Abstract
Background
Recent studies revealed that long non-coding RNAs (lncRNA) play crucial roles in cancer initiation and progression. However, the function and underlying mechanism of lncRNAs in triple-negative breast cancer (TNBC) are little investigated.
Methods
qRT-PCR was used to investigate LINC00096 expression in TNBC tissues and cells. Function assays were used to test the effects of LINC00096 on TNBC cells progression. In addition, luciferase reporter and qRT-PCR assays were used to determine the underlying mechanism of LINC00096 on TNBC progression.
Results
In our present study, we identify LINC00096 as one of the most upregulated lncRNA in TNBC progression by using microarray screening. High LINC00096 expression was obviously related to advanced tumor stage, metastasis, poor prognosis of patients. Loss-of-function assays showed that LINC00096 suppression reduced TNBC cells proliferation and invasive abilities in vitro. Mechanistically, we demonstrated that LINC00096 directly interacted with miR-383-5p, subsequently acted as a miRNA sponge to increase RBM3 expression.
Conclusion
In the present study, we indicated that LINC00096 might promote the proliferation and invasion through regulating the miR-383-5p/RBM3 pathway in TNBC, which providing a novel therapeutic target for cancer treatment.
Keywords: LINC00096, miR-383-5p, RBM3, triple-negative breast cancer
Introduction
Breast cancer (BC) is the most pervasive carcinoma and one of the primary causes of mortality in women worldwide.1,2 Triple-negative breast cancer (TNBC) is characterized by a lack of ER, PR, and Her2, and is the most aggressive subtype of all BC cases.3 Although the treatments of BC have developed considerably in recent years, the recurrence and 5-year survival rate are still unsatisfactory, especially in TNCB.4,5 Thus, it is necessary to explore the underlying molecular mechanism of TNBC progression.
Long non-coding RNAs (lncRNAs) are a family of non-coding RNA (ncRNA) which more than 200 nucleotides (nt) in length without translation (protein-coding) ability.6 Recently, increasing evidence revealed that lncRNAs could play critical functions in numbers of biological processes via post-transcriptional, transcriptional, and even by epigenetic alterations, especially in cancer progression.7 For example, Cao et al showed that GAS5 expression was significantly decreased and associated with advanced clinical features and poor overall survival in patients with cervical cancer.8 Li et al found that lncRNA CASC2 reduced gastric cancer cells proliferation via regulating MAPK signaling pathway.9 In TNBC studies, Li et al found that lncRNA GAS5 was downregulated in TNBC and overexpression of GAS5 could promote the chemosensitivity and apoptosis.10 Collina et al found that lncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of TNBC.11 Although a large number of lncRNAs have been explored in tumor progression, the underlying mechanisms of lncRNAs in TNBC progression are little investigated.
Emerging evidence showed that lncRNAs might serve as a competitive endogenous RNAs (ceRNAs) to sponge miRNAs and then reduced their functions.12 For example, Ma et al showed that lncRNA CCAT1 promoted gallbladder cancer development via negative modulation of miR-218-5p.13 Chen et al found that lncRNA TTN-AS1 promoted cervical cancer cell growth and metastasis through miR-573/E2F3 axis.14 In the current study, we identified a new differentially-expressed lncRNA (LINC00096), which was upregulated in TNBC and promoted the proliferation and invasion in vitro. Mechanistically, we demonstrated that LINC00096 might accelerate TNBC progression by regulating the miR-383-5p/RBM3 axis.
Materials and Methods
Patient Samples
A total of 90 paired TNBC tissues and adjacent non-tumor tissues were collected from patients who underwent surgery in Huaihe Hospital of Henan University. No patients had received radiotherapy or chemotherapy before surgery. All tissue samples were frozen in liquid nitrogen and stored at −80°C for subsequent experiments. All patients read and signed the informed consent forms and this study was approved by the Research Ethic Committee of Huaihe Hospital of Henan University
Microarray Analysis
Total RNA was extracted from 2 paired TNBC tissues. Tissue samples preparation and microarray hybridization were performed according to the instructions. LncRNA microarray assay (Agilent-045142) was performed by Biotechnology Corporation (Shanghai, China). The analysis was performed as described previously.15 The threshold cutoff was a fold change ≥2.0 and a P value ≤0.05 for downregulated and upregulated lncRNAs.16
Cell Culture and Transfection
Human TNBC cells (BT-549, MDA-MB-231, MDA-MB-134, MDA-MB-157 and CAL-51) and human breast epithelial cells (MCF-10A) were purchased from cell bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin under a humidified environment with 5% CO2 at 37°C.
