Abstract
Background
Tea-oil tree (Camellia oleifera) is a unique edible-oil tree in China, and anthracnose occurs in wherever it is cultivated, causing great economic losses each year. We have previously identified the Ascomycete fungus Colletotrichum fructicola as the major pathogen of anthracnose in Ca. oleifera. The purpose of this study was to characterize the biological function of Snf1 protein, a key component of the AMPK (AMP-activated protein kinase) pathway, for the molecular pathogenic-mechanisms of C. fructicola.
Results
We characterized CfSnf1 as the homolog of Saccharomyces cerevisiae Snf1. Targeted CfSNF1 gene deletion revealed that CfSnf1 is involved in the utilization of specific carbon sources, conidiation, and stress responses. We further found that the ΔCfSnf1 mutant was not pathogenic to Ca. oleifera, resulting from its defect in appressorium formation. In addition, we provided evidence showing crosstalk between the AMPK and the cAMP/PKA pathways for the first time in filamentous fungi.
Conclusion
This study indicate that CfSnf1 is a critical factor in the development and pathogenicity of C. fructicola and, therefore, a potential fungicide target for anthracnose control.
Keywords: Conidiation, Appressorium formation, Pathogenicity, C. fructicola
Background
Tea-oil tree (Camellia oleifera) is a commercial shrub native to China and has been widely cultivated throughout southern China for more than 2000 years with abundant edible oil in its seeds [1]. Owing to its lower cholesterol concentrations and the ability to decrease lipid concentration and prevent hypertension and arteriosclerosis, tea oil is considered as an excellent source of oil for human health, similar to that of olive oil [2, 3]. In China, Tea-oil tree covers more than 30,000 km2 and produces 250,000 tons of edible oil each year, however, it is still unable to meet the consumer demand for tea oil. One of the major limiting factors is the serious diseases occurred in Tea-oil tree.
Anthracnose is the most devastating disease in Tea-oil tree and happens in wherever it is cultivated [4]. The buds, fruits, and leaves of Tea-oil tree all are susceptible to the disease, causing the wilt or even fall of the plant tissues. Anthracnose typically results in 10%~ 30% reduction of tea oil each year, and the severely affected areas often experience more than 50% of tea oil losses. Our previous studies demonstrated that there are at least five pathogens of anthracnose in Tea-oil tree, namely: Colletotrichum fructicola, Colletotrichum siamense, Colletotrichum gloeosporioides, Colletotrichum camelliae, and Colletotrichum karstii. Among them, C. fructicola showed the widest distribution and highest pathogenicity, acting as the major pathogen [4, 5].
C. fructicola was first identified in coffee berries in northern Thailand in 2009 and has been found to cause diseases in more than 50 plants, including apple, pear, strawberry etc; and was distributed broadly on five continents [6–9]. However, despite its ecological and economic importance, very little is known about its molecular pathogenesis. The recent transcriptomic analysis showed that there are many putative pathogenic-related genes in C. fructicola, and most of them are significantly up-regulated in early infectious stages [6, 10]. However, the functions of these putative pathogenic-related genes remains confirmed.
Colletotrichum is among the top 10 fungal pathogens of plants, with infections primarily start with conidia, the asexual spores [11]. Briefly, conidia attach onto the host surface and germinate to produce germ tubes, the end of which will develop into specialized infection structures named appressoria. The enormous turgor pressure of the appressoria help the fungi to penetrate and colonize host tissues, causing disease lesions on plants. Each infected tissue may produce thousands of conidia that when released, can initiate a new disease cycle on new plant tissues [12, 13]. When first attaching on the host surface, the conidia probably lack nutrients and energy, thus changes in metabolic processes for pathogens are needed in order to grow and infect. There are at least three signaling pathways involved in nutrient metabolism in organisms, namely AMPK (AMP-activated protein kinase), cAMP-PKA (cAMP dependent protein kinase A), and TOR (target of rapamycin) [14, 15]. Among them, AMPK and cAMP-PKA are mainly involved in the carbon metabolisms and TOR responses to the nitrogen/ amino acid metabolisms. Previous studies have systematically demonstrated that the cAMP-PKA and TOR pathways regulate pathogenicity through host surface sensing in filamentous fungi [16–18]. However, little is known about the roles of AMPK pathway in filamentous fungi.
AMPK pathway is also called Snf1 (sucrose non-fermenting 1) pathway in Saccharomyces cerevisiae, which is a heterotrimeric complex composed of the catalytic α subunit Snf1, a regulatory β subunit (one of the Gal83, Sip1 and Sip2) and the γ subunit Snf4 [19]. The serine/threonine protein kinase Snf1 is at the center of the heterotrimeric complex and is required for the adaptation of yeast cells to glucose limitation [20]. In high glucose concentrations Snf1 is inactivated. In contrast, when glucose is limited, the threonine residue of Snf1 is phosphorylated and activated [21]. The active Snf1 regulates the expression of more than 400 genes involved in adaptation to nutrient stress [22]. Besides the roles in carbon metabolism, more and more studies have revealed that Snf1 plays important roles in autophagy, aging, biofilm formation, mitochondrial homeostasis and the response to various environmental stresses [20, 23–25]. To date, Snf1 homologs have been identified in a small number of filamentous fungi and found to play critical roles in the pathogenicity of Magnaporthe oryzae, Verticillium dahliae, and Cochliobolus carbonum [26–28]. Despite the importance, no information is available regarding the roles of such proteins in forest fungal pathogens including C. fructicola. Here, we identified the protein kinase CfSnf1 from C. fructicola and characterized its functions.
