ABSTRACT
The study was conducted to estimate the prevalence of Escherichia coli (E. coli) in sub-clinically mastitic (SCM) animals, and in wild and migratory birds which may act as reservoir disseminating such pathogen. Farm hygiene, management and milking procedures were listed through a questionnaire. Thirty lactating cows and 15 lactating buffaloes from five small-scale dairy farms were randomly selected and screened for subclinical mastitis (SCM) using California Mastitis Test (CMT) and somatic cell count (SCC). In addition, 80 teat skin swabs, 5 drinking water samples and 38 wild and migratory bird faecal matter were also collected. All samples were processed for E. coli isolation by culturing on Levine’s Eosin Methylene Blue (L-EMB) agar, followed by purification and biochemical identification. Positive samples were subjected to molecular identification and serotyping. In addition, the presence of extended-spectrum beta-lactamase (ESBL) and carbapenemase-producing E. coli have been reported by antimicrobial sensitivity testing. Escherichia coli were isolated from 7.7%, 50% and 50% of the positive CMT cows’ quarters, cows’ composite and buffaloes’ composite milk samples, respectively. In addition, 14% of cows’ teats, 20% of water samples, 70% of faecal matter from wild bird, and 33.3% of faecal matter from migratory waterfowls were carrying E. coli. Serotyping, antibiotic-resistant pattern and phylogenetic analysis have pointed the bearable implication of milking hygiene and wild birds in disseminating E. coli strains causing intramammary infections.
KEYWORDS: Sub-clinical mastitis, E. coli, buffalo, cow, wild birds, antibiotic-resistance
1. Introduction
Bovine mastitis continues to be a major infection that affects a high proportion of dairy populations worldwide with a negative economic impact on dairy farmers and the dairy industry [1–3]. Furthermore, it possesses a public health concern [4], besides it is considered the single most prevalent cause for antibacterial use in lactating dairy animals [5]. The types of mastitis pathogens are either contagious or environmental organisms [6]. Coliform bacteria are a common aetiology of bovine mastitis. Until recently, Escherichia coli (E. coli) is the most predominant species and is responsible for more than 80% of coliform mastitis cases [3,7,8] and categorized as a Gram-negative opportunistic environmental bacterium. Mastitis caused by E. coli is usually sporadic with clinical signs range from mild to very severe or even fatal forms [9]. Properties and virulence factors specific to E. coli strains causing mastitis are still ill-defined, although a high prevalence of phylogroup A was observed in mastitis caused by E. coli [10–13]. E. coli is mainly a commensal bacterium in the intestinal tract of animals, birds and humans [14]. Although the establishment of biosecurity measures to prevent the entry of infectious agents into dairy farms, dairy herd health still faces several serious threats, regarding open space, which allows uncontrolled access of wild birds into yards, facilities and even feed storage areas [15]. However, few studies focused on wild birds rooming nearby dairy farms [16]. Migratory and non-migratory wild birds serve as reservoirs of coliform bacteria, like E. coli, inclosing antimicrobial-resistance genes [17,18]. Water and food contact were suggested to be the primary routes of transmission of resistant bacteria between human or animal and wild birds [19]. Antibiotics are very remarkable for the treatment of bacterial infections in animals and humans [20]. The FDA reported that resistance in E. coli is steadily the highest for antimicrobial agents used in human and veterinary medicine [21]. Extended-spectrum beta-lactamase (ESBL) producing E. coli is mostly insensitive to lots of commonly used antibiotics causing an increase in the use of last-resort antimicrobial drugs (i.e. carbapenems) during treatment.
The aim of this study was to understand the possible role of wild and migratory birds in carrying and spreading mastitis-causing E. coli strains to the dairy farm environment which could represent potential veterinary and public health hazards through infecting the animals and contaminating the milk with antimicrobial-resistant avian strains, in a coincidence of poor milking hygiene.
2. Materials and methods
2.1. Collection of samples
2.1.1. Milk samples
Milk samples were collected from five dairy farms located in Cairo, Giza, and El-Ismailia governorates in Egypt during the period from August 2017 to January 2018. A total of 15 composite milk samples from 15 lactating cows and 15 composite milk samples from 15 lactating buffaloes were collected. In addition, 60 quarter milk samples from other 15 lactating cows were collected. All animals were apparently healthy.
Milk samples were collected into sterile vials after washing, drying and swabbing of teat ends with 70% ethyl alcohol, and discarding of the first 3–4 streams of milk. Milk samples were subjected to California Mastitis Test (CMT) according to the American Public Health Association (APHA) [22] and somatic cell counting according to Zecconi et al. [23].
2.1.2. Teat swabs
A total of 80 teat skin swabs were collected from 25 cows and 15 buffaloes (40 Hind and 40 fore teat swabs) by a cotton-tipped stick through rotating the cotton-tip on the teat barrel, as described by Piccinini et al. [24].
2.1.3. Drinking water samples
Five water samples (50 ml from each farm) were collected from the drinking troughs of the lactating animals into disposable sterilized test tubes. All samples were stored in an ice box until transported to the laboratory.
2.1.4. Wild and migratory birds faecal matter
A total number of 38 faecal samples were collected from 20 resident wild birds (12 laughing doves “Spilopelia senegalensis” and 8 hooded crows “Corvus cornix”) and 18 migratory waterfowls (10 eurasian coots “Fulica atra” and eight northern shovelers “Anas clypeata”) in Giza, Cairo, El-Ismailia and Port-said governorates in Egypt.
Port-Said is one of the Canal Zone governorates and considered a main stop for migratory waterfowls seeking food and rest during the long migration in the spring and fall seasons.
Bird traps were used to capture wild birds around dairy farms and migratory waterfowls during winter migration. Once trapped, faecal swabs were taken, and the birds were released. The swabs were then placed in 2 ml sterile saline (0.9% NaCl) and stored in ice box until transported to the laboratory.
Protocols for the collection of samples were conducted in accordance with the applicable legislation of the Institutional Animal Care and Use Committee, of the Faculty of Veterinary Medicine, Cairo University, Egypt (VetCU10102019087).
