Abstract
Background
Diarrhoea is a major cause of death in neonate pigs and most of the viruses that cause it are RNA viruses. Next Generation Sequencing (NGS) deeply characterize the genetic diversity among rapidly mutating virus populations at the interspecific as well as the intraspecific level. The diversity of RNA viruses present in faeces of neonatal piglets suffering from diarrhoea in 47 farms, plus 4 samples from non-diarrhoeic piglets has been evaluated by NGS. Samples were selected among the cases submitted to the Veterinary Diagnostic Laboratories of Infectious Diseases of the Universitat Autònoma de Barcelona (Barcelona, Spain) and Universidad de León (León, Spain).
Results
The analyses identified the presence of 12 virus species corresponding to 8 genera of RNA viruses. Most samples were co-infected by several viruses. Kobuvirus and Rotavirus were more commonly reported, with Sapovirus, Astrovirus 3, 4 and 5, Enterovirus G, Porcine epidemic diarrhoea virus, Pasivirus and Posavirus being less frequently detected. Most sequences showed a low identity with the sequences deposited in GenBank, allowing us to propose several new VP4 and VP7 genotypes for Rotavirus B and Rotavirus C.
Conclusions
Among the cases analysed, Rotaviruses were the main aetiological agents of diarrhoea in neonate pigs. Besides, in a small number of cases Kobuvirus and Sapovirus may also have an aetiological role. Even most animals were co-infected in early life, the association with enteric disease among the other examined viruses was unclear. The NGS method applied successfully characterized the RNA virome present in faeces and detected a high level of unreported intraspecific diversity.
Keywords: RNA virus, Piglets, Diarrhoea, Next-generation sequencing
Background
Diarrhoea in neonatal piglets is a common problem in many pig herds. Usually, this condition is seen either in the offspring of gilts or as an epidemic problem affecting litters from sows of any parity. In the first case, the problem is usually attributed to an inadequate acclimation protocol for gilts while in the latter, the most often cause is the introduction of a new enteric pathogen. These patterns reflect the immune status of sows against the causing agents and the transfer of colostral and lactogenic immunity to piglets. The agents involved in neonatal diarrhoea of piglets are diverse and include bacterial (Escherichia coli, Clostridium perfringens, etc.) and viral pathogens (coronaviruses, rotaviruses, etc.). Less commonly neonates can suffer from diarrhoea caused by parasites [1]. Most of the viruses causing outbreaks of diarrhoea in neonate piglets are RNA viruses such as Porcine epidemic diarrhoea virus (PEDV), Transmissible gastroenteritis virus (TGEV), Porcine deltacoronavirus (PDCoV) or Rotavirus A, B, C or H (RVA, RVB, RVC, RVH) [2–9]. Among them, Rotavirus A, B and C are a major cause of diarrhoea in pigs worldwide, having a significant impact in health and productivity, plus the potential of zoonotic transmission to humans [7]. The virus is transmitted by the faecal-oral route and large number of viral particles is excreted during Rotavirus infection. In contrast, other RNA viruses including Kobuvirus, Astrovirus, Sapovirus, Sapelovirus, Teschovirus, and Torovirus, have been detected in pig faeces but its role as causative agents of neonatal diarrhoea has not so far been fully elucidated [10–14]. For instance, porcine Kobuvirus (KobuV) has been found frequently (16–99%) in faecal samples of pigs worldwide [15]. In some cases, the presence of the virus was associated with diarrhoea, but in others no significant differences between infection rates in healthy or diarrhoeic animals was found. Frequently KobuV were present concomitantly with other viruses making difficult a precise assessment of its role as agent of disease [12, 16].
Next Generation Sequencing (NGS) allows for an in-depth characterization of the genetic diversity among rapidly mutating virus populations by directly sequencing viral strains at the interspecific as well as the intraspecific level [17, 18]. The technology has been successfully applied to identify the diversity in the viral genome of pigs’ faeces [15, 19–21]. The aim of the present study is to explore the diversity of RNA viruses present in faeces of neonatal piglets suffering from diarrhoea in Spain, applying a tailor-made NGS protocol using the total RNA extracted, without any amplification step, where the results obtained represent not only the RNA viruses present in a sample, but also their relative abundances.
Results
The results reported among the 47 diarrhoeic samples analysed include representatives of 12 virus species corresponding to 8 genera of RNA viruses (Additional file 1): Kobuvirus, Rotavirus (RVA, RVB and RVC), Sapovirus (SAV), Mamastrovirus (Porcine Astrovirus types 3 - AstV3 -, 4 - AstV4 - and 5 – AstV5 -), Alphacoronavirus (PEDV), Enterovirus (Enterovirus G, EntVG), Pasivirus (PasiV) and Posavirus (PosaV). The most commonly reported RNA virus was KobuV, followed by RVA, RVC, SAV, RVB and AstV3 (N values in Fig. 1). Among the 9 diarrhoeic samples where a single virus was detected, RVA was present in 5 samples and KobuV was present in 4. Thirty-eight samples (81%) presented more than one virus and in one sample seven RNA viruses were identified: KobuV, RVC, AstV 3, 4 and 5, SAV and PEDV. Rotavirus (either RVA, RVB or RVC) plus KobuV was the combination present in 12 out of the 14 (86%) samples infected with 2 viruses. RVA, RVB, RVC or a combination of two or three Rotavirus were also present in 41 out of the 47 (87%) diarrhoeic samples analysed. For the 6 Rotavirus-negative samples KobuV was present in all 6; being the single virus present in 4 of them. In the other two samples Astrovirus 3, 4, and 5, SAV, PasiV, PosaV and EntVG were detected (Additional file 1).
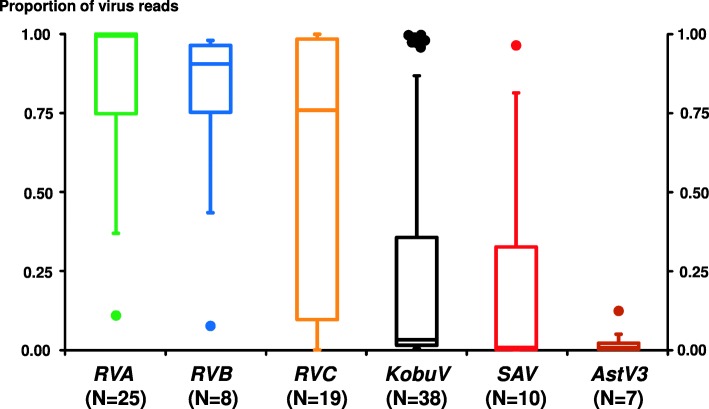
Fig. 1.