Results
Identification and phylogenetic analysis of CfSnf1 in C. fructicola
Using S. cerevisiae Snf1 sequence as the trace, we acquired its single homolog in C. fructicola genome database by a BLAST_P search. Then, we submitted its sequence to NCBI database (https://www.ncbi.nlm.nih.gov/) (GenBank accession number MN094751) and named it CfSnf1. CfSnf1 was predicted to encode 741 amino acids and phylogenetic analysis revealed that CfSnf1 shows sequence conservation among other fungi Snf1 proteins; CfSnf1 shows higher amino acid sequence homology with that of C. gloeosporioides (96% identify and 96% similarity) and lesser homology with S. cerevisiae Snf1 (still 43% identify and 56% similarity) (Fig. 1a). This result indicates that Snf1 proteins are well conserved in fungi.
Fig. 1.
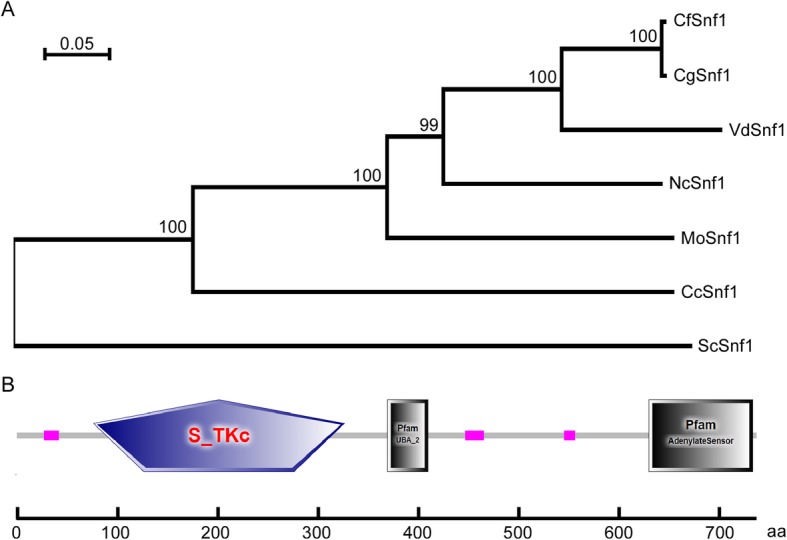
Phylogenetic analysis and domain prediction of CfSnf1. (a) The Snf1 proteins from diverse fungi were aligned using the CLUSTAL_W and the neighbour-joining tree was constructed by MEGA 5.05 with 1000 bootstrap replicates. The organisms and GenBank accession numbers are shown as follows: CfSnf1 (C. fructicola, MN094751); CgSnf1 (C. gloeosporioides, EQB56372.1); VdSnf1 (V. dahliae, RXG48310.1); NcSnf1 (N. crassa, XP_958665.3); MoSnf1 (M. oryzae, XP_003718181.1); CcSnf1 (C. carbonum, AF159253); ScSnf1 (S. cerevisiae, NP_010765.3). (b) The domains of CfSnf1 were predicted. The purple pentagon indicates the S_TKc (Serine/Threonine protein kinases, catalytic) domain, the gray quadrilaterals represent the UBA (ubiquitin-associated) domain and the Adenylate Sensor domain, and pink boxes refer to three low complexity regions
The domain prediction using the SMART website (http://smart.embl-heidelberg.de/) suggested that CfSnf1 contains a S_TKc (Serine/Threonine protein kinases, catalytic) domain, a UBA (ubiquitin-associated) domain, an Adenylate Sensor domain, and three low complexity regions (Fig. 1b).
Targeted deletion of CfSNF1 gene in C. fructicola
To characterize the functions of CfSnf1, a CfSNF1 gene-deletion construct was generated according to the homologous recombination principle by replacing the coding region with a hygromycin-resistance cassette (HPH) (Additional file 1: Figure S1a). Putative transformants were screened on hygromycin media, then verified by PCR amplification, and we thus obtained two deletion mutants ΔCfSnf1#3 and ΔCfSnf1#5 (Additional file 1: Figure S1b). Since these two mutants showed the same biological phenotypes, we selected ΔCfSnf1#3 for further analysis. To complement the mutant strain, the genomic DNA sequence of CfSNF1 containing a 1.5-kb promoter was retransformed to the ΔCfSnf1#3 mutant and restored all of the mutant defects.
