ABSTRACT
Tumour cells release diverse populations of extracellular vesicles (EVs) ranging in size, molecular cargo, and function. We sought to characterize mRNA and protein content of EV subpopulations released by human glioblastoma (GBM) cells expressing a mutant form of epidermal growth factor receptor (U87EGFRvIII) in vitro and in vivo with respect to size, morphology and the presence of tumour cargo. The two EV subpopulations purified from GBM U87EGFRvIII cancer cells, non-cancer human umbilical vein endothelial cells (HUVEC; control) and serum of U87EGFRvIII glioma-bearing mice using differential centrifugation (EVs that sediment at 10,000 × g or 100,000 × g are termed large EVs and small EVs, respectively) were characterized using transmission electron microscopy (TEM), confocal microscopy, nanoparticle tracking analysis (NTA), flow cytometry, immunofluorescence (IF), quantitative-polymerase chain reaction (qPCR), droplet digital polymerase chain reaction (ddPCR) and micro-nuclear magnetic resonance (μNMR). We report that both U87EGFRvIII and HUVEC release a similar number of small EVs, but U87EGFRvIII glioma cells alone release a higher number of large EVs compared to non-cancer HUVEC. The EGFRvIII mRNA from the two EV subpopulations from U87EGFRvIII glioma cells was comparable, while the EGFR protein (wild type + vIII) levels are significantly higher in large EVs. Similarly, EGFRvIII mRNA in large and small EVs isolated from the serum of U87EGFRvIII glioma-bearing mice is comparable, while the EGFR protein (wild type + vIII) levels are significantly higher in large EVs. Here we report for the first time a direct comparison of large and small EVs released by glioma U87EGFRvIII cells and from serum of U87EGFRvIII glioma-bearing mice. Both large and small EVs contain tumour-specific EGFRvIII mRNA and proteins and combining these platforms may be beneficial in detecting rare mutant events in circulating biofluids.
KEYWORDS: Extracellular vesicles, large extracellular vesicles, small extracellular vesicles, biomarkers, glioma
Introduction
Extracellular vesicles (EVs) are small membrane enclosed particles released by both cancer and non-cancer cells into the extracellular space. Once they leave the cells, EVs travel through extracellular matrices and reach various biofluids including blood. They can therefore be isolated from biological fluids and analysed for molecular signatures of the primary tumour. EVs contain proteins, RNA and DNA that can inform us of the molecular status of their respective producer cell. EVs are thought to be a potential means by which tumour cells can communicate and influence their microenvironment to promote tumour progression and dissemination, via altering immune responses, increasing cell proliferation and promoting angiogenesis, matrix remodelling, and, ultimately, metastasis [1,2]. EVs are explored as biomarkers for tumour RNA profiling and mutation detection at the time of initial diagnosis, longitudinally for patient follow up, and also for evaluation of tumour response to therapy [3–6].
EVs are markedly heterogeneous and it is increasingly recognized that each cell type secretes a unique mixture of different EV subpopulations that vary in size, content and function [7–13]. A commonly studied subfraction of EVs are small EVs, also called exosomes (~100 nm diameter) [14,15]. Glioblastoma-specific small EVs are demonstrated to contain several of the transcripts of parental tumour cells and can be used to detect EGFR variant III (EGFRvIII) mRNA transcripts and other tumour-specific mRNA and miRNAs from various biofluids [11,16–18]. Recent studies demonstrated that prostate cancer cells which exhibit amoeboid behaviour release large EVs, also called large oncosomes (>1000 nm diameter) [12,19–22], into the medium which result from pinching off of non-apoptotic blebs from the plasma membrane. These large EVs represent a distinct subclass of EVs, containing tumour-specific mRNA, miRNA and proteins that could be used as potential biomarkers for tumour diagnosis and monitoring. Large EVs have been associated with disease progression in the context of prostate cancer cells, but remain largely uncharacterized in GBM cells [12,19–22]. Hence, we sought to characterize large and small EVs in GBM cells, in vitro and in vivo. Currently, differential centrifugation is the most widely used technique to separate vesicles of different size or density [20,23]. Small EVs are conventionally sedimented at 100,000 × g spin using high-speed ultracentrifugation after a 2800 × g clearing step, while large EVs have been shown to sediment at a lower speed of 10,000 × g [21,24]. EVs that sediment at 100,000 × g or 10,000 × g are termed small EVs or large EVs, respectively. In this study, we performed sequential centrifugation (10,000 × g followed by 100,000 × g) for all EV isolations.
EGFRvIII mRNA and protein status can provide GBM-specific liquid biopsy based monitoring that can help not only diagnose but also evaluate tumour progression [8]. Since most groups focus on either large or small EVs for biomarker studies, we focused on understanding the differences between the two EV subfractions in terms of size, mRNA and protein content. We first sought to investigate if GBM cells release large EVs in vitro and in vivo. We then directly compared the two EV subpopulations (large and small EVs) derived from a glioma cell line (U87EGFRvIII) and a control, non-cancer cell line (HUVEC) for EGFRvIII mRNA and protein. We later quantified EGFRvIII mRNA and protein in both large and small EVs isolated from serum samples of U87EGFRvIII glioma-bearing xenograft models. Our findings suggest that both large and small EVs contain comparable quantities of EGFRvIII mRNA while large EVs contain higher levels of EGFR protein (EGFRwt + EGFRvIII) and combining both these EV subpopulations could result in higher rates of mutation detection.