Lentiviral vector suppressing LINC00096 (sh-LINC00096), miR-383-5p mimics and RBM3 overexpression vectors were constructed by Sangon (Shanghai, China). The transfection was conducted via Lipofectamine 2000 reagents according to the manufacturer’s instructions
Cell Proliferation Assay
The CCK-8 assay (Beyotime, China) was used for evaluation of cell viability. Briefly, transfected cells were cultured in 96-well plates (2×103cells/well). Subsequent to incubation at the indicated times, each well was added with 10 μL. CCK-8 reagents for 2 h culture at 37°C. The absorbance at a wavelength of 490 nm was analyzed using Microplate Reader (Bio-Rad, CA, USA).
Colony Formation Assay
For evaluation of colony formation, 1000 cells per well were seeded into 6-well plates, followed by culturing for 7 days. Then colonies were washed with PBS, fixed with 4% paraformaldehyde for 10 min, and stained with 0.1% crystal violet for 10 min at room temperature.
Transwell Assay
Cell invasion ability was analyzed using the Matrigel pre-coated chamber (Corning, NY, USA). 2 × 104 cells were cultured to the upper chamber in RPMI-1640 without FBS, and RPMI-1640 containing 10% FBS was added to the lower chamber. 48 h after incubation, the non-invasive cells were removed, while the invasive cells were fixed with paraformaldehyde, stained with crystal violet, and photographed under a microscope (Nikon, Japan).
Quantitative Real-Time PCR
Total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instruction. QRT-PCR was performed with SYBR green PCR kit (TaKaRa, China) on the Applied Biosystems StepOne Real-Time PCR Systems. All signals from LINC00096 and RBM3 were conducted in triplicate and normalized to GAPDH. MiR-383-5p qRT-PCR kit was purchased from GenePharma and normalized to U6 snRNA. The relative mRNA expression was calculated using 2−ΔΔCt.
Western Blot
Total proteins from cells were extracted using RIPA lysis buffer (Abcam, USA) and protein concentrations were measured with BCA protein assay. Proteins were denatured and electrophoresis was performed using 12% SDS-PAGE. Proteins were transferred to PVDF membranes, followed by blocking in 5% non-fat milk. After that, the membranes were incubated with primary antibodies overnight at 4 °C. Then the membranes were incubated with specific HRP-conjugated secondary at room temperature for 1 h. Protein bands were visualized with an enhanced chemiluminescence (ECL, Sigma, USA).
Dual-Luciferase Reporter Assay
The LINC00096 cDNA sequence contains the binding site for miR-383-5p was inserted into pmiR-GLO vector to obtain a wild-type LINC00096 (LINC00096-Wt). Mutant LINC00096 (LINC00096-Mut) was constructed using site-direct mutagenesis kit (Takara). To investigate the association between miR-383-5p and RBM3, 3ʹ-UTR of RBM3 contains miR-383-5p binding site was cloned into pmiR-GLO. Cells were transfected with LINC00096-Wt or RBM3-Wt and miR-383-5p mimics using Lipofectamine 2000. After 48h of transfection, luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocols.
Immunohistochemistry Staining
The experimental methods are based on the previous study.17
Statistical Analysis
All analyses were performed with SPSS 19.0. Data were depicted as mean ± standard deviation (SD). Student’s t-test was used for comparison between two groups and one-way ANOVA for more than two groups. p< 0.05 was considered to be statistically significant.