CfSnf1 is involved in the utilization of specific carbon sources and aerial hyphae growth
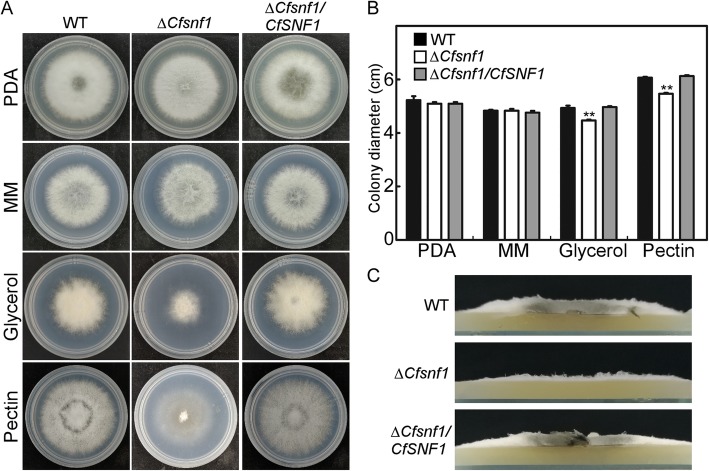
Snf1 proteins have been demonstrated to be important for the utilization of specific carbon sources in V. dahliae and M. oryzae [26, 27]. To investigate the functions of CfSnf1 in utilization of carbon sources, Wide-Type (WT), ΔCfSnf1 mutant, and complemented strain ΔCfSnf1/CfSNF1 were cultured in the plates of PDA (Potato Dextrose Agar), MM (Minimal Medium), and MM supplemented with 1% Glycerol or 1% Pectin as sole carbon source for 4 days. All the strains showed similar growth levels on PDA and MM, while ΔCfSnf1 mutant showed significantly reduced growth rates to WT and ΔCfSnf1/CfSNF1 on MM supplemented with Glycerol or Pectin (Fig. 2a and b). Meanwhile, we also found that ΔCfSnf1 mutant exhibit a flat colony compared with the fluffy colony of WT and ΔCfSnf1/CfSNF1, owing to its reduced aerial hyphal growth (Fig. 2a and c).
Fig. 2.
CfSnf1 is involved in the utilization of specific carbon sources and aerial hyphae growth. (a) Growth of WT, ΔCfSnf1 mutant, and complemented strain ΔCfSnf1/CfSNF1 on the PDA, MM and MM plates supplemented with 1% Glycerol or 1% Pectin as sole carbon source for 4 days at 28 °C. (b) Colony diameters were measured and statistically analyzed by Duncan analysis from three replicates. Asterisks indicate significant differences (p < 0.01). (c) Aerial hyphae growth is reduced in ΔCfSnf1 mutant. Strains were cultured in PDA for 4 days and colony side views are shown from three replicates
CfSnf1 is important in asexual development
Like most fungal pathogens, asexual conidia are important for the disease cycle and infection in Colletotrichum [29, 30]. To examine the role of CfSnf1 in conidiation, WT, ΔCfSnf1 mutant, and ΔCfSnf1/CfSNF1 were cultured in liquid shaking PDB for 4 days. WT and ΔCfSnf1/CfSNF1 showed more than 220 × 104 conidia per milliliter, comparing with the less than 5 × 104 conidia per milliliter for ΔCfSnf1 mutant (Fig. 3). This result demonstrates that CfSnf1 is important in conidiation.
Fig. 3.
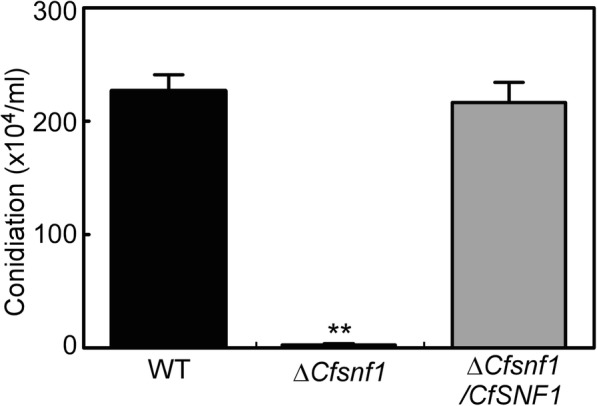
CfSnf1 is important in asexual development. The conidia produced by WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 cultured in PDB medium for 4 days were collected and quantified, then analyzed by Duncan analysis (p < 0.01) from three replicates. Three independent experiments yielded similar results
CfSnf1 is essential for pathogenicity
As a plant-pathogenic fungus, we concentrated our interests on the role of pathogenicity for CfSnf1 in C. fructicola. Mycelial plugs of the WT, ΔCfSnf1 mutant, and ΔCfSnf1/CfSNF1 were inoculated onto the intact Ca. oleifera leaves. After 4 days incubation, ΔCfSnf1 mutant caused no lesions, in contrast to the large and typical lesions produced by the WT and ΔCfSnf1/CfSNF1 (Fig. 4a left panel). To examine whether CfSnf1 plays roles in infectious growth, we further carried out the pathogenicity assay on wounded leaves. The result showed that ΔCfSnf1 mutant still causes no lesions, compared with the typical lesions of WT and ΔCfSnf1/CfSNF1 (Fig. 4a right panel). This result indicates that CfSnf1 is essential for pathogenicity.
Fig. 4.

CfSnf1 is essential for pathogenicity. (a) Mycelial plugs of the WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 were inoculated onto unwounded or wounded C. oleifera leaves with three replicates, and photographed at 4 days post inoculation (dpi), CK: controls. (b) Colonies of the strains with three replicates were cultured on top of cellophane membranes placed on PDA plates (Before). After 3 days incubation, the cellophane membranes with fungal dishes were removed, and the plates were further cultured for 3 days and photographed (After)
To further analyze the pathogenicity defect of ΔCfSnf1 mutant, we performed the cellophane penetration assay. Colonies of the strains were first cultured on top of cellophane membranes placed on PDA plates for 3 days incubation, then the cellophane membranes with fungal dishes were removed. After the further incubation for 3 days, we found ΔCfSnf1 mutant could not penetrate the cellophane, compared with the mycelial growth of WT and ΔCfSnf1/CfSNF1 on the plates (Fig. 4b). This result suggests that CfSnf1 is required for penetration.