The box-and-whisker plots show, for the 47 samples from diarrhoeic animals, the distribution of the proportion of reads for a given mammalian viral species over the total number of mammalian viral reads (viral reads in a sample/total viral reads). Minimum, maximum, median, 25 and 75 quartiles are shown. Only virus showing read proportions larger than 0.001 are represented. Virus abbreviations: Rotavirus A, RVA; Rotavirus B, RVB; Rotavirus C, RVC; Kobuvirus, KobuV; Sapovirus, SAV; Astrovirus 3, AstV3
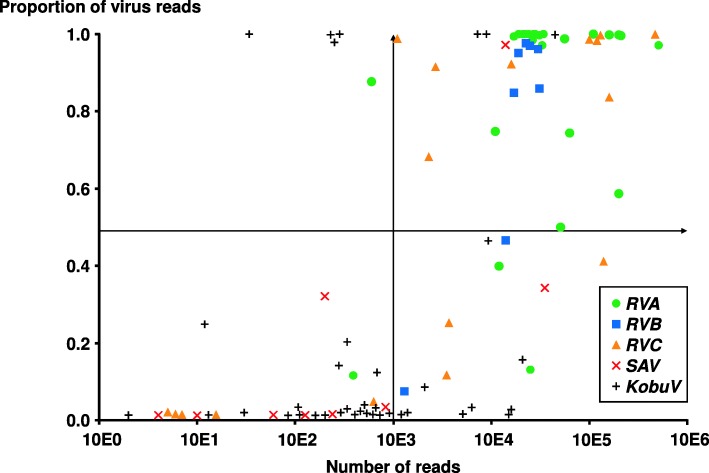
The NGS approach used allowed the comparison of the relative frequencies of each viral species in the examined samples. Thus, when the number of reads for a given virus was compared to the total number of reads attributable to a mammalian RNA virus in the sample, it was evident that Rotaviruses, particularly RVA and RVB, clearly outnumbered any other virus that could be present (Fig. 1). In only nine cases, KobuV or SAV reads were predominant among viral reads (8 and 1 cases, respectively). In all 47 cases, one virus represented at least 45% of the total viral reads. Interestingly, when mixed rotavirus infections were detected (RVA-RVC or RVB-RVC), the proportion of one virus clearly predominated against the other. When coupled with the viral load, the relative frequencies per virus (Fig. 2) showed that for a majority of cases (40/47, 85.1%), the predominant virus in the sample represented read proportions higher than 0.5 and viral reads figures larger than 10E3. Nearly all of them (36/40, 90.0%) corresponded to Rotavirus – either RVA (21), RVB (6) or RVC (9) –, with KobuV and SAV being much less frequent (3 and 1 cases, respectively). Besides, few cases (4 KobuV and 1 RVA) represented simultaneously high proportions with viral reads below 10E3.
Fig. 2.
Scatter plot graph representing the number of viral reads indexed per virus and per sample (X-axis), versus the proportion that those viral reads represents over the total number of mammalian virus reads indexed (Y-axis) in each one of the 47 diarrhoeic animals analysed. Only Rotavirus A, B and C, KobuV and SAV were considered
In the four non-diarrhoeic samples, the number of viral reads obtained and indexed was lower compared to those of the diarrhoeic samples, none of the mammalian virus detected surpasses 10E0 to 10E1 reads (Additional file 1). In contrast, a large number of reads were indexed against RNA viruses of plants (i.e. Fabavirus, Bromovirus, Fijivirus, Luteovirus) and fungi (i.e. Hypovirus, Alphapartivirus).
The filtering results included both whole genome (complete) and partial (less than 90% of the genome covered) sequences of the virus listed above (Additional file 1). Despite KobuV was more commonly reported than RVA, the number of full-length genome sequences obtained was higher in RVA (n = 23) than in KobuV (n = 17). The full-length sequences obtained – 17 KobuV, 23 RVA, 11 RVC, 8 RVB, 4 SAV and 2 AstV3 – were deposited in GenBank with the Accession Numbers MH238075 to MH238338, MK936372 to MK936426, MK953017 to MK953236, and MK962320 to MK962342. Considering the segmented nature of the Rotavirus genome, every segment for every genome was uploaded separately. For the remaining RNA viruses, every complete genome was uploaded in a single file.
Regarding the phylogenetic analysis of KobuV (Fig. 3), all Spanish sequences formed a monophyletic cluster where a Hungarian sequence was also included. The average nucleotide identity among Spanish isolates was 90.0%. It is worth noting that the depth of the terminal branches in the phylogenetic tree was very high compared to the inner branches, indicating a low number of shared mutations.
