Abstract
Previous studies have shown that OVATE family proteins (OFPs) participate in various aspects of plant growth and development. How OFPs affect leaf chlorophyll accumulation and leaf senescence has not been reported yet. Here, we found that overexpression of SlOFP20 in tomato not only impacted plant architecture but also enhanced the leaf chlorophyll accumulation and retarded leaf senescence. Gene expression analysis of SlGLK1, SlGLK2, and HY5, encoding transcription factors that are putatively involved in chloroplast development and chlorophyll levels, were significantly up-regulated in SlOFP20-OE lines. Both chlorophyll biosynthesis and degradation genes were distinctly regulated in transgenic plants. Moreover, SlOFP20-OE plants accumulated more starch and soluble sugar than wild-type plants, indicating that an increased chlorophyll content conferred some higher photosynthetic performance in SlOFP20-OE plants. Furthermore, The levels of leaf senescence-related indexes, such as hydrogen peroxide, malondialdehyde, and antioxidant enzymes activities, were differently altered, too. SlOFP20 overexpression repressed the expression of senescence-related genes, SAG12, RAV1, and WRKY53. Moreover, abscisic acid and ethylene synthesis genes were down-regulated in transgenic lines. These results provide new insights into how SlOFP20 regulates chlorophyll accumulation and leaf senescence.
Keywords: OVATE family protein, SlOFP20, chlorophyll, sugar metabolism, leaf senescence
Introduction
Chlorophylls, in complex with their binding proteins, serve primary functions in photosynthesis through trapping light energy and transferring it to the reaction centers of photosystems (Meskauskiene et al., 2001; Goslings et al., 2004). Many factors affect the accumulation of chlorophyll, such as environmental conditions, the structural integrity of the chloroplast, plant hormones, as well as the expression levels of transcription factors and structural genes within the chlorophyll biosynthesis pathway (Meng et al., 2018). To date, some transcription factors have been reported to control chloroplast development and chlorophyll synthesis. For example, the transcription factor SlGLK2 determines plastid and chlorophyll levels via enhancing photosynthesis gene expression and chloroplast development (Fitter et al., 2002; Powell et al., 2012; Nguyen et al., 2014). TKN2 and TKN4, two Class I KNOTTED1-LIKE HOMEOBOX (KNOX) proteins, induce the expression of SlGLK2 and SlAPRR2-LIKE genes to promote fruit chloroplast development in tomato (Nadakuduti et al., 2014). Triple mutant plants of GRAS gene family members, namely SCL6-II, SCL6-III, and SCL6-IV, exhibit increased chlorophyll accumulation in leaves (Wang et al., 2010). Moreover, phytohormones participate in chloroplast development and chlorophyll synthesis. For instance, cytokinins (CKs) affect chloroplast function through regulating photosynthetic performance (Cortleven and Schmulling, 2015) and triggering chloroplast-related genes (Cortleven et al., 2016). The abscisic acid-deficient mutant, high-pigment 3, accumulates more carotenoids and chlorophylls in leaves and fruits (Galpaz et al., 2008). Previous research has suggested that detached roots activate chlorophyll biosynthesis through a reduction in auxin signaling, reflecting the repressive effect of auxin on root greening (Kobayashi et al., 2012).
Leaf senescence occurs at the final stage of leaf development and involves a series of changes at the molecular, cellular and phenotypic levels. Senescence is initiated by characteristic degenerative processes, such as chlorophyll degradation and macromolecule breakdown, and particularly the recycling of nutrients to actively growing tissues or storage organs (Gan and Amasino, 1997). Developmentally regulated metabolic pathways are typically influenced by transcription factors (Yin et al., 2016). For instance, suppression of SlNAP2 expression in tomato delays leaf senescence but boosts fruit yield and sugar content (Ma et al., 2018). However, the wrky54/wrky70 double mutant distinctly shows premature senescence and chlorophyll degradation (Besseau et al., 2012). In addition, phytohormones can either induce or inhibit leaf senescence. Ethylene-insensitive mutants, such as etr1-1 (Grbić and Bleecker, 1995) and ein2/ore3 (Oh et al., 1997), exhibit a delayed senescence phenotype. Exogenous application of abscisic acid (ABA) rapidly induces the senescence syndrome and expression of several senescence-associated genes (SAGs) (Gepstein and Thimann, 1980; Weaver et al., 1998). Increased CKs production could slow down leaf senescence, whereas reduced endogenous CK levels may result in premature senescence (Zhang et al., 2010; Qin et al., 2011).
Brassinosteroids (BRs) are steroid hormones of plants that were identified in the 1970s because of their strong growth-promoting capacities (Mitchell et al., 1970; Grove et al., 1979). BRs regulate cell elongation, cell division, and cell differentiation and function throughout plant development in various developmental programs, including seedling development in the light and dark, adult shoot and root growth, flowering, fruit development, and senescence (Clouse, 2011). Extensive studies using genetic, molecular, and proteomic approaches have been applied to reveal how BRs modulate a wide range of plant growth and development processes. However, the effects of BRs on tomato growth and development are largely unknown, as well as the underlying molecular mechanism. Recently, ectopic expression of BZR1-1D, encoding a transcription factor in BR signaling, enhances carotenoid accumulation and fruit quality attributes in tomato (Liu et al., 2014). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato (Li et al., 2016). BR enhances chilling tolerance through a signaling cascade involving RBOH1 (RESPIRATORY BURST OXIDASE HOMOLOG1), GRX (GLUTAREDOXIN) genes, and 2-Cys Prx (2-cysteine peroxiredoxin) in tomato (Xia et al., 2018).
The OVATE gene was originally identified as a primary quantitative trait locus that controls fruit shape in tomato (Liu et al., 2002). OVATE family proteins (OFPs) control multiple aspects of plant growth and development. It has been suggested that AtOFP1 functions as a transcriptional repressor and regulates cell elongation (Wang et al., 2007). AtOFP5 negatively regulates the activity of a BLH1-KNAT3 complex during early embryo sac development (Pagnussat et al., 2007). More recently, co-expression of MuMADS1 and MaOFP1 in the ovate mutant could compensate for fruit shape and inferior qualities (Liu et al., 2018). Overexpression of OsOFP19 caused a semi-dwarf stature with thicker leaves and stronger culms and roots (Yang et al., 2018).
Here, we report the functional characterization of tomato SlOFP20, an OFP family member that is a homolog of the Arabidopsis AtOFP1 and rice OsOFP19. Some quite recent research indicates that SlOFP20 control fruit shape, and down-regulation of SlOFP20 in WT tomato does not generate visible phenotypes (Wu et al., 2018). In our study, overexpression of SlOFP20 produced pleiotropic phenotypes. We focused on the accumulation of chlorophyll and delay of leaf senescence caused by SlOFP20 overexpression. Our data expand our understanding of the functions of OFPs in the regulation mechanisms of chlorophyll accumulation and leaf senescence.
Materials and Methods
Plant Materials and Growth Conditions
The WT tomato (Solanum lycopersicon Mill. cv. Ailsa Craig) and T2 SlOFP20-OE transgenic plants were used in our research and grown in a greenhouse with the following conditions (16-h-day/8-h-night cycle, 28°C/18°C day/night temperature). To examine the response of SlOFP20 to phytohormones, 50 µM gibberellic acid (GA3), 50 µM indole-3-acetic acid, 100 µM ABA (0.0264 g ABA was dissolved in a small amount of methanol, followed by adding distilled water to 1 l), 50 µM 1-aminocyclopropane-1-carboxylate (ACC) or distilled water (control) were used to treat 35-day-old WT tomato seedlings. For each case, individual plants were used for each treatment with three biological replicates. After treatments for 0, 1, 2, 4, 8, 12, and 24 h, the third leaves from the top of the WT tomato seedlings were collected. For salinity stress treatment, the roots of tomato seedlings were immersed in a solution with 200 mM sodium chloride for 0, 1, 2, 4, 8, 12, and 24 h, then leaves and roots from the treated seedlings and control plants were collected. For dehydration experiments, the whole tomato seedlings were carefully pulled out of the pots, washed gently with water to remove soil and left on a piece of dry filter paper under dim light at 25 ± 1°C. For low temperature experiments, the whole potted tomato seedlings were incubated at 4°C for 0, 1, 4, 8, 12, and 24 h, after which the leaves were harvested. All these samples were instantly frozen with liquid nitrogen and kept at –80°C.