Materials and methods
Cell line
The human GBM cell line U87EGFRvIII was obtained from the American Type Culture Collection (Manassas, VA). U87EGFRvIII glioma cells were stably engineered to express the deleted form of EGFR – EGFRvIII (for further details, please see [25]) and cultured in high glucose Dulbecco’s modified essential medium (DMEM, Life Technologies, Grand Island, NY), which contained 10% foetal bovine serum (FBS; Sigma, St. Louis, MO) and penicillin/streptomycin (10 IU ml−1 and 10 µg ml−1, respectively; Cellgro, Manassas, VA). HUVEC were provided by Drs. Francis W. Luscinskas and Kay Case, Cell Core Facility, Brigham and Women’s Hospital (supported by NIH P01 HI36028). HUVEC were cultured in gelatin-coated flasks using endothelial basal medium (Lonza, Allendale, NJ) supplemented with human epidermal growth factor (hEGF), hydrocortisone, and media with growth factors (GA-1000; Singlequots from Lonza). All in vitro experiments were performed with a cell confluency of 50–70% to minimize cell death. All the cell lines are periodically verified for mycoplasma contamination using commercial mycoplasma PCR (PCR Mycoplasma Detection Kit, Applied Biological Materials Incorporated, Richmond, British Columbia).
Xenograft tumour models
Ten adult nude mice (nu/nu NCI) were each injected subcutaneously in both flanks with 5 × 106 human GBM U87EGFRvIII cells (Figure 2(a)). Tumours were allowed to grow for 1 month, mice were deeply anaesthetized, and blood was drawn via cardiac puncture. Tumour mass at euthanization for each mouse was recorded as follows: 1–2.1 g; 2–1.5 g; 3–0.7 g; 4–0.8 g; 5–0.5 g; 6–1.1 g; 7–1.2 g; 8–0.9 g; 9–1.5; 10–1.2 g. Approximately, 1 ml of total blood was collected from each mouse and allowed to clot at room temperature for 30 min prior to being centrifuged at room temperature for 10 min at 1300 × g. Serum was transferred to a clean Eppendorf tube and stored at −80°C until processing. All samples were then thawed once and centrifuged at 10,000 × g for 30 min, yielding a pellet which was labelled “large EVs”. Then, the supernatant was ultracentrifuged at 100,000 × g for 1 h and the resulting pellet was labelled “small EVs”.
Figure 2.
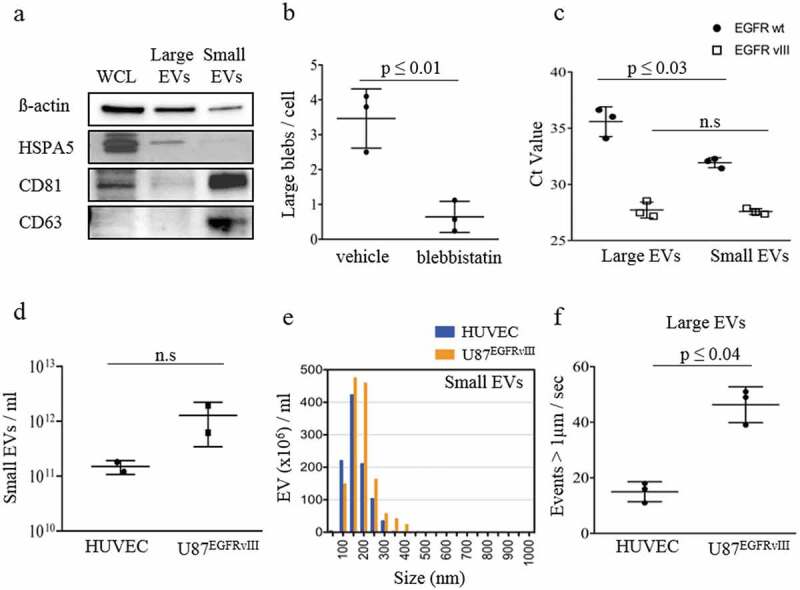
Characterization of large and small EV cargo released from GBM U87EGFRvIII cells.