Fig. 3.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the complete genomes for Kobuvirus. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains the 51 complete Kobuvirus genomes available in GenBank plus the 17 genomes – marked in green and with a circle – reported in this work
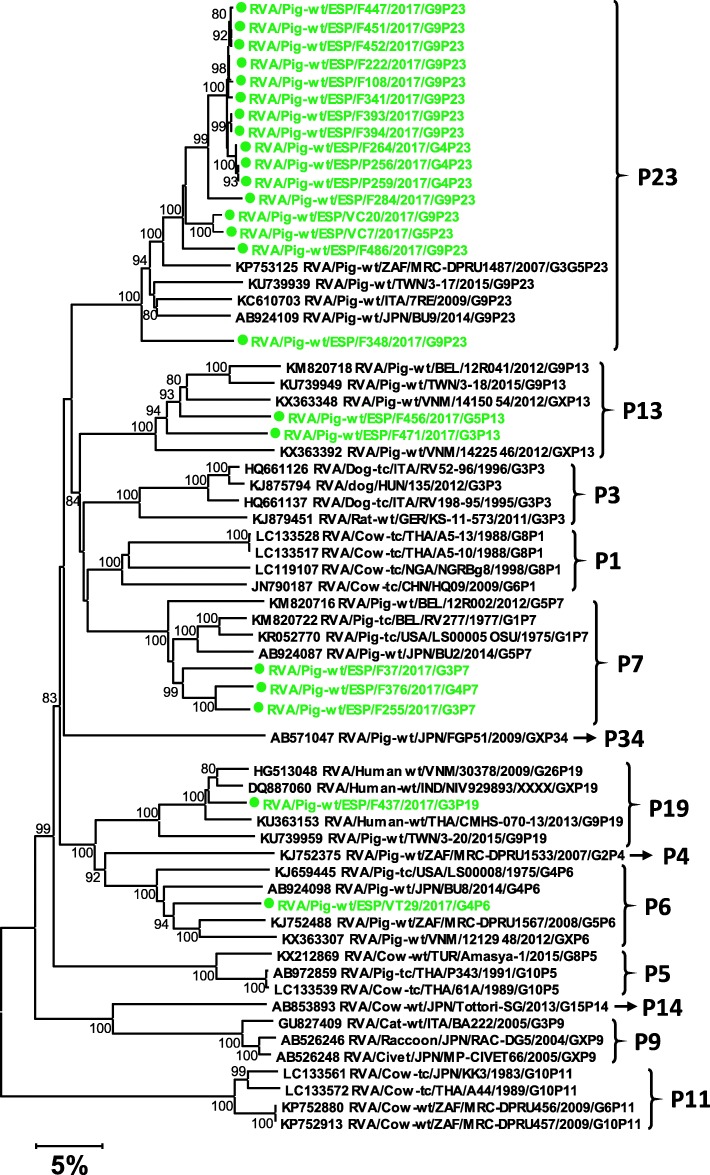
The genotyping of the 23 RVA genomes obtained indicated that the most common combination was G9P [22] (12 cases; 52%), with G4P [22] (3 cases; 13%) and G3P [7] (2 cases; 9%) being much less frequent. The genotypes G3P [13], G3P [19], G4P [7], G5P [13], G5P [22] and G4P [6] were reported only once (Fig. 4). The nucleotide identities reported for the RVA segments compared to the existing GenBank sequences ranged between 89 and 98%. In contrast, for the 8 RVB complete genomes characterized in this work (Fig. 5), the overall nucleotide identities (among the 11 segments of the RVB genome) with the closest GenBank sequences ranged from between 74 and 90%. In fact, according to the proposed nucleotide cut-off values for VP7 (80%), two undescribed VP7 genotypes were reported in nearly all (7 out of 8) the RVB positive samples. Similarly, for VP4, 3 samples could harbour a new RVB genotype (tree not shown). The RVB genotypes reported and its frequency would be: GX2P [4] (3, 37.5%), GX1P[X] (2, 25%), GX1P [4] (2, 25%), and G12P[X] (1, 12.5%).
Fig. 4.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the VP7 segment for Rotavirus A. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains 41 reference sequences representing RVA strains isolated in different species, included for comparison purposes, plus the 23 sequences – marked in green and with a circle – reported in this work. The division of genotypes is based on the 80% nucleotide cut-off values proposed by the RotaC 2.0 genotyping tool [23], and the RVA genotype determination tool of Virus Pathogen Resource [24]
Fig. 5.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the VP7 segment for Rotavirus B. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains 58 reference sequences representing the 26 RVB VP7 genotypes described plus the 8 sequences – marked with a circle – reported in this work. The division of genotypes is based on the 80% nucleotide cut-off value proposed by [25]; accordingly, two new genotypes (labelled as GX1 and GX2 in the Figure) are tentatively proposed
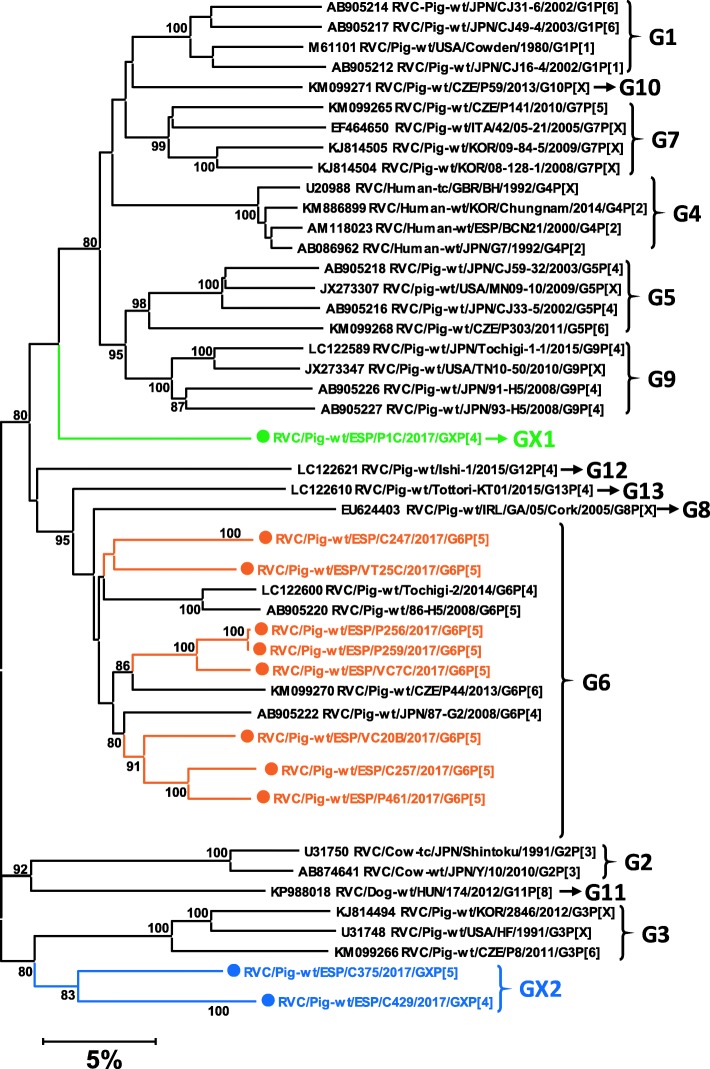
For RVC (Fig. 6), the nucleotide identities with the sequences available in GenBank ranged between 83 and 91%. The most common genotype was G6P [5] (8 samples), while the remaining 3 genomes would present two new RVC VP7 genotypes in combination with P [4] – one with GX1P [4] and one with GX2P [4] – and one GX2P [5].