Construction of SlOFP20 Over-Expression Vector and Plant Transformation
To obtain SlOFP20 overexpression transgenic lines, the full-length SlOFP20 coding sequence was amplified using WT tomato complementary DNA with SlOFP20-F and SlOFP20-R primers, which tailed with Xba I and Sac I restriction sites at the 5′end, respectively. The amplified products were digested and cloned into the plant binary vector pBI121 placed under the control of CaMV 35S promoter. The resulting expression vector was transferred into Agrobacterium LBA4404 strain, and the positive LBA4404 strain was transferred into WT tomato cotyledon explants. Transformed lines were selected for kanamycin resistance and then analyzed by polymerase chain reaction (PCR) to determine the presence of T-DNA using the primers NPTII-F/R.
Gene Expression Analysis
Gene expression analysis was performed according to our previous report (Zhou et al., 2018). Primers used for quantitative real-time (RT)-PCR are listed in Supplementary Table S1.
Stoma Morphology and Anatomical Analyses of the Leaves
The lower epidermis from WT and OE3 mature leaves were torn off and observed under a Nikon E100 microscope. The middle parts of WT and OE3 mature leaves were cut and fixed by FAA [70% ethanol/acetic acid/formaldehyde (18:1:1)]. Then the dehydration, fixation, sectioning, dewaxing and staining by safranin and fast green (Bryan, 1955) were performed to prepare the cross sections of WT and OE3 transgenic plant leaves, which were visualized under a Nikon E100 microscope and photographed.
Quantitative Analysis of Chlorophyll Content
Chlorophyll content was determined in mature leaves of WT and SlOFP20-OE plants according to the methods described by Arnon (1949).
Sugar Content Analysis
Leaves from WT and SlOFP20-OE plants were collected and immersed in 95% ethanol to destain for at least 24 h, then the thoroughly bleached leaves were stained with an I2/KI solution for 20 min. For measurement of starch and soluble sugar contents, 0.5 g fresh leaves of WT and SlOFP20-OE transgenic lines were pounded to powder with liquid nitrogen, extracted with 5 ml 80% ethanol for 30 min in a water-bath at 80°C, and then centrifuged 3,500 g for 10 min at room temperature. The supernatant was transferred to a 50 ml centrifuge tube, which was used to measure soluble sugar. These steps were repeated twice. Two milliliter distilled water was added to the residue, which then placed in a boiling water bath for 15 min. After cooling, added 2 ml 9.2 M perchloric acid, stirred for 15 min, mixed with 4 ml distilled water, and centrifuged 4,000 g for 10 min, the supernatant was transferred to a 50 ml centrifuge tube, 4.2 M perchloric acid was added to the residue again, stirred for 15 min, mixed with 5 ml distilled water, centrifuged 4,000 g for 10 min, the supernatant was merged into the centrifuge tube, which was used to measure starch content. The anthrone–sulfuric acid method was used to measure starch and soluble sugar contents (Leng et al., 2016). Three independent experiments were carried out and results expressed as mean ± SD of all replicates.
Analysis of Hydrogen Peroxide Content, Malondialdehyde Content, and Antioxidant Enzyme Activities
Hydrogen peroxide (H2O2) content was determined according to the method of Sergiev et al. (1997). Malondialdehyde (MDA) content was estimated according to Zhu et al. (2015). The activity of superoxide dismutase (SOD) was determined according to the methods described by Beyer and Fridovich (1987). The activity of catalase (CAT) was determined according to the methods described by Rao et al. (1996). The activity of peroxidase (POD) was determined according to the methods described by Morohashi (2002).
Statistical Analysis
All the experiments included three independent repeats. The significant difference between WT and SlOFP20-OE lines was analyzed by Student's t-test (P < 0.05).
Results
The Response of SlOFP20 to Various Hormone and Stress Treatments
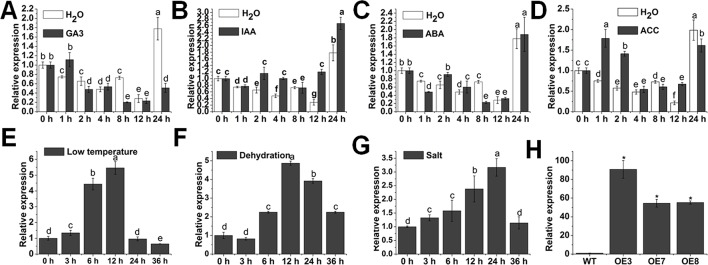
Phytohormones act as crucial regulators to coordinate multiple developmental processes and responses to environmental stresses (Bari and Jones, 2009). To obtain some clues of whether SlOFP20 is involved in hormone signaling, we investigated the expression patterns of SlOFP20 under different hormone treatments by quantitative RT-PCR technology (qRT-PCR). SlOFP20 was significantly repressed by GA3 treatment at 8 and 24 h (Figure 1A). For indole-3-acetic acid, the transcript level of SlOFP20 was induced from 2 to 4 h and 12 to 24 h, and no noticeable change at 8 h (Figure 1B). When treated with ABA, the expression of SlOFP20 had no significant change until 8 h, and then declined sharply at 8 h, reached a similar level from 12 to 24 h. (Figure 1C) The SlOFP20 messenger RNA (mRNA) was accumulated from 1 to 2 h and at 12 h after ACC treatment, and no apparent change at other time points (Figure 1D). These results indicated that SlOFP20 may play an important role in plant hormone response and signal transduction. Rice OFP 6 has been suggested to confer resistance to drought and cold stresses (Ma et al., 2017). Except for this, few studies have been focused on the functional characterization of OFPs referring to stress response. To know whether SlOFP20 participates in response to various abiotic stresses, qRT-PCR was used to detect the expression patterns of SlOFP20 under different stress treatments. Under low-temperature conditions, the expression of SlOFP20 was distinctly increased and peaked at 12 h and then declined (Figure 1E). SlOFP20 mRNA was remarkably raised after dehydration treatments and peaked at 12 h, and then fell (Figure 1F). SlOFP20 was significantly induced after salt treatments and peaked at 24 h and then decreased (Figure 1G). These results give a further hint that SlOFP20 may take part in response to abiotic stress in tomato.
Figure 1.
Expression patterns of SlOFP20. The expression level of SlOFP20 in response to the phytohormones gibberellic acid (A), indole-3-acetic acid (B), abscisic acid (C), and 1-aminocyclopropane-1-carboxylate (D). The expression of SlOFP20 under low temperature (E), dehydration (F), and salt stress (G). Data are the means ± SD of three independent experiments. Significant differences (P < 0.05) are denoted by different letters. Relative expression profiles of SlOFP20 between WT and SlOFP20-OE lines. Data are the means ± SD of three independent experiments. Asterisks indicate a significant difference (P < 0.05).
To understand the biological function of SlOFP20, the plasmid 35S:SlOFP20 was introduced into WT tomato AC++, the expression levels of SlOFP20 in T2 transgenic lines were detected by qRT-PCR. The expression of SlOFP20 was 90-, 54-, and 55-fold higher in the overexpression T2 lines OE3, OE7, and OE8, respectively, compared with WT plants (Figure 1H).
Ectopic Expression of SlOFP20 May Lead to Typical Brassinosteroids-Insensitive and Gibberellic Acid-Deficient Phenotypes
A gain-of-function mutant of AtOFP1 exhibited decreased lengths in all aerial organs, which was partially attributed to the deficiency in gibberellin biosynthesis (Wang et al., 2007). Overexpression of OsOFP19 in rice modified plant architecture, including semi-dwarf stature with thicker leaves, stronger culms and roots and grain shape, by integrating the cell division pattern and BR signaling (Yang et al., 2018). In our study, we also observed that overexpression of SlOFP20 in WT tomato resulted in a series of phenotypes related to plant vegetative growth.
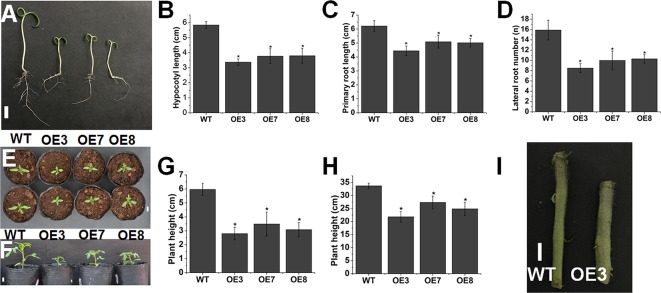
Compared with WT tomato seedlings, overexpression of SlOFP20 led to decreased length of the primary root, hypocotyl and cotyledon and reduced number of lateral roots (Figures 2A–E). We also found kidney-shaped cotyledons in SlOFP20-OE transgenic plants (data not shown). In addition, overexpression of SlOFP20 caused a dwarf phenotype (Figure 2F), we measured the plant height of WT and SlOFP20-OE transgenic lines at 30 and 60 days after sowing in pots under the same conditions, and the height of SlOFP20-OE transgenic plants was much shorter than WT tomato plants (Figures 2G, H). We also observed that the stem of SlOFP20-OE transgenic plants was much thicker when compared with WT tomato plants (Figure 2I).