(a) Western blot analysis of β-actin, HSPA5, CD81 and CD63 in whole cell lysate (WCL), large and small EV fractions isolated from U87EGFRvIII cells demonstrated WCL enriched in β-actin, HSPA5, CD81; large EVs enriched in HSPA5and small EVs enriched in CD81, CD63. (b) Quantitative analysis of large blebs released per cell in U87EGFRvIII cells treated with blebbistatin (a known inhibitor of bleb formation) or vehicle (mock) yielded a significant decrease in large EV release. (c) EGFRwt and EGFRvIII mRNA quantification using qRT-PCR demonstrated significantly higher amounts of EGFRwt mRNA in small EVs and similar amounts of EGFRvIII mRNA in both large and small EVs. (d) Nanoparticle tracking analysis (NTA) of particle counts of small EVs derived from HUVEC and U87EGFRvIII cells represent a similar number of small EVs released from both the cell lines. (e) Size distribution of small EVs derived from HUVEC and U87EGFRvIII cells depicted a similar size distribution of small EVs released from both the cell lines. (f) Flow-cytometry analysis of large EVs derived from HUVEC and U87EGFRvIII cells depicting higher large EV release from U87EGFRvIII cells. The results are presented as the mean ± SD ((f) n = 3; (d) n = 2).
Extracellular vesicles isolation
U87EGFRvIII cells were grown in 15-cm plates (~20 million cells/plate) in 20 ml media containing 5% EV-depleted FBS. For EV depletion, FBS was ultracentrifuged (100,000 × g) for 16 h at 4°C and the supernatant filtered under sterile conditions using a 0.22 µm filter (Millex-GV, PVDF; Millipore, Billerica, MA). Conditioned media was collected after 48 h and processed as follows: 300 × g speed spin at 4°C for 10 min, followed by transferring the supernatant to a clean Falcon tube and centrifuging again at 2,000 × g. The supernatant was removed and re-centrifuged at 10,000 × g for 30 min; the remaining pellet was resuspended in 100 µl of PBS and stored on ice for 1 h. This is labelled “large EVs’. The supernatant was transferred to Beckman polyallomer tubes and ultracentrifuged at 100,000 × g for 1 h at 4°C. The pellet containing predominantly small EVs was resuspended in 100 µl of PBS and labelled “small EVs”. A fixed angle rotor 70 Ti was used to isolate EVs from conditioned media. A MLA-55 fixed angle rotor was used to isolate EVs from serum of glioma-bearing mice.
RNA extraction and qRT-PCR
The large and small EV pellets were lysed in 700 µl Qiazol lysis buffer. The miRNeasy kit (Qiagen, Valencia CA) was used to isolate RNA from EV subfractions, as per the manufacturer’s recommendations. DNase digestion was performed on-column before assessing the quality and quantity of the extracted RNA using ThermoFisher Nanodrop (for accurate RNA quantification) and Agilent’s Bioanalyzer RNA 6000 pico (for sizing, quantification and quality control of the RNA). Equal volumes (14 µl) of RNA from large and small EV pellets were used as input for the cDNA reactions using SuperScript VILO (Invitrogen, Carlsbad, CA). All qRT-PCR reactions were performed in 25 µl reaction volumes using fast TaqMan MasterMix (Applied Biosystems, Foster City, CA). Amplification was performed using ABI PRISM 7500 (Applied Biosystems) set to the following conditions: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s; and 60°C for 1 min on standard mode. Primers used in this study were provided by Applied Biosystems and corresponded to the following sequences. EGFRvIII primers were as follows: EGFRvIII Forw: CTGCTGGCTGCGCTCTG; EGFRvIII Rev: GTGATCTGTCACCACATAATTACCTTTC; EGFR probe: TTCCTCCAGAGCCCGACT. EGFRwt primers were as follows: EGFR Forw: TATGTCCTCATTGCCCTCAACA; EGFR Rev: CTGATGATCTGCAGGTTTTCCA; EGFR Probe: AAGGAATTCGCTCCACTG. TaqMan ThermoFisher assay GAPDH-Hs03929097_g1 (within one exon; ThermoFisher).
Droplet digital PCR (ddPCR)
A volume of 5 µl of cDNA was used as input in duplicate reactions. About 20,000 droplets were generated using the AutoDroplet generator (Bio-Rad, Hercules, CA). PCR conditions were as follows: 95 °C for 10 min, 39 cycles at 94 °C for 30 s and 61 °C for 1 min. The last stage was 98 °C for 10 min followed by 4 °C. Droplets were analysed using the Droplet Reader (Bio-Rad). Gates were set to exclude all events from the cDNA no-template control sample. All events above the no-template control gates were considered positive. Concentrations were calculated in auto mode using the Bio-Rad software.
Nanoparticle tracking analysis (NTA)
EVs were purified by ultracentrifugation and quantified using the Nanosight LM10 (Malvern, Framingham, MA) [26,27]. Samples were diluted in PBS 1× (1:20) from the concentrated pellet. Each sample was recorded three times for 30 s, with manual monitoring of temperature and cameral level set to 14. Analysis was performed using the NTA v3.1 software, with detection threshold set to 7 (Malvern, Framingham, MA). NTA EV concentration was expressed in EVs/ml and EV size in mean values.