CfSnf1 is localized to cytoplasm
To detect the subcellular localization of CfSnf1, we fused a green fluorescent protein (GFP) tag to the C-terminus of CfSnf1 and introduced it into the ΔCfSnf1 mutant. Strong GFP signals were uniformly distributed throughout the cytoplasm, but not in vacuoles, both in hyphae and conidia (Fig. 5). This result suggests the expression and cytoplasm-localized pattern of CfSnf1 in hyphae and conidia.
Fig. 5.
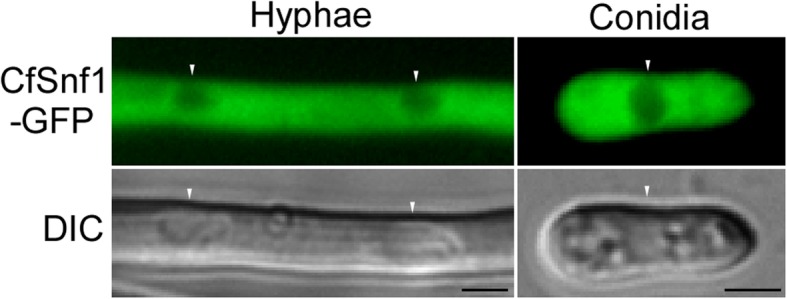
CfSnf1 is localized to cytoplasm. The localization pattern of CfSnf1 in the hyphae and conidia. Three independent biological experiments were carried out with three replicates each time. Arrows indicate vacuoles. DIC: differential interference contrast. Bar = 5 μm
CfSnf1 is involved in response to osmotic stress
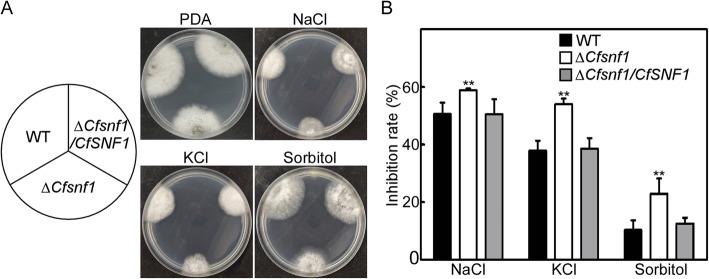
For normal growth and infection, fungi must undergo many types of stresses in nature, such as osmotic stress, oxidative stress and cell wall integrity stress etc. Studies in yeast concluded that ScSnf1 plays critical roles in response to various environmental stresses [20, 31, 32]. Here we investigated the roles of CfSnf1 in the response to environmental stresses. We first cultured the WT, ΔCfSnf1 mutant, and ΔCfSnf1/CfSNF1 on PDA plates supplemented with osmotic stress (1 M NaCl, 1 M KCl, and 1 M sorbitol) for 4 days. We found that ΔCfSnf1 mutant showed significant higher inhibition rates than that of WT and ΔCfSnf1/CfSNF1 in all three osmotic stresses (Fig. 6a and b). These results indicate that CfSnf1 is involved in response to osmotic stress. Then, we examined the sensitivity of the strains to two cell wall integrity inhibitors, Congo red (CR) and sodium dodecyl sulfate (SDS), and oxidative stress H2O2. Consistently, all the strains showed similar inhibition rates on CR, SDS and H2O2 plates (Additional file 2:Fig. S2).
Fig. 6.
CfSnf1 is involved in response to osmotic stress. (a) The WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 strains were cultured onto PDA plates with different osmotic stresses (NaCl, KCl, and Sorbitol) at 28 °C for 4 days and photographed. (b) Statistical analysis of inhibition rates of the strains to osmotic stress. Three independent biological experiments were carried out with three replicates each time, and similar results were obtained among the biological replicates. Error bars represent SD of three replicates and asterisks represent significant differences (p < 0.01)
CfSnf1 is required for appressorium formation
Based on the fact that ΔCfSnf1 mutant caused no lesions on both unwounded and wounded tea-oil tree leaves and its non-penetration on cellophane membranes, we wondered whether the mutant could not produce any functional appressorium. To further elucidate the mechanisms of the abolished pathogenicity in ΔCfSnf1 mutant, the conidia of WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 were inoculated onto hydrophobic artificial surfaces to induce the appressorium formation. At 4 h post inoculation (hpi), all the strains exhibited more than 80% conidial germination rate and showed no significant difference among them. However, the germ tubes of ΔCfSnf1 were elongated abnormally and no appressoria were developed at 24 hpi, in contrast with the normal germ tubes and more than 80% appressorium formation rate in WT and ΔCfSnf1/CfSNF1 (Fig. 7a and b). This result indicates that CfSnf1 is required for appressorium formation.
Fig. 7.