Results
Expression Profiles of lncRNAs in TNBC
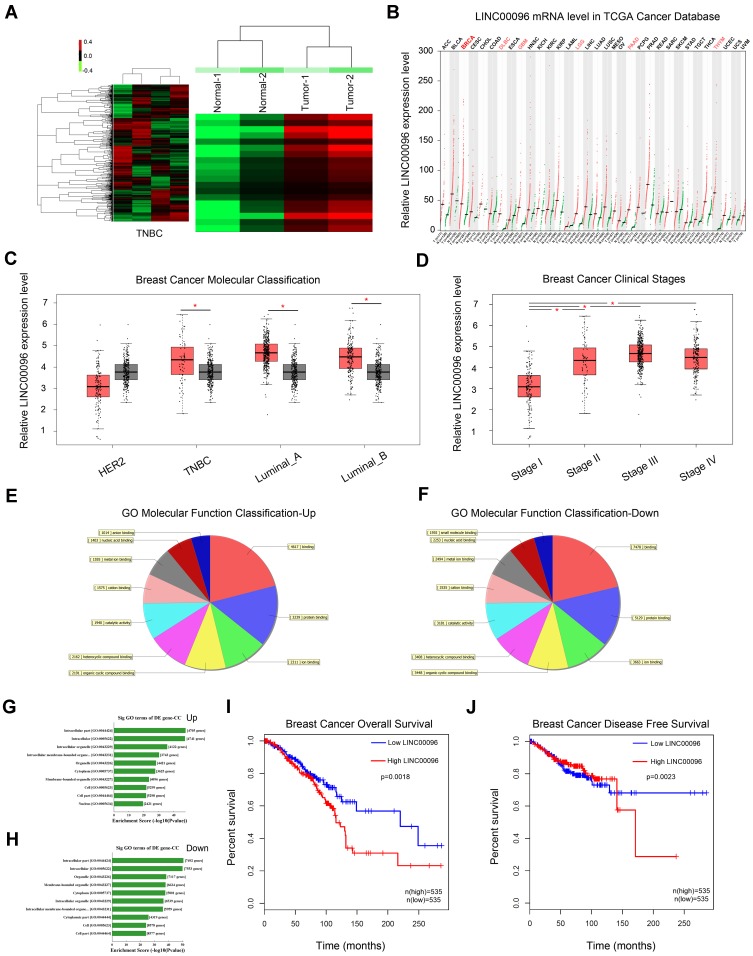
LncRNA microarray analysis was used to identify the unique lncRNAs involved in TNBC progression. Based on fold change ≥2.0 and P<0.05, 631 upregulated lncRNAs and 355 downregulated lncRNAs were identified (Figure 1A). We showed the top 20 significantly dysregulated lncRNA transcripts in Table 1, and LINC00096 (the mostly upregulated lncRNAs) was selected for further study. Next, LINC00096 expression was explored in TCGA database. Results showed that the expression of LINC00096 was obviously upregulated in tumors, including breast cancer (Figure 1B). LINC00096 expression was further confirmed in its subtype and clinical stage (Figure1C and D). As shown in Figure 1E–H, molecular function and cellular components using gene ontology (GO) analysis based on abundant lncRNAs found in TNBC tissues were subsequently performed. Subsequently, we found that high LINC00096 expression was associated with poor overall survival and disease-free survival rate in patients (Figure 1I and J). Thus, these data indicated that LINC00096 was involved in TNBC progression.
Figure 1.
Screening and expression of LINC01124 in TNBC tissues. (A) Aberrantly expression of lncRNAs was evaluated in 2 pairs of TNBC tissues though lncRNA expression microarray. (B) LINC00096 expression in TCGA Cancer database. (C and D) LINC00096 expression was expression in its subtype (TNBC, Luminal_A, Luminal_B) and clinical stage of TNBC patients. (E–H) The top 10 upregulated and downregulated GO functions of the molecular function domains (E and F) and cellular component (G and H). (I and J) High LINC00096 expression was associated with poor prognosis. *P<0.05.
Table 1.
Significantly Aberrantly Expressed IncRNAs Determined by Microarray (Up-Regulation/Down-regulation Top 20)
| Upregulated | Downregulated | ||||
|---|---|---|---|---|---|
| lncRNAs | Log2 Fold Change(T/N)* | P-Values | lncRNAs | Log2 Fold Change(T/N) | P-Values |
| H_RG012D21.9 | 7.412 | 0.000353215 | ENST00000448318 | −8.213 | 0.006014172 |
| ENST00000418938 | 7.257 | 0.009300485 | AI621345 | −7.562 | 0.028098678 |
| BC070168 | 7.081 | 0.048587215 | AI347010 | −7.341 | 0.045254085 |
| ENST00000453128 | 6.621 | 0.047495328 | NR_001543 | −7.126 | 0.042348313 |
| CR587433 | 6.237 | 0.014162886 | ENST00000512505 | –6.314 | 0.011188389 |
| ENST00000442607 | 6.102 | 0.012060102 | ENST00000458329 | −6.218 | 0.018865758 |
| CV340376 | 5.943 | 0.016635862 | NR_026805 | −6.013 | 0.041336894 |
| ENST00000499483 | 5.361 | 0.007502416 | uc010zaz.1 | −5.743 | 0.041422996 |
| ENST00000514936 | 5.137 | 0.044007764 | BG897081 | −5.312 | 0.036204094 |
| BY797754 | 5.036 | 0.033521155 | ENST00000377547 | −5.085 | 0.039384513 |
| ENST00000445923 | 4.622 | 0.042554580 | ENST00000513362 | −4.781 | 0.044832756 |
| ENST00000436848 | 4.504 | 0.027469783 | ENST00000395181 | −4.427 | 0.011413775 |
| BC140261 | 4.499 | 0.040541542 | AK126669 | −3.879 | 0.046313446 |
| ENST00469321 | 3.342 | 0.000757127 | BX485243 | −3.660 | 0.017514558 |
| NR_029455 | 3.262 | 0.024922906 | ENST00000430364 | −3.398 | 0.016387004 |
| CD656032 | 2.780 | 0.024085074 | DB264073 | −3.246 | 0.047435468 |
| NR_033658 | 2.341 | 0.040642026 | NR_026052 | −2.947 | 0.009538409 |
| ENST00000340510 | 1.734 | 0.018036641 | BC013423 | −2.453 | 0.008560159 |
| ENST00000426884 | 1.589 | 0.028023339 | AK123278 | −1.652 | 0.004500777 |
| AK097341 | 1.263 | 0.007748291 | ENST00000500040 | −1.341 | 0.044767683 |
Notes: *T/N: breast tumor samples relative to corresponding non-tumor samples. H_RG012D21.9: NCRNA00096, LINC00096.
