Abstract.
Fifty-two febrile patients living in Barquisimeto, Venezuela, were screened for arbovirus infection by virus culture during an outbreak of what was thought to be Zika virus infection. We report identification of Mayaro virus (MAYV) on culture of plasma from one patient, an 18-year-old woman with acute febrile illness, arthralgias, and psoriasiform rash. The strain was sequenced and was found to be most closely related to a 1999 strain from French Guiana, which, in turn, was related to two 2014 strains from Haiti. By contrast, previously reported outbreak-related MAYV strains from a sylvatic area approximately 80 miles from where the case patient lived were most closely related to Peruvian isolates. The two strain groups show evidence of having diverged genetically approximately 100 years ago.
INTRODUCTION
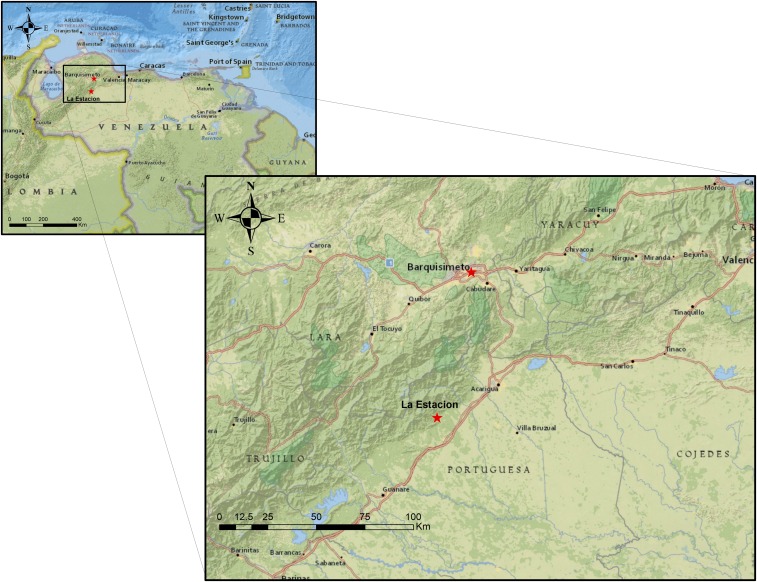
Mayaro virus (MAYV), first isolated from forest workers in Trinidad and Tobago in 1954,1 is a single-stranded positive-sense RNA virus in the genus Alphavirus and family Togaviridae. Signs of Mayaro febrile illness include fever (lasting 3–5 days), headache, retro-orbital pain, arthralgia, myalgia, vomiting, diarrhea, and rashes,2–4 similar to findings reported for chikungunya fever. Severe joint pain has been reported to last for months and up to 1 year.4,5 Since 1954, outbreaks of Mayaro fever have been detected in sites in South America as well as in parts of Central America and the Caribbean,6 mostly among forest workers and hunters in rural areas.2–4,7,8 One of the largest MAYV outbreaks to date occurred in 2010 in the village of La Estación, Portuguesa State, Venezuela.7 That village is located in a former tropical forest which had recently been converted for coffee production and other farming. The village is approximately 125 km (78 mi) from Barquisimeto, the 4th largest city in Venezuela with a population of close to one million, and the home of the patient described in this report (Figure 1).
Figure 1.
Locations of 2010 outbreak (La Estación) of Mayaro virus infection and of case patient in current report (Barquisimeto). The city of Barquisimeto (elevation: 566 m) is approximately 80 miles from La Estación (elevation: 450 m). Source: Esri. “National Geographic” (basemap). “National Geographic World Map”. Created December 13, 2011; updated April 24, 2019. https://services.arcgisonline.com/ArcGIS/rest/services/NatGeo_World_Map/MapServer (July 10, 2019). This figure appears in color at www.ajtmh.org.