Transmission electron microscopy (TEM)
A 10 µl droplet of 4% paraformaldehyde fixed large and small EV suspension was transferred onto 200 mesh Formvar/carbon-coated nickel grids and allowed to adsorb 15 min. Suspensions were diluted, as needed, in phosphate-buffered saline. The grids containing adsorbed EVs were blotted and contrast-stained in 2% methyl cellulose (tylose)-uranyl acetate solution, then blotted on filter paper and air dried prior to analysis. Examination of preparations was done using a JEOL JEM 1011 transmission electron microscope at 80 kV. Images were collected using an AMT digital imaging system with proprietary image capture software (Advanced Microscopy Techniques, Danvers, MA).
Blebbistatin treatment and cholera toxin B labelling
Cells were treated with blebbistatin (50 µM) for 24 h or vehicle and then incubated with the cholera toxin B subunit directly conjugated with FITC label (CTxB-FITC) (Sigma) and imaged using the Axioplan 2 microscope (Zeiss, Oberkochen, Germany), as described in the previous studies [12,19,22].
Western blotting
Cells and EV pellets were lysed in RIPA buffer on ice for 15 min. The protein lysates were centrifuged at 12,000 × g for 15 min at 4°C. Protein lysates from cells and EVs were quantified using the Bradford Protein Assay (Bio-Rad, Hercules, CA). Samples were resolved by SDS-PAGE and then analysed using the following antibodies: rabbit polyclonal CD63 (1:1000; H-193; Santa Cruz), mouse monoclonal CD81 (1:1000; M38; Abcam) and rabbit monoclonal heat shock protein family A HSPA5 (1:1000; Abcam) and mouse monoclonal beta-actin (1:5000; AC-15; Sigma).
Flow cytometry analysis of large EVs
Following purification, large EVs were washed in PBS and their size was analysed using a LSRII Flow Cytometer (BD) as previously described [12,19,22]. For each sample, a minimum of 3000 events were recorded. Data analysis was performed using the FlowJo software (Treestar); only events that resulted >1 µm/s were taken into account.
EV preparation and labelling for μNMR
Isolated EVs were resuspended in PBS and labelled for µNMR analysis. Briefly, EVs were mixed with trans-cyclooctene (TCO)-modified antibodies. The antibody-targeted EVs were then coupled with magnetic nanoparticles (MNP) derivatized with 1,2,4,5-tetrazine (TZ), mixed within the microfluidic device and processed for μNMR measurements. All experiments were performed with TCO-modified isotype control antibodies to determine non-specific background binding. EV protein expression profiles were normalized by their CD63 expression to account for variations in EV numbers.
μNMR measurement
We performed μNMR measurements using a previously described miniaturized NMR relaxometer [17,28]. The operating magnetic field, generated by a portable permanent magnet, was 0.5 T. The R2 relaxation was measured on 1 µl sample volumes using Carr-Purcell-Meiboom-Gill pulse sequences with the following parameters: echo time, 4 ms; repetition time, 1 s; the number of 180° pulses per scan, 50; the number of scans, 8. All measurements were done in triplicate and data are displayed as mean ± SEM.
Statistical analysis
Statistical analyses were performed using unpaired two-tailed Student’s t-test in GraphPad Prism 8 software and p < 0.05 was considered statistically significant. The results are presented as the mean ± SD.
Results
Glioma cells release large and small extracellular vesicles
Large and small EVs were isolated from conditioned media derived from HUVEC and U87EGFRvIII glioma cells. Electron microscopy images of the two EV subpopulations from both HUVEC (Figure 1(a) and Supplementary Figure S1) and U87EGFRvIII cells (Figure 1(b) and Supplementary Figure S2) were obtained using TEM. Small EVs ranged approximately between 50 and 300 nm in diameter and displayed a lipid bilayer membrane and cup-shaped EVs. Large EVs were >1 µm in diameter and showed a definable structure and not aggregates of small EVs (Figure 1(a,b)). Cultured U87EGFRvIII glioma cells were stained with cholera toxin B (CTxB), which labels lipid membranes and allows for clear visualization of membrane blebs from which large EVs are known to originate [20,21]. Cells were visualized using confocal microscopy 10 min after labelling. Abundant large EVs were observed originating from the plasma membrane into the extracellular space (Figure 1(c)), with instances of large EVs enclosing small EVs (Figure 1(d,e)).
Figure 1.