LINC00096 Expression Is Upregulated in TNBC
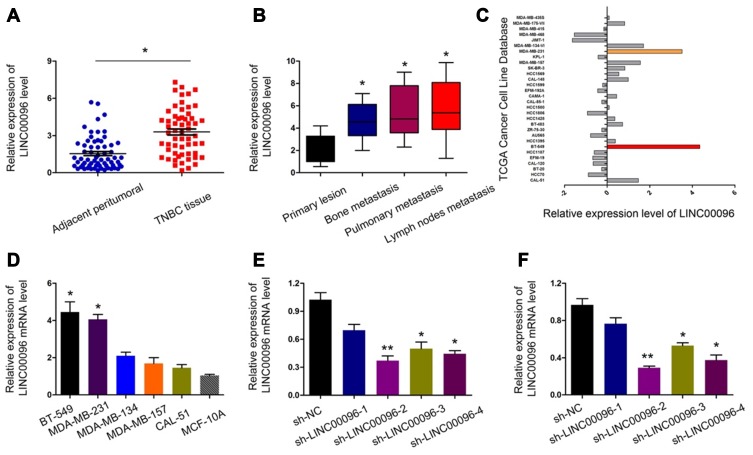
Next, we determined LINC00096 expression in TNBC tissues and cell lines. qRT-PCR showed that the expression of LINC00096 was significantly increased in TNBC tissues and associated with tumor metastasis (Figure 2A and B). In addition, correlation analysis showed that high LINC00096 expression was associated with advanced tumor stage, histological differentiation, metastasis and Ki-67 levels (Table 2). Subsequently, qRT-PCR showed that LINC00096 expression was significantly increased in TNBC cells (BT-549, MDA-MB-231, MDA-MB-134, MDA-MB-157 and CAL-51) compared to MCF-10A cells (Figure 2D). And the data were further confirmed by TCGA Cancer Cell Line database (Figure 2C). These results showed that LINC0009 might play critical roles in TNBC progression.
Figure 2.
LINC00096 was upregulated in TNBC. (A) qRT-PCR showed that LINC00096 expression was significantly increased in TNBC tissues. (B) High LINC00096 expression was positively associated with metastasis. (C) LINC00096 expression in TCGA Cancer Cell Line database. (D) qRT-PCR showed that LINC00096 expression was significantly increased in TNBC cells compared to MCF-10A cells. (E and F) Knockdown of LINC00096 by shRNA in BT-549 and MDA-MB-231 cells. *P<0.05, **P<0.01.
Table 2.
Association of LINC00096 Expression with Clinicopathological Parameters of Breast Cancer Patients
| Characteristic | No. | LINC00096 Relative Expression Level | χ2 | P Value | |
|---|---|---|---|---|---|
| High (%) | Low (%) | ||||
| Age (y) | 0.216 | 0.739 | |||
| ≤50 | 48 (53.3) | 26 (28.9) | 22 (24.4) | ||
| >50 | 42 (46.7) | 24 (26.7) | 18 (20.0) | ||
| Tumor stage | 4.768 | 0.021* | |||
| I-II stage | 39 (43.3) | 14 (15.6) | 25 (27.8) | ||
| III-IV stage | 51 (56.7) | 38 (42.2) | 13 (14.4) | ||
| Lymphatic metastasis | 8.963 | 0.001* | |||
| Positive | 34 (37.8) | 26 (28.9) | 8 (8.9) | ||
| Negative | 56 (62.2) | 25 (27.8) | 31 (34.4) | ||
| Tumor size | 0.431 | 0.073 | |||
| ≤5cm | 43 (47.8) | 23 (25.6) | 20 (22.2) | ||
| >5cm | 47 (52.2) | 29 (32.2) | 18 (20.0) | ||
| Distant metastasis | 2.543 | 0.034* | |||
| M0 | 69 (76.7) | 32 (35.6) | 37 (41.1) | ||
| M1 | 21 (23.3) | 17 (18.9) | 4 (4.4) | ||
| Histological differentiation | 1.076 | 0.026* | |||
| Low-grade | 23 (25.6) | 12 (13.3) | 11 (12.2) | ||
| Middle-grade | 35 (38.9) | 25 (27.8) | 10 (11.1) | ||
| High-grade | 32 (35.5) | 21 (23.3) | 11 (12.2) | ||
| Ki-67 level | 2.963 | 0.047* | |||
| ≤10 | 57 (63.3) | 25 (27.8) | 32 (35.6) | ||
| >10 | 33 (36.7) | 26 (28.9) | 7 (7.8) | ||
Note: *P<0.05.