In February 2016, physicians in Barquisimeto reported a spike in febrile illnesses associated with the outbreak of Zika fever in the region. Access to diagnostic services within the country was limited as a severe economic crisis was occurring at the time. To identify the etiologic agent(s) of the febrile illness, a collaborative study was established between infectious disease physicians at the Instituto Diagnóstico Barquisimeto (IDB) Biomedical Research Institute and arbovirus researchers at the University of Florida (UF) in Gainesville, FL. Protocols were developed and implemented for the collection of diagnostic specimens (blood and urine) from patients seen for acute undifferentiated febrile illness, together with maculopapular rash, conjunctivitis, myalgias, arthralgias, or other possible signs of infection with Zika virus (ZIKV).
From February through July 2016, blood and urine specimens were obtained from patients who met the criteria for acute undifferentiated febrile illness; samples from 52 patients were screened by virus culture at the UF. The protocol for sample collection was approved by the IDB Biomedical Research Institute Institutional Review Board and the UF Institutional Review Board and written informed consent was obtained from the patient. Reports of isolation of ZIKV, dengue virus serotype 2, and Madariaga virus arising from this collaborative study have been previously published.9–11 Herein, we report detection and isolation of MAYV from a woman with no history of recent travel outside of the city of Barquisimeto.
CASE DESCRIPTION
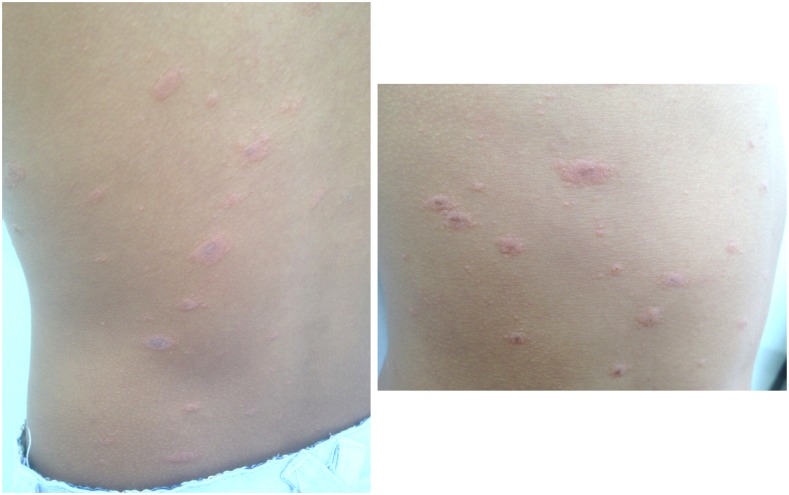
An 18-year-old woman presented as an outpatient for evaluation of fever, chills, rash, and small joint tenderness. Her temperature was 40°C. She also complained of malaise and fatigue. Her past medical history was unremarkable, and she denied any travel history, pet, or tick exposure. Three days prior, with the onset of fever, she had developed a pruritic maculopapular rash, which evolved into erythematous fine scaling oval-shaped macules and plaques distributed mainly in the trunk and proximal aspects of the limbs (Figure 2). She had no conjunctivitis, retro-ocular pain, or myalgias. On examination, the patient had a psoriasiform eruption reaction pattern with the presence of multiple small pink slightly scaly macules and plaques highly suggestive of pityriasis rosea, which resolved uneventfully after 5 days. She denied any associated chest pain, cough, or changes in her stool or urine, and any family or personal history of psoriasis. A skin biopsy was not performed.
Figure 2.
Patient rash. Generalized round to oval erythematous scaly macules and plaques ranging from 1.0 to 3.0 cm in diameter distributed mainly in the trunk and proximal aspects of the limbs. This figure appears in color at www.ajtmh.org.