Figure 2.
Overexpression of SlOFP20 affected plant growth. (A) Seven-day-old seedlings of WT and SlOFP20-OE lines. Bar = 1 cm. (B) Hypocotyl lengths of 7-day-old WT and SlOFP20-OE seedlings. (C) The length of primary root in WT and SlOFP20-OE 7-day-old seedlings. (D) The number of lateral roots in WT and SlOFP20-OE 7-day-old seedlings. Error bars show the standard error between three biological replicates with 10 plants for each replicate performed. Asterisks indicate a significant difference (P < 0.05). (E) Top view of cotyledons from 7-day-old WT and SlOFP20-OE lines. (F) Image of 30-day-old WT and SlOFP20-OE plants. Bar = 1 cm. (G, H).Plant height of WT and SlOFP20-OE plants at 30 days (G) and 60 days (H) after sowing. Error bars show the standard error between three biological replicates with eight plants for each replicate performed. (I) Stems of WT and OE3 plants. Bar = 1 cm.
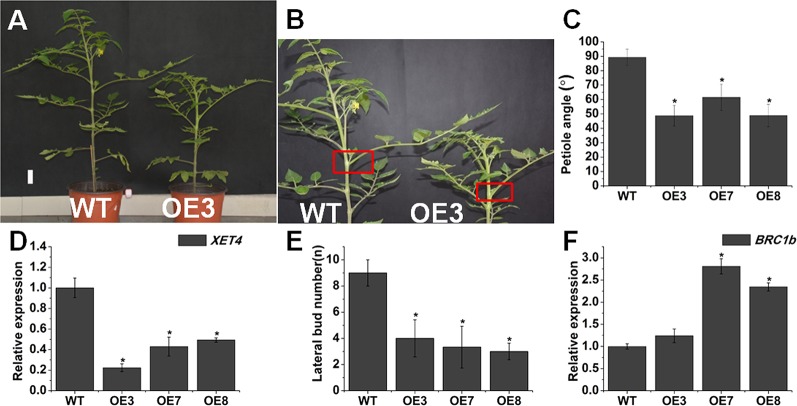
The changes of endogenous BR level and BR signaling affect leaf angle and lateral bud initiation (Li et al., 2016). SlOFP20-OE plants also displayed upright leaves. The 9th leaf petiole angle of SlOFP20-OE plants was clearly reduced, in comparison with WT tomato plants (Figures 3A–C). We inferred that SlOFP20 may affect leaf inclination through transcriptional regulation of the relevant genes. OsXTR1, which encodes a xyloglucan endotransglycosylase, is a cell wall-loosening enzyme necessary for cell elongation (Duan et al., 2006). Compared with WT tomato plants, the expression of XET4, the homologous gene of OsXTR1 in tomato, was significantly down-regulated in SlOFP20-OE plants. Furthermore, SlOFP20-OE transgenic plants had few branches throughout the life cycle. Compared with WT tomato plants, the number of branches reduced by at least 50% in overexpression lines (Figure 3E). SlBRC1b acts as a suppressor of shoot branching in tomato (Martín-Trillo et al., 2011). Gene expression analysis suggested that the expression of BRC1b was increased in SlOFP20 overexpression lines when compared with WT plants (Figure 3F).
Figure 3.
Overexpression of SlOFP20 restricts leaf inclination and lateral bud outgrowth. (A) Picture of 60-day-old WT and OE3 plants. Bar = 1 cm. (B) The red boxes indicated the petiole angle of the 9th leaf. (C) The petiole angle was determined at 60 days in the ninth leaf with eight plants after sowing. Error bars show the standard error between three biological replicates. Asterisks indicate a significant difference (P < 0.05). (D) The expression level of XET4 in WT and SlOFP20-OE lines. (E) Lateral bud numbers of WT and transgenic plants. At least eight plants were measured for WT and each transgenic line. Error bars show the standard error between three biological replicates. Asterisks indicate a significant difference (P < 0.05). (F) Expression analysis of BRClb in WT and transgenic plants. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
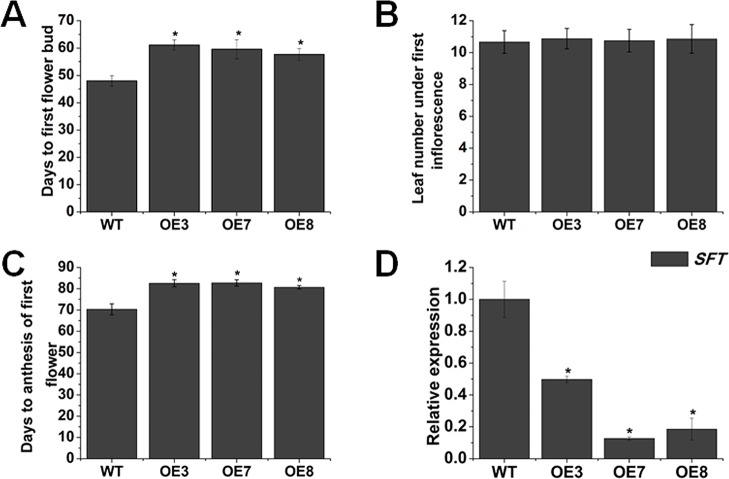
Nearly all BR-deficient and BR-insensitive mutants display late-flowering phenotype (Li et al., 2010). Here, we also observed that overexpression of SlOFP20 obviously restrained the initiation of flower buds. To quantify this phenotype in more detail, the related parameters, which are usually used to assess the flowering time, were measured. For days to first flower bud emergence, the SlOFP20-OE transgenic plants generated flower buds much later than those of WT plants. Flower buds arose at least 9 days later in overexpression lines than WT tomato plants in our growth conditions (Figure 4A). In addition, we also measured days to anthesis of first flower (Figure 4C), which were consistent with the days to first flower bud. However, another parameter, leaf numbers below the first inflorescence, showed no obvious discrepancy between WT and SlOFP20-OE transgenic plants (Figure 4B). SFT is a central regulator to control flowering time in tomato (Lifschitz and Eshed, 2006; Shalit et al., 2009). We supposed that overexpression of SlOFP20 delayed flowering through the SFT pathway. Thus, the expression level of SFT was determined, which was significantly reduced in SlOFP20-OE plants when compared with WT tomato plants (Figure 4D).
Figure 4.
Overexpression of SlOFP20 restrains the onset of flowering. Analysis of the days to first flower bud (A), leaf number under first inflorescence (B) and (C) days to anthesis of first flower of WT and transgenic plants. Error bars show the standard error between three biological replicates with eight plants for each replicate performed. (D) Expression analysis of SFT in WT and transgenic plants. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
Moreover, 10 µM 24-epi BR (eBR) was used to treat WT and SlOFP20-OE plants, and as a result, the leaf angle of WT plants was significantly increased, while that of the OE plants was only slightly increased. This implies that SlOFP20-OE plants displayed a decreased sensitivity to eBR treatment (Supplementary Figure S1). Furthermore, exogenous application of 50 µM GA3 could partially restore defects in plant height (Supplementary Figure S2). Combining the results of our current paper with previous results (Wang et al., 2007; Yang et al., 2018), we suppose that ectopic expression of SlOFP20 may exhibit typical BRs-insensitive and GA-deficient phenotypes.