LINC00096 Inhibition Reduces TNBC Cells Proliferation and Invasion
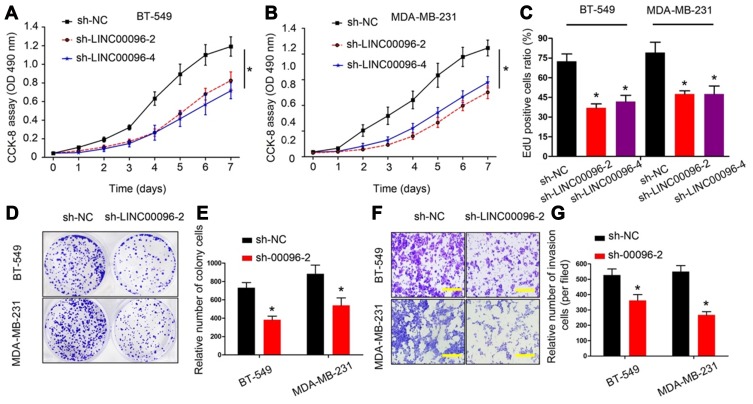
To further identify the roles of LINC0009 in TNBC, we firstly transfected sh-LINC00096#1/2/3/4 into BT-549 and MDA-MB-231 cells, and the interference efficiency was determined by qRT-PCR (Figure 2E and F). CCK-8 and EdU assays revealed that BT-549 and MDA-MB-231 cells transfected with sh-LINC0009 showed a poor cell viability compared with sh-NC group (Figure 3A–C). Colony formation assay suggested that LINC0009 inhibition reduced the colony numbers of BT-549 and MDA-MB-231 cells compared to sh-NC group (Figure 3D and E). In addition, transwell assay showed that LINC0009 inhibition significantly reduced the number of invaded BT-549 and MDA-MB-231 cells compared to sh-NC group (Figure 3F and G).
Figure 3.
LINC00096 inhibition reduced TNBC cells proliferation and invasion. (A–C) The effects of LINC00096 on TNBC cells viabilities were determined by CCK-8 and EdU assays. (D and E) Colony formation assay showed that LINC00096 inhibition reduced the colony numbers of BT-549 and MDA-MB-231 cells. (F and G) Transwell assay showed that LINC00096 inhibition reduced BT-549 and MDA-MB-231 cells invasion ability in vitro. *P<0.05.
LINC00096 Direct Binding to miR-383-5p
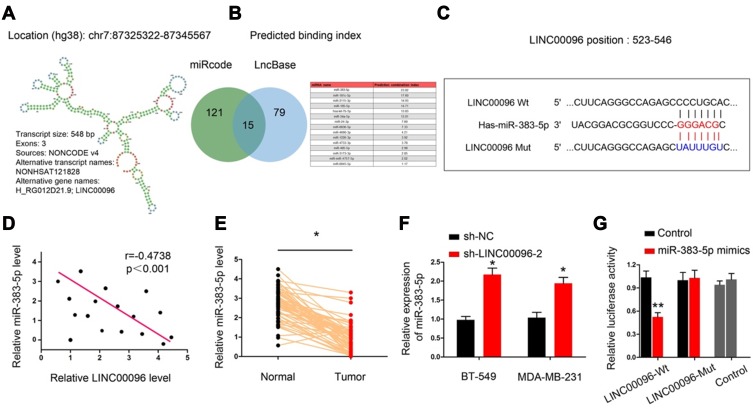
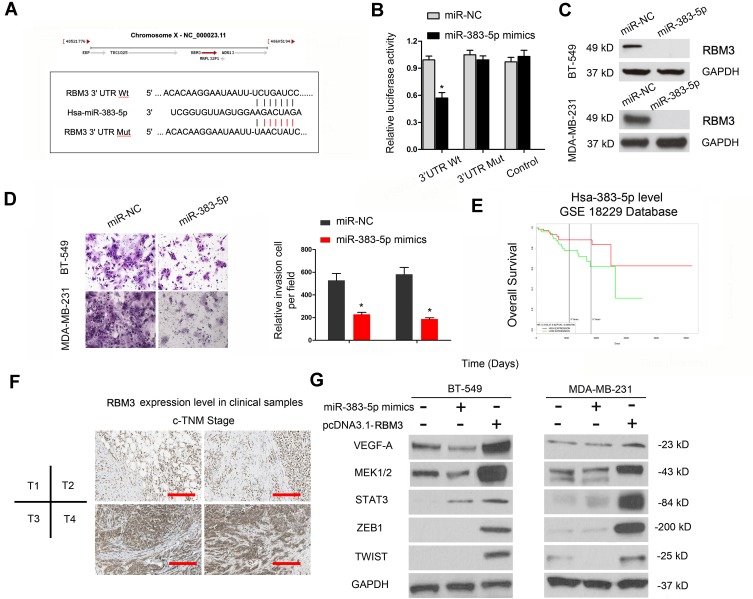
Previous studies showed lncRNA could serve as miRNA “sponge” to affect the activity of miRNA.18 Thus, we determined the regulatory roles of LINC00096 at the post-transcriptional level. Results showed that miR-383-5p ranked top among potential targets (miRcode, and LncBase), the secondary structure of LINC00096 and possible binding sites were shown in Figure 4A–C. QRT-PCR analysis showed that miR-383-5p expression was significantly decreased and negatively correlated with LINC00096 expression in TNBC tissues (Figure 4D and E). sh-LINC00096 significantly increased miR-383-5p expression in BT-549 and MDA-MB-231 cells (Figure 4F). Luciferase reporter assay revealed that miR-383-5p mimics reduced the luciferase activity of LINC00096-Wt vectors (Figure 4G). These data indicated that LINC00096 might act as a sponge for miR-383-5p in TNBC progression.