Fig. 6.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the VP7 segment for Rotavirus C. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains 34 reference sequences representing the 11 RVC VP7 genotypes described plus the 17 sequences – marked with a circle – reported in this work. The division of genotypes is based on the 85–86% nucleotide cut-off value proposed by [26]; accordingly, two new genotypes (labelled as GX1 and GX2 in the Figure) are tentatively proposed
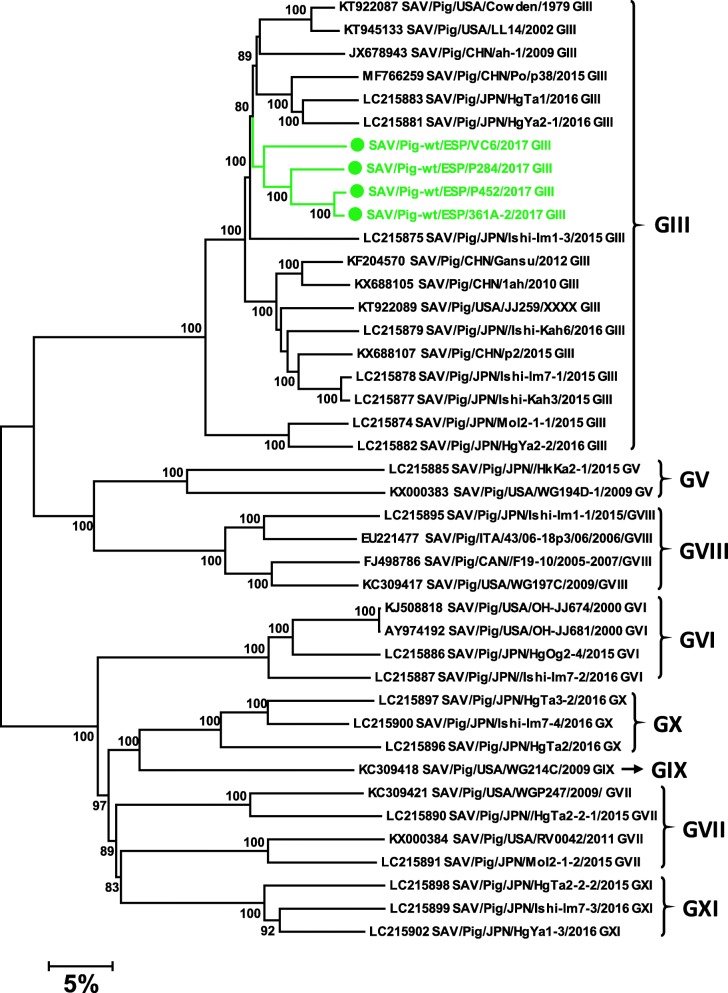
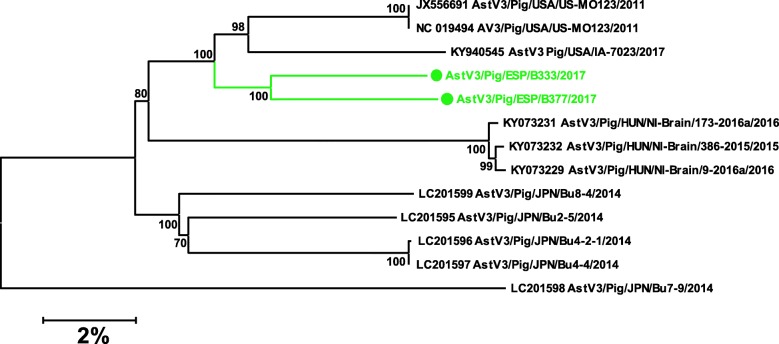
For other viruses, nucleotide identities with the closest sequences deposited were low: AstV3 (91%), AstV4 (89%), SAV (87%), EntVG (82%) and PasiV (86%) except for PosaV (96%), AstV5 (97%) and PEDV (99%) which were very similar to other European sequences reported. Figures 7 and 8 show the phylogenetic trees for SAV and AstV3, respectively.
Fig. 7.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the complete genomes of Porcine Sapovirus. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains 37 reference genomes representing the 8 Sapovirus genogroups described so far in swine plus the 4 complete sequences – marked with a circle – reported in this work. The division of genotypes is based on the classification proposed by [27]
Fig. 8.
Neighbor-joining phylogenetic tree based on the p-distance among the nucleotide sequences of the complete genomes of Porcine Astrovirus 3. Along the branches, percentage of bootstrap values based on 10,000 replicates. Only values equal or larger than 80% are shown. The dataset contains the 11 complete genomes available so far in swine plus the 2 complete sequences – marked with a circle – reported in this work
The de novo assembler and virus detection analyses performed did not detect contigs that could be related to the presence of an unknown RNA virus, as all of them where positively indexed against the Reference RNA sequences dataset and/or the RefSeq Genome Database available at the BLAST resource of the NCBI website.
Discussion
Diarrhoea is the most common infectious cause of neonatal death in pigs [28]. Prevention of this condition can be achieved by immunization of the sow with the aim of ensuring transfer of immunoglobulin A in colostrum and milk. Indeed, outbreaks of infectious diarrhoea in neonates are associated to the lack of specific immunity in the sow.
Classically, infectious neonatal diarrhoea has been seen as a problem caused by single aetiological agents. However, with the development of molecular techniques, a growing number of evidences indicated that many agents can be found in the faeces of affected animals [19, 21]. The problem now is to distinguish which of those agents primary cause diarrhoea, which others act as secondary or associated agents and which of them are part of the enteric microbiome without involvement in the disease.
In this work, we examined the RNA virome present in faecal samples from 47 cases of diarrhoea plus four non-diarrhoeic negative controls in which specific bacteriological agents were excluded. The approach taken using NGS did not include any previous enrichment or PCR step; hence, the results obtained represented not only the RNA viruses present in a sample analysed but also their relative abundances. This may contribute to our understanding of the role of each virus in a given case.