Full blood count and chemistry test results were unremarkable except for mild leukopenia (3.34 × 103/µL) and lymphocytosis (58%), mildly elevated erythrocyte sedimentation rate (25 mm/hours [nL for female < 50 years: 0–12]), and transaminase elevation (aspartate aminotransferase: 119 mg/dL [nL < 40 mg/dL] and alanine aminotransferase: 56 mg/dL [nL < 40 mg/dL]). Antistreptolysin O titers returned within normal limits (180 IU/mL [nL < 200 IU/L]). Peripheral (thick and thin) blood smears were negative for hemoparasites. Serologic test results included cytomegalovirus IgG (+) and IgM (−), Epstein–Barr virus IgG (−) and IgM (−), DENV IgG (−) and IgM (−), and parvovirus (−). The serology test used to detect DENV antibodies was the OneStep IgG/IgM Ab Rapid Test SD (Standard Diagnostics, Inc., Kyonggi-do, Korea).
METHODS
Approximately 1.5 mL of patient plasma and urine were stored in liquid nitrogen within 1 hour of collection. One cryovial of each specimen was shipped on dry ice by express courier to the UF and stored at −80°C. Mammalian cell lines LLC-MK2 (CCL-7), MRC-5 (CCL-171), and Vero E6 (CRL-1586), obtained from the American Type Culture Collection (ATCC, Manassas, VA), were propagated as monolayers at 37°C and 5% CO2 using advanced Dulbecco’s essential medium (aDMEM) and 10% fetal bovine serum (FBS) as previously described.12
Subconfluent cells in 75-cm2 U-shaped canted neck cell culture flasks (with vented cap and tissue culture–treated surface [Corning®, Corning, NY]) were inoculated with 50 µL aliquots of the plasma. Twelve hours after inoculation, the cell growth medium was replenished by removing the spent medium and replacing it with reduced-serum aDMEM containing 3% FBS. Two days postinoculation, whereupon virus-induced cytopathic effects (CPEs) were observed, aliquots of spent medium from the inoculated cells were tested by reverse transcription polymerase chain reaction (RT-PCR) for the presence of alpha- and flavivirus genomic RNAs (vRNAs) as described previously.12,13
Reverse transcription polymerase chain reaction screens for ZIKV, dengue virus serotypes 1–4, MAYV, and other vRNAs.
Virus genomic RNAs (vRNAs) were extracted from virions in spent cell growth medium using the QIAamp Viral RNA Mini Kit (Qiagen Inc., Valencia, CA). SUPERase·In™ RNase inhibitor (Ambion™; Invitrogen, Carlsbad, CA) was then added to a final concentration of 0.25 U/µL. Reverse transcription polymerase chain reaction tests were then conducted on the vRNAs to determine the presence of common arthropod-borne alpha- and flaviviruses. For the detection of ZIKV and DENV1–4 vRNAs, a Superscript™ III Platinum™ One-step qRT-PCR kit was used with 3 µL of purified vRNAs. Primers and probe were ZIKV 1086, ZIKV 1162c and ZIKV 1107-FAM,14 and CDC DENV1–4 using DENV serotype–specific forward and reverse primers and probes, respectively.15 Samples were tested by real-time RT-PCR for ZIKV and for all four DENV serotypes in a CFX96 C1000 Real-Time System (BioRAD, Hercules, CA).
To determine whether the infectious agent was an alphavirus or a flavivirus, duplex RT-PCR was completed using primers and procedures modified from de Morais Bronzoni et al.16 First, cDNA was synthesized using Omniscript Reverse Transcriptase (Qiagen Inc.), 8 µL of purified vRNAs, and reverse primers (cM3W for alphavirus and FG2 for flavivirus). Amplification of cDNA was then completed using OneTaq DNA polymerase (New England Biolabs, Ipswich, MA), 1 µL of cDNA, cM3W (reverse) and M2W (forward) primers for alphavirus, and FG1 (forward) and FG2 (reverse) primers for flaviviruses. For detection of MAYV, nMAY (+)-specific forward primer was used to amplify 1 µL cDNA from the alphavirus RT-PCR product. All PCR amplicons in this assay were visualized on a 2% agarose gel with ethidium bromide.
Phylogenetic analyses.