Figure 4.
LINC00096 directly interacted with miR-383-5p in TNBC. (A) Chromosome location information and secondary structure of LINC00096. (B and C) The potential binding site between LINC00096 and miR-383-5p. (D) miR-383-5p expression was negatively correlated with LINC00096 expression in TNBC tissues. (E) miR-383-5p expression was significantly decreased in TNBC tissues. (F) sh-LINC00096 significantly increased miR-383-5p expression in BT-549 and MDA-MB-231 cells. (G) miR-383-5p mimics decreased luciferase activity of LINC00096-Wt group. *P<0.05, **P<0.01.
LINC00096 Regulated RBM3 by Absorbing miR-383-5p
Based on the above data, we further explored the entire regulation mechanism underlying LINC00096. By bioinformatics exploration, we sought out the interplay between miR-383-5p and RBM3 (Figure 5A). Luciferase reporter assay showed that miR-383-5p mimics reduced the luciferase activity of RBM3-Wt group (Figure 5B). Western blot revealed that miR-383-5p mimics reduced RBM3 expression in BT-549 and MDA-MB-231 cells (Figure 5C).
Figure 5.
LINC00096 regulated RBM3 by absorbing miR-383-5p. (A) The potential binding site between miR-383-5p and RBM3. (B) miR-383-5p mimics decreased luciferase activity of RBM3-Wt group. (C) miR-383-5p mimics reduced RBM3 protein expression in BT-549 and MDA-MB-231 cells. (D) miR-383-5p mimics reduced TNBC cells invasion ability. (E) TCGA database revealed that low miR-383-5p expression was correlated with poor overall survival in TNBC patients. (F) RBM3 protein expression in TNBC tissues was detected by IHC. (G) The effects of miR-383-5p mimics or RBM3 overexpression on EMT progression. *P<0.05.