In the samples examined, the predominance of Rotaviruses reads reinforce the notion of these viruses as primary agents of neonatal diarrhoea; although, occasionally, KobuV and SAV may have a role in this process. The examination of the relative proportions of viral reads versus the number of reads showed that for any given case always one virus predominated, representing more than 45% of the mammalian viral reads obtained. This value can be tentatively proposed as a cut-off for the assignment of an etiological agent. In contrast, for all the other viruses examined, the relative abundance of their genomes would be more consistent with a subclinical infection. Whether this could be the consequence of a limited virulence, because of some level of passive immunity, or other causes, cannot be addressed in the present study.
Since the NGS filtering method used in the present study could only detect known agents, additional analyses to screen for potential viral motifs were performed. The de novo assembly and virus detection analyses were not able to identify any contig that could be related to the presence of an unknown RNA virus.
Interestingly, in the non-diarrhoeic samples analysed the number of mammalian reads was very scarce, but a higher number of plant and fungi viruses was reported. This is somewhat surprising, since the examined animal were suckling piglets (less than 1 week of age) that do not eat feedstuff. It is difficult to explain the origin of those viruses, since samples were taken from the rectum and thus, environmental contamination is little likely.
Our results agree with a number of studies that reported different Rotavirus species as agents of neonatal diarrhoea (reviewed in [7]). One interesting observation from the present study is that coinfections or RVA-RVC and RVB-RVC was very common, suggesting the need for simultaneous testing all three agents in cases of neonatal diarrhoea. Regarding KobuV, our results also agree with an increased prevalence of this agent observed in cases of diarrhoea in suckling piglets worldwide: Brazil [22], Korea [29] and Vietnam [30]; despite several studies have observed non-significant differences in KobuV infection between diarrheic and healthy piglets in several European countries [12, 16, 31]. As mentioned above, two different patterns were observed in cases where KobuV was present: one with high proportion of KobuV reads and high number of viral reads, and others where these two circumstances were not fulfilled. The first pattern could correspond to clinical cases caused by KobuV, while the second could correspond to secondary or concomitant infections by this agent. A similar pattern would apply to SAV. Regarding the other agents, since they were always in combination and their reads were not preeminent, its role must be seen as secondary at best. For instance, no differences in Astrovirus prevalence between diarrheic and non-diarrheic piglets were reported [32, 33], nor for Enterovirus prevalence between healthy and diarrheic pigs [30]. Interestingly, PEDV was seldom found in our samples and when found only traces could be detected. Prior to this, a higher frequency for this pathogen could be expected. However, while PEDV has caused recently severe epidemics in America [18], in Europe the incidence seems to be much lower [34]. Of note is the total absence of TGEV, a virus which was widespread in Europe, but has consistently declined, or even disappeared, after the apparition and fast spread worldwide in the nineties of Porcine Respiratory Coronavirus [35].
In any case, it is somewhat surprising to detect that many different viruses (up to seven in a single sample) in so young animals, even though the pattern was already observed in diarrhoeic piglets [21]. While it is easy to understand that the introduction of a new enteric virus in the farm will result in its rapid spread and the eventual development of an epidemic, it is more difficult to understand how so many viruses can be present in the maternities without generating a herd immunity resulting in colostral and lactogenic protection of suckling piglets. One possibility is that, most often, maternal immunity could be enough to limit the replication of those agents but not to completely prevent the infection; namely only provided partial immunity. In other cases, on farms reporting disease even in sows, RVA was present. This is compatible with its assumed role as primary agent. We have recently described the introduction of a new RVA strain that rapidly spread across pig farms in Spain and that was also present in the samples of this study as well [36].
It is worth noting that the phylogenetic analyses for all the examined viruses indicated the existence of local clusters except for RVA (discussed above). This, together with the depth of the corresponding branches in the trees, suggested a long evolutionary history on a local basis. Certainly, we cannot discard the existence of other clusters for any of the reported viruses. However, the detection of several new genotypes for RVB and RVC plus the relative low nucleotide identity with other available sequences in GenBank reinforces the idea of local evolution at least for these two viruses. Moreover, with the results obtained we propose to define for RVB one new VP4 (P [6]) and two new VP7 (G27 and G28) genotypes; as well as genotypes G14 and G15 for RVC. The geographical pattern observed in the RVB phylogenetic trees agrees with the notion that RVB genotypes may be specific for host species and region [25]. Regarding KobuV, no differences in the clustering pattern were observed between those strains coming from cases in which KobuV was predominant or not. This suggests that phylogenetic clustering probably is not predictive of virulence for this virus and most probably, other causes (i.e. immunity) are more relevant with regards to the clinical expression of the disease. Similarly, for SAV no differences were seen, and all isolates clustered together within genogroup III, the most commonly reported porcine SAV [37].
Conclusions
The results of this study suggested that based on the abundance among the cases analysed, Rotaviruses were the main RNA viruses involved in neonatal diarrhoea in pigs. Although Kobuvirus was the most common virus detected, probably it was the primary agent of diarrhoea only in a small number of cases. Similarly, Sapovirus would be responsible of a single diarrhoea case. For the other examined viruses, the results indicated that many animals were infected in early life but the association with enteric disease was unclear. The NGS approach applied permit not only the detection RNA viruses present in a sample, but also to determine their relative abundances. This approach can be used for a more comprehensive diagnosis of neonatal diarrhoea and is useful for the examination of the RNA virome in faeces.
Methods
The study was conducted with samples selected from cases of neonatal diarrhoea (less than 7 days old) routinely submitted for diagnosis during 2017 to the Veterinary Diagnostic Laboratories of Infectious Diseases of the Universitat Autònoma de Barcelona (UAB, Barcelona, Spain) and Universidad de León (UL, León, Spain). Four samples from healthy non-diarrhoeic piglets were also included to be used as negative controls. In the cases studied, a bacteriological cause for the diarrhoea had been excluded after a microbiological culture for aerobes and anaerobes and molecular analyses (PCR for E. coli virulence factors and C. perfringens and C. difficile toxins). Only cases apparently unrelated (different farms with no evident connection) were included. Finally, 47 faecal samples, each one representing an outbreak of neonatal diarrhoea in 47 farms from Catalonia, Castile and Leon, Aragon, Galicia and Valencia were analysed (Additional file 1).