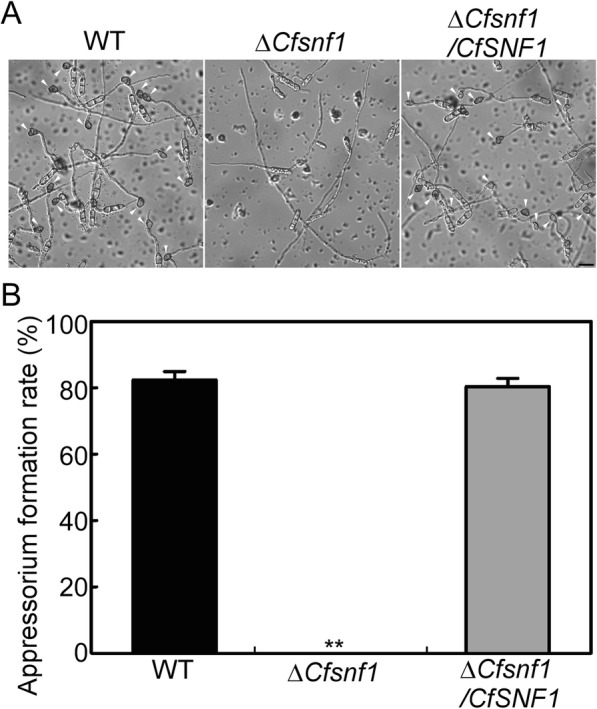
CfSnf1 is required for appressorium formation. (a) Conidia collected from the WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 strains were inoculated onto hydrophobic artificial surfaces at 24 h post inoculation (hpi) and photographed. Arrows indicate appressoria. (b) The percentage of appressorium formation was statistically analyzed. Error bars represent SD and asterisks indicate statistically significant differences (p < 0.01) from three replicates. Bar = 10 μm
cAMP treatment partially restores the appressorium formation defect of ΔCfSnf1 mutant
Studies in M. oryzae, which acts as the model for studying the mechanisms of fungal pathogenesis and host-microbe interactions, have revealed that the loss of cAMP/PKA signaling pathway abolishes appressorium formation and appressorium formation can be rescued in strains carrying deletions upstream of cAMP/PKA by cAMP treatment [31, 33, 34]. Therefore, we performed cAMP treatment for ΔCfSnf1, when inducing appressorium formation. The result showed that 88% of ΔCfSnf1 conidia germinated but do not formed appressoria and the rest conidia were not germinated, however, about 20% of ΔCfSnf1 conidia formed appressoria when cAMP was added (Fig. 8). This result reveals that cAMP treatment could partially restore the appressorium formation defect of ΔCfSnf1 mutant.
Fig. 8.
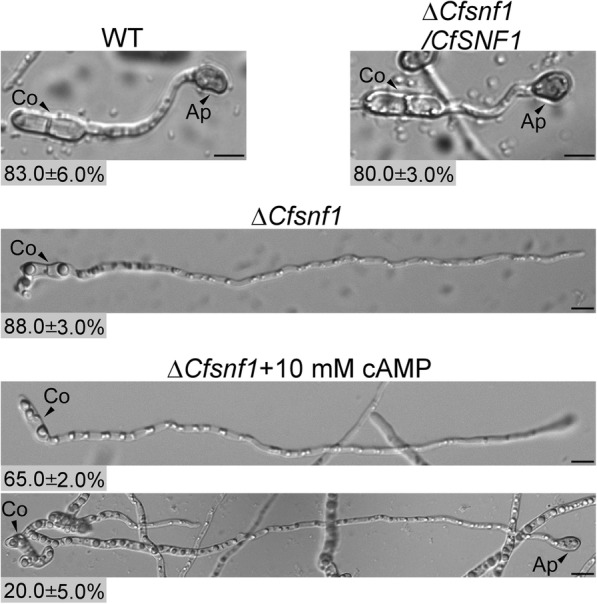
cAMP treatment partially restored the appressorium formation of ΔCfSnf1 mutant. Appressorium formation rates were determined at 24 hpi from 100 spores per hydrophilic slide for WT, ΔCfSnf1, ΔCfSnf1/CfSNF1 and ΔCfSnf1 with 10 mM cAMP treatment, repeated in triplicate with three replicates each time. Co: Conidia; Ap: Appressorium. Bar = 10 μm
Discussion
In the present study, we characterized CfSnf1 as the homolog of S. cerevisiae Snf1 in C. fructicola. We found that CfSnf1 plays critical roles in the utilization of specific carbon sources, conidiation, appressorium formation, stress responses and pathogenicity in C. fructicola.
Targeted CfSNF1 gene deletion mutant ΔCfSnf1 showed similar mycelial growth rate as the WT in PDA and MM plates, but showed decreased growth rate compared to WT on MM with Glycerol or Pectin as sole carbon source. Considering the fact that PDA and MM medium use glucose for carbon source and the yeast Snf1 is inactivated in high glucose concentrations and activated in low glucose [21], we speculate that CfSnf1 also mainly functions in low glucose condition in C. fructicola. Moreover, the homologs of Snf1 in V. dahliae and C. carbonum are also required for the utilization of specific carbon sources [26, 27], which is consistent with our study. These findings indicate that Snf1 proteins shared conserved mechanisms during carbon utilization.
Asexual conidia play critical roles in the disease cycle and infection of Colletotrichum [29, 30]. The ΔCfSnf1 mutant produced dramatically decreased conidiation, which is consistent with the studies in M. oryzae [28, 35]. However, the ΔCcSnf1 mutant showed normal conidiation in C. carbonum and the ΔVdSnf1 mutant even produced more conidia in V. dahliae [26, 27]. These differences in conidiation by SNF1 mutants of the fungal pathogens reflect distinct functions of Snf1 proteins during asexual development in different fungal species.
The studies in yeast revealed the roles of Snf1 in response to various environmental stresses through the activation of downstream proteins [20, 36]. Our study demonstrated that CfSnf1 is involved in response to osmotic stress but plays no role in the response to cell wall integrity stress and oxidative stress in C. fructicola. Such differences probably contribute to the specificity of CfSnf1 in stress responses. Furthermore, since fungi must undergo many types of stresses in nature for normal infection, the function of CfSnf1 in osmotic stress response might foretell its role in pathogenicity.