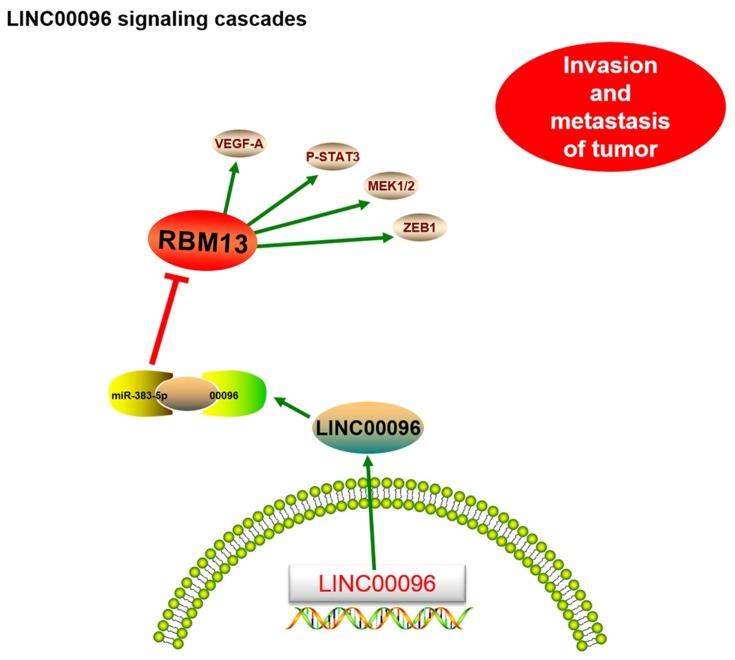
Next, we explored miR-383-5p and RBM3 roles in TNBC cells. Transwell assay showed that miR-383-5p mimics significantly reduced BT-549 and MDA-MB-231 cells invasion ability in vitro (Figure 5D). Low miR-383-5p expression was correlated with poor overall survival of TNBC patients (Figure 5E). In addition, IHC showed that RBM3 expression was significantly increased in TNBC patients and positively correlated with TNM stage (Figure 5F). Subsequently, we detected the effects of overexpression of miR-383-5p or RBM3 on epithelial-mesenchymal transition (EMT; VEGF, MEK1/2, STAT3, ZEB1, and TWIST) progression.19,20 Western blot assay indicated miR-383-5p mimics significantly reduced EMT processes, while RBM3 overexpression promoted EMT processes (Figure 5G). Thus, the present study suggested that LINC00096 might regulat EMT processes via regulating the miR-383-5p/RBM3 axis in TNBC (Figure 6).
Figure 6.
Schematic diagram of LINC00096 in TNBC progression.
Discussion
TNBC is a group of hormone-deficient breast cancer associated with very aggressive tumor biology and a poor prognosis.3 Recently, lots of studies showed that lncRNA could play critical roles in breast cancer progression. For instance, Shi et al revealed that lncRNA-ATB could promote trastuzumab resistance and invasion-metastasis cascade in breast cancer.17 Zuo et al showed that lncRNA MALAT1 promoted TNBC cells proliferation and invasion by targeting miR-129-5p.21 Wang et al suggested that lncRNA HCP5 promoted TNBC progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p.22
In our study, we identified a novel lncRNA LINC00096 via lncRNA microarray. Subsequently, we showed that the expression of LINC00096 was significantly upregulated and correlated with advanced clinical features of TNBC patients. Kaplan-Meier analysis suggested that high LINC00096 expression was associated with poor prognosis in TNBC patients. Moreover, function assays showed that LINC00096 suppression reduced TNBC cells proliferation, colony formation, and invasion abilities in vitro. Thus, we suggested that LINC00096 might serve as an oncogenic lncRNA in TNBC tumorigenesis.
MicroRNA is a class of small ncRNAs which acts as either tumor suppressors or oncogenes and implicate a variety of cellular processes, such as cell proliferation, invasion, apoptosis, and differentiation.23 miR-383-5p play important roles in tumorigenesis. For example, Jiang et al found that miR-383-5p reduced ovarian cancer cells proliferation and enhanced chemo-sensitivity via regulating TRIM27 expression.24 Wang et al showed that AKR1B10 modulated regulated by miR-383-5p, promoted HCC progression.25 However, the roles of miR-383-5p in TNBC remain unclear. In our study, miR-383-5p was predicted as a target of LINC00096. MiR-383-5p expression was decreased and negatively correlated poor overall survival of TNBC patients. Overexpression of miR-383-5p reduced EMT progression. In addition, we confirmed that LINC00096 could directly bind to miR-383-5p in TNBC cells. Overall, we showed that LINC00096 could act as a sponge for miR-383-5p in TNBC progression.
RNA-binding motif protein 3 (RBM3) is an RNA and DNA-binding protein that in response to various types of cellular functions, such as cell cycle regulation, invasion.26 Recent studies showed that RBM3 could play critical roles in tumor progression. For example, Jonsson et al showed that high RBM3 expression was associated with biochemical recurrence and disease progression in patients with melanoma cancer.27 Venugopal et al found that RBM3 increased β‐catenin signaling to increase stem cell characteristics in colorectal cancer cells.28 Moreover, Chen et al showed that RBM3 acted as an oncogene upregulated ARPC2 and contributed to breast cancer progression.29 In our study, bioinformatic prediction and experimental verification showed that RBM3 could act as a direct target of miR-383-5p in TNBC progression. IHC revealed that RBM3 expression was significantly increased and positively correlated with TNM stage of TNBC patients. In addition, Western blot showed that RBM3 overexpression significantly increased EMT related genes expression in TNBC.