Faecal samples were kept frozen at − 80 °C until used. When needed, samples were thawed and diluted 1:5 in sterile distilled water. Then, to remove debris, diluted samples were sequentially clarified for 2 min at 2000 g, 5 min at 5000 g and 10 min at 10,000 g. Total RNA extraction was performed using the TRIzol reagent (ThermoFischer Scientific) following the manufacturer’s instructions. For each reaction, 250 μL of the diluted faeces was used.
The assessment of the diversity of viral RNA within each sample was directly characterised from the total RNA extracted, without any previous amplification step (no primers were required), applying a NGS approach previously applied to faeces [36]. Briefly, the procedures included: (i) the construction of a genomic library for Illumina NGS sequencing from the total RNA extracted, using a commercial protocol and reagents (Protocol for use with Purified mRNA or rRNA Depleted RNA and NEBNext® Ultra™ II RNA Library Prep Kit for Illumina®, New England Biolabs); ii) the NGS runs using an Illumina MiSeq platform available at the Genomics and Bioinformatics Service of the UAB and a read length of 250 bp; iii) the trimming of low quality reads (those showing a QC score < 20 as determined by FastQC©software, Babraham informatics), using Trimmomatic© [38]; iv) the taxonomic classification of the quality reads against a pre-built database containing the viral genomes available at GenBank with Kraken [39]; v) the initial filtering of quality reads against a reference file containing the concatenated sequences of a panel of RNA viruses described in faeces (Additional file 2); vi) the mapping of quality reads against the complete genomes of the RNA virus identified in the steps iv) and v), with the Burrows-Wheeler Aligner [40], applying the BWA-MEM algorithm for long reads [41]; and vii) the assembly of a consensus sequence in fasta format for every sample and RNA species identified, using the program QUASR [42]. For the software programs used in steps iii), iv), vi) and vii) - FastQC©software, Kraken, Burrows-Wheeler Aligner and QUASR, respectively -, the default parameters of every program were applied. Next, a database for every RNA species identified was constructed by downloading the available complete genome sequences from GenBank. Finally, the sequences obtained were aligned and phylogenetically compared with the datasets constructed for every complete genome, or segment in the segmented genomes identified. The phylogenetic relationships among sequences were analysed using the software MEGA7 [43], by means of a Neighbor-joining (NJ) algorithm, using the matrix of pairwise p-distances and 10,000 bootstrap replicates to estimate the confidence of the internal branches of the trees.
In the cases where a presumptive aetiological agent could not be clearly established, further analysis to detect unknown RNA viruses were undertaken with the program rnaSPAdes [44], an RNAseq de novo assembler included in the package SPADES [45]. Also, the RNAseq outputs were analysed with VirFind, a web-based bioinformatic pipeline specifically designed for virus detection and discovery [46]. In both cases, the default parameters provided by the programs were used.
Supplementary information
Additional file 1: Summary of the 51 samples analyzed: name, origin, RNA viruses detected and number of reads indexed.
Additional file 2: Summary of the RNA viruses and Accession Numbers of the strains used to filter the quality reads in the step iv) of the NGS approach, used together with step iii), to identify the RNA virus species present in a given sample.
Acknowledgements
Our grateful thanks to the participating veterinarians that kindly provided the field samples.
Abbreviations
- AstV3
Porcine Astrovirus type 3
- AstV4
Porcine Astrovirus type 4
- AstV5
Porcine Astrovirus type 5
- BWA-MEM
Burrows-Wheeler Alignment tool with Maximal Exact Matches
- EntVG
Enterovirus G
- KobuV
Kobuvirus
- NGS
Next Generation Sequencing
- PasiV
Pasivirus
- PCR
Polymerase Chain Reaction
- PDCoV
Porcine deltacoronavirus
- PEDV
Porcine epidemic diarrhoea virus
- PosaV
Posavirus
- QC
Quality Control score
- RNA
Ribonucleic acid
- RVA
Rotavirus A
- RVB
Rotavirus B
- RVC
Rotavirus C
- RVH
Rotavirus H
- SAV
Sapovirus
- TGEV
Transmissible gastroenteritis virus
Authors’ contributions
AV, LD, PJGN, MM and HP performed the microbiological cultures. MC, ID, GM, PJGN, AC, MM and EM selected the samples. MC, ID and AV carried out the laboratory work. MC, ID, AV and GF performed the NGS filtering. MC, ID, GM and EM build up the datasets and performed the phylogenetic analyses. MC, ID, MM and EM discussed the results and wrote the manuscript. All authors revised and approved the manuscript.
Funding
This work was partially supported by the National Institute of Research and Agricultural and Food Technology (INIA, reference E-RTA2015–0003-C02–01 and E-RTA2015–0003-C02–02). Both projects partially fund laboratory analyses and reagents. A. Vidal was supported by a PIF grant from the Universitat Autònoma de Barcelona. M. Cortey was funded by Spanish Ministry of Economy and Competitiveness, program Ramón y Cajal (grant RyC-2015-17154). The role of the funding bodies not included the design of the study, the sample collection or the analysis, neither the interpretation of data, nor the writing of the manuscript.
Availability of data and materials
The complete genome sequences obtained and analysed during the current study are available in the National Center for Biotechnology Information database repository, (https://www.ncbi.nlm.nih.gov/) with the Accession Numbers MH238075 to MH238338, MK936372 to MK936426, MK953017 to MK953236, and MK962320 to MK962342.
Ethics approval and consent to participate
Samples submitted for diagnosis to the Veterinary Diagnostic Laboratories of Infectious Diseases at the Universitat Autònoma de Barcelona and Universidad de León comply with institutional, national, and international guidelines regarding ethics on research.
Regarding the paper “High levels of unreported intraspecific diversity among RNA viruses in faeces of neonatal piglets with diarrhoea” (authors: Martí Cortey, Ivan Díaz, Anna Vidal, Gerard Martín-Valls, Giovanni Franzo, Pedro José Gómez de Nova, Laila Darwich, Héctor Puente, Ana Carvajal, Marga Martín, Enric Mateu) has been done on the basis of clinical testing. According to Directive 63/2010 of the European Parliament and of the Council, on the protection of animals used for scientific purposes -Article 1, point 5(b) “non-experimental clinical veterinary practices”- the procedures carried out to obtain data are not within the scope of named directive. Hence, no authorization from the ethics committee on animal and human experimentation from the UAB and UL was necessary.