Alignments for MAYV full genome sequences were obtained using multiple alignment fast Fourier transform (MAFFT).17 Because MAYV has exhibited recombination in the past,18 we checked for recombination using the algorithms implemented in the RDP4 software19 specifying for linear genome. The program DAMBE720 was used to check for nucleotide substitution saturation, whereas IQ-TREE21 was used to evaluate the presence of phylogenetic signal by likelihood mapping22 and to infer maximum likelihood phylogeny of the isolates based on the best-fit model chosen according to Bayesian Information Criterion.21,23 Statistical robustness for internal branching order in the phylogeny was assessed by UFBoot—Ultrafast Bootstrap (BB) Approximation (2,000 replicates) implemented in IQ-TREE—and strong statistical support along the branches was defined as BB > 90%.24
Correlation between root-to-tip genetic divergence and date of sampling was conducted in TempEst25 to assess clock signal in the dataset prior Bayesian phylogenetic analysis. Time-scaled phylogenetic trees were reconstructed using the Bayesian phylogenetic inference framework available in BEAST v.1.8.4.26 Markov chain Monte Carlo samplers were run until achieving for 100 million generations to ensure proper mixing of the Markov chain, assessed by calculating the effective sampling size of the parameter estimates. The Hasegawa, Kishino, and Yano substitution model was used with empirical base frequencies and gamma distribution of site-specific rate heterogeneity. The fit of the strict or relaxed uncorrelated molecular clock models and a constant size or Bayesian Skyline Plot27 demographic models were tested. Marginal likelihood estimates (MLE) for Bayesian model testing were obtained using path sampling and stepping-stone sampling methods.28 The strength of evidence against the null hypothesis (H0) was evaluated via MLE comparison with the more complex model (HA), referred to as the Bayes Factor (BF), wherein lnBF < 2 indicates no evidence against H0, 2–6—weak evidence, 6–10—strong evidence, and > 10 indicates very strong evidence. The best model was strict clock and constant demographic size. Maximum clade credibility tree was inferred from the posterior distribution of trees using TreeAnnotator specifying a burn-in of 10% and median node heights and edited graphically in FigTree v1.4.4.
RESULTS
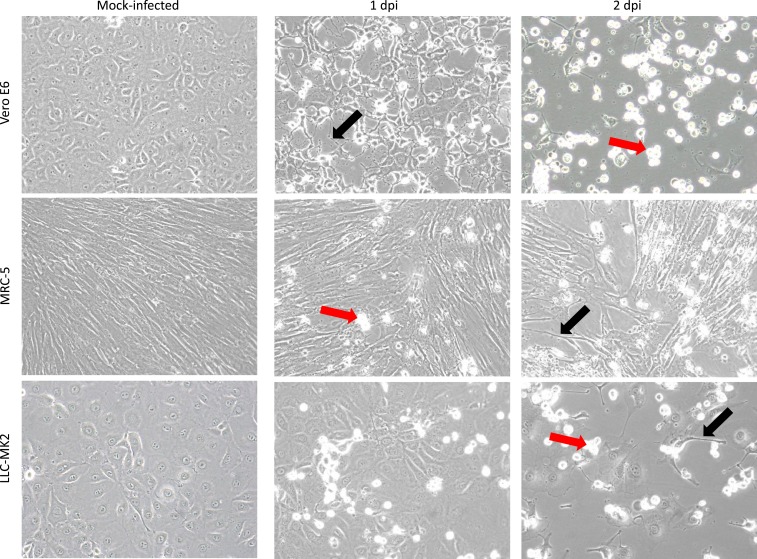
One day postinoculation, CPEs were observed in Vero E6, LLC-MK2, and MRC-5 cells that had been inoculated with plasma. Two days postinoculation, some dead cells had detached from the growth surface, resulting in gaps in the monolayer, and some of the remaining attached cells were undergoing apoptosis (Figure 3). Real-time RT-PCR tests of RNA purified from spent cell growth medium were negative for ZIKV and DENV1-4 vRNAs. The duplex RT-PCR test for alphavirus and flavivirus vRNAs yielded an ∼434-base pair PCR amplicon, suggesting that the sample was positive for alphavirus vRNA. Subsequent PCR of cDNA obtained from the duplex RT-PCR test further suggested that the virus present was MAYV (Supplemental Figure 1). The MAYV vRNAs were Sanger-sequenced using a gene walking approach with overlapping primers as previously described,12 and the virus sequence was deposited in GenBank (Accession #MK288026).