The ΔCfSnf1 mutant were non-pathogenic on intact Tea-oil leaves, indicating the penetrating defect of the mutant and its non-penetration on cellophane membranes confirmed this. Furthermore, the lack of pathogenicity on wounded Tea-oil leaves also revealed the host-colonizing defect of ΔCfSnf1 mutant. We reasoned that the abolished pathogenicity of ΔCfSnf1 mutant was directly due to the defect in appressorium formation, which is essential for the penetration and host-colonization of Colletotrichum [29, 30]. In addition, the defect of appressorium formation in ΔCfSnf1 mutant was partially restored by cAMP, which is a positive determinant in cAMP/PKA pathway [31]. Thus, we predict a relationship between Snf1 and cAMP/PKA pathway in C. fructicola, which has also been reported but not clearly clarified in yeast and mammalian cells [21, 37]. Though the related mechanism is unknown and needs to be further investigated, at least, we establish the first crosstalk between Snf1 and cAMP/PKA pathway in filamentous fungi.
Conclusion
This study concluded that CfSnf1 is involved in utilization of specific carbon sources, conidiation, and stress responses in C. fructicola. CfSnf1 also functions as a key regulator for appressorium formation that are crucial for the pathogenicity of the fungus.
Methods
Strains and culture conditions
C. fructicola CFLH16 was used as the wild-type strain. All strains were cultured on PDA (200 g peeled potato, 20 g dextrose, and 15 g agar in 1 L ddH2O) plates at 28 °C in the darkness, unless indicated otherwise. The strains were cultured in liquid PDA medium in darkness shaking at 28 °C for 2 days and collected for the extraction of genomic DNA.
Phylogenetic tree construction and domain prediction
The Snf1 proteins of C. fructicola, C. gloeosporioides, V. dahliae, Neurospora crassa, M. oryzae, C. carbonum, and S. cerevisiae were acquired from the NCBI database (https://www.ncbi.nlm.nih.gov/). The phylogenetic tree was constructed by MEGA 5.05 programs using neighbor-joining method with 1000 bootstrap replicates. The domain of CfSnf1 was predicted by the SMART website (http://smart.embl-heidelberg.de/).
Gene deletion and complementation assays
CfSNF1 targeted gene deletion was performed by one-step replacement strategy [38]. Two about 1.0-kb DNA fragments flanking the CfSNF1 gene and 1.4-kb HPH gene were amplified using primes (Additional file 3: Table. S1), then the two flanking sequences were ligated to the flanks of HPH, respectively, by overlap PCR. The PCR products were further cloned into the pMD19-T vector. After sequencing, the 3.4-kb fragments, which contain the flanking sequences and hygromycin cassette, were amplified and transformed into the protoplasts of wild-type strain. The transformants were selected by hygromycin and screened by PCR. For complementation assays, approximately 1.5-kb native promoter and the full-length of CfSNF1 were amplified and ligated to the pYF11 vector (bleomycin resistance). After sequencing, the fused-pYF11 plasmids were transformed into the protoplasts of ΔCfSnf1 mutant for complementation.
Growth assays on different carbon sources
The strains were cultured on PDA, MM (6 g NaNO3, 0.52 g KCl, 0.152 g MgSO4·7H2O, 1.52 g KH2PO4, 0.01 g VB1, 1 ml 1000 × trace elements, 10 g Glucose, and 15 g agar in 1 L ddH2O), and MM with the Glucose was substituted by 1% Glycerol or 1% Pectin. After 4 days incubation, the colony diameters were measured and statistically analyzed.
Stress response assays
The strains were cultured on PDA and PDA added with different stresses, including osmotic stress (1 M NaCl, 1 M KCl, and 1 M Sorbitol), cell wall integrity stress (0.01% SDS and 400 μg/ml CR), and oxidative stress (10 mM H2O2). After 4 days incubation, the colony diameters were measured and the inhibition rates were statistically analyzed.
Conidiation and appressorium formation assays
For conidiation, the strains were cultured in liquid PDB (Potato Dextrose Broth) for 4 days, then filtered with three layers of lens paper and the conidia were collected and statistically analyzed. For appressorium formation, the collected conidia were resuspended to a concentration of 1 × 105 spores/mL and inoculated onto hydrophobic artificial surfaces for germination and appressorium formation. For cAMP treatment, the conidial suspensions of ΔCfSnf1 mutant were added with 10 mM cAMP and then were induced for appressorium formation.
Pathogenicity and penetration assays
For pathogenicity assay, the mycelial plugs of the strains were inoculated onto the leaf margin of the detached Tea-oil tree leaves. The inoculated leaves were kept in a humidity plate with a 12 h light and 12 h dark cycle for 4 days and then photographed. The penetration assay on cellophane membranes was performed as previously described [39, 40] with minor modifications, colonies of the strains were first cultured on top of cellophane membranes placed on PDA plates. After 3 days incubation, the cellophane membranes with fungal dishes were removed, and the plates were further cultured for 3 days.
Localization observation
The green fluorescent protein (GFP) tag was fused to the C-terminus of CfSnf1 and transformed into the ΔCfSnf1 mutant. The fluorescence of the hyphae and conidia was observed under microscope.