At the moment of submission to the Veterinary Diagnostic Laboratories, clients were informed with a written consent that the data obtained from piglet faeces’ may be used for research purposes and published.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Martí Cortey, Email: marti.cortey@uab.cat.
Ivan Díaz, Email: ivan.diaz@irta.cat.
Anna Vidal, Email: anna.vidal@uab.cat.
Gerard Martín-Valls, Email: gerard.martin@uab.cat.
Giovanni Franzo, Email: giovanni.franzo@unipd.it.
Pedro José Gómez de Nova, Email: pedrojgdn@gmail.com.
Laila Darwich, Email: laila.darwich@uab.cat.
Héctor Puente, Email: hpuenf00@estudiantes.unileon.es.
Ana Carvajal, Email: ana.carvajal@unileon.es.
Marga Martín, Email: marga.martin@uab.cat.
Enric Mateu, Email: enric.mateu@uab.cat.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12917-019-2204-2.
References
- 1.Chan G, Farzan A, DeLay J, McEwen B, Prescott JF, Friendship RM. A retrospective study on the etiological diagnoses of diarrhea in neonatal piglets in Ontario, Canada, between 2001 and 2010. Can J Vet Res. 2013;77(4):254–260. [PMC free article] [PubMed] [Google Scholar]
- 2.Kongsted H, Pedersen K, Hjulsager CK, Larsen LE, Pedersen KS, Jorsal SE, Bækbo P. Diarrhoea in neonatal piglets: a case control study on microbiological findings. Porcine Health Manag. 2018;4:17. doi: 10.1186/s40813-018-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marthaler D, Rossow K, Culhane M, Collins J, Goyal S, Ciarlet M, Matthijnssens J. Identification, phylogenetic analysis and classification of porcine group C rotavirus VP7 sequences from the United States and Canada. Virology. 2013;446:189–198. doi: 10.1016/j.virol.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Marthaler D, Homwong N, Rossow K, Culhane M, Goyal S, Collins J, Matthijnssens J, Ciarlet M. Rapid detection and high occurrence of porcine rotavirus a, B, and C by RT-qPCR in diagnostic samples. J Virol Methods. 2014;209:30–34. doi: 10.1016/j.jviromet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Mesonero-Escuredo S, Strutzberg-Minder K, Casanovas C, Segalés J. Viral and bacterial investigations on the aetiology of recurrent pig neonatal diarrhoea cases in Spain. Porcine Health Manag. 2018;4:5. doi: 10.1186/s40813-018-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz VL, Bersano JG, Carvalho AF, Catroxo MH, Chiebao DP, Gregori F, Miyashiro S, Nassar AF, Oliveira TM, Ogata RA, Scarcelli EP, Tonietti PO. Case-control study of pathogens involved in piglet diarrhea. BMC Res Notes. 2016;9:22. doi: 10.1186/s13104-015-1751-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlasova AN, Amimo JO, Saif LJ. Porcine rotaviruses: epidemiology, immune responses and control strategies. Viruses. 2017;9:E48. doi: 10.3390/v9030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakuda M, Ide T, Sasaki J, Komoto S, Ishii J, Sanekata T, Taniguchi K. Porcine rotavirus closely related to novel group of human rotaviruses. Emerg Infect Dis. 2011;17:1491–1493. doi: 10.3201/eid1708.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Zhang Y, Byrum B. Development and evaluation of a duplex real-time RT-PCR for detection and differentiation of virulent and variant strains of porcine epidemic diarrhea viruses from the United States. J Virol Methods. 2014;207:154–157. doi: 10.1016/j.jviromet.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anbalagan S, Peterson J, Wassman B, Elston J, Schwartz K. Genome sequence of Torovirus identified from a pig with porcine epidemic diarrhea virus from the United States. Genome Announc. 2014;2:e01291–e01214. doi: 10.1128/genomeA.01291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goecke NB, Hjulsager CK, Kongsted H, Boye M, Rasmussen S, Granberg F, Fischer TK, Midgley SE, Rasmussen LD, Angen Ø, Nielsen JP, Jorsal SE, Larsen LE. No evidence of enteric viral involvement in the new neonatal porcine diarrhoea syndrome in Danish pigs. BMC Vet Res. 2017;13:315. doi: 10.1186/s12917-017-1239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theuns S, Vanmechelen B, Bernaert Q, Deboutte W, Vandenhole M, Beller L, Matthijnssens J, Maes P, Nauwynck HJ. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Sci Rep. 2018;8:9830. [DOI] [PMC free article] [PubMed]
- 13.Lauritsen KT, Hansen MS, Johnsen CK, Jungersen G, Bottiger B. Repeated examination of natural sapovirus infections in pig litters raised under experimental conditions. Acta Vet Scand. 2015;57:60–65. doi: 10.1186/s13028-015-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leme RA, Silva DR, Lorenzetti E, Moraes DA, Alfieri AF, Alfieri AA. Longitudinal survey of Teschovirus a, Sapelovirus a, and Enterovirus G fecal excretion in suckling and weaned pigs. Braz J Microbiol. 2019;50:321–327. doi: 10.1007/s42770-018-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Zhu L, Zhou Y, Xu Z, Guo W, Yang W. Molecular and phylogenetic analysis of the porcine kobuvirus VP1 region using infected pigs from Sichuan Province. China Virology J. 2013;10:281. doi: 10.1186/1743-422X-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackova A, Sliz I, Mandelik R, Salamunova S, Novotny J, Kolesarova M, Vlasakova M, Vilcek S. Porcine kobuvirus 1 in healthy and diarrheic pigs: genetic detection and characterization of virus and co-infection with rotavirus a. Infect Genet Evol. 2017;49:73–77. doi: 10.1016/j.meegid.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Leung P, Eltahla AA, Lloyd AR, Bull RA, Luciani F. Understanding the complex evolution of rapidly mutating viruses with deep sequencing: beyond the analysis of viral diversity. Virus Res. 2017;239:43–54. doi: 10.1016/j.virusres.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Posada-Cespedes Susana, Seifert David, Beerenwinkel Niko. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Research. 2017;239:17–32. doi: 10.1016/j.virusres.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q, Wang L, Zheng Y, Zhang J, Guo B, Yoon KJ, Gauger PC, Harmon KM, Main RG, Li G. Metagenomic analysis of the RNA fraction of the fecal virome indicates high diversity in pigs infected by porcine endemic diarrhea virus in the United States. Virol J. 2018;15:95. doi: 10.1186/s12985-018-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachsenröder J, Twardziok S, Hammerl JA, Janczyk P, Wrede P, Hertwig S, Johne R. Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PLoS One. 2012;7:e34631. doi: 10.1371/journal.pone.0034631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Tang C, Yue H, Ren Y, Song Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J Gen Virol. 2014;95:1603–1611. doi: 10.1099/vir.0.063743-0. [DOI] [PubMed] [Google Scholar]
- 22.Barry AF, Ribeiro J, Alfieri AF, Van der Poel WHM, Alfieri AA. First detection of kobuvirus in farm animals in Brazil and the Netherlands. Infect Genet Evol. 2011;11:1811–1814. doi: 10.1016/j.meegid.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Maes P, Matthijnssens J, Rahman M, VanRanst M. RotaC: a web-based tool for the complete genome classification of group a rotaviruses. BMC Microbiol. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.RVA genotype determination tool: Virus Pathogen Resource. https://www.viprbrc.org (2019). Accessed 2 April 2019.