2.2. Isolation and identification of coliform and E. coli
Loopfuls from positive CMT milk samples were enriched into nutrient broth and incubated aerobically at 37°C for 24 h. The enriched milk samples, teat swabs and loopfuls from water samples were streaked on Violet Red Bile agar (VRB) for isolation of coliforms. Additionally, these samples beside the faecal samples from wild and migratory birds were plated on Levine’s Eosin Methylene Blue plates (L-EMB) for isolation of E. coli and incubated at 37ºC for 24–48 h. Suspected colonies of E. coli which appeared as dark centred, flat colonies with metallic green lustre were picked up and cultured on agar slants and incubated at 35°C for 18 h and subjected to further identification. Biochemical identification of E. coli was carried out according to BAM and Quinn et al. [25,26].
A total of eight representative E. coli isolates from buffalo’s milk, cow’s milk, teat skin swabs, drinking water, Laughing dove, Hooded crow, Eurasian coot, Northern shoveler faecal swabs that were isolated from the same locality were selected and serologically identified by slide agglutination test for O-antigen group screening using E. coli antisera (Denka Seiken Co., LTD. Chuo-Ku,Tokyo, Japan) according to Kok et al. [27].
2.3. Molecular identification of E. coli
All E. coli isolates were extracted using DNA Mini Kit (Qiagen, Hombrechtikon, Switzerland) according to the manufacturer’s protocol. For molecular identification of E. coli, Primers for 16S rRNA gene of E. coli were selected according to Wang et al. [28]. The reaction was carried out using a total volume of 25 μl contain 3 μl of DNA as a template, 5 pmol of each primer and 5μl of 1X PCR master mix (Jena bioscience, GmbH, Germany). The PCR mixtures were then subjected to the following cycling conditions: 50°C (2 min, 1 cycle); 95°C (5min, 1 cycle); 40 cycles of 95°C (45 s), 50°C (1 min), and 72°C (1 min); and 72°C (7 min, 1 cycle) in a thermal cycler (Perkin-Elmer, Waltham, USA).
2.4. Antibiotic sensitivity test
The antimicrobial sensitivity for the fore mentioned eight serotyped isolates was evaluated using eight antimicrobials that represent three groups: Carbapenems (Imipenem, Ertapenem, Meropenem), Cephalosporins (Ceftazidime, Cefotaxime, Ceftriaxone, Cefpodoxime) and Macrolides (Azithromycin).
ESBL production was confirmed by the increase of ≥5 mm in the zone of inhibition for Cefotaxime/Clavulanic acid (30/10 mcg) and Ceftazidime/Clavulanic acid (30/10 mcg) discs compared to cefotaxime or ceftazidime alone. The interpretation was done according to Clinical and Laboratory Standards Institute (CLSI) [29].
2.5. Sequence analysis
The amplified 16S rRNA fragments of these eight isolates were purified using the QIAquick gel extraction kit (Qiagen, Hombrechtikon, Switzerland) according to the manufacturer’s instructions and sequenced at Promega Lab Technology (Madison, USA) by using the forward and reverse primers listed in table (Table 1). The sequences of the 16S rRNA gene have been deposited in the GenBank database under the accession numbers mentioned in Table 4. Genes sequenced in this study were compared with the sequences available in public domain using NCBI BLAST server. The 16S rRNA gene sequences were aligned using CLUSTALW in BioEdit version 7.0.1.4. Phylogenetic analysis was performed with MEGA version 7, using the neighbour-joining approach. The bootstrap consensus tree was estimated from 1000 replicates.
Table 1.
Primer sequences 16 s Escherichia coli.
| Primer name (Target gene) |
Oligonucleotide sequence (5–3`)* |
Amplicon size (bp) |
|---|---|---|
| E16S (16S rRNA) | F: CCCCCTGGACGAAGACTGAC | 401 bp |
| R: ACCGCTGGCAACAAAGGATA |
Table 4.
Serotyping of E. coli isolates and their antibiogram.
| Dairy farms |
Resident birds |
Migratory birds |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Samples | Cow milk | Buffalo milk | teat skin | Drinking water | Laughing dove | Hooded crow | Eurasian coot | Northern shoveler | |
| E. coli serotypes | O166 | O146 | O146 | O1 | O166 | O78 | O18 | O158 | |
| Accession number | MK503655 | MK503954 | MK503972 | MK503975 | MK503974 | MK503973 | MK503976 | MK503978 | |
| Antibiotics (mcg*) | |||||||||
| Carbapenems | Imipenem (10) | S | S | I | S | I | I | I | S |
| Ertapenem (10) | S | S | R | R | I | I | I | S | |
| Meropenem (10) | S | S | R | I | S | S | S | S | |
| Cephalosporins | Ceftazidime (30) | I | I | S | R | R | R | R | I |
| (Cephotaxime) (30) | R | R | R | R | R | R | R | R | |
| Ceftriaxone (30) | R | S | R | R | R | R | R | S | |
| Cefpodoxime (10) | R | R | R | R | R | R | R | R | |
| Macrolides | Azithromycin (15) | I | I | S | R | R | R | R | I |
| ESBL producer | Ceftazidime/Clavulanic acid (30/10) | - | - | - | - | - | ESBL | - | - |
| Cefotaxime/Clavulanic acid (30/10) | - | ESBL | - | ESBL | ESBL | ESBL | ESBL | - | |
* mcg: Micrograms.
2.6. Statistical analysis
Results were analysed using PASW Statistics, Version 18.0 software (SPSS Inc., Chicago, IL, USA). Somatic cell counts were compared by independent sample T-test and expressed as mean ± standard error. Frequencies of isolation were tested with the Chi-square (x2) (provided that at least 80% of the cells have an expected frequency of 5 or greater, and that no cell has an expected frequency smaller than 1.0), otherwise, the Fisher’s Exact test and the Fisher-Freeman-Halton Exact test (it is the Fisher’s Exact test for contingency tables larger than 2 × 2) were used. A P-value <0.05 was considered statistically significant.