Figure 3.
Isolation of Mayaro virus in cell culture. Mock-infected Vero E6, MRC-5, and LLC-MK2 cells are shown 2 days postseed in the left panels. Cells inoculated with plasma from patient HI_112 are shown 1 dpi and 2 dpi in the middle and right panels, respectively. Refractile floating dead cells are evident (red arrows) and virus-induced cytopathic effects include apoptosis and appearance of cells with shrunken cytoplasm (black arrows). Original images at ×200 magnification. This figure appears in color at www.ajtmh.org.
Phylogenetic analyses.
We first assessed the presence of phylogenetic signal and absence of nucleotide substitution saturation (Supplemental Figure 2), as well as the absence of recombination based on full genome alignments. We then explored the phylogenetic relationship of the 2016 MAYV isolate from Venezuela with that of all other MAYV isolates available in GenBank, based on the nonrecombinant regions of the genome, as previously described,18 to be able to compare the genome of MAYV Venezuela with the recombinant genomes belonging to genotype L.18
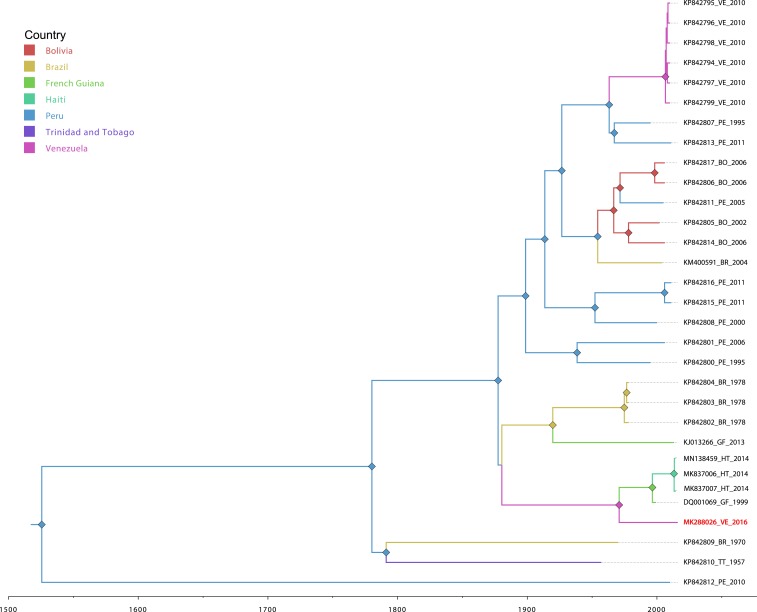
We performed a molecular clock analysis to date the divergence of the strain with previous isolates collected in Venezuela during the outbreak in 2010 (Figure 4). To get an accurate dating of the most recent common ancestor (MRCA) of the 2016 MAYV isolate, we included only genotype D full genome sequences in our analysis. The analysis showed that the MAYV isolate diverged about 100 years ago from the 2010 Venezuelan isolates, with the time of the MRCA (tMRCA) dating back to 1914 (with a 95% high posterior density (HPD) interval of 1883–1936). The tMRCA for the 2016 MAYV isolate and the most closely related MAYV isolates to this isolate (DQ001069, isolated in 1999 in French Guiana, and MK837006 and MK837007, isolated in 2014 in Haiti29) was more recent, dating back to 1980 (with a 95% HPD interval of 1971–1988). The tMRCA for subsequent divergence of the lineages leading to the French Guiana strain and the Haitian strains dated to 1996.
Figure 4.