Overexpression of SlOFP20 in Tomato Promotes Chlorophyll Accumulation in Leaves
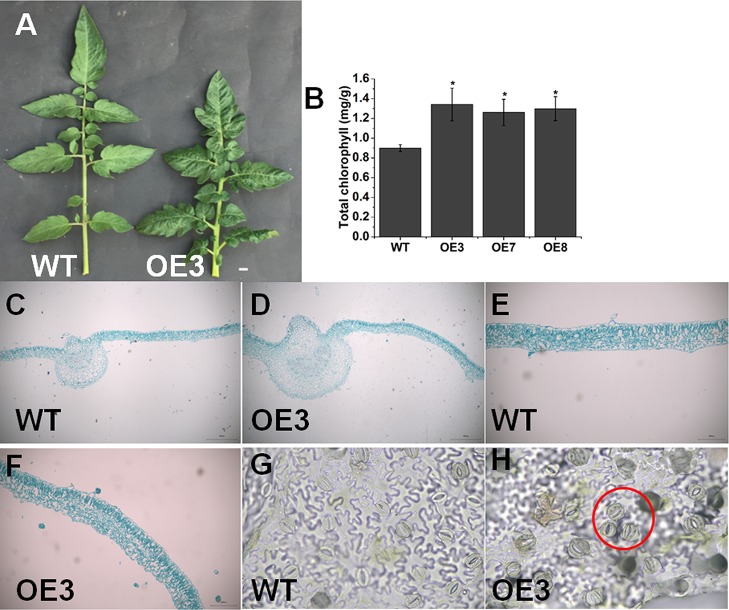
We found that SlOFP20-OE plants exhibited shorter compound leaves and darker green when compared with WT plants (Figure 5A). The chlorophyll contents of leaves were measured, indicating that overexpression lines had more chlorophyll compared to WT plants (Figure 5B). Moreover, SlOFP20-OE transgenic lines showed thicker leaves, and the anatomical study confirmed the increased thickness of leaf blade in SlOFP20-OE lines relative to WT plants (Figures 5C–F). We also performed hand sectioning to observe stomata morphology and stomata clustering event was found in SlOFP20-OE transgenic lines (Figures 5G, H), which was closely related to BR deficiency and BR signaling deficiency (Kim et al., 2012).
Figure 5.
Overexpression of SlOFP20 alters leaf growth. (A) Comparison of the leaves of WT and OE3 plants. Bar = 1 cm. (B) Total chlorophyll content in mature leaves of WT and transgenic plants. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05). (C–F) The leaf structure of WT and OE3 mature leaves. Bar = 100 μm. Stoma morphology of WT (G) and OE3 (H) leaves. Bar = 100 μm.
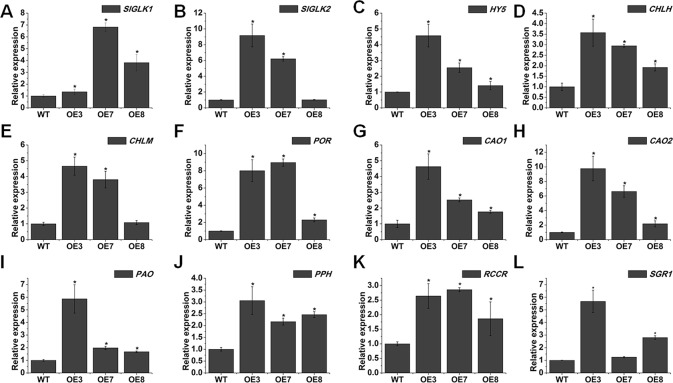
To reveal the possible molecular mechanism of chlorophyll accumulation in SlOFP20-OE dark green leaves, we analyzed the expression levels of genes related to chlorophyll accumulation and chloroplast development in mature leaves of WT and SlOFP20-OE lines. GLK transcription factors are involved in regulating the chloroplast and chlorophyll levels (Waters et al., 2008). There are two GLKs existing in tomato, namely SlGLK1 and SlGLK2. The expression of both was significantly increased in SlOFP20-OE plants (Figures 6A, B). We also found the upregulation of LeHY5 in SlOFP20-OE transgenic plants (Figure 6C). Because LeHY5-RNAi plants show defects in light responses, including inhibited seedling photomorphogenesis, loss of thylakoid organization, and reduced carotenoid accumulation (Liu et al., 2004), the observed phenotype in SlOFP20-OE transgenic plants could be reasonable.
Figure 6.
Overexpression of SlOFP20 alters the expression of chloroplast development- and chlorophyll metabolism-related genes. (A–C) Quantitative RT-PCR analysis of the expression levels of SlGLK1, SlGLK2, and HY5. (D–H) Expression analysis of genes involved in the chlorophyll biosynthesis. in the mature leaves of WT and transgenic plants (I–L) qRT-PCR analysis of chlorophyll degradation-related genes in the mature leaves of WT and transgenic plants Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
To clarify whether SlOFP20 affected chlorophyll metabolism, transcript levels of chlorophyll biosynthesis genes were detected. Five structural genes in the chlorophyll synthesis pathway, magnesium chelatase H subunit (CHLH), Mg protoporphyrin IX methyltransferase (CHLM), protochlorophyllide reductase (POR), and chlorophyllide a oxygenase (CAO1 and CAO2), were dramatically up-regulated in SlOFP20 transgenic lines (Figures 6D–H).
To determine whether SlOFP20 is involved in chlorophyll degradation, the expression levels of three chlorophyll degradation genes, pheophytin pheophorbide hydrolase (PPH), pheophorbide an oxygenase (PAO), and red chlcatabolite reductase (RCCR) were examined. All of them were significantly increased in SlOFP20-OE plants when compared with WT plants (Figures 6I–K). SGR1 is a chlorophyll degradation-related gene, and silencing of SGR1 reduces chlorophyll degradation in tomato leaves and fruits (Hu et al., 2011). The expression level of SGR1 was remarkably enhanced in SlOFP20-OE lines (Figure 6L).
Overexpression of SlOFP20 Affects Sugar Metabolism
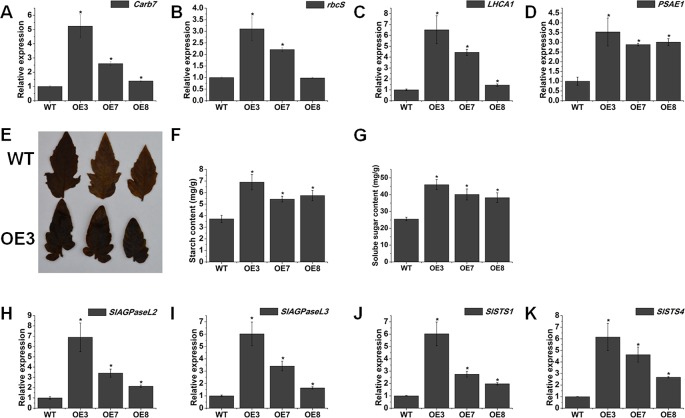
We wondered whether the increased chlorophyll content could confer higher photosynthetic performance in the SlOFP20-OE plants. To this end, we checked the expression level of photosynthesis-associated genes cab7 and rbcS (John et al., 1995), and showed that their mRNAs were present at much higher levels in SlOFP20-OE mature leaves compared with WT mature leaves (Figures 7A, B). The mRNA accumulation of LHCA1 and PSAE1 which encode chlorophyll a/b-binding polypeptides and photosystem I reaction center subunit IV A respectively, was distinctively enhanced in SlOFP20-OE as compared to the WT (Figures 7C, D). We also detected the accumulation of sugars in the leaves of WT and SlOFP20-OE plants, which are the main products of chloroplast activity and photosynthesis. I2/KI staining was used to determine the accumulation of starch in the leaves of WT and SlOFP20-OE plants, indicating that SlOFP20-OE plants leaves accumulated much more starch than those of WT plants leaves (Figure 7E). Moreover, we measured the starch and soluble sugar content in the leaves of WT and SlOFP20-OE plants. These results suggested that SlOFP20-OE plants had significantly higher starch and soluble sugar contents when compared with WT plants (Figures 7F, G). To gain more insight into the mechanism of sugar metabolism in the SlOFP20-OE transgenic plants, we examined the expression patterns of starch biosynthesis genes. SlAGPaseL2 and AGPaseL3 encode the most important enzymes in starch synthesis that catalyse the first step of the reaction. SlSTS1 and SlSTS4 encode the starch synthases which catalyse the second step of starch synthesis. These results showed that all of these genes were distinctly induced in the mature leaves of SlOFP20-OE plants (Figures 7H–K).
Figure 7.
Ectopic expression of SlOFP20 affects sugar metabolism. (A–D) The transcript levels of photosynthesis-related genes. (E) Starch I2/KI staining results for the WT and OE3 leaves. (F, G) Contents of starch and soluble sugar. (H–K) Expression analysis of genes involved in starch synthesis. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
Transgenic Plants Overexpressing SlOFP20 Display Prolonged Natural Senescence
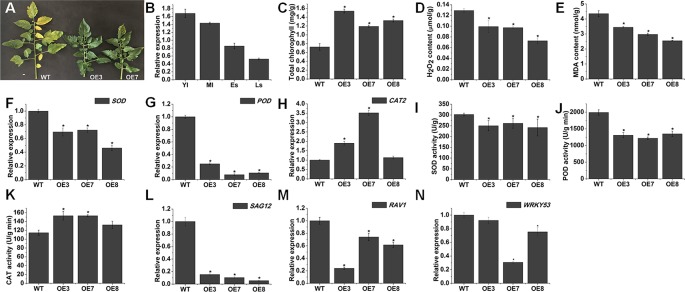
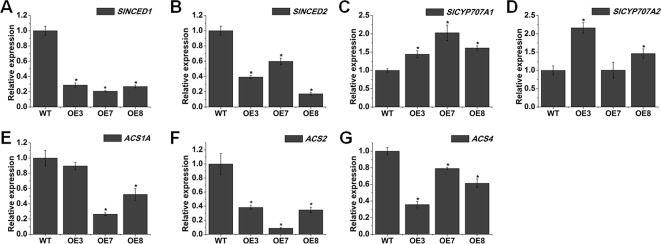
We found some delayed senescence in SlOFP20-OE transgenic lines when compared with WT plants during the whole life cycle (Figure 8A). To evaluate the contribution of SlOFP20 in leaf senescence, we examined the expression level of SlOFP20 at different developmental stages of WT tomato leaves, indicating that the mRNA accumulation of SlOFP20 was gradually reduced since the beginning of senescence (Figure 8B). The visible appearance of leaf senescence is the degradation of chlorophyll, which is used as an index to assess leaf senescence. As shown in Figure 8C, there was a 1.64, 2.12, and 1.83 times higher total chlorophyll content in the leaves of OE3, OE7, and OE8 transgenic lines, respectively, when compared to WT leaves. To further explore this phenotype, we measure a series of physiological indicators related to leaf senescence. Enhanced production of reactive oxygen species (ROS) as well as MDA, a marker for lipid peroxidation, is characteristic of naturally senescent leaves (Dhindsa et al., 1981; Khanna-Chopra, 2012; Wang et al., 2013). The results suggested that the leaves of SlOFP20-OE lines had much less H2O2 and MDA contents than WT leaves (Figures 8D, E). Moreover, we also examined the mRNA accumulation of oxidative stress-related genes, which encode SOD, POD, and CAT, respectively. The expression levels of SOD and POD were significantly reduced in SlOFP20-OE lines when compared with WT (Figures 8F, G). However, the expression level of CAT2 in SlOFP20-OE plants was higher than in WT plants (Figure 8H). In addition, the activities of SOD, POD, and CAT were measured, the results suggested that the activities of SOD and POD were significantly lower in transgenic lines than in WT plants (Figures 8I, J). However, a significantly higher CAT activity was measured in transgenic lines (Figure 8K). To determine whether overexpression of SlOFP20 mediates the expressions of senescence-mediated genes, SAG12 was evaluated as a marker gene of leaf senescence (Weaver et al., 1998). RAV1 acts as a positive regulator of leaf senescence in Arabidopsis (Woo et al., 2010). WRKY53 acts in a complicated transcription factor signaling network regulating senescence specific gene expression (Miao et al., 2004). The results showed that the expression level of SAG12, RAV1, and WRKY53 was down-regulated in SlOFP20-OE lines (Figures 8L–N). Leaf senescence is closely linked to differential phytohormones signaling, such as ABA and ethylene. To understand whether SlOFP20 modulates leaf senescence via phytohormone pathways, we analyzed the expression level of ABA biosynthesis genes (SlNCED1 and SlNCED2) and degradation genes (SlCYP707A1 and SlCYP707A2). Gene expression analysis revealed that the transcript abundance of SlNCED1 and SlNCED2 in SlOFP20-OE lines was much less than WT plants (Figures 9A, B). In contrast, both SlCYP707A1 and SlCYP707A2 were up-regulated in SlOFP20-OE lines (Figures 9C, D). We further evaluated ethylene synthesis genes, including ACS1A, ACS2 and ACS4, and the results suggested that expression of all these three genes was substantially repressed in SlOFP20-OE lines when compared to WT plants (Figures 9E–G).
Figure 8.
SlOFP20 overexpression delays leaf senescence. (A) Phenotype of delayed leaf senescence in SlOFP20-OE lines. (B) The expression levels of SlOFP20 in the leaves at different developmental stages: young leaves (YL), mature leaves (ML), senescent leaves (SL), and late senescent leaves (LS). (C, D) Comparison of H2O2 and MDA Contents in the senescent leaves of WT and SlOFP20-OE plants. (F–H) Comparison of transcript levels of SOD, POD, and CAT2 in the senescent leaves of WT and transgenic plants. (I–K). Comparison of SOD, POD, and CAT activities in the senescent leaves of WT and transgenic plants. (L–N) Expression of senescence-related genes (SAG12, RAV1, and WRKY53) in the senescent leaves of WT and transgenic plants. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
Figure 9.
Overexpression of SlOFP20 leads to the changes of abscisic acid (ABA)- and ethylene-related genes. (A, B) The expression of ABA biosynthesis genes (SlNCED1 and SlNCED2) between WT and SlOFP20-OE lines. (C, D) The expression of ABA degradation genes (SlCYP707A1 and SlCYP707A2) between WT and SlOFP20-OE lines. (E–G) The expression of ethylene biosynthesis genes (ACS1A, ACS2, and ACS4) between WT and SlOFP20-OE lines. Each value represents the mean ± SE of three biological replicates. Asterisks indicate a significant difference (P < 0.05).
Discussion
OVATE domain-containing proteins belong to a plant-specific and poorly characterized family of plant regulatory proteins (Hackbusch et al., 2005). Recent studies reveal that OFPs participate in a series of plant growth and developmental processes through modulating hormone signaling. AtOFP1 overexpression in Arabidopsis generates dwarf phenotype through weakening GA synthesis (Wang et al., 2007). The function of OsOFP8 in plant growth and development is at least partly through the BR signaling pathway in rice (Yang et al., 2016). Rice OFP 6 controls lateral root growth and initiation via mediating auxin (Ma et al., 2017). OsOFP19 negatively modulates the BR response to adjust plant growth and development (Yang et al., 2018). These findings strongly support the fact that OFPs are involved in mediating hormone signaling, BR and GA in particular. BR and GA are the two most important hormones that determine plant height by regulating cell elongation. Mutants deficient in either BR or GA display a dwarf stature. In our study, we observed that overexpression of SlOFP20 in WT tomato significantly reduced plant height (Figure 2F). The diminished plant size in AtOFP1 and OsOFP19 overexpression lines could partially be attributed to the deficiency in gibberellin biosynthesis and the reduced BR response, respectively. Exogenous application of eBR suppresses the expression of SlOFP20 (unpublished data), as well as GA3 (Figure 1A). Therefore, we speculated that the multiple phenotypes caused by SlOFP20 overexpression may be closely related to BR and GA signaling.
Here, we found that overexpression of SlOFP20 led to reduced cotyledon size (Figure 2E) and hypocotyl length (Figures 2A, B), as well as plant height (Figure 2F). Moreover, decreased length of primary root and reduced number of lateral roots were observed, in agreement with the earlier observations as to some partial deficiency in key BR biosynthetic gene DWARF (Li et al., 2016). The deficiency of BR biosynthesis or signaling results in upright leaves. LEAF and TILLER ANGLE INCREASED CONTROLLER (LIC) acts as a negative regulator in BR response, and its gain-of-function mutant displays upright leaves (Zhang et al., 2012). BR level is connected with the bud outgrowth and DWARF overexpression obviously increased the number of lateral branches (Li et al., 2016). SlOFP20-OE plants showed obvious erect leaves (Figures 3A, B), the leaf angles of SlOFP20-OE lines were much smaller than WT plants (Figure 3C), and the expression level of XET4 positively related to leaf inclination was significantly reduced in SlOFP20-OE lines (Figure 3D). Moreover, we also examined the lateral bud number in WT and SlOFP20-OE plants, the results suggested that the lateral bud number in SlOFP20-OE lines was clearly reduced (Figure 3E), and the transcript accumulation of BRC1b gene, which inhibits the outgrowth of lateral branches, was significantly enhanced in SlOFP20-OE lines (Figure 3F). Late-flowering phenotypes are found in BR-deficient or BR-insensitive mutants in Arabidopsis (Domagalska et al., 2007), and mutant plants that are deficient in GA also exhibit a late-flowering phenotype (Davière and Achard, 2013). Based on our observation, overexpression of SlOFP20 delayed the flowering process, further statistical analysis confirmed this phenotype (Figures 4A–C), and the expression level of SFT, which encodes a major player in flowering time control of the day-neutral plant tomato, was distinctly inhibited in SlOFP20-OE lines (Figure 4D). Furthermore, we also determined the expression levels of BR- and GA- related genes, and the results suggested that these genes were significantly affected in SlOFP20-OE lines (unpublished data). In addition, our yeast two-hybrid experiments confirmed that SlOFP20, KNOX1, and GRAS41 can form a complex to directly repress BR signaling, which was consistent with the previous finding that OsOFP19, OSH1, and DLT can form a complex that finely modulates the balance between plant growth and development and BR signaling (Yang et al., 2018). SlOFP20, KNOX1, and GRAS41 are homologous genes of OsOFP19, KNOX1, and DLT from rice (unpublished data). SlOFP20-OE plants displayed greatly reduced sensitivity to eBR treatment (Supplementary Figure S1), and exogenous GA3 can partially restore defects in cell elongation in plants overexpressing SlOFP20 (Supplementary Figure S2), which is consistent with previous observations (Wang et al., 2007; Yang et al., 2018). Based on this, we would like to conclude that ectopic expression of SlOFP20 results in typical BRs-insensitive and GA-deficient phenotypes. The phenotypes produced by SlOFP20 overexpression resembled those of AtOFP1-OE and OsOFP19-OE plants, indicating that these homologous genes share similar functions in different plant species, most possibly as well as underlying molecular mechanisms. Besides, we found that overexpression of SlOFP20 in tomato plants enhanced the accumulation of chlorophyll in leaves (Figure 5A), and retarded leaf senescence (Figure 8A), which have not been studied in other OFPs.
Our knowledge of the complex regulatory mechanisms of chloroplast development and chlorophyll metabolism is very limited. In this report, we speculated that SlOFP20 may positively regulate chloroplast development and chlorophyll metabolism. SlOFP20-OE lines showed significantly higher levels of chlorophyll content when compared with WT plants. To investigate the mechanism of chlorophyll accumulation and chloroplast development in the mature leaves of SlOFP20-OE lines, we analyzed gene expression levels in the mature leaves of SlOFP20-OE lines and WT plants. HY5 and GLKs promote the expression of many genes related to chloroplast development (Kobayashi et al., 2012; Powell et al., 2012; Pogson et al., 2015). The expression levels of HY5, SlGLK1 and SlGLK2 were distinctly enhanced by overexpressing SlOFP20 in WT tomato (Figures 6A–C). Moreover, we also explored the effects of SlOFP20 overexpression on chlorophyll metabolism. These five chlorophyll biosynthetic genes, LHCH, LHCM, POR, CAO1, and CAO2 were sharply boosted in transgenic lines (Figures 6D–H), whereas the chlorophyll degradation genes, PPH, PAO, RCCR, and SGR1 were also significantly increased (Figures 6I–L), implying that the accumulation of chlorophyll in SlOFP20-OE plants may activate the chlorophyll degradation pathways. A number of studies have suggested that chlorophyll content may be a positive correlation with photosynthetic capacity. For example, overexpression of SlARF10 results in a dark-green phenotype and positively affected photosynthesis in both leaves and fruit (Yuan et al., 2018). The enhanced chlorophyll content in SlARF4 down-regulated fruits correlates with a higher photochemical efficiency compared with WT fruits (Sagar et al., 2013). The basic helix-loop-helix transcription factor PHYTOCHROME INTERACTING FACTOR3 (PIF3) negatively regulates light responses, repressing light-mediated cotyledon expansion, opening, and chlorophyll biosynthesis in the dark (Liu et al., 2013). We speculate that the increased chlorophyll content may upgrade the photosynthesis in SlOFP20-OE lines. Therefore, the expression levels of two photosynthetic-related genes, cab7 and rbcS, were checked, the results suggested that both of them were increased in SlOFP20-OE lines (Figures 7A, B). LHCA1 and PSAE1 which encode chlorophyll a/b-binding polypeptides and photosystem I reaction center subunit IV A respectively, the mRNA accumulation of them was distinctly enhanced in SlOFP20-OE when compared to WT (Figures 7C, D). Moreover, we also measured sugar content in the mature leaves of SlOFP20-OE and WT plants, as the main products of photosynthesis. As expected, SlOFP20-OE plants accumulated more starch and soluble sugar content than WT plants (Figures 7E–G). We also examined the expression patterns of starch biosynthesis genes, including SlAGPaseL2, AGPaseL3, SlSTS1, and SlSTS4, indicating that overexpression of SlOFP20 increased the transcripts of all four genes in mature leaves (Figures 7H–K). These results suggested that SlOFP20 may play an important role in chlorophyll and sugar metabolism.
Leaf senescence represents the final stage of leaf development. Its time of onset is thus a major determinant of crop yield and quality. Therefore, plasticity in the timing of leaf senescence and the delicate balance between the onset and extent of leaf senescence are essential for ecological success and crop yield (Ma et al., 2018). In this study, transgenic plants with enhanced expression of SlOFP20 exhibit a delay of leaf senescence during typical age-dependent senescence (Figure 8A). The expression SlOFP20 of in leaves collected at different developmental stages declined along with the onset of leaf senescence (Figure 8B), implying that it may play a negative regulatory role in this process. Consistent with the delayed-senescence phenotype, SlOFP20-OE plants retained more total Chl levels than WT plants during senescence (Figure 8C). Leaf senescence induces the expression level of many SAGs, such as hormone signaling, transcriptional regulation, and chlorophyll catabolism (Buchanan-Wollaston et al., 2003; Lim et al., 2007; Li et al., 2014; Li et al., 2017). The senescence specificity of SAG12, encoding a cysteine protease, makes this gene as a molecular marker to study the senescence process (Noh and Amasino, 1999). WRKY53 overexpression, RNAi and knock-out lines showed accelerated and delayed senescence phenotypes, respectively, and exhibited altered expression levels of the target genes (Miao et al., 2004). Constitutive and inducible overexpression of RAV1 caused premature leaf senescence (Woo et al., 2010). SAG12, WRKY53, and RAV1 were down-regulated in SlOFP20-OE lines when compared with WT plants, which was consistent with the phenotype of delayed senescence (Figures 8L–N). Reactive oxygen species are continuously produced in plants as products of aerobic metabolism (Maurino and Flügge, 2008). Excess ROS accumulation leads to oxidative damage to thylakoid membranes and other cellular components (Maurino and Flügge, 2008). Previous studies showed that early leaf senescence is usually associated with excessive ROS (Han et al., 2014; Ren et al., 2018; Wang et al., 2018). Compared with SlOFP20-OE plants, WT plants accumulated more H2O2 (Figure 8D). Furthermore, the content of MDA, a product of ROS-induced membrane lipid peroxidation, was also increased in WT plants when compared with SlOFP20-OE plants (Figure 8E). ROS can lead to the activation of antioxidative enzymes in plants, including SOD, APX, CAT, POD, and GST, to alleviate oxidative damage (Apel and Hirt, 2004). Plant senescence can lead to the synthesis of antioxidative enzymes to remove ROS (Miller et al., 2010). Rice dwarf and early-senescence leaf1 (del1) exhibits early leaf senescence with the accumulation of ROS, and the activities of SOD and POD are increased in del1 plants (Leng et al., 2017). The activities of SOD and POD in SlOFP20-OE plants were lower than those of WT plants (Figures 8I, J). The activity of CAT was increased in SlOFP20-OE plants (Figure 8K). Moreover, the changes in transcript levels of SOD, POD, and CAT2 agreed with the changes in activity (Figures 8F–H). Furthermore, numerous studies confirmed that ABA and ethylene play vital roles in leaf senescence (Gao et al., 2016; Ren et al., 2018; Tang et al., 2018). Here, we measured the transcript accumulation of ABA biosynthesis genes (SlNCED1 and SlNCED2) and degradation genes (SlCYP707A1 and SlCYP707A2). SlNCED1 and SlNCED2 were down-regulated in SlOFP20-OE plants (Figures 9A, B), and the SlCYP707A1 and SlCYP707A2 were up-regulated (Figures 9C, D). We also determined the mRNA levels of ethylene synthesis genes, including ACS1A, ACS2, and ACS4, and their expression levels were decreased in SlOFP20-OE plants (Figures 9E–G). Therefore, we inferred that overexpression of SlOFP20 delays leaf senescence in relation to ABA and ethylene synthesis.