3. Results
3.1. Field observation
Dairy farms included in this study raised from 10 to 100 animals. Animal breeds were Holstein Friesian cows and native buffaloes. Cows age ranged from 6 to 10 years, while buffaloes were around 12 years old. Animals were housed in open yards surrounded with a fence, partially covered with sheds and muddy floors and each yard contains one common water trough for drinking. Each yard is occupied by 10 to 25 lactating cows or buffaloes. All animals were automatically milked twice daily either in-stanchion (10 animals) or in milk parlours (40 to 100 animals). Pre-milking practice was restricted to udder washing and wiping without pre-milking sanitizing teat dip. Only, two farms applied post-milking teat dipping.
3.2. California mastitis test (CMT) and milk somatic cell count (SCC)
California mastitis test (CMT) reacted positive with 26 (43.3%) quarters milk samples of cows, 14 (93.3%) composite milk samples of cows, and 4 (26.7%) composite milk samples of buffalos (Figure 1), revealing significant association of subclinical mastitis to cows rather than buffaloes, x2 (1, N = 30) = 13.889, p< 0.001.
Figure 1.
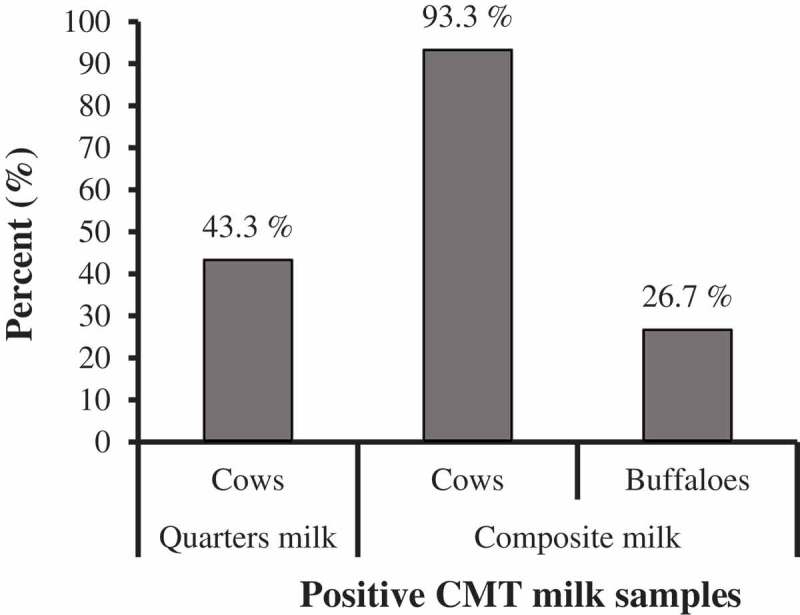
Prevalence of subclinical mastitis in the examined quarter and composite milk samples from cows and buffaloes using California Mastitis Test (CMT).
Results demonstrated that somatic cell count (SCC) in cows’ quarter milk samples was 8.5 × 105 (±1.9 × 105, ±95% C.I.) cells/ml significantly greater than that in composite milk samples (t(69.79) = 8.838, p< 0.001) (Table 2). While, somatic cell count (SCC) in composite milk samples of cows was 30 × 104 (±1.9 × 105, ±95% C.I.) cells/ml numerically but not significantly greater than that of buffaloes (t(15) = 0.333, p= 0.744) (Table 2). According to the Egyptian standards of raw milk [30], subclinical mastitis is considered when SCC exceeds 500 × 103 cells/ml. Consequently, 88.3% of examined cow quarters recorded unacceptable SCC, as well as, 46.7% of cows’ composite and 50% of buffaloes’ composite milk samples (Figure 2). Subclinical mastitis in cows was significantly more likely to be detected in quarter milk samples than in composite ones (p =0.002, Fisher’s exact test, FET).
Table 2.
SCC, coliforms and E. coli in cows’ and buffaloes’ positive CMT milk samples.
| Types of milk samples | Total no. | +ve CMT | Minimum SCC (Cell/ml) | Maximum SCC (Cell/ml) | Mean SCC ±SE (Cell/ml) | Coliforms No. (%) |
E. coli No. (%) |
|---|---|---|---|---|---|---|---|
| Quarters – cows | 60 | 26 | 336 × 103 | 2 × 106 | 1.368 × 106 ± 0.085 × 106 a | 26 (100%) | 2 (7.7%) b |
| Composite | |||||||
| Cows | 15 | 14 | 350 × 103 | 817 × 103 | 515 × 103 ± 45 × 103 b | 13 (92.9%) | 7 (50%) a |
| Buffaloes | 15 | 4 | 312 × 103 | 639 × 103 | 485 × 103 ± 68 × 103 | 4 (100%) | 2 (50%) |
a,b Different superscripts indicate significant difference (p < 0.05)
Figure 2.
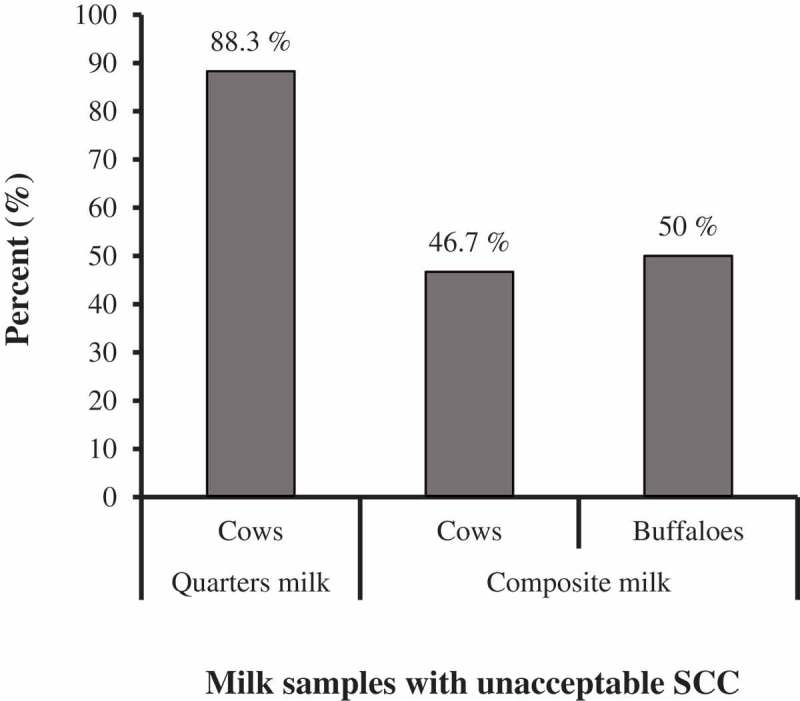
Prevalence of unacceptable milk SCC in examined quarters and composite milk from cows and buffaloes, according to Egyptian standards of raw milk, 2010.