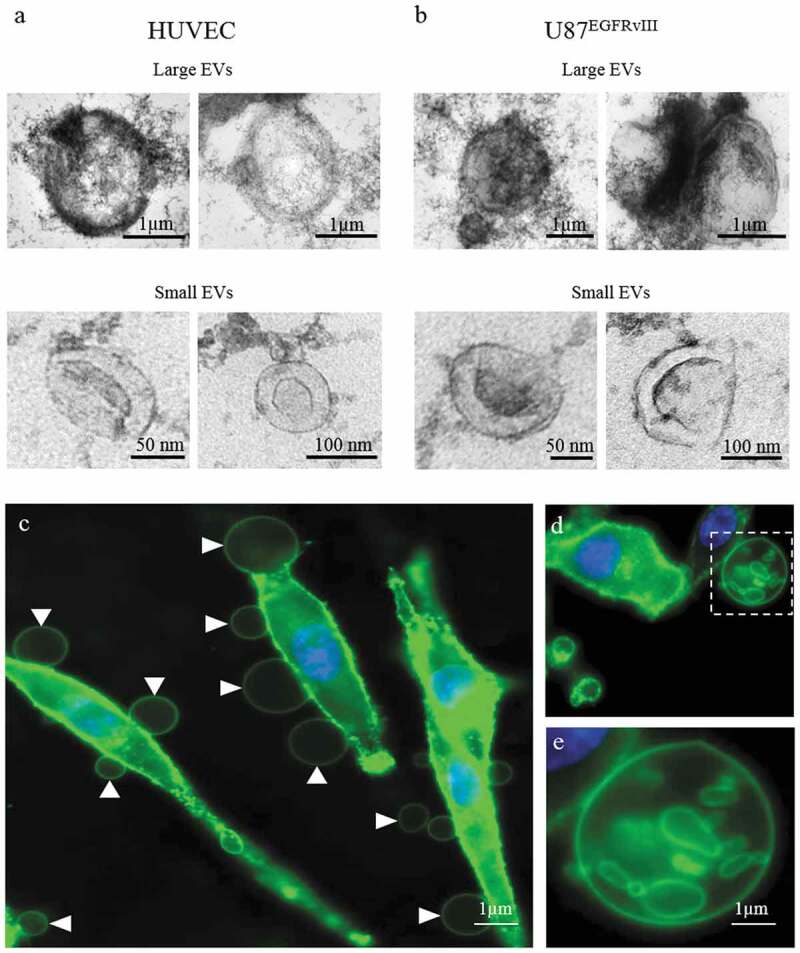
Structural visualization of large and small EVs released from HUVEC and GBM U87EGFRvIII cells.
Transmission electron microscopy (TEM) images of large and small EVs derived from (a) HUVEC and (b) U87EGFRvIII cells. (c) Confocal images of U87EGFRvIII cells stained with CTxB-FITC showing large EVs (arrowhead) blebbing from the plasma membrane. (d, e) Magnified snapshot of a large EV enclosing small EVs.
We isolated large and small EVs from U87EGFRvIII glioma cells using ultracentrifugation (see Methods). To confirm the nature of our EV subfractions, we performed western blot analysis for β-actin, HSPA5, CD81, CD63 in whole cell lysate (WCL), large and small EV fractions from U87EGFRvIII cells (10 µg of protein per lane). β-actin is a ubiquitous cytoskeletal protein, involved in cellular motility, structure and integrity, abundant in all eukaryotic cells. HSPA5 is a protein that affects glutamine metabolism [29] and is reported to be enriched in Large EVs [21]. Tetraspanins like CD81 and CD63 are enriched in small EVs and have been traditionally used as markers for small EVs [30]. We observed that large EVs contained higher levels of HSPA5, while small EVs were enriched in the canonical small EV markers, CD63 and CD81, which were very low in the large EV fraction. Of note, large EVs contained slightly higher levels of β-actin than small EVs. WCL contained high levels of β-actin, HSPA5 and CD81, but relatively low levels of CD63 (Figure 2(a); Supplementary Figure S3). This result confirmed that EVs isolated using 10,000 × g and 100,000 × g spin from U87EGFRvIII glioma cells were enriched in large EVs and small EVs, respectively.
Inhibition of cellular blebbing using blebbistatin induced a significant drop in large EV formation as compared to mock (vehicle only) treated cells (p ≤ 0.01; Figure 2(b)). This result suggests that, similar to prostate cancer, large EV formation in GBM cells is regulated by Myosin II and therefore in line with an amoeboid phenotype. The rate of blebbing was determined as the number of blebs released by a single cell over a minute.
We also quantified the EGFRwt and EGFRvIII mRNA levels within large and small EVs isolated from U87EGFRvIII glioma cells and show that both EV subfractions contained similar amount of EGFRvIII levels while EGFRwt levels were significantly higher in small EVs (p ≤ 0.03) compared to large EVs. Data were normalized to RNA input and GAPDH (p ≤ 0.03; Figure 2(c)).
Normal HUVEC cells and GBM cells release EVs that range in size
Nanoparticle tracking analysis (NTA) was used to study the counts and the size distribution of the small EV populations derived from both GBM U87EGFRvIII and HUVEC. NTA data indicate that both U87EGFRvIII and HUVEC release similar numbers of small EVs (Figure 2(d)) with similar size distributions (Figure 2(e); Supplementary Figure S4). However, due to the limitations of NTA in counting large EVs, we also employed flow cytometry to compare EVs that sediment at 10,000 × g from HUVEC and U87EGFRvIII cells. Large EVs were collected in the same volume of filtered PBS (200 µl) and processed for flow analysis. The analysis of events >1 µm normalized per time units, in seconds, showed that U87EGFRvIII glioma cells released a significantly higher number of large EVs compared to HUVEC cells (p ≤ 0.04; Figure 2(f)).