Supplementary information
Additional files 1: Figure S1. Generation of the CfSNF1 gene deletion mutants in C. fructicola. (A) Strategy for the construction of gene replacement fragment. ~ 1.0-kb left fragment (LF) and right fragment (RF) flanking the CfSNF1 gene and 1.4-kb HPH gene from pCX62 vector were amplified, then the LF and RF were ligated to the flanks of HPH, respectively, by overlap PCR. The PCR products were further cloned into the pMD19-T vector and obtained the vector of pMD-CfSNF1-KO. After sequencing, the 3.4-kb fragments, which contain the LF, RF and HPH, were amplified for gene replacement. (B) Schematic illustration for deletion strategy of CfSNF1 gene. (C) Validation of the gene deletion mutants by PCR amplified with primers 1 (NBF/NBR) and primers 2 (BWF/HPHR). M: marker; +: positive control; −: negative control; #3 and #5: mutants.
Additional files 2: Figure S2. CfSnf1 is not involved in the tolerance of cell wall stress and oxidative stress. (A) The WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 strains were cultured onto PDA plates with different cell wall stresses (CR and SDS) and oxidative stress (H2O2) at 28 °C for 4 days and photographed. (B) Statistical analysis of inhibition rates of the strains to cell wall stresses and oxidative stress. Error bars represent SD of three replicates.
Additional files 3: Table S1. Primers used in this study.
Acknowledgements
Not applicable.
Abbreviations
- CR
Congo red
- PCR
Polymerase chain reaction
- SDS
Sodium dodecyl sulfate
Authors’ contributions
SZ, YG, and SL carried out the experiment and drafted the manuscript. GZ, JL, and HL participated in the design and coordination. JX help with the English writing. All authors read and approved the final manuscript.
Funding
This research was funded by Natural Science Foundation of China (grant/award number 31570641). The funding source had no role in the data collection, design and analysis of the study.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12863-019-0796-y.
References
- 1.Chen YZ, Wang BM, Chen JJ, Wang XN, Wang R, Peng SF, Chen LS, Ma L, Luo J. Identification of Rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front Plant Sci. 2015;6:189. doi: 10.3389/fpls.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di, T.M., Yang, S.L., Du, F.Y., Zhao, L., Li, X.H., Xia, T., and Zhang, X.F. (2018). Oleiferasaponin A2, a novel saponin from Camellia oleifera Abel. Seeds, inhibits lipid accumulation of HepG2 cells through regulating fatty acid metabolism. Molecules 23: pii E3296. [DOI] [PMC free article] [PubMed]
- 3.Feas, X., Estevinho, L.M., Salinero, C., Vela, P., Sainz, M.J., Vazquez-Tato, M.P., and Seijas, J.A. (2013). Triacylglyceride, antioxidant and antimicrobial features of virgin Camellia oleifera, C reticulata and C sasanqua Oils Molecules 18: 4573–4587. [DOI] [PMC free article] [PubMed]
- 4.Li H, Zhou GY, Liu JA, Xu JP. Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PLoS One. 2016;11:e0156841. doi: 10.1371/journal.pone.0156841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang SQ, Li H. First report of leaf anthracnose caused by colletotrichum karstii on tea-oil trees (Camellia oleifera) in China. Plant Dis. 2018;102:674–675. doi: 10.1094/PDIS-08-17-1195-PDN. [DOI] [Google Scholar]
- 6.Liang X, Shang S, Dong Q, Wang B, Zhang R, Gleason ML, Sun G. Transcriptomic analysis reveals candidate genes regulating development and host interactions of Colletotrichum fructicola. BMC Genomics. 2018;19:557. doi: 10.1186/s12864-018-4934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prihastuti H, Cai L, Chen H, McKenzie EHC, Hyde KD. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89–109. [Google Scholar]
- 8.Rockenbach MF, Velho AC, Goncalves AE, Mondino PE, Alaniz SM, Stadnik MJ. Genetic structure of Colletotrichum fructicola associated to apple bitter rot and glomerella leaf spot in southern Brazil and Uruguay. Phytopathol. 2016;106:774–781. doi: 10.1094/PHYTO-09-15-0222-R. [DOI] [PubMed] [Google Scholar]
- 9.Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012:115–80. [DOI] [PMC free article] [PubMed]
- 10.Baroncelli R, Amby DB, Zapparata A, Sarrocco S, Vannacci G, Le Floch G, Harrison RJ, Holub E, Sukno SA, Sreenivasaprasad S, et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics. 2016;17:555. doi: 10.1186/s12864-016-2917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JY, Fang YL, Wang P, Ye JR, Huang L. Pleiotropic roles of ChSat4 in asexual development, cell wall integrity maintenance, and pathogenicity in Colletotrichum higginsianum. Front Microbiol. 2018;9:2311. doi: 10.3389/fmicb.2018.02311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shashkova, S., Welkenhuysen, N., and Hohmann, S. (2015). Molecular communication: crosstalk between the Snf1 and other signaling pathways. FEMS yeast res 15: fov026. [DOI] [PubMed]