- 25.Marthaler D, Rossow K, Gramer M, Collins J, Goyal S, Tsunemitsu H, Kuga K, Suzuki T, Ciarlet M, Matthijnssens J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology. 2012;433:85–96. doi: 10.1016/j.virol.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niira K, Ito M, Masuda T, Saitou T, Abe T, Komoto S, Sato M, Yamasato H, Kishimoto M, Naoi Y, Sano K, Tuchiaka S, Okada T, Omatsu T, Furuya T, Aoki H, Katayama Y, Oba M, Shirai J, Taniguchi K, Mizutani T, Nagai M. Whole genome sequences of Japanese porcine species C rotaviruses reveal a high diversity of genotypes of individual genes and will contribute to a comprehensive, generally accepted classification system. Infect Genet Evol. 2010;44:106–113. doi: 10.1016/j.meegid.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 27.Scheuer KA, Oka T, Hoet AE, Gebreyes WA, Molla BZ, Saif LJ, Q. Prevalence of porcine noroviruses, molecular characterization of emerging porcine sapoviruses from finisher swine in the United States, and unified classification scheme for sapoviruses. J Clin Microbiol. 2013;51:2344–23453. doi: 10.1128/JCM.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cutler RS, Fahy VA, Cronin GM, Spicer EM. Chapter 62: Prewening mortality. In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ, editors. Diseases of swine. 9. Oxford: Blackwell Publishing; 2006. pp. 993–1010. [Google Scholar]
- 29.Park SJ, Kim HK, Moon HJ, Song DS, Rho SM, Han JY, Nguyen VG, Park BK. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch Virol. 2010;155:1803–1811. doi: 10.1007/s00705-010-0774-1. [DOI] [PubMed] [Google Scholar]
- 30.Van Dung N, Anh PH, Van Cuong N, Hoa NT, Carrique-Mas J, Hien VB, Sharp C, Rabaa M, Berto A, Campbell J, Baker S, Farrar J, Woolhouse ME, Bryant JE, Simmonds P. Large-scale screening and characterization of enteroviruses and kobuviruses infecting pigs in Vietnam. J Gen Virol. 2016;97:378–388. doi: 10.1099/jgv.0.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Ullman K, Chowdry V, Reining M, Benyeda Z, Baule C, Juremalm M, Wallgren P, Schwarz L, Zhou E, Pina-Pedrero S, Hennig-Pauka I, Segales J, Liu L. Molecular investigations on the prevalence and viral load of enteric viruses in pigs from five European countries. Vet Microbiol. 2016;182:75–78. doi: 10.1016/j.vetmic.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumthip K, Khamrin P, Saikruang W, Kongkaew A, Vachirachewin R, Ushijima H, Maneekarn N. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch Virol. 2018;163:1823–1829. doi: 10.1007/s00705-018-3806-x. [DOI] [PubMed] [Google Scholar]
- 33.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals – molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvajal A, Argüello H, Martínez-Lobo FJ, Costillas S, Miranda R, de Nova PJ G, Rubio P. Porcine epidemic diarrhoea: new insights into an old disease. Porcine Health Manag. 2015;1:12. doi: 10.1186/s40813-015-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, Knutson TP, Rossow S, Saif LJ, Marthaler DG. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci Report. 2019;9:3953. [DOI] [PMC free article] [PubMed]
- 36.Vidal A, Clilverd H, Cortey M, Martín-Valls GE, Franzo G, Darwich L, Martín M, Mateu E. Full-genome characterization by deep sequencing of rotavirus a isolates from outbreaks of neonatal diarrhoea in pigs in Spain. Vet Microbiol. 2018;227:12–19. doi: 10.1016/j.vetmic.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Marthaler D, Fredrickson R, Gauger PC, Zhang J, Burrough ER, Petznick T, Li G. Genetically divergent porcine sapovirus identified in pigs, United States. Transbound Emerg Dis. 2019;00:1–11. doi: 10.1111/tbed.13337. [DOI] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26:489–495. doi: 10.1093/bioinformatics/btq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience. 2019;8:1–13. doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho T, Tzanetakis IE. Development of a virus detection and discovery pipeline using next generation sequencing. Virology. 2014;471–473:54–60. doi: 10.1016/j.virol.2014.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Summary of the 51 samples analyzed: name, origin, RNA viruses detected and number of reads indexed.
Additional file 2: Summary of the RNA viruses and Accession Numbers of the strains used to filter the quality reads in the step iv) of the NGS approach, used together with step iii), to identify the RNA virus species present in a given sample.
Data Availability Statement
The complete genome sequences obtained and analysed during the current study are available in the National Center for Biotechnology Information database repository, (https://www.ncbi.nlm.nih.gov/) with the Accession Numbers MH238075 to MH238338, MK936372 to MK936426, MK953017 to MK953236, and MK962320 to MK962342.