Abstract
Objective:
A study was conducted to identify metabolic-related effects of benzo[a]pyrene (BaP) on human lung epithelial cells and validate these findings using human sera.
Methods:
Human lung epithelial cells were treated with BaP, and extracts were analyzed with a global metabolome-wide association study (MWAS) to test for pathways and metabolites altered relative to vehicle controls.
Results:
MWAS results showed that BaP metabolites were among the top metabolites differing between BaP-treated cells and controls. Pathway enrichment analysis further confirmed that fatty acid, lipid, and mitochondrial pathways were altered by BaP. Human sera analysis showed that lipids varied with BaP concentration. BaP associations with amino acid metabolism were found in both models.
Conclusions:
These findings show that BaP has broad metabolic effects, and suggest that air pollution exacerbates disease processes by altered mitochondrial and amino acid metabolism.
Keywords: Bio-monitoring, environmental toxicology, exposome, high-resolution metabolomics, HRM, MWAS
INTRODUCTION
Benzo[a]pyrene (BaP) is a well-studied polycyclic aromatic hydrocarbon (PAH) that is a widespread environmental pollutant in diesel exhaust, cigarette smoke and other sources with incomplete combustion of organic matter. BaP is metabolized through cytochrome P450 pathways, forming reactive epoxides and diol-epoxides, which bind to the DNA and contribute to environmental mutagenesis and carcinogenesis (1-4). The primary route of BaP exposure for smokers is through inhalation, and the effects of BaP exposure on the pulmonary system have been well characterized (5). For non-smokers, the primary route of BaP exposure is through the diet, and this is also well characterized (6-8). Studies show that in addition to tumor initiation (9) and lung cancer (10, 11), BaP exposure potentiates lung inflammation (12, 13) and asthma (14). These effects on multiple disease processes suggest that BaP has adverse impacts on multiple biologic pathways.
Mechanistic studies in human lung cell models (e.g., A549 adenocarcinoma human alveolar basal epithelial cells) support the interpretation that BaP has pleiotropic effects on pulmonary function and diseases (15-18). BaP promotes cell migration and invasion as well as time-dependent alterations in cytochrome 1B1 mRNA levels (16, 19). BaP increases formation of reactive epoxide and diol epoxide metabolites by increasing CYP1A1 and CYP1B1 expression (20). Barhoumi et al. showed that fatty acids, such as docosahexaenoic acid, affected BaP uptake and perturbed metabolism (15).
Zhang et al (21) used metabolomics to study effects of BaP as a component of particulate matter in air pollution particles (PM2.5) in A549 cells and in vivo in rats. They found that BaP promoted pulmonary injury by increasing phospholipase A2 (PLA2) activity. These results are consistent with our earlier findings that BaP levels in serum were associated with microRNA’s, lipid and fatty acid metabolism (22-24). Our studies also showed, however, that changes in carnitine metabolism, sulfur amino acid metabolism, xenobiotic metabolism and butanoate metabolism also varied with BaP (22). This study of human serum indicated that BaP has metabolic effects in addition to those caused by increased phospholipase activity.
In the present study, we used a high-resolution metabolomics platform that complements the previous human metabolomics (21) and rat and cellular lipidomics analyses (25) to further examine metabolic effects of BaP in human serum and cultured lung cells. The results support the prior evidence that BaP alters lipid metabolism and shows that BaP causes additional disruptions in mitochondrial metabolism and amino acid metabolism.
METHODS
Chemicals
Acetonitrile (HPLC grade), formic acid (HPLC grade), benzo[a]pyrene (HPLC grade), and water (HPLC grade) were from Sigma–Aldrich (St. Louis, MO). Stable isotopic chemicals for internal standards (26) were obtained from Cambridge Isotope Laboratories, Inc. (Andover, Pennsylvania). BaP was prepared by dissolving in acetonitrile to make 3 mM stock solution. BaP was then spiked into 2 mL of media to give a final concentration of 3 μM.
Cell Culture and BaP treatment
A549 human alveolar epithelial adenocarcinoma cells were obtained from ATCC (Manassas, VA) and cultured in humidified incubator (5% CO2, 37°C) in complete media (10% fetal bovine serum (FBS) in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) with glutamine (Mediatech, Manassas, VA) until 90% confluence. Cells were incubated with 3 μM BaP or acetonitrile (vehicle control) for 24 h in low serum containing medium (0.5% FBS in DMEM/F12). Culture media was removed, and cells were washed three times with ice-cold phosphate buffered saline (PBS). Cells were lysed and extracted by addition of 150 μL of lysis solution [2:1, acetonitrile (ACN): water] containing a mixture of isotopic standards (26). Samples were allowed to stand on ice for 30 min, centrifuged at 13,000 x g for 10 min to remove protein, and supernatants were analyzed by liquid chromatography coupled to ultra-high-resolution mass spectrometry (LC-HRMS) as described below.