Bayesian phylogeographic analysis of Mayaro virus genotype D strains. Time-scaled phylogenetic maximum clade credibility tree inferred using the constant demographic prior, strict molecular clock and asymmetric phylogeographic diffusion models, implemented in BEAST v1.8.4. Branches are colored according to geographical location, and diamonds represent branches supported by posterior probability > 0.9 and are colored based on the ancestral state reconstruction for the country of origin. This figure appears in color at www.ajtmh.org.
DISCUSSION
Infections with MAYV are likely under-recognized because other closely related arboviruses, such as chikungunya virus (CHIKV), DENV, and ZIKV, cause clinical illness with a significant overlap of signs and symptoms.30 Our findings indicate that MAYV was circulating in Barquisimeto during the epidemic of Zika disease in 2016. Although the complaint of arthralgias in our patient was consistent with an alphavirus infection (CHIKV and MAYV), a definitive diagnosis requiring virus-specific screening which, in this case, was performed by virus culture, with confirmatory PCR and sequencing. These findings highlight some of the ongoing problems in arbovirus diagnosis in endemic regions. When highly specific PCR-based tests are used, only the target-specific pathogens are detected (i.e., if you are not looking for MAYV, you will not find MAYV). If multiplex PCR methods are used, sensitivity decreases; when universal (virus group specific) tests are used, both specificity and sensitivity are affected. Taking all of the aforementioned disadvantages into consideration, at a research level, our laboratory attempts virus isolation in cell culture as an adjunct for PCR-based tests. We would hope that specific PCR assays for MAYV will become increasingly available for use in areas of potential endemicity; however, our work underscores the continuing value of traditional culture techniques.
At a clinical level, the case was also of interest because of the nature of the associated rash. Among a wide constellation of triggers and predisposing factors, there is increasing evidence that infection can play a role in initiating or exacerbating psoriasiform rashes.31 Epstein–Barr virus, varicella zoster virus, cytomegalovirus, human papillomavirus, herpes simplex virus, and more recently ZIKV have all been associated with the onset of psoriasis.32 Alphavirus-associated psoriasis has also been previously reported in patients infected with CHIKV.33,34 Although we lack biopsy confirmation of the diagnosis, our observations suggest that MAYV can trigger a psoriasiform dermatosis clinically mimicking pityriasis rosea, an acute, self-limiting papulosquamous skin disorder, which has been linked to endogenous systemic reactivation of human herpesvirus (HHV)-6 and/or HHV-7 and also to virus and vaccine-induced (influenza A [H1N1], hepatitis B–mediated) reactions.35,36
Although MAYV was present in Barquisimeto, we did not see evidence of a major MAYV epidemic: in culturing more than 50 samples from patients with acute undifferentiated febrile illness during this time period, we identified only one MAYV case. Given that the patient had no history of recent travel outside of Barquisimeto, findings would be consistent with low-level transmission and possible endemicity within the city. Mayaro virus has been associated with a variety of other reservoir hosts, including birds and reptiles, and urban-dwelling mosquitoes such as Aedes aegypti and Aedes albopictus appear to be competent vectors.37 This pattern contrasts with findings in a large MAYV outbreak in La Estación Village, Portuguesa State, Venezuela, in 2010, in which 77 potential cases were identified, 19 of which were laboratory confirmed. Genome sequence data were available for MAYV isolates from six patients: all were within a single clade, which was most closely related to 1995 and 2011 MAYV strains from Peru7 (Figure 4). The village is located in a former tropical forest that had recently been converted for coffee production and other farming; several monkey species and competent MAYV vectors (i.e., Haemagogus mosquitoes) were described as being present within its immediate vicinity.7 The apparent high attack rate is suggestive of virus introduction into a MAYV-naive population, linked with enzootic circulation in a recently cleared forest area, and contrasting with what may be endemic, urban transmission within Barquisimeto.