In conclusion, we identified a new lncRNA LINC00096, which was upregulated in TNBC and promoted the proliferation and invasion in vitro. Mechanistically, the LINC00096/miR-383-5p/RBM3 axis might play an important role in TNBC progression, which providing a novel therapeutic target for cancer treatment.
Acknowledgments
We appreciate each authors assistance with the experiments and constructive comments on the manuscript.
Ethics Approval and Consent to Participate
Ethical approval was given by the Ethics Committee of Huaihe Hospital of Henan University.
Data Sharing Statement
The dataset supporting the conclusions of this article is included within the article.
Disclosure
All the authors declare that they have no conflict of interest.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.McPherson K, Steel CM, Dixon JM. Breast cancer—epidemiology, risk factors, and genetics. BMJ. 2000;321(7261):624–628. doi: 10.1136/bmj.321.7261.624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133–140. doi: 10.1016/S1470-2045(00)00254-0 [DOI] [PubMed] [Google Scholar]
- 4.Tao ZQ, Shi A, Lu C, et al. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72(2):333–338. doi: 10.1007/s12013-014-0459-6 [DOI] [PubMed] [Google Scholar]
- 5.DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31–42. doi: 10.3322/caac.21320 [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 7.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10(1):38. doi: 10.1186/1476-4598-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao S, Liu W, Li F, et al. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7(10):6776. [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, Xue WJ, Feng Y, et al. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8(8):3522. [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Li L, Yuan H, et al. Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle. 2019;18(16):1965–1975. doi: 10.1080/15384101.2019.1635870 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Collina F, Aquino G, Brogna M, et al. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of triple negative breast cancer. J Cancer. 2019;10(9):2018. doi: 10.7150/jca.29670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6(1):e1583. doi: 10.1038/cddis.2014.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P, Wang R, Yue Q, et al. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun. 2018;503(4):2956–2962. doi: 10.1016/j.bbrc.2018.08.077 [DOI] [PubMed] [Google Scholar]
- 15.Sun D, Yu Z, Fang X, et al. LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017;18(10):1801–1816. doi: 10.15252/embr.201643668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min W, Dai D, Wang J, et al. Long noncoding RNA miR210HG as a potential biomarker for the diagnosis of glioma. PLoS One. 2016;11(9):e0160451. doi: 10.1371/journal.pone.0160451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652. doi: 10.18632/oncotarget.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta Gene Regul Mech. 2016;1859(1):169–176. doi: 10.1016/j.bbagrm.2015.06.015 [DOI] [PubMed] [Google Scholar]
- 19.Soini Y, Tuhkanen H, Sironen R, et al. Transcription factors zeb1, twist and snai1 in breast carcinoma. BMC Cancer. 2011;11(1):73. doi: 10.1186/1471-2407-11-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang D, Chen H, Zhu JY, et al. Epithelial–mesenchymal transition of ovarian cancer cells is sustained by Rac1 through simultaneous activation of MEK1/2 and Src signaling pathways. Oncogene. 2017;36(11):1546. doi: 10.1038/onc.2016.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo Y, Li Y, Zhou Z, et al. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–928. doi: 10.1016/j.biopha.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Luan T, Zhou S, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR‐219a‐5p. Cancer Med. 2019;8(9):4389–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834. doi: 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 24.Jiang J, Xie C, Liu Y, et al. Up-regulation of miR-383-5p suppresses proliferation and enhances chemosensitivity in ovarian cancer cells by targeting TRIM27. Biomed Pharmacother. 2019;109:595–601. doi: 10.1016/j.biopha.2018.10.148 [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Zhou Y, Fei X, et al. Biostatistics mining associated method identifies AKR1B10 enhancing hepatocellular carcinoma cell growth and degenerated by miR-383-5p. Sci Rep. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukong KE, Chang K, Khandjian EW, et al. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004 [DOI] [PubMed] [Google Scholar]
- 27.Jonsson L, Bergman J, Nodin B, et al. Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: an analysis of 215 cases from the Malmö Diet and Cancer Study. J Transl Med. 2011;9(1):114. doi: 10.1186/1479-5876-9-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venugopal A, Subramaniam D, Balmaceda J, et al. RNA binding protein RBM3 increases β‐catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol Carcinog. 2016;55(11):1503–1516. doi: 10.1002/mc.v55.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen P, Yue X, Xiong H, et al. RBM3 upregulates ARPC2 by binding the 3ʹUTR and contributes to breast cancer progression. Int J Oncol. 2019;54(4):1387–1397. doi: 10.3892/ijo.2019.4698 [DOI] [PubMed] [Google Scholar]