3.3. Prevalence of coliforms and E. coli
3.3.1. Milk samples
Bacteriological examination of positive CMT milk resulted in isolation of Coliforms from 26 (100%) quarter milk samples, 13 (92.9%) composite milk of cows, and 4 (100%) composite milk of buffaloes (p= 0.409; Fisher-Freeman-Halton Exact test = 2.193). Further examination of positive CMT milk samples resulted in isolation of E. coli from 2 (7.7%) quarter milk samples, 7 (50%) composite milk of cows, and 2 (50%) composite milk of buffaloes (Table 2). Escherichia coli (E. coli) was significantly more likely to be isolated from composite milk samples than from quarter milk samples in cows (p =0.004, Fisher’s exact test, FET).
3.3.2. Teat skin swabs and drinking water
Coliforms were isolated from 19 (38%) and 6 (20%) teats of cows and buffaloes, respectively; a difference that was not statistically significant, x2 (1, N = 80) = 2.828, p= 0.093. Further examination resulted in the isolation of E. coli from 7 (14%) of cow’s teat skins, but could not be isolated from buffaloes’ teat skins, as listed in Table 3. Coliforms and E. coli were isolated from the one-water sample (20%).
Table 3.
Prevalence of coliforms and E. coli on teat skin swabs, drinking water samples and birds’ faecal swabs.
| Samples | Total no. | Coliforms No. (%) |
E. coli No. (%) |
|---|---|---|---|
| Cows: | |||
|
25 | 10 (40%) | 3 (12%) |
|
25 | 9 (36%) | 4 (16%) |
|
50 | 19 (38%) a | 7 (14%) |
| Buffaloes: | |||
|
15 | 4 (26.67%) | 0 |
|
15 | 2 (13.33%) | 0 |
|
30 | 6 (20%) a | 0 |
| Drinking water: | 5 | 1 (20%) | 1 (20%) |
| Resident wild birds: | |||
|
12 | 8 (66.6%) | |
|
8 | 6 (75%) | |
|
20 | 14 (70%) a | |
| Migratory waterfowls: | |||
|
10 | 3 (30%) | |
|
8 | 3 (37.5%) | |
|
18 | 6 (33.3%) b |
a,b Different superscripts indicate significant difference (p < 0.05)
3.3.3. Faecal samples of resident wild birds and migratory waterfowls
Bacteriological examination of 38 faecal samples from wild birds and migratory waterfowls resulted in isolation of E. coli from 14 (70%) of resident wild birds (laughing doves “Spilopelia senegalensis” and hooded crows “Corvus cornix”) and 6 (33.3%) of migratory waterfowls (eurasian coot “Fulica atra” and northern shoveler “Anas clypeata”) (Table 3). Isolation rate of E. coli from faeces of wild birds was significantly higher than that from migratory waterfowls, x2 (1, N = 38) = 5.109, p= 0.024.
All E. coli isolates were molecularly confirmed by PCR using Primers for 16S rRNA gene of E. coli.
3.4. Serotyping of E. coli
Results in Table 4 revealed different E. coli serotypes. E. coli O166 serotype was isolated from both cow’s milk and laughing dove’s faecal swab, and O146 serotype was isolated from both buffalo’s milk and teat skin. Serotypes O1, O78, O18 and O158 were isolated from drinking water, faecal swab of hooded crow, faecal swab of eurasian coot and of northern shoveler; respectively.
3.5. Antibiotic sensitivity of E. coli isolates
The antibiogram demonstrated that the Carbapenems group of antibiotics showed the greatest efficacy against most of the E. coli isolates. Resistance to Carbapenems group appeared from the environmental E. coli serotypes isolated from teat skin and drinking water. All the eight E. coli isolates displayed resistance against both Cefotaxime and Cefpodoxime antibiotics.
3.6. Sequence analysis
Phylogenetic analysis of 16S rRNA gene sequences of these eight E coli isolates showed100% homology between isolates from northern shoveler, eurasian coot, laughing dove, hooded crow and water as shown now in the phylogenetic tree (Figure 3).
Figure 3.
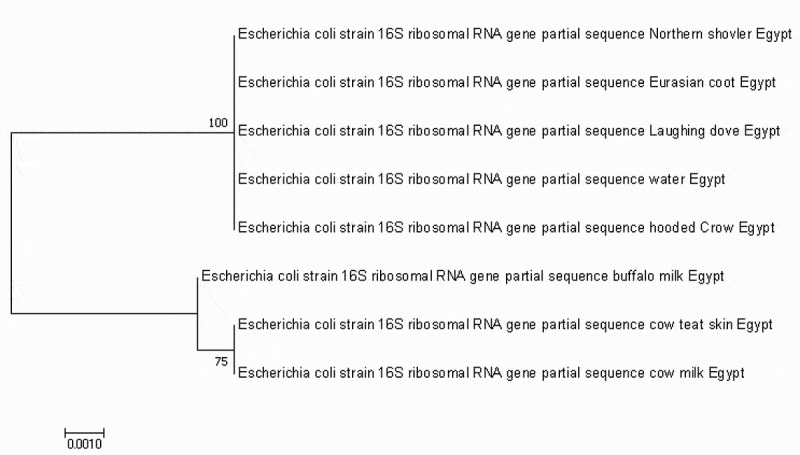
Phylogenetic analysis using neighbour-joining method based on the partial sequence of 16s rRNA. The bootstrap consensus tree demonstrates the evolutionary history of the obtained E. coli strain. The tree was constructed by Mega 7 software.