In summary, our data suggest that SlOFP20 overexpression exert manifold effects on tomato growth and development through coordinating BR and GA signaling. In addition, we found that overexpression of SlOFP20 promotes the accumulation of chlorophyll and sugar, and restrains the beginning of leaf senescence, which has not been reported in OFPs. Knockdown of BRASSINOSTEROID INSENSITIVE1 (BRI1), coding for a BR receptor, results in reduced BR signaling and a dwarf stature with upright and dark-green leaves (Kir et al., 2015). BRs could promote ethylene biosynthesis via regulation of ACC synthase 5 (ACS5) and ACC oxidase activities (Hansen et al., 2009). BRs could also induce ABA biosynthesis (Xia et al., 2014; Zhou et al., 2014). The expression of ethylene biosynthesis genes and ABA biosynthesis genes was inhibited in SlOFP20-OE plants. We infer that these two phenotypes may be closely linked with the reduced BR response in SlOFP20-OE lines. Therefore, it is meaningful to decipher how SlOFP20 harmonizes the crosstalk between different hormones to control plant growth and development in the future.
Data Availability Statement
All datasets for this study are included in the article/ Supplementary Material.
Author Contributions
GC, QX, and ZH designed and managed the research work and improved the manuscript. SZ, XC, FL, PF, and GH performed the experiments. SZ wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (no. 31872121), and the National Natural Science Foundation of Chongqing of China (cstc2018jcyjAX0458).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01510/full#supplementary-material
References
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 (1), 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 (4), 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63 (7), 2667–2679. 10.1093/jxb/err450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer W. F., Jr., Fridovich I. (1987). Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal. Biochem. 161 (2), 559–566. 10.1016/0003-2697(87)90489-1 [DOI] [PubMed] [Google Scholar]
- Bryan J. H. D. (1955). Differential staining with a mixture of safranin and fast green FCF. Stain Technol. 30 (4), 153–157. 10.3109/10520295509114456 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., et al. (2003). The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnol. J. 1 (1), 3–22. 10.1046/j.1467-7652.2003.00004.x [DOI] [PubMed] [Google Scholar]
- Clouse S. D. (2011). Brassinosteroids. Arabidopsis Book 9, e0151–e0151. 10.1199/tab.0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Schmulling T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66 (16), 4999–5013. 10.1093/jxb/erv132 [DOI] [PubMed] [Google Scholar]
- Cortleven A., Marg I., Yamburenko M. V., Schlicke H., Hill K., Grimm B., et al. , (2016). Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 172 (1), 464–478. 10.1104/pp.16.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J. M., Achard P. (2013). Gibberellin signaling in plants. Development 140 (6), 1147–1151. 10.1242/dev.087650 [DOI] [PubMed] [Google Scholar]
- Dhindsa R. S., Plumb-dhindsa P., Thorpe T. A. (1981). Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 32 (1), 93–101. 10.1093/jxb/32.1.93 [DOI] [Google Scholar]
- Domagalska M. A., Schomburg F. M., Amasino R. M., Vierstra R. D., Nagy F., Davis S. J. (2007). Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134 (15), 2841–2850. 10.1242/dev.02866 [DOI] [PubMed] [Google Scholar]
- Duan K., Li L., Hu P., Xu S. P., Xu Z. H., Xue H. W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47 (4), 519–531. 10.1111/j.1365-313X.2006.02804.x [DOI] [PubMed] [Google Scholar]
- Fitter D. W., Martin D. J., Copley M. J., Scotland R. W., Langdale J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31 (6), 713–727. 10.1046/j.1365-313x.2002.01390.x [DOI] [PubMed] [Google Scholar]
- Galpaz N., Wang Q., Menda N., Zamir D., Hirschberg J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53 (5), 717–730. 10.1111/j.1365-313X.2007.03362.x [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113 (2), 313. 10.1104/pp.113.2.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Gao J., Zhu X., Song Y., Li Z., Ren G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9 (9), 1272–1285. 10.1016/j.molp.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Gepstein S., Thimann K. V. (1980). Changes in the abscisic acid content of oat leaves during senescence. Proc. Natl. Acad. Sci. U.S.A. 77 (4), 2050–2053. 10.1073/pnas.77.4.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslings D., Meskauskiene R., Kim C., Lee K. P., Nater M., Apel K. (2004). Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 40 (6), 957–967. 10.1111/j.1365-313X.2004.02262.x [DOI] [PubMed] [Google Scholar]
- Grbić V., Bleecker A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8 (4), 595–602. 10.1046/j.1365-313X.1995.8040595.x [DOI] [Google Scholar]
- Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N., Worley J. F., Warthen J. D., et al. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281 (5728), 216–217. 10.1038/281216a0 [DOI] [Google Scholar]
- Hackbusch J., Richter K., Müller J., Salamini F., Uhrig J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102 (13), 4908–4912. 10.1073/pnas.0501181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim C. Y., Lee J., Lee S. K., Jeon J. S. (2014). OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 37 (7), 532–539. 10.14348/molcells.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Chae H. S., Kieber J. J. (2009). Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 57 (4), 606–614. 10.1111/j.1365-313X.2008.03711.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. L., Deng L., Yan B., Pan Y., Luo M., Chen X. Q., et al. (2011). Silencing of the LeSGR1 gene in tomato inhibits chlorophyll degradation and exhibits a stay-green phenotype. Biol. Plantarum 55 (1), 27–34. 10.1007/s10535-011-0004-z [DOI] [Google Scholar]
- John I., Drake R., Farrell A., Cooper W., Lee P., Horton P., et al. (1995). Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J. 7 (3), 483–490. 10.1046/j.1365-313X.1995.7030483.x [DOI] [Google Scholar]
- Khanna-Chopra R. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249 (3), 469–481. 10.1007/s00709-011-0308-z [DOI] [PubMed] [Google Scholar]
- Kim T. W., Michniewicz M., Bergmann D. C., Wang Z. Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482 (7385), 419–422. 10.1038/nature10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir G., Ye H., Nelissen H., Neelakandan A. K., Kusnandar A. S., Luo A., et al. (2015). RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 169 (1), 826–839. 10.1104/pp.15.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Baba S., Obayashi T., Sato M., Toyooka K., Keranen M., et al. (2012). Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24 (3), 1081–1095. 10.1105/tpc.111.092254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F., Sunl S., Jing Y., Wang F., Wei Q., Wang X., et al. (2016). A rapid and sensitive method for determination of trace amounts of glucose by anthrone-sulfuric acid method. Bulg. Chem. Commun. 48 (1), 109–113. [Google Scholar]
- Leng Y., Yang Y., Ren D., Huang L., Dai L., Wang Y., et al. (2017). A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 174 (2), 1151–1166. 10.1104/pp.16.01625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Y., Chen S., An L. (2010). Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 61 (15), 4221–4230. 10.1093/jxb/erq241 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Peng J., Guo H., Luo J. (2014). LSD 2.0: An update of the leaf senescence database. Nucleic Acids Res. 42 (D1), D1200–D1205. 10.1093/nar/gkt1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Chen X. J., Guo X., Yin L. L., Ahammed G. J., Xu C. J., et al. (2016). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14 (3), 1021–1033. 10.1111/pbi.12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Jiang Z., Peng J., Jin J., et al. (2017). Construction of the leaf senescence database and functional assessment of senescence-associated genes. Methods Mol. Biol. 1533, 315–333. 10.1007/978-1-4939-6658-5_19 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57 (13), 3405–3414. 10.1093/jxb/erl106 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Gil Nam H. (2007). Leaf Senescence. Annu. Rev. Plant Biol. 58 (1), 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Liu J., Van Eck J., Cong B., Tanksley S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U. S. A. 99 (20), 13302–13306. 10.1073/pnas.162485999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Roof S., Ye Z., Barry C., van Tuinen A., Vrebalov J., et al. (2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. U. S. A. 101 (26), 9897–9902. 10.1073/pnas.0400935101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen C. Y., Wang K. C., Luo M., Tai R., Yuan L., et al. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25 (4), 1258–1273. 10.1105/tpc.113.109710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Jia C., Zhang M., Chen D., Chen S., Guo R., et al. (2014). Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol. J. 12 (1), 105–115. 10.1111/pbi.12121 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang J., Wang J., Zhang J., Miao H., Jia C., et al. (2018). MuMADS1 and MaOFP1 regulate fruit quality in a tomato ovate mutant. Plant Biotechnol. J. 16 (5), 989–1001. 10.1111/pbi.12843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Yang C., He Y., Tian Z., Li J. (2017). Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J. Exp. Bot. 68 (17), 4885–4898. 10.1093/jxb/erx309 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Y., Tureckova V., Xue G. P., Fernie A. R., Mueller-Roeber B., et al. (2018). The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 177 (3), 1286–1302. 10.1104/pp.18.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E. G., Serra F., Marcel F., Rodríguez-Buey M. L., Schmitz G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67 (4), 701–714. 10.1111/j.1365-313X.2011.04629.x [DOI] [PubMed] [Google Scholar]