Geographically, Barquisimeto and La Estación village are approximately 80 miles apart by road, albeit separated by the Terepaima mountain range (Figure 1). However, in contrast to the La Estación village isolate, the Barquisimeto isolate clustered most closely with isolates from French Guiana and, in turn, Haiti, with molecular clock analysis suggesting that the La Estación village and Barquisimeto isolates diverged genetically some 100 years ago. Given the relatively limited number of full genome sequences available for MAYV (Figure 4), it is difficult to fully reconstruct the pattern of movement of the virus within the Amazon Basin and other regions of northern South America. It is possible that the La Estación strain was a recent introduction into Venezuela; alternatively, the two strains could have evolved separately, albeit in close proximity, for 100 years In this context, the rate of evolution of MAYV appears to be relatively slow, given the 17-year gap (2016 and 1999) between the very similar Barquisimeto and French Guiana strains; similar observations have been made by other investigators.7,18 Taken together, our data underscore the range, potential for movement, and relative antiquity of the virus in the region and highlight its potential for ongoing spread through the Americas.
Supplemental figures
Acknowledgment:
We thank Sadie Ryan for providing the map included as Figure 1.
Note: Supplemental figures appear at www.ajtmh.org.
REFERENCES
- 1.Anderson CR, Downs WG, Wattley GH, Ahin NW, Reese AA, 1957. Mayaro virus: a new human disease agent. II. Isolation from blood of patients in Trinidad, B.W.I. Am J Trop Med Hyg 6: 1012–1016. [DOI] [PubMed] [Google Scholar]
- 2.Pinheiro F, LeDuc J, 1998. Mayaro virus disease. Monath TP, ed. The Arboviruses: Epidemiology and Ecology. Boca Raton, FL: CRC Press, 137–150. [Google Scholar]
- 3.Tesh RB, Watts DM, Russell KL, Damodaran C, Calampa C, Cabezas C, 1999. Mayaro virus disease: an emerging mosquito-borne zoonosis in tropical South America. Clin Infect Dis 28: 67–73. [DOI] [PubMed] [Google Scholar]
- 4.Halsey ES, Siles C, Guevara C, Vilcarromero S, Jhonston EJ, Ramal C, 2013. Mayaro virus infection, Amazon basin region, Peru, 2010–2013. Emerg Infect Dis 19: 1839–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor SF, Patel PR, Herold TJ, 2005. Recurrent arthralgias in a patient with previous Mayaro fever infection. South Med J 98: 484–485. [DOI] [PubMed] [Google Scholar]
- 6.Acosta-Ampudia Y, Monsalve DM, Rodríguez Y, Pacheco Y, Anaya JM, Ramírez-Santana C, 2018. Mayaro: an emerging viral threat? Emerg Microbes Infect 7: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auguste AJ, et al. 2015. Evolutionary and ecological characterization of Mayaro virus strains isolated during an outbreak, Venezuela, 2010. Emerg Infect Dis 21: 1742–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñoz MJ, Navarro JC, 2012. Mayaro: a re-emerging arbovirus in Venezuela and Latin America [article in Spanish]. Biomedica 32: 286–302. [DOI] [PubMed] [Google Scholar]
- 9.Blohm GM, et al. 2018. Evidence for mother-to-child transmission of Zika virus through breast milk. Clin Infect Dis 66: 1120–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blohm GM, Paniz-Mondolfi AE, Márquez-Colmenarez MC, Loeb JC, Pacheco CA, Pulliam JRC, Morris JG, Jr., 2018. Complete genome sequence of dengue virus serotype 2, Asian/American genotype, isolated from the urine of a Venezuelan child with hemorrhagic fever in 2016. Genome Announc 6: e00529– 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blohm GM, Lednicky JA, White SK, Mavian CN, Márquez-Colmenarez MC, González-García KP, Salemi M, Morris JG, Jr., Paniz-Mondolfi AE, 2018. Madariaga virus: identification of a lineage III strain in a Venezuelan child with acute undifferentiated febrile illness, in the setting of a possible equine epizootic. Clin Infect Dis 67: 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lednicky JA, et al. 2016. Zika virus outbreak in Haiti in 2014: molecular and clinical data. PLoS Negl Trop Dis 10: e0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lednicky JA, et al. 2016. Mayaro virus in a child with acute febrile illness, Haiti, 2015. Emerg Infect Dis 22: 2000–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR, 2007. Genetic and serologic properties of Zika virus associated with an epidemic, Yap state, Micronesia. Emerg Infect Dis 14: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, Medina F, Colón C, Margolis H, Muñoz-Jordán JL, 2013. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 7: e2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Morais Bronzoni RV, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Moraes Figueiredo LT, 2005. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol 43: 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K, Standley DM, 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32: 1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavian C, Rife BD, Dollar JJ, Cella E, Ciccozzi M, Prosperi MC, Lednicky JA, Morris JG, Jr., Capua I, Salemi M, 2017. Emergence of recombinant Mayaro virus strains from the Amazon basin. Sci Rep 7: 8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia X, 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol 35: 1550–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ, 2015. IQ-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt HA, Strimmer K, Vingron M, von Haeseler A, 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504. [DOI] [PubMed] [Google Scholar]
- 23.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ, 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44: W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minh BQ, Thi Nguyen MA, von Haeseler A, 2013. Ultrafast approximation for phylogenetic Bootstrap. Mol Biol Evol 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rambaut A, Lam TT, Carvalho LM, Pybus OG, 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Patho-O-Gen). Virus Evol 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Suchard MA, Xie D, Rambaut A, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall MD, Woolhouse ME, Rambaut A, 2016. The effects of sampling strategy on the quality of reconstruction of viral population dynamics using Bayesian skyline family coalescent methods: a simulation study. Virus Evol 2: vew003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV, 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blohm G, et al. 2019. Mayaro as a Caribbean traveler: evidence for multiple introductions and transmisión of the virus in Haiti. Int J Infect Dis. Available at: [DOI]
- 30.Paniz-Mondolfi AE, Rodríguez-Morales AJ, Blohm GM, Márquez-Colmenarez MC, Villamil-Gomez WE, 2016. ChikDenMaZika syndrome: the challenge of diagnosing arboviral infections in the midst of concurrent epidemics. Ann Clin Microbiol Antimicrob 15: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry L, Baker BS, 2007. Triggering psoriasis: the role of infections and medications. Clin Dermatol 25: 606–615. [DOI] [PubMed] [Google Scholar]
- 32.Paniz-Mondolfi AE, Hernandez Perez M, Blohm GM, Márquez-Colmenarez MC, Mogollon Mendoza A, Hernandez-Pereira CE, Escalona MA, Lodeiro Colatosti A, Rothe DeArocha J, Rodriguez Morales AJ, 2017. Generalized pustular psoriasis triggered by Zika virus infection. Clin Exp Dermatol 43: 171–174. [DOI] [PubMed] [Google Scholar]
- 33.Seetharam KA, Sridevi K, 2011. Chikungunya infection: a new trigger for psoriasis. J Dermatol 38: 1033–1034. [DOI] [PubMed] [Google Scholar]
- 34.Kumar R, Sharma MK, Jain SK, Yadav SK, Singhal AK, 2017. Cutaneous manifestations of chikungunya fever: observations from an outbreak at a tertiary care hospital in southeast Rajasthan, India. Indian Dermatol Online J 8: 336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li A, Li P, Li Y, Li W, 2018. Recurrent pityriasis rosea: a case report. Hum Vaccin Immunother 14: 1024–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drago F, Ciccarese G, Rebora A, Broccolo F, Parodi A, 2016. Pityriasis rosea: a comprehensive classification. Dermatology 232: 431–437. [DOI] [PubMed] [Google Scholar]
- 37.Wiggins K, Eastmond B, Alto BW, 2018. Transmission potential of Mayaro virus in Florida Aedes aegypti and Aedes albopictus mosquitoes. Med Vet Entomol 32: 436–442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.