4. Discussion
The Small-scale dairy farming is a significant economic sector for the enhancement of agriculture and livelihoods in developing countries [31,32]. It begins from 2, 15 up to 20 or even 50 milking animals [33–35]. Mastitis is a multi-factorial production disease and possess a threat for small-scale producers as its prevalence exceeded 50% according to FAO, 2014 report [36]. In subclinical mastitis, milk and udder appear normal, but there is decreased milk production, positive reactivity with CMT, high milk somatic cell count, presence of pathogens in the milk, and altered milk composition [37–39]. California Mastitis Test (CMT) is a common screening test used for detection of subclinical mastitis and it revealed that 43.3% of quarter milk samples were affected with subclinical mastitis. These results agreed with that reported by Sori et al. [40] (40.6%) and Ayano et al. [41] (43.02%). A higher prevalence (60%) was reported by Ayano et al. [42], and a lower prevalence ranged from 18.14% to 23.25% were reported by Bangar et al. [43], Patel and Trivedi [44], and Singh et al. [45]. On the other hand, somatic cell count (SCC) is the gold standard for confirming subclinical mastitis [46]. Healthy udder has milk SCC ranges from 50,000 and 100,000 cells/ml, up to 200,000 cells/mL, while in subclinical mastitis the SCC exceeds 200,000 cells/ml [47]. In Egypt, subclinical mastitis is confirmed when SCC exceeds 500,000 cells/ml [30]. In Table 2, the average SCC of quarter milk samples and composite milk samples were similar to the average value (9.24 ± 2.0 × 105 cells/ml) reported by Skrzypek et al. [48] and average (44.8 ± 0.26 x 104 cell/ml) reported by Mohamed et al. [49], respectively.
Coliforms are environmental mastitis-causing pathogens, they are primarily present in moisture, mud, faeces and other organic matter present around the animals. Escherichia coli is the most commonly isolated coliform species from intramammary infections and clinical mastitis; besides it has a significant public health importance as it causes diarrhoea in human [40,50,51]. Prevalence of coliform infections (Table 2) disagreed with Hawari and Al-Dabbas [52] who detected coliforms in the milk of 31.9% of the mastitic quarters. On the other side, El-Khodery and Osman [53] detected coliforms in 80.36% of the affected buffaloes, but Sayed [54] recorded lower coliform prevalence (21.78%) in the examined milk samples. Escherichia coli was isolated form 7.7% quarter milk samples which is lower than that reported by Verma et al. [55] (21.28%). Escherichia coli has been detected in 50% of composite milk of buffaloes and cows which agreed with Mansour et al. [56] who found E. coli in 47% of machine-milked samples.
Poor hygiene, husbandry and milking technique could predispose to environmental mastitis as well as milk contamination [57]. Gram-negative bacteria pass into the mammary gland through teat canal [50]. Previous studies stated that regular teat dipping for controlling of mastitis is not a common practice at small-scale farms, which agrees with the findings of our study [58]. Escherichia coli was detected on the skin of 14% of cows’ teats as shown in Table 3, which poses a critical threat to the animals as well as consumer health as concluded by Galiero [59]. Teat skin act as the primary reservoir of microbes that can infect milk during milking [60]. Moreover, Jones [61] showed that the incidence of clinical mastitis was related to bacterial populations on teat ends.
As E. coli is a ubiquitous organism, it colonizes the gut of birds as well as mammals [62]. Recent studies assumed that wild birds play a crucial role in spreading of foodborne bacteria among dairy farms [63,64]. The practice of feeding dairy animals in open yards and accumulation of their manure lead to the attraction of wild birds into dairy farms [65]. Results in Table 3 showed that faecal matter of 70% of wild resident birds and 33.3% of migratory waterfowls contained E. coli. Similar studies isolated identical E. coli strains from excreta of wild birds collected from two dairy farms with 32.5 km distance apart, on the same sampling time [64,66].
Serotyping of E. coli isolates from different sources in the same locality revealed that O-146 and O-166 serotypes were the most frequent strains (Table 4). The O-146 type was isolated from both buffalo milk and teat skin and this indicated that contamination of teat skin with environmental pathogens possess a risk for occurrence of mastitis [61]. The same serotype was isolated by Wenz [12] from milk samples of cows with acute coliform mastitis and by Wang et al. [67] from ducks in China. Escherichia coli O-166 serotype was isolated from both cow milk and dove excreta which shows the probable role of wild birds in acting as reservoir and spreader of mastitis-causing pathogens among dairy farms [65–67]. Furthermore, the same serotype was previously reported to cause an outbreak of gastroenteritis in humans in Japan [68], which displays the significance of this serotype on the public health. Escherichia coli serotypes O-1, O-18, O-78 and O-158 which were isolated from drinking water, migratory ducks (coot), wild birds (crows) and migratory shoveler duck, respectively, were known as the most frequent Avian Pathogenic and Avian Faecal E. coli (APEC and AFEC) serotypes infecting chickens, turkeys, ducks or other birds [67,69–71]. Moreover, O-18, O-78 and O-158 were previously isolated by Wenz et al. [12] from milk samples of cows suffering acute coliform mastitis with varying systemic disease severity.