- Maurino V. G., Flügge U. I. (2008). Experimental systems to assess the effects of reactive oxygen species in plant tissues. Plant Signal Behav. 3 (11), 923–928. 10.4161/psb.7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Fan Z., Zhang Q., Wang C., Gao Y., Deng Y., et al. (2018). BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. 94 (6), 1126–1140. 10.1111/tpj.13924 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., Op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98 (22), 12826–12831. 10.1073/pnas.221252798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55 (6), 853–867. 10.1007/s11103-004-2142-6 [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33 (4), 453–467. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Mitchell J. W., Mandava N., Worley J. F., Plimmer J. R., Smith M. V. (1970). Brassins-a new family of plant hormones from rape pollen. Nature 225 (5237), 1065–1066. 10.1038/2251065a0 [DOI] [PubMed] [Google Scholar]
- Morohashi Y. (2002). Peroxidase activity develops in the micropylar endosperm of tomato seeds prior to radicle protrusion. J. Exp. Bot. 53 (374), 1643–1650. 10.1093/jxb/erf012 [DOI] [PubMed] [Google Scholar]
- Nadakuduti S. S., Holdsworth W. L., Klein C. L., Barry C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78 (6), 1022–1033. 10.1111/tpj.12529 [DOI] [PubMed] [Google Scholar]
- Nguyen C. V., Vrebalov J. T., Gapper N. E., Zheng Y., Zhong S., Fei Z., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26 (2), 585–601. 10.1105/tpc.113.118794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y. S., Amasino R. M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41 (2), 181–194. 10.1023/a:1006342412688 [DOI] [PubMed] [Google Scholar]
- Oh S. A., Park J. H., Lee G. I., Paek K. H., Park S. K., Nam H. G. (1997). Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 12 (3), 527–535. 10.1046/j.1365-313x.1997.00527.x [DOI] [PubMed] [Google Scholar]
- Pagnussat G. C., Yu H. J., Sundaresan V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19 (11), 3578–3592. 10.1105/tpc.107.054890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B. J., Ganguly D., Albrecht-Borth V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847 (9), 1017–1024. 10.1016/j.bbabio.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Powell A. L., Nguyen C. V., Hill T., Cheng K. L., Figueroa-Balderas R., Aktas H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336 (6089), 1711–1715. 10.1126/science.1222218 [DOI] [PubMed] [Google Scholar]
- Qin H., Gu Q., Zhang J., Sun L., Kuppu S., Zhang Y., et al. (2011). Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 52 (11), 1904–1914. 10.1093/pcp/pcr125 [DOI] [PubMed] [Google Scholar]
- Rao M. V., Paliyath G., Ormrod D. P. (1996). Ultraviolet-B- and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 110 (1), 125–136. 10.1104/pp.110.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Wang J., Zhao M., Gong X., Wang S., Wang G., et al. (2018). Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247 (1), 53–68. 10.1007/s00425-017-2770-0 [DOI] [PubMed] [Google Scholar]
- Sagar M., Chervin C., Mila I., Hao Y., Roustan J. P., Benichou M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161 (3), 1362–1374. 10.1104/pp.113.213843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev I., Alexieva V., Karanov E. (1997). Effect of spermine, atrazine and combination between them on some endogenous protective systems and stress markers in plants. Compt. Rend Acad. Bulg. Sci. 51, 121–124. [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J. P., Bowman J. L., Eshed Y., et al. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. U.S.A. 106 (20), 8392–8397. 10.1073/pnas.0810810106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Li L., Yan T., Fu X., Shi P., Shen Q., et al. (2018). AaEIN3 mediates the downregulation of artemisinin biosynthesis by ethylene signaling through promoting leaf senescence in Artemisia annua. Front. Plant Sci. 9, 413. 10.3389/fpls.2018.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chang Y., Guo J., Chen J. G. (2007). Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50 (5), 858–872. 10.1111/j.1365-313X.2007.03096.x [DOI] [PubMed] [Google Scholar]
- Wang L., Mai Y. X., Zhang Y. C., Luo Q., Yang H. Q. (2010). MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant 3 (5), 794–806. 10.1093/mp/ssq042 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lin A., Loake G. J., Chu C. (2013). H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. J. Integr. Plant Biol. 55 (3), 202–208. 10.1111/jipb.12032 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang T., Peng H., Luo S., Tan J., Jiang K., et al. (2018). Rice Premature Leaf Senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front. Plant Sci. 9, 560. 10.3389/fpls.2018.00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Moylan E. C., Langdale J. A. (2008). GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56 (3), 432–444. 10.1111/j.1365-313X.2008.03616.x [DOI] [PubMed] [Google Scholar]
- Weaver L. M., Gan S., Quirino B., Amasino R. M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37 (3), 455–469. 10.1023/A:1005934428906 [DOI] [PubMed] [Google Scholar]
- Woo H. R., Kim J. H., Kim J., Kim J., Lee U., Song I. J., et al. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61 (14), 3947–3957. 10.1093/jxb/erq206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang B., Keyhaninejad N., Rodríguez G. R., Kim H. J., Chakrabarti M., et al. (2018). A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 9 (1), 4734. 10.1038/s41467-018-07216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. J., Fang P. P., Guo X., Qian X. J., Zhou J., Shi K., et al. (2018). Brassinosteroid-mediated apoplastic H2O2-glutaredoxin 12/14 cascade regulates antioxidant capacity in response to chilling in tomato. Plant Cell Environ. 41 (5), 1052–1064. 10.1111/pce.13052 [DOI] [PubMed] [Google Scholar]
- Xia X.J., Gao C.J., Song L.X., Zhou Y.H., Shi K., Yu J. Q. (2014). Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ. 37 (9), 2036–2050. 10.1111/pce.12275 [DOI] [PubMed] [Google Scholar]
- Yang C., Shen W., He Y., Tian Z., Li J. (2016). OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PloS Genet. 12 (6), e1006118. 10.1371/journal.pgen.1006118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Ma Y., He Y., Tian Z., Li J. (2018). OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 93 (3), 489–501. 10.1111/tpj.13793 [DOI] [PubMed] [Google Scholar]
- Yin X. R., Xie X. L., Xia X. J., Yu J. Q., Ferguson I. B., Giovannoni J. J., et al. (2016). Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 86 (5), 403–412. 10.1111/tpj.13178 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Mei L., Wu M., Wei W., Shan W., Gong Z., et al. (2018). SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 69 (22), 5507–5518. 10.1093/jxb/ery328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang W. Q., Zhang G. L., Kaminek M., Dobrev P., Xu J., et al. (2010). Senescence-inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in Cassava. J. Integr. Plant Biol. 52 (7), 653–669. 10.1111/j.1744-7909.2010.00956.x [DOI] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., et al. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PloS Genet. 8 (4), e1002686. 10.1371/journal.pgen.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Li X., Xia X. J., Zhou Y. H., Shi K., et al. (2014). H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 65 (15), 4371–4383. 10.1093/jxb/eru217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Hu Z., Li F., Yu X., Naeem M., Zhang Y., et al. (2018). Manipulation of plant architecture and flowering time by down-regulation of the GRAS transcription factor SlGRAS26 in Solanum lycopersicum. Plant Sci. 271, 81–93. 10.1016/j.plantsci.2018.03.017 [DOI] [PubMed] [Google Scholar]
- Zhu M., Chen G., Dong T., Wang L., Zhang J., Zhao Z., et al. (2015). SlDEAD31, a putative DEAD-Box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato. PloS One 10 (8), e0133849. 10.1371/journal.pone.0133849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets for this study are included in the article/ Supplementary Material.