. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–17. [DOI] [PubMed] [Google Scholar]
- [2].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [3].Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shah R, Patel T, Freedman JE.. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379:958–966. [DOI] [PubMed] [Google Scholar]
- [5].Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111:14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Durcin M, Fleury A, Taillebois E, et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J Extracell Vesicles. 2017;6:1305677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Clos-Garcia M, Loizaga-Iriarte A, Zuniga-Garcia P, et al. Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J Extracell Vesicles. 2018;7:1470442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Whitham M, Parker BL, Friedrichsen M, et al. Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab. 2018;27:237–251. [DOI] [PubMed] [Google Scholar]
- [9].Alderton GK. Diagnosis: fishing for exosomes. Nat Rev Cancer. 2015;15:453. [DOI] [PubMed] [Google Scholar]
- [10].Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. [DOI] [PubMed] [Google Scholar]
- [11].Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mateescu B, Kowal EJ, van Balkom BW, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6:1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;8:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:3.22.1–3.22.29 Chapter 3:Unit 3 22 [DOI] [PubMed] [Google Scholar]
- [16].Gardiner C, Di Vizio D, Sahoo S, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rider MA, Hurwitz SN, Meckes DG Jr.. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. 2016;6:23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Welch JL, Madison MN, Margolick JB, et al. Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep. 2017;7:45034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Boing AN, van der Pol E, Grootemaat AE, et al. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014;3:23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Welton JL, Brennan P, Gurney M, et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array. J Extracell Vesicles. 2016;5:31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lobb RJ, Becker M, Wen SW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang TT, Anderson KW, Turko IV. Assessment of extracellular vesicles purity using proteomic standards. Anal Chem. 2017;89:11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yoo CE, Kim G, Kim M, et al. A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Anal Biochem. 2012;431:96–98. [DOI] [PubMed] [Google Scholar]
- [24].Fang XN, Duan YK, Adkins GB, et al. Highly efficient exosome isolation and protein analysis by an integrated nanomaterial-based platform. Anal Chem. 2018;90:2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee K, Shao H, Weissleder R, et al. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9:2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wunsch BH, Smith JT, Gifford SM, et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat Nanotechnol. 2016;11:936–940. [DOI] [PubMed] [Google Scholar]
- [27].Wu M, Ouyang Y, Wang Z, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA. 2017;114:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Karttunen J, Heiskanen M, Navarro-Ferrandis V, et al. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J Extracell Vesicles. 2018;8:1555410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Oeyen E, Van Mol K, Baggerman G, et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles. 2018;7:1490143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li P, Kaslan M, Lee SH, et al. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shao H, Im H, Castro CM, et al. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakata R, Shimada H, Fernandez GE, et al. Contribution of neuroblastoma-derived exosomes to the production of pro-tumorigenic signals by bone marrow mesenchymal stromal cells. J Extracell Vesicles. 2017;6:1332941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Buschmann D, Kirchner B, Hermann S, et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles. 2018;7:1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sodar BW, Kittel A, Paloczi K, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep. 2016;6:24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Simonsen JB. What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ Res. 2017;121:920–922. [DOI] [PubMed] [Google Scholar]
- [36].Andreu Z, Rivas E, Sanguino-Pascual A, et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J Extracell Vesicles. 2016;5:31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Helwa I, Cai JW, Drewry MD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. Plos One. 2017;12:0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ding M, Wang C, Lu XL, et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal Bioanal Chem. 2018;410:3805–3814. [DOI] [PubMed] [Google Scholar]
- [39].Martins TS, Catita J, Rosa IM, et al. Exosome isolation from distinct biofluids using precipitation and column-based approaches. Plos One. 2018;13:e0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stranska R, Gysbrechts L, Wouters J, et al. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med. 2018;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].van der Pol E, Coumans FA, Grootemaat AE, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182–1192. [DOI] [PubMed] [Google Scholar]
- [42].Zabeo D, Cvjetkovic A, Lässer C, et al. Exosomes purified from a single cell type have diverse morphology. J Extracell Vesicles. 2017;7:1329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tian Y, Ma L, Gong M, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671–680. [DOI] [PubMed] [Google Scholar]
- [44].Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles. 2013;2:19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Abramowicz A, Widlak P, Pietrowska M. Proteomic analysis of exosomal cargo: the challenge of high purity vesicle isolation. Mol Biosyst. 2016;12:1407–1419. [DOI] [PubMed] [Google Scholar]
- [47].Zhu S, Ma L, Wang S, et al. Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles. ACS Nano. 2014;8:10998–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ma L, Zhu S, Tian Y, et al. Label-free analysis of single viruses with a resolution comparable to that of electron microscopy and the throughput of flow cytometry. Angew Chem Int Ed Engl. 2016;55:10239–10243. [DOI] [PubMed] [Google Scholar]
- [49].Yang L, Zhu S, Hang W, et al. Development of an ultrasensitive dual-channel flow cytometer for the individual analysis of nanosized particles and biomolecules. Anal Chem. 2009;81:2555–2563. [DOI] [PubMed] [Google Scholar]
- [50].Zhu SB, Yang LL, Long Y, et al. Size differentiation and absolute quantification of gold nanoparticles via single particle detection with a laboratory-built high-sensitivity flow cytometer. J Am Chem Soc. 2010;132:12176–12178. [DOI] [PubMed] [Google Scholar]
- [51].Laven P. MiePlot (A computer program for scattering of light from a sphere using Mie theory and the Debye series); 2011. [cited 2016 October19] Available from: http://www.philiplaven.com
- [52].Zhang W, Tian Y, Hu X, et al. Light-scattering sizing of single submicron particles by high-sensitivity flow cytometry. Anal Chem. 2018;90:12768–12775. [DOI] [PubMed] [Google Scholar]
- [53].Arraud N, Linares R, Tan S, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12:614–627. [DOI] [PubMed] [Google Scholar]
- [54].Hergenreider E, Heydt S, Treguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. [DOI] [PubMed] [Google Scholar]
- [55].Osteikoetxea X, Sodar B, Nemeth A, et al. Differential detergent sensitivity of extracellular vesicle subpopulations. Org Biomol Chem. 2015;13:9775–9782. [DOI] [PubMed] [Google Scholar]
- [56].Karimi N, Cvjetkovic A, Jang SC, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75:2873–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Linares R, Tan S, Gounou C, et al. High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles. 2015;4:29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lacroix R, Judicone C, Poncelet P, et al. Impact of pre-analytical parameters on the measurement of circulating microparticles: towards standardization of protocol. J Thromb Haemost. 2012;10:437–446. [DOI] [PubMed] [Google Scholar]
- [60].Coumans FAW, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120:1632–1648. [DOI] [PubMed] [Google Scholar]
- [61].Woo HK, Sunkara V, Park J, et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017;11:1360–1370. [DOI] [PubMed] [Google Scholar]
- [62].Takov K, Yellon DM, Davidson SM. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J Extracell Vesicles. 2019;8:1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7:1311–1326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Laven P. MiePlot (A computer program for scattering of light from a sphere using Mie theory and the Debye series); 2011. [cited 2016 October19] Available from: http://www.philiplaven.com