The antibiotic resistance of E. coli is of a particular concern because it is the most common Gram-negative pathogen in humans [72]. In addition, resistant E. coli strains could transfer and acquire antibiotic-resistance elements to and from other bacteria [73]. In the current study, the serotyped E. coli isolates showed resistance against most of Cephalosporins group of antibiotics (Table 4). On contrary, all E. coli isolates showed considerable sensitivity to Carbapenems group of antibiotics except that isolated from teat skin and water (Table 4). Carbapenems, like imipenem or meropenem, are considered the “last line of defense” in the treatment of infections caused by multi-resistant Gram-negative bacteria. Previous studies stated that bacteria isolated from wild and migratory bird populations acquired resistance although they are not directly affected by antibiotic practices [74]. The number of studies describing the prevalence of ESBL-producing Enterobacteriaceae has increased rapidly around the world [75]. Out of the eight E. coli isolates, five ESBL producing E. coli isolates were detected (two from wild resident birds, one from migratory waterfowls, one from buffalo milk, and one from drinking water). In recent years, ESBL-producing Enterobacteriaceae have been recovered from different sources in the community, including cattle, chickens, and raw milk [76–78]. Interestingly, ESBL-producing E. coli in wildlife was firstly reported shortly after their isolation from livestock farming which could suggest the spread of ESBL-E. coli into the environment through manure [79], and subsequently spread to different ecological niches [80].
These results were also confirmed by sequence analysis of 16S rRNA gene of these eight isolates. The phylogenetic analysis revealed that the sequence from northern shoveler, eurasian coot, laughing dove, hooded crow and water were identical to each other suggesting the possible role of migratory birds to contaminate water sources in the farm since they were found in the same clade.
5. Conclusion
The study showed that faecal matter of wild and migratory birds could be considered as a potential risk factor for disseminating pathogenic and/or resistant E. coli in a dairy farm environment, leading to contaminating teat skin of lactating animals, triggering mastitis and/or milk contamination, and threatening animal and human health. Frequent removal of manure could lessen wild bird density inside dairy farms. Milking hygiene practices, as; washing worker hands, rinsing udder and teats in sanitizing solution, then drying them, followed by effective germicidal teat dip could reduce pathogenic bacterial load on the udder skin and prevent mastitis-causing pathogens from populating the environment or teat skin of the animals. Further research should be carried out to explore other environmental risk factors responsible for intramammary infections within dairy herds.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Bradley A. Bovine mastitis: an evolving disease. Vet J. 2002;164:116–128. [DOI] [PubMed] [Google Scholar]
- [2].Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res. 2003;34:475–491. [DOI] [PubMed] [Google Scholar]
- [3].Roussel P, Porcherie A, ReÂpeÂrant-Ferter M, et al. Escherichia coli mastitis strains: in vitro phenotypes and severity of infection in vivo. PLoS One. 2017;12:e0178285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kivaria FM, Noordhuizen JPTM, Kapaga AM. Evaluation of the hygienic quality and associated public health hazards of raw milk marketed by smallholder dairy producers in the Dar es Salaam region, Tanzania. Trop Anim Health Prod. 2006;38:185–194. [DOI] [PubMed] [Google Scholar]
- [5].Erskine RJ, Walker RD, Bolin CA, et al. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J Dairy Sci. 2002;85:1111–1118. [DOI] [PubMed] [Google Scholar]
- [6].Blowey RW, Edmondson PW. Mastitis control in dairy herds. Ipswich, England: Farming Press; 1995. [Google Scholar]
- [7].Bradley AJ, Leach KA, Breen JE, et al. Survey of the incidence and etiology of mastitis in dairy farms in England and Wales. Vet Rec. 2007;160:253–258. [DOI] [PubMed] [Google Scholar]
- [8].Botrel MA, Haenni M, Morignat E, et al. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhone-Alpes, France. Foodborne Pathog Dis. 2010;7:479–487. [DOI] [PubMed] [Google Scholar]
- [9].Shpigel NY, Elazar S, Rosenshine I. Mammary pathogenic Escherichia coli. Curr Opin Microbiol. 2008;11:60–65. [DOI] [PubMed] [Google Scholar]
- [10].Bean A, Williamson J, Cursons RT. Virulence genes of Escherichia coli strains isolated from mastitic milk. J Vet Med B Infect Dis Vet Public Health. 2004;51:285–287. [DOI] [PubMed] [Google Scholar]
- [11].Suojala L, Pohjanvirta T, Simojoki H, et al. Phylogeny, virulence factors and antimicrobial susceptibility of Escherichia coli isolated in clinical bovine mastitis. Vet Microbiol. 2011;147:383–388. [DOI] [PubMed] [Google Scholar]
- [12].Wenz JR, Barrington GM, Garry FB, et al. Escherichia coli isolates’ serotypes, genotypes, and virulence genes and clinical coliform mastitis severity. J Dairy Sci. 2006;89:3408–3412. [DOI] [PubMed] [Google Scholar]
- [13].Schukken YH, Gunther J, Fitzpatrick J, et al. Host-response patterns of intramammary infections in dairy cows. Vet Immunol Immunopathol. 2011;144:270–289. [DOI] [PubMed] [Google Scholar]
- [14].Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stanković B, Hristov S, Bojkovski J, et al. The possibility of dairy farms isolation assessment - biosecurity aspect. Biotechnol Anim Husbandry. 2011;27:1425–1431. [Google Scholar]
- [16].Hald B, Skov MN, Nielsen EM, et al. Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Vet Scand. 2016;58:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dolejska M, Bierosova B, Kohoutova L, et al. Antibiotic- resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta- lactamases in surface water and sympatric black-headed gulls. J Appl Microbiol. 2009;106:1941–1950. [DOI] [PubMed] [Google Scholar]
- [18].Shobrak MY, Abo-Amer AE. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Braz J Microbiol. 2015;45:1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cole D, Drum DJ, Stalknecht DE, et al. Free-living Canada geese and antimicrobial resistance. Emerg Infect Dis. 2005;11:935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Adzitey F, Saba CK, Teye GA. Antibiotic susceptibility of Escherichia coli isolated from milk and hands of milkers in Nyankpala community of Ghana. Curr Res Dairy Sci. 2016;8:6–11. [Google Scholar]
- [21].FDA National antimicrobial resistance monitoring system.2014. http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoring System/default.htm.
- [22].APHA Standard methods for examination of dairy products. 17th ed. New York, USA: American Public Health Association; 2004. [Google Scholar]
- [23].Zecconi A, Cesaris L, Liandris E, et al. Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathog. 2006;40:177–183. [DOI] [PubMed] [Google Scholar]
- [24].Piccinini R, Cesaris L, Dapra` V, et al. The role of teat skin contamination in the epidemiology of Staphylococcus aureus intramammary infections. J Dairy Res. 2009;76:36–41. [DOI] [PubMed] [Google Scholar]
- [25].BAM Bacteriological analytical manual, food and drug administration center for food safety and applied nutrition, Chapter 4. Enumeration of Escherichia coli and the coliform bacteria. 2002.
- [26].Quinn PJ, Markey BK, Carter ME, et al. Veterinary microbiology and microbial diseases. 1st ed. USA: Wiley-Blackwell Science; 2002. p. 544 ISBN-13: 978-0632055258. [Google Scholar]
- [27].Kok T, Worswich D, Gowans E. Some serological techniques for Microbial and viral infections In: Fraser J,A, Marmion B, Simmons A, editors. Mackie and Mccartney practical medical microbiology, collee, 14th India: Elsevier; 1996. ISBN: 9788131203934 179–204. [Google Scholar]
- [28].Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence Factors, the Genes defining the O157: h7serotype, and components of the type 2 shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40:3613–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Performance CLSI. Standards for antimicrobial susceptibility testing. 26th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. CLSI supplement M100S. [Google Scholar]
- [30].Egyptian standards of raw milk Raw milk specifications, Egyptian organization for standardization and quality control. No 7123. Egypt: Ministry of Industry; 2010. [Google Scholar]
- [31].McDermott JJ, Randolph TF, Staal SJ. The economics of optimal health and productivity in smallholder livestock systems in developing countries. Revue Scientifique Et Technique De L’Office International Des Epizooties. 1999;18:399–424. [DOI] [PubMed] [Google Scholar]
- [32].FAO (Food and Agriculture Organization of the United Nations). Milk for health and wealth. Prepared by J. Henriksen. FAO Diversification Booklet. 2009;6:70. [Google Scholar]
- [33].Devendra C. Smallholder dairy production systems in developing countries: characteristics, potential and opportunities for improvement – review. Asian-Australas J Anim Sci. 2001;14:104–113. [Google Scholar]
- [34].Moran J . Improved management requirements for high quality dairy cows on small holder farms in Indonesia. Padang (West Sumatra): International Dairy Seminar, Uni of Andalas; 2009.
- [35].Phiri BJ, Benshop J, French NP. Systematic review of causes and factors associated with morbidity and mortality on smallholder dairy farms in Eastern and Southern Africa. Prev Vet Med. 2010;94:1–8. [DOI] [PubMed] [Google Scholar]
- [36].FAO Impact of mastitis in small scale dairy production systems. Animal Production and Health Working Paper. No. 13. Rome; 2014. [Google Scholar]
- [37].Tyler JW, Cullor JS. Mammary gland health and disorders In: Smith BP, editor. Large animal internal medicine. 3rd London: Blackwell Science; 1990:1019–1022. [Google Scholar]
- [38].Mungube ED, Tenghagen BA, Regassa F, et al. Reduced milk production in udder quarters with subclinical mastitis and associated economic losses in crossbred dairy cows in Ethiopia. Trop Anim Health Prod. 2005;37:503–512. [DOI] [PubMed] [Google Scholar]
- [39].Batavani RA, Asri S, Naebzadeh H. The effect of subclinical mastitis on milk composition in dairy cows. Iran J Vet Res. University of Shiraz. 2007; 8:205–211. [Google Scholar]
- [40].Sori H, Zerihun A, Abdicho S. Dairy cattle mastitis in and around Sebeta, Ethiopia. Int J Appl Res Vet Med. 2005;3:332–338. [Google Scholar]
- [41].Ayano AA, Hiriko F, Simyalew AM, et al. Prevalence of subclinical mastitis in lactating cows in selected commercial dairy farms of Holeta district. J Vet Med Anim Health. 2013;5:67–72. [Google Scholar]
- [42].García-Sánchez F, Sánchez-Santana T, López-Vigoa O, et al. Prevalence of subclinical mastitis and associated microorganisms. Pastos Y Forrajes. 2018;41(1):33+. [Google Scholar]
- [43].Bangar YC, Singh B, Dohare AK, et al. A systematic review and meta-analysis of prevalence of subclinical mastitis in dairy cows in India. Trop Anim Health Prod. 2015;47:291–297. [DOI] [PubMed] [Google Scholar]
- [44].Patel YG, Trivedi MM. Quarter-wise prevalence of subclinical mastitis in crossbred 31 cows. Trends Biosci. 2015;8:4727–4729. [Google Scholar]
- [45].Singh A, Bansal BK, Singh RS, et al. Prevalence and diagnosis of subclinical mastitis in sahiwal dairy cows. Int J Livestock Res. 2018;8:337–345. [Google Scholar]
- [46].Hegde R, Isloor S, Prabhu KN, et al. Incidence of subclinical mastitis and prevalence of major mastitis pathogens in organized farms and unorganized sectors. Indian J Microbiol. 2013;53:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Skrzypek R, Wojtowski J, Fahr RD. Factors affecting somatic cell count in cow bulk tank milk- A case study from Poland. J Vet Med A. 2004;51:127–131. [DOI] [PubMed] [Google Scholar]
- [48].Mohamed AA, Wahba AK, Faisal RA. Some bacteriological and biochemical studies on subclinical mastitis in dairy cows. NY Sci. 2013;6:71–79. [Google Scholar]
- [49].Sri Balaji N, Saravanan R, Senthilkumar A, et al. Effect of subclinical mastitis on somatic cell count and milk profile changes in dairy cows. Int J Sci Environ Technol. 2016;5:4427–4431. [Google Scholar]
- [50].Hogan J, Smith KL. Coliform mastitis. Vet Res. 2003;34:507–519. [DOI] [PubMed] [Google Scholar]
- [51].Galiero G. The control of environmental mastitis Bubalus bubalis. 2002;I:26–28. [Google Scholar]