Large and small EVs from serum of glioma-bearing xenograft models contain EGFRvIII mRNA
Xenograft mouse models were used to study small and large EVs released from tumours in vivo. Nude mice were injected subcutaneously in both flanks with U87EGFRvIII cells (Figure 3(a)). Tumours were allowed to grow for 1 month. Blood was collected while mice were under deep anaesthesia, and serum samples were used to isolate both large and small EVs. We observed a significantly higher amount of total RNA in large EVs compared to small EVs (p ≤ 0.05; Figure 3(b)), in line with our previous observations [21]. Total RNA from large and small EV subpopulations represent the total extracellular RNA cargo isolated from 10,000 × g and 100,000 × g pellets, respectively (Figure 3(b)). Droplet digital PCR analysis of EGFRvIII mRNA reveals comparable copy numbers in both large and small EVs (Figure 3(c)).
Figure 3.
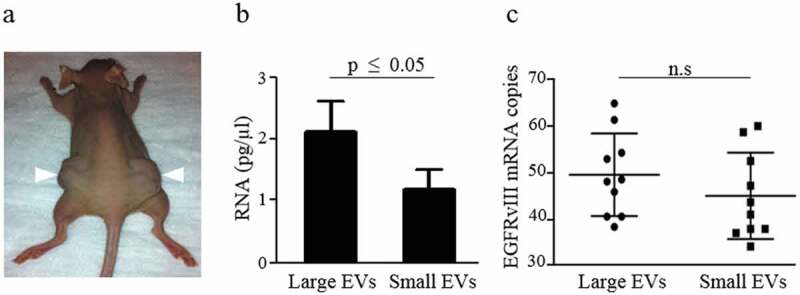
Characterization of large and small EVs released from glioma-bearing mice.
(a) A representative mouse injected with human GBM U87EGFRvIII cells at the time of sacrifice. Arrowheads indicate the site of injection of tumour cells. (b) Total RNA quantification using Agilent Bioanalyzer RNA 6000 pico analysis demonstrated significantly higher total RNA from large EVs. (c) EGFRvIII mRNA quantification using droplet digital PCR demonstrated similar amounts of EGFRvIII mRNA in both large and small EVs. The results are presented as the mean ± SD (n = 3; (c) n = 10).
Protein measurements of tumour biomarker using µNMR
We used a highly sensitive protein analytic technology, micro-nuclear magnetic resonance (µNMR), that enables rapid and sensitive biomarker detection from small amounts of input EVs (1 µl of EV samples). The EVs are targeted with immunospecific MNP and detected by miniature µNMR system [28,31,32]. The presence of MNP-labelled EVs speeds up the decay of NMR signals, as local magnetic fields from MNPs perturb coherent relaxation of 1H spins [17,33,34] (Figure 4(a)). CD63 and a combined EGFR protein (EGFRwt + EGFRvIII) levels were measured in large and small EVs from U87EGFRvIII-conditioned cell media. The results show CD63 protein enrichment in small EVs in comparison with large EVs (p ≤ 0.01), whereas the levels of EGFR protein (EGFRwt + EGFRvIII) in large EVs was significantly higher than in small EVs (p ≤ 0.04; Figure 4(b)). We also measured the EGFR protein (EGFRwt + EGFRvIII) levels in large and small EVs isolated from the serum of U87EGFRvIII glioma-bearing mice (p ≤ 0.00002; Figure 4(c)). The data, similar to the U87EGFRvIII cell derived EVs, showed significantly higher levels of EGFR (EGFRwt + EGFRvIII) protein levels in large EVs compared to small EVs (p ≤ 0.00002). Ratios of EGFR protein (EGFRwt + EGFRvIII) tumour marker levels in large and small EVs from glioma cell lines (1.4) and glioma-bearing xenograft models (1.5) were comparable (Figure 4(b,c)). This raises the possibility of the presence of specific patterns of protein distribution in large and small EVs.
Figure 4.