- 16.Liu WD, Zhou XY, Li GT, Li L, Kong LG, Wang CF, Zhang HF, Xu JR. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011;7:e1001261. doi: 10.1371/journal.ppat.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu FW, Gu Q, Yun YZ, Yin YN, Xu JR, Shim WB, Ma ZH. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014;203:219–232. doi: 10.1111/nph.12776. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Kim Y, Park G, Xu JR. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 20.Hong SP, Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- 21.Coccetti P, Nicastro R, Tripodi F. Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microb Cell. 2018;5:482–494. doi: 10.15698/mic2018.11.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young ET, Zhang C, Shokat KM, Parua PK, Braun KA. The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae. J Biol Chem. 2012;287:29021–29034. doi: 10.1074/jbc.M112.380147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchin S, Vyas VK, Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol. 2002;22:3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Y, Tsuchiyama S, Yang C, Bulteau AL, He C, Robison B, Tsuchiya M, Miller D, Briones V, Tar K, et al. Proteasomes, Sir2, and Hxk2 form an interconnected aging network that impinges on the AMPK/Snf1-regulated transcriptional repressor Mig1. PLoS Genet. 2015;11:e1004968. doi: 10.1371/journal.pgen.1004968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonukari NJ, Scott-Craig JS, Walton JD. The Cochliobolus carbonum SNF1 gene is required for cell wall-degrading enzyme expression and virulence on maize. Plant Cell. 2000;12:237–248. doi: 10.1105/tpc.12.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzima AK, Paplomatas EJ, Rauyaree P, Ospina-Giraldo MD, Kang S. VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol Plant Microbe In. 2011;24:129–142. doi: 10.1094/MPMI-09-09-0217. [DOI] [PubMed] [Google Scholar]
- 28.Yi M, Park JH, Ahn JH, Lee YH. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol. 2008:1172–81. [DOI] [PubMed]
- 29.Fukada F, Kodama S, Nishiuchi T, Kajikawa N, Kubo Y. Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 2019;222:1909–1923. doi: 10.1111/nph.15728. [DOI] [PubMed] [Google Scholar]
- 30.Fukada F, Kubo Y. Colletotrichum orbiculare regulates cell cycle G1/S progression via a two-component GAP and a GTPase to establish plant infection. Plant Cell. 2015;27:2530–2544. doi: 10.1105/tpc.15.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Zhou X, Xu JR. Genetic control of infection-related development in Magnaporthe oryzae. Curr Opin Microbiol. 2012;15:678–684. doi: 10.1016/j.mib.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Zheng X, Zhang Z. The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol. 2016;17:796–804. doi: 10.1111/mpp.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Zhong K, Yin Z, Hu J, Wang W, Li L, Zhang H, Zheng X, Wang P, Zhang Z. The seven transmembrane domain protein MoRgs7 functions in surface perception and undergoes coronin MoCrn1-dependent endocytosis in complex with Galpha subunit MoMagA to promote cAMP signaling and appressorium formation in Magnaporthe oryzae. PLoS Pathog. 2019;15:e1007382. doi: 10.1371/journal.ppat.1007382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marroquin-Guzman M, Sun G, Wilson RA. Glucose-ABL1-TOR signaling modulates cell cycle tuning to control terminal appressorial cell differentiation. PLoS Genet. 2017;13:e1006557. doi: 10.1371/journal.pgen.1006557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng XQ, Chen GQ, Liu XH, Dong B, Shi HB, Lu JP, Lin F. Crosstalk between SNF1 pathway and the peroxisome-mediated lipid metabolism in Magnaporthe oryzae. PLoS One. 2014;9:e103124. doi: 10.1371/journal.pone.0103124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrer-Dalmau J, Randez-Gil F, Marquina M, Prieto JA, Casamayor A. Protein kinase Snf1 is involved in the proper regulation of the unfolded protein response in Saccharomyces cerevisiae. Biochem J. 2015;468:33–47. doi: 10.1042/BJ20140734. [DOI] [PubMed] [Google Scholar]
- 37.Djouder N, Tuerk RD, Suter M, Salvioni P, Thali RF, Scholz R, Vaahtomeri K, Auchli Y, Rechsteiner H, Brunisholz RA, et al. PKA phosphorylates and inactivates AMPKα to promote efficient lipolysis. EMBO J. 2010;29:469–481. doi: 10.1038/emboj.2009.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, Chen Y, Wang J, Qi Z, Wang Z, et al. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011;7:e1002450. doi: 10.1371/journal.ppat.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Q, Chen Y, Liu Y, Zhang C, Ma Z. The transmembrane protein FgSho1 regulates fungal development and pathogenicity via the MAPK module Ste50-Ste11-Ste7 in Fusarium graminearum. New Phytol. 2015;206:315–328. doi: 10.1111/nph.13158. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A. A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell. 2010;22:2459–2475. doi: 10.1105/tpc.110.075937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional files 1: Figure S1. Generation of the CfSNF1 gene deletion mutants in C. fructicola. (A) Strategy for the construction of gene replacement fragment. ~ 1.0-kb left fragment (LF) and right fragment (RF) flanking the CfSNF1 gene and 1.4-kb HPH gene from pCX62 vector were amplified, then the LF and RF were ligated to the flanks of HPH, respectively, by overlap PCR. The PCR products were further cloned into the pMD19-T vector and obtained the vector of pMD-CfSNF1-KO. After sequencing, the 3.4-kb fragments, which contain the LF, RF and HPH, were amplified for gene replacement. (B) Schematic illustration for deletion strategy of CfSNF1 gene. (C) Validation of the gene deletion mutants by PCR amplified with primers 1 (NBF/NBR) and primers 2 (BWF/HPHR). M: marker; +: positive control; −: negative control; #3 and #5: mutants.
Additional files 2: Figure S2. CfSnf1 is not involved in the tolerance of cell wall stress and oxidative stress. (A) The WT, ΔCfSnf1 and ΔCfSnf1/CfSNF1 strains were cultured onto PDA plates with different cell wall stresses (CR and SDS) and oxidative stress (H2O2) at 28 °C for 4 days and photographed. (B) Statistical analysis of inhibition rates of the strains to cell wall stresses and oxidative stress. Error bars represent SD of three replicates.
Additional files 3: Table S1. Primers used in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary additional files.