- [52].Hawari AD, Al-Dabbas F. Prevalence and distribution of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Jordan. Am J Anim Vet Sci. 2008;3:36–39. [Google Scholar]
- [53].El-Khodery SA, Osman SA. Acute coliform mastitis in buffaloes (Bubalus bubalis): clinical findings and treatment outcomes. Trop Anim Health Prod. 2008;40:93–99. [DOI] [PubMed] [Google Scholar]
- [54].Sayed SM. A contribution on Coliforms causing mastitis in cows with reference to serotypes and virulence factors of E. coli isolates. Assiut Univ Bull Environ Res. 2014;17:85–95. [Google Scholar]
- [55].Verma H, Rawat S, Sharma N, et al. Prevalence, bacterial etiology and antibiotic susceptibility pattern of bovine mastitis in Meerut. J Entomol Zool Stud. 2018;6:706–709. [Google Scholar]
- [56].Mansour MA, Ayoub MA, Abd El Aal SF, et al. Prevalence of virulent Escherichia coli in cow milk in Sharkia Governorate. Egypt Zag Vet J. 2013;41:57–65. [Google Scholar]
- [57].Maniruzzaman M, Khan MFR, Amin MM, et al. Isolation and identifaction of bacterial flora from milk of apparently health buffalo- cows. Int J Biol Res. 2010;1:13–16. [Google Scholar]
- [58].Sudhan NA, Sharma N Mastitis: An important production disease of dairy animals. 1st Ed. Gurgoan (India): Sarva Manav Vikash Samiti; 2010:72–88. [Google Scholar]
- [59].Palaha R, Chaudhary N, Kumar H. Detection of Escherichia coli from the udder of the dairy farm buffaloes in Phagwara region, Punjab, India. Vet World. 2012;5:522–525. [Google Scholar]
- [60].Vacheyrou M, Normand A-C, Guyot P, et al. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int J Food Microbiol. 2011;146:253–262. [DOI] [PubMed] [Google Scholar]
- [61].Neave FK, Dood FH, Kingwill RG. A method of controlling udder disease. Vet Rec. 1966;78:521–523. [DOI] [PubMed] [Google Scholar]
- [62].Goldberg TL, Gillespie TR, Rwego IB, et al. Forest fragmentation as cause of bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerg Infect Dis. 2008;14:1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].LeJeune JT, Homan J, Linz G, et al. Role of the European starling in the transmission of E. coli O157 on dairy farms. In: Proceedings 23rd Vertebrate Pest Conference, Davis, University of California; 2008; pp. 31–34. [Google Scholar]
- [64].Cernicchiaro N, Pearl DL, McEwen SA, et al. Association of wild bird density and farm management factors with the prevalence of E. coli O157: h7in dairy herds in Ohio (2007–2009). Zoonoses Public Health. 2012;59:320–329. [DOI] [PubMed] [Google Scholar]
- [65].Medhanie GA, Pearl DL, McEwen SA, et al. Dairy cattle management factors that influence on-farm density of European starlings in Ohio, 2007–2009. Prev Vet Med. 2015;120:162–168. [DOI] [PubMed] [Google Scholar]
- [66].Van Donkersgoed J, Berg J, Potter A, et al. Environmental sources and transmission of Escherichia coli O157 in feedlot cattle. Can Vet J. 2001;42:714–720. [PMC free article] [PubMed] [Google Scholar]
- [67].Wang Y, Cheng T, Xuehui YU, et al. Distribution of serotypes and virulence- associated genes in pathogenic Escherichia coli isolated from ducks. Avian Pathol. 2010;39:297–302. [DOI] [PubMed] [Google Scholar]
- [68].Nishikawa Y, Ogasawara J, Helander A, et al. An outbreak of gastroenteritis in Japan due to Escherichia coli O166. Emerg Infect Dis. 1999;5:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ewers C, Janssen T, Kiessling S, et al. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet Microbiol. 2004;104:91–101. [DOI] [PubMed] [Google Scholar]
- [70].Rodriguez-Siek KE, Giddings CW, Doetkott C, et al. Characterizing the APEC pathotype. Vet Res. 2005;36:241–256. [DOI] [PubMed] [Google Scholar]
- [71].Vandekerchove D, Vandemaele F, Adriaensen C, et al. Virulence-associated traits in avian Escherichia coli: comparison between isolates from colibacillosis-affected and clinically healthy layer flocks. Vet Microbiol. 2005;108:75–87. [DOI] [PubMed] [Google Scholar]
- [72].Salvadori M, Coleman BL, Louie M, et al. Consumption of antimicrobial-resistant Escherichia coli-contaminated well water: human health impact. PSI Clin Res. 2004;1:6–25. [Google Scholar]
- [73].Österblad M, Hadanen A, Manninen R, et al. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. J Antimicrob Chemother. 2000;44:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Literak I, Dolejska M, Janoszowska D, et al. Antibiotic- resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in Waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol. 2010;76:8126–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tham J. Extended-spectrum beta-lactamase-producing enterobacteriaceae: epidemiology, risk factors, and duration of carriage. Sweden: Department of Clinical Sciences/Malmö Infectious Disease Research Unit/Lund University; 2012. [Google Scholar]
- [76].Jakobsen L, Kurbasic A, Skjot-Rasmussen L, et al. Escherichia coli isolates from broiler chicken meat, broiler chickens, pork, and pigs share phylogroups and antimicrobial resistance with community-dwelling humans and patients with urinary tract infection. Foodborne Pathog Dis. 2010;7:537–547. [DOI] [PubMed] [Google Scholar]
- [77].Ramchandani M, Manges AR, DebRoy C, et al. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin Infect Dis. 2005;40:251–257. [DOI] [PubMed] [Google Scholar]
- [78].Vincent C, Boerlin P, Daignault D, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis. 2010;16:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kummerer K. Antibiotics in the aquatic environment – a review – part I. Chemosphere. 2009;75:417–434. [DOI] [PubMed] [Google Scholar]
- [80].O’Brien TF. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin Infect Dis. 2002;34:S78–S84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- FDA National antimicrobial resistance monitoring system.2014. http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoring System/default.htm.