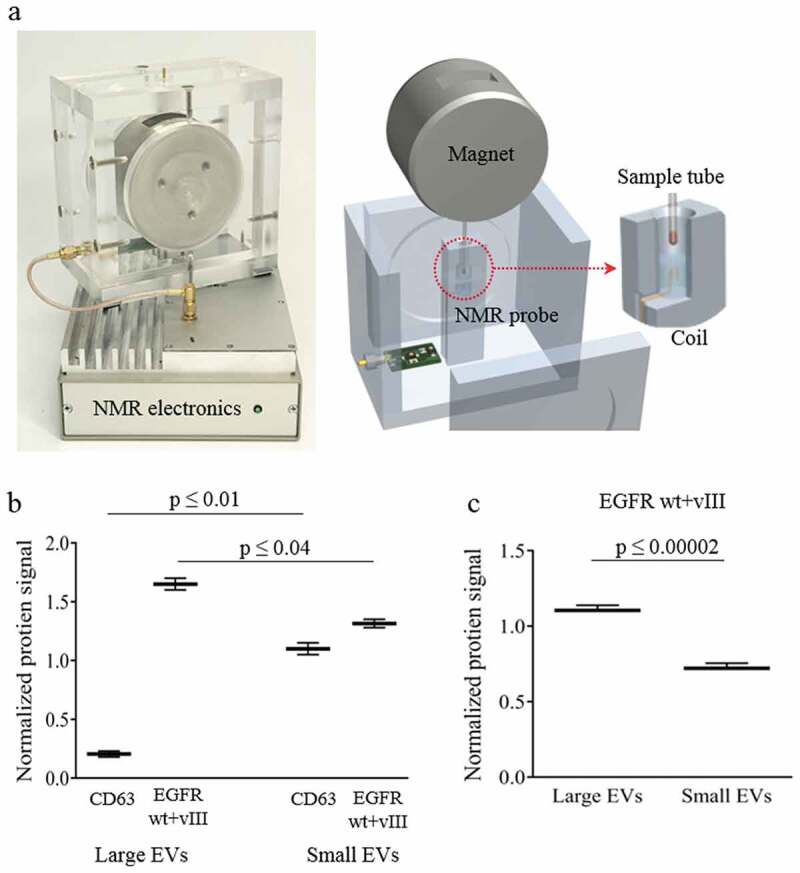
Protein quantification of large and small EVs released from glioma cells and glioma-bearing mice.
(a) Diagnostic micro-nuclear magnetic resonance (µNMR) for large and small EV protein quantification. (b) µNMR protein analysis of CD63 and EGFR (wt and vIII) protein expression levels in large and small EVs isolated from U87EGFRvIII cells, shows enrichment of CD63 in small EVs and enrichment of EGFR (wt and vIII) protein levels in large EVs. (c) µNMR protein analysis of EGFR (wt and vIII) protein expression levels in large and small EVs isolated from serum of U87EGFRvIII glioma-bearing mice shows higher levels of EGFR (wt and vIII) protein in large EVs. (Note: EGFR antibody detects both wt and mutant protein.) The results are presented as the mean ± SD (n = 3).
Discussion
EVs are released from all cells in varying sizes and with different contents. In this report, we aimed to characterize two different subpopulations of EVs released from a cancer cell line (GBM U87EGFRvIII) and normal non-cancer cell line (HUVEC). We show that GBM cells release both large and small EVs, while normal brain endothelial cells mostly release small EVs. Prostate cancer cells were previously described to release large EVs containing tumour-specific biomarkers [20,21,23,24]. Studies in prostate cancer have also shown that large EVs mediate intercellular transfer of functional miRNA [22]. Several studies in the past decade have shown that small EVs mediate transfer of several bioactive molecules including miRNA [11,35,36], indicating that they may play an important role in tumour progression and spreading malignancy [37]. Considering the heterogeneity in the EV subpopulations, our study explores the small and large EV subfractions in the context of size distribution, mRNA and protein content.
Our findings indicate that the canonical EGFR biomarker and the deleted variant III (EGFRvIII) mRNA and proteins are also readily detected in both subpopulations of EVs secreted from GBM U87EGFRvIII cells. Interestingly, the EGFRvIII mRNA levels were comparable in large and small EVs isolated from U87EGFRvIII cells, while large EVs has higher levels of EGFR protein (EGFRwt + EGFRvIII) compared to small EVs. These in vitro findings were consistent with the in vivo findings, where analysis of EGFRvIII mRNA and proteins from large and small EVs derived from the serum of glioma-bearing xenograft models, showed similar levels of EGFRvIII mRNA in both EV subpopulations and higher levels of EGFR protein (EGFRwt + EGFRvIII) in large EVs. This suggests that both large and small EVs are relevant for biomarker studies and should be included when analysing circulating biomarkers, particularly in cancer. It is important to ensure that biofluid collection protocols are inclusive of the large EVs found in the circulation as they may increase the number of tumour-specific signals that often times may be too low to detect, with consequent increase in the signal-to-background ratio and lead to more robust biomarker assays.
We also used micro-nuclear magnetic resonance, an advanced technology with high sensitivity and robust specificity to detect tumour protein markers from low levels of protein input. Our study combines the analysis of RNA and protein, in two distinct subpopulations of EVs which are relevant in tumours. We find comparable levels of the EGFRvIII mRNA in both populations in vitro and in vivo studies and hence both large and small EVs are a good representative signature of the parent tumour cells. In contrast, large EVs showed higher levels of EGFR protein (EGFRwt + EGFRvIII) compared to small EVs in both in vitro and in vivo studies, suggesting that more emphasis should be placed in studying large EV protein cargo. We and others have previously shown that mRNA and protein markers are found in small EVs [11,38–42] and large EVs [12,19–22] but this is the first study that compares the levels of the same tumour marker at the mRNA and protein levels in both these EV subpopulations, using complementary technologies. Large EVs, by definition enclose larger volume of mRNA and protein cargo, therefore, it is highly likely that copy numbers of mRNA of interest may be higher in a single large EV as compared to a small EV and future studies will address this hypothesis. We also found similar ratios of protein tumour marker in large and small EVs from cell lines (1.4) and serum samples from xenograft models injected with human GBM cells (1.5) suggesting that there may be a specific pattern of protein distribution in large and small EVs. It is important to note that membrane debris that sediment with large EVs could be a confounding factor for the higher protein amounts that were detected in the large EVs in both glioma cells and glioma-bearing mice.
Our study aims to understand the differences in EV mRNA and protein cargo in large and small EVs from glioma cells in vitro and in vivo. Understanding differences in RNA cargo and protein content in EV subpopulations will help gain insights into the EV subfractions that are the best representative of the primary tumour at diagnosis as well as over time. These general insights into the heterogeneity of EV subpopulations and their cargoes are pivotal in increasing the utility of EVs as circulating biomarkers.
Supplementary Material
Acknowledgments
We thank Dr. Johan Skog for valuable insight, Sabrina Roy for helpful discussions.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Declaration of interest statement
None of the authors declare any conflict of interest.
Supplementary Material
Supplementary data for this article can be accessed here.
References
- [1].Wang J, Bettegowda C.. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J Mol Diagn. 2017;19:24–10. [DOI] [PubMed] [Google Scholar]
- [2].Shankar GM, Balaj L, Stott SL, et al. Liquid biopsy for brain tumors. Expert Rev Mol Diagn. 2017;17:943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. [DOI] [PubMed] [Google Scholar]
- [7].Inda -M-D-M, Bonavia R, Mukasa A, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Bio. 2008;10:619–624. [DOI] [PubMed] [Google Scholar]
- [9].Andaloussi EL, Mäger S, Breakefield I. X. O. & Wood M. J. A.. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. [DOI] [PubMed] [Google Scholar]
- [10].Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Bio. 2008;10:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Di Vizio D, Kim J, Hager MH, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].van der Vos KE, Balaj L, Skog J, et al. Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol Neurobiol. 2011;31:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quezada C, Torres Á, Niechi I, et al. Role of extracellular vesicles in glioma progression. Mol Aspects Med. 2018;60:38–51. [DOI] [PubMed] [Google Scholar]
- [16].Zachariah MA, Oliveira-Costa JP, Carter BS, et al. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018;20:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reátegui E, van der Vos KE, Lai CP, et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun. 2018;9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Figueroa JM, Skog J, Akers J, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19:1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hager MH, Morley S, Bielenberg DR, et al. DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med. 2012;4:743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Di Vizio D, Morello M, Dudley AC, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181:1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Minciacchi VR, You S, Spinelli C, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Morello M, Minciacchi V, de Candia P, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12:3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meehan B, Rak J, Di Vizio D. Oncosomes - large and small: what are they, where they came from? J Extracell Vesicles. 2016;5:33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7:1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Grandi P, Fernandez J, Szentirmai O, et al. Targeting HSV-1 virions for specific binding to epidermal growth factor receptor-vIII-bearing tumor cells. Cancer Gene Ther. 2010;17:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gardiner C, Ferreira YJ, Dragovic RA, et al. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Balaj L, Atai NA, Chen W, et al. Heparin affinity purification of extracellular vesicles. Sci Rep. 2015;5:10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Z, Wang Y, Wu H, et al. GRP78 enhances the glutamine metabolism to support cell survival from glucose deficiency by modulating the β-catenin signaling. Oncotarget. 2014;5:5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Min C, Shao H, Liong M, et al. Mechanism of magnetic relaxation switching sensing. ACS Nano. 2012;6:6821–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yoon T-J, Lee H, Shao H, et al. Multicore assemblies potentiate magnetic properties of biomagnetic nanoparticles. Adv Mater. 2011;23:4793–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peck TL, Magin RL, Lauterbur PC. Design and analysis of microcoils for NMR microscopy. J Magn Reson B. 1995;108:114–124. [DOI] [PubMed] [Google Scholar]
- [34].Lee H, Sun E, Ham D, et al. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang J-K, Yang J-P, Tong J, et al. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J Neurooncol. 2017;131:255–265. [DOI] [PubMed] [Google Scholar]
- [36].Thuringer D, Chanteloup G, Boucher J, et al. Modulation of the inwardly rectifying potassium channel Kir4.1 by the pro-invasive miR-5096 in glioblastoma cells. Oncotarget. 2017;8:37681–37693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Le MTN, Hamar P, Guo C, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Demory Beckler M, Higginbotham JN, Franklin JL, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Graner MW, Alzate O, Dechkovskaia AM, et al. Proteomic and immunologic analyses of brain tumor exosomes. Faseb J. 2009;23:1541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Crescitelli R, Lässer C, Szabó TG, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell Vesicles. 2013;2. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xiao D, Ohlendorf J, Chen Y, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One. 2012;7: e46874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li CCY, Eaton SA, Young PE, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