Serum Samples
Thirty non-identified human serum samples from the Department of Defense Serum Repository were processed as described by Walker et al. (22)
Liquid chromatography-high-resolution mass spectrometry
We followed methods described by Walker et al. in analyzing the A549 extracts with a shortened run time (22). A549 extracts were analyzed by same procedure with a shortened run time in which the first 1-min consisted of 5% solution A (2% formic acid), 60% water, 35% ACN, followed by a 2-min linear gradient to 5% solution A, 0% water, 95% ACN and final 2-min at 5% A, 95% ACN. Data were collected continuously during chromatographic separations and stored as .raw files. A quality control pooled reference plasma sample (Q-Std3) was included at the beginning and end of each batch of 25 samples for normalization and post hoc quantification (27).
Mass spectral data extraction and pre-processing
Mass spectral files in .raw format were converted to .cdf files using XCalibur file converter software (Thermo Fisher, Waltham, MA). These files were extracted using apLCMSv6.3.3 (28), and xMSanalyzerv2.0.7 (29) to provide data tables containing mass spectral features defined by mass-to-charge ratio (m/z), retention time, and ion abundance.
Feature Selection and Pathway Analysis
For A549 cells, triplicate technical replicates were averaged, and cell lysates from replicate cultures were then averaged, and only m/z features with at least 80% non-missing values in either of the groups and more than 50% non-missing values across all samples were retained, (n=7 for BaP exposed group and n=9 for vehicle control). After filtering based on missing values, data were log2 transformed and quantile normalized (30). Selection of differentially expressed m/z features was performed based on one-way ANOVA using the limma package in R (31). Benjamini-Hochberg false discovery method was used for multiple hypothesis testing correction at FDR<0.2 threshold (32). Visualization of the data, which was based on similarity in expression, was performed using unsupervised two-way hierarchal clustering analysis (HCA) utilizing the hclust() function in R to determine the clustering pattern of selected m/z features and samples. Principal component analysis (PCA) was performed using the pca() function implemented in R package pcaMethods.
For the human serum samples, metabolic features correlated with serum BaP concentration were selected using linear regression analysis and criteria described by Walker et al. (22) Two-way hierarchal cluster analysis was performed with the hclust() function in R. Pathway enrichment analysis was performed using mummichog (33). For this analysis, features differing at P < 0.05 were selected to protect against type 2 error, and permutation testing (P < 0.05) was used in pathway enrichment analysis to protect against type 1 error (34).
RESULTS
Effect of BaP on A549 lung cell metabolome
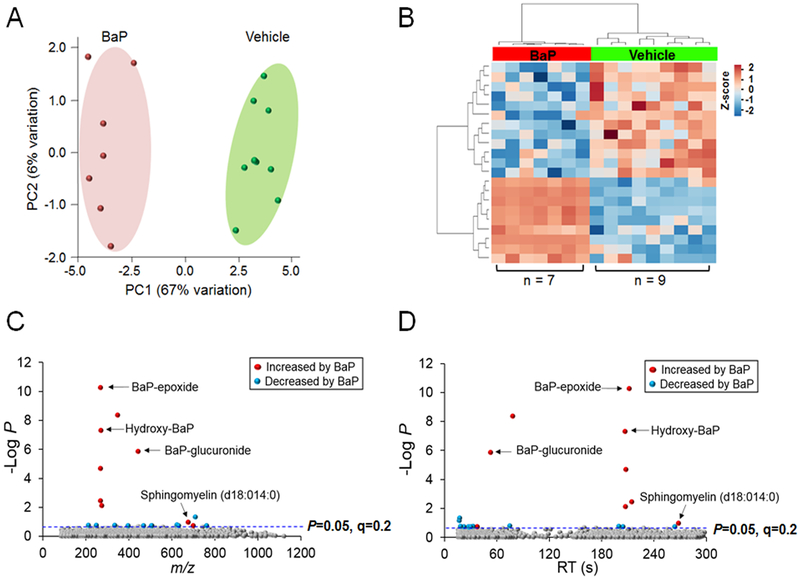
Experiments were performed with A549 lung cells treated with 3 μM BaP for 24 h because prior research showed that this concentration is non-toxic and that protein expression changes were maximal at this time point (21). To understand lung metabolic responses to BaP exposure, we performed LQ-HRMS on cells treated with BaP and compared results to identically treated vehicle controls. After data extraction and quality filtering using apLCMS and xMSanalyzer, 3,217 mass spectral features were used for analyses (Supplementary Table 1). Principal component analysis (PCA) showed separation by the first two principal components, PC1 (67%) and PC2 (6%) (Fig 1A). While 360 features differed in abundance at P < 0.05 (without FDR correction), only 21 features differed after using FDR corrected P < 0.2 (Fig 1B). Of these, nine were increased while twelve were decreased by BaP (Fig 1B); detailed information of these metabolites is provided in Table 1.
Figure 1. Metabolic separation and annotation of metabolites after exposure to BaP in A549 cells.
A. PCA plot showing separation of the vehicle control group (shown in green) and BaP exposed group (shown in red), through the 1st (67% variation) and 2nd (6% variation) principal components. B. Unsupervised hierarchal clustering heatmap indicate that intensity of 21 features drive the separation between vehicle control and BaP exposed groups. C. Type I Manhattan plot of m/z features plotted against the −log P. Shown in gray are the 3,217 features identified after filtering and normalization. 21 features were found to be different between the two groups at FDR < 0.2 as indicated by the blue dotted line. Shown in red were features identified to be increased after BaP exposure (9/21) which include BaP-epoxide (m/z 267.0813, 212 s), hydroxy-BaP (BaP-OH) (m/z 269.0881, 207 s), BaP-glucuronide (m/z 443.1134, 53 s) and sphingomyelin (d18:140) (m/z 675.54379, 268 s), while features which were decreased by BaP are shown in blue (12/21). D. Type II Manhattan plot using time plotted against −log P. The more polar features elute earlier than the more non-polar metabolites as seen by BaP-glucuronide versus hydroxy-BaP which elute at a later time due to the methods and column described above in the methods. n=7 for BaP exposed group and n=9 for vehicle control.
Table 1.
Annotation of 21 metabolic features (Level 5 identity by criteria of Schymanski et al, 2014) altered by BaP in A549 cells (FDR 0.2, n ≥ 7) using HMDB, KEGG and Metlin. Fold changes were calculated from mean intensities; BaP-increased abundance (positive), BaP-decreased abundance (negative).
| m/z | RT, s | Name | Adduct | Fold change, BaP/vehicle |
|
|---|---|---|---|---|---|
| 1 | 210.9368 | 144 | Dichlorobenzoic acid | M+Na-2H | −1.8 |
| 2 | 248.0716 | 200 | Dubamine | M-H | −3.6 |
| 3 | 267.0014 | 208 | Cyclobrassinone | M+Cl | 26 |
| 4 | 267.0813 | 212 | BaP-epoxide | M-H | 1099 |
| 5 | 267.1571 | 215 | No Match | 13 | |
| 6 | 269.0881 | 207 | Hydroxy-BaP | M-H | 28 |
| 7 | 273.0378 | 208 | Bis(24-dihydroxyphenyl)ethanedione | M-H | 16 |
| 8 | 333.0556 | 19 | 7-Hydroxymethyl-12-methylbenzaanthracene sulfate | M-H2O-H | −3.1 |
| 9 | 347.0380 | 78 | 2-O-(alpha-D-Glucopyranosyl)-3-phospho-D-glycerate;(2R)-2-(alpha-D-Glucopyranosyloxy)-3-(phosphonooxy)propanoate | M-H | |
| 10 | 395.9023 | 30 | No Match | −2.1 | |
| 11 | 443.1134 | 53 | BaP-glucuronide | M-H | 844 |
| 12 | 468.2634 | 204 | Benzethonium chloride | M+Na-2H | −3.2 |
| 13 | 504.6294 | 28 | No Match | −2.1 | |
| 14 | 506.6275 | 28 | No Match | −1.6 | |
| 15 | 623.9064 | 75 | No Match | −2.7 | |
| 16 | 630.57607 | 22 | No Match | −3.2 | |
| 17 | 675.5438 | 268 | Sphingomyelin (d18:014:0) | M-H | 4.3 |
| 18 | 698.0841 | 37 | No Match | 2.8 | |
| 19 | 709.1226 | 17 | Cyanidin3-(4"-malonyl-2"-glucuronosylglucoside) | M-H | −2.6 |
| 20 | 761.5995 | 264 | Sphingomyelin (d18:022:3) | M-H2O-H | −1.9 |
| 21 | 1232.6274 | 17 | No Match | −2.0 |
Thirteen of the 21 features that differed at FDR < 0.2 matched to metabolites in the Human Metabolome Database (HMDB), the Kyoto Encyclopedia of Genes and Genomes (KEGG), or the METLIN™ metabolite database, while eight were not matched, indicated as “No match” (Table 1). Three of these 21 metabolic features were BaP metabolites with significant fold increase in BaP treated cells (Fig 1C); these include BaP-epoxide (m/z 267.0813, 212 s), hydroxy-BaP (BaP-OH) (m/z 269.0881, 207 s), and BaP-glucuronide (m/z 443.1134, 53 s) (Fig 1C, 1D, Table 1). A Type II Manhattan plot (-log10 P as a function of RT on C18 chromatography) showed that these BaP metabolites separated based on polarity; the more polar BaP-glucuronide eluted earlier with more polar metabolites while BaP-epoxide and BaP-OH eluted later with more non-polar metabolites (Fig 1D).
Consistent with findings of Zhang et al, (21) two features matching sphingomyelin species were also altered in abundance by BaP (fold change ≥ 1.9) and eluted out later (264s, 268s) based on their non-polar lipid properties (Fig 1D). The other annotated features included the PAH metabolite 7-hydroxymethyl-12-methylbenz(a)anthracene sulfate, dichlorobenzoate, hydroxybenzyl, benzethonium chloride, and other several features (dubamine, cyclobrassinone, cyanidin-malonyl-glucuronosylglucoside, and 2-O-(glucopyranosyl)-phospho-glycerate) that are annotated based upon accurate mass (Level 5 identification by criteria of Schymanski et al. (35).)
Pathway Enrichment Analysis of BaP exposure in A549 cells
Prior research showed that BaP perturbed lipid metabolism through effects on phospholipase A2 (21) and that changes in carnitine, butyrate, sulfur amino acid and xenobiotic metabolism were associated with BaP in human serum (22). To test for global pathway effects of BaP on metabolism in A549 cells, we used mummichog (33) to test for pathway enrichment among all of the 360 features selected at P < 0.05 (168 increased, 192 decreased by BaP; Supplemental Table 2). The results of mummichog pathway enrichment analysis showed that 4 metabolic pathways affected by BaP (P < 0.05, Supplemental Table 3) were 1) CoA biosynthesis and catabolism (P = 0.02), 2) arginine and proline metabolism (P = 0.03), 3) fatty acid biosynthesis (P = 0.03) and 4) glycerophospholipid metabolism (P = 0.04) (Fig 2). Detailed information of 26 features enriched in 4 pathways is provided in Supplemental Table 3. Results of KEGG pathway analysis (http://www.genome.jp/kegg/tool/map_pathway2.html) of the 360 features also identified pathways for energy metabolism, carbohydrate and lipid metabolism, consistent with the mummichog analysis (data not shown).
Figure 2. Pathway enrichment analysis of A549 cells after exposure to BaP.
Pathway enrichment rate analysis of A549 cells after BaP exposure compared to vehicle control. A total of (4/119) enriched pathways were determined (Filled gray bars indicate significance less than P < 0.05, and the cutoff is indicated by the dotted line). These pathways include CoA biosynthesis and catabolism (3/4), arginine and proline metabolism (9/33), fatty acid biosynthesis (8/29) and glycerophospholipids (6/34). Numbers in the parentheses indicate the overlap of the metabolites as matched through KEGG database matching using m/z.
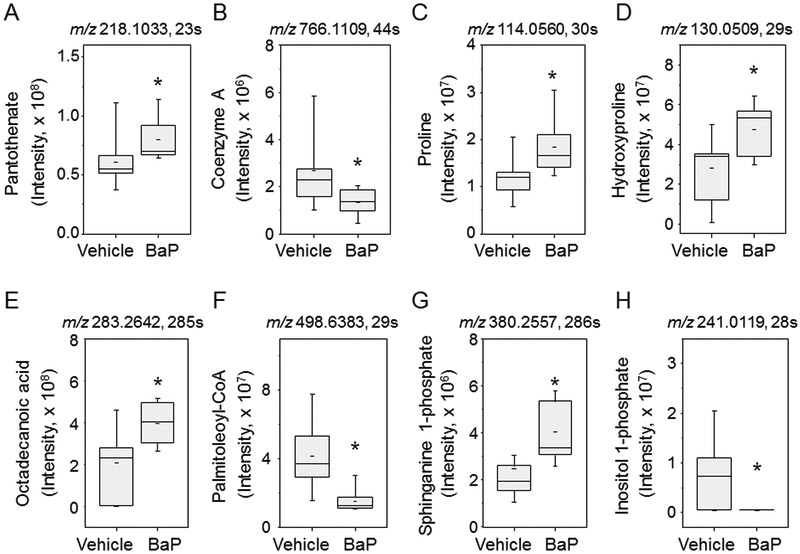
Selected metabolites from these pathways are shown by Whisker plots in Fig 3. For CoA biosynthesis and catabolism, panthothenate (3A) was increased and coenzyme A (3B) was decreased. In arginine and proline metabolism, both proline (3C) and hydroxyproline (3D) were increased. In fatty acid biosynthesis and metabolism pathways, the free fatty acid, octadecanoic acid (3E) was increased and the acyl-CoA, palmitoleoyl-CoA (3F), was decreased. Lastly, in glycerophospholipid metabolism, sphinganine 1-phosphate (3G) was increased and inositol 1-phosphate (3H) was decreased. Collectively, the pathway enrichment analyses and targeted examination of individual metabolites showed that BaP has multiple effects on metabolism in A549 cells.
Figure 3. Comparison of selected metabolites between the BaP and vehicle control exposed groups.
Two metabolites were selected from each metabolic pathway from mummichog pathway enrichment analysis. Data shown are mean and standard deviation for spectral intensities of BaP and vehicle groups. CoA biosynthesis and catabolism: A. Pantothenate (m/z 218.1033, 23 s) and B. Coenzyme A (m/z 766.1109, 44 s). Arginine and proline metabolism: C. Proline (m/z 114.0560 30 s) and D. hydroxyproline (m/z 130.05099, 29 s). Fatty acid biosynthesis: E. octadecanoic acid (m/z 283.2642, 285 s) and F. palmitoleoyl-CoA (m/z 498.6383, 29 s). Glycerophospholipids: G. sphinganine 1-phosphate (m/z 380.2557, 286 s) and H. inositol 1-phosphate (m/z 241.0119, 28 s).
Metabolic associations with BaP in human sera
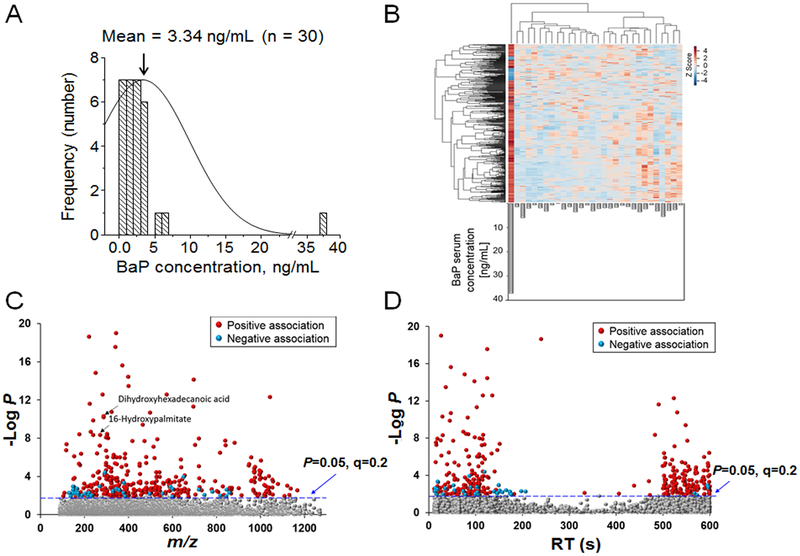
Prior analyses with LC-HRMS using C18 chromatography with positive electrospray ionization showed that carnitine, butyrate, sulfur amino acid and xenobiotic metabolism pathways were associated with BaP in 30 human serum samples from the DoDSR (22). To test for additional metabolic associations with BaP, we used C18 chromatography with negative electrospray ionization to reanalyze these 30 DoDSR samples. The data showing distribution of BaP levels are replotted in Fig 4A. Mean BaP concentration was 3.34 ng/mL (13.6 nM), with the highest concentration being 37.3 ng/mL (148 nM). After data were averaged, normalized, and filtered, 6,123 features were used for analysis (Supplemental Table 4). Four hundred features correlated with BaP at FDR < 0.2 (Fig 4B). Of these 400 features, 44 were negatively associated and 356 were positively associated with BaP (Fig 4C, D; Supplemental Table 5). Many of the features annotated using KEGG play a role in lipid and fatty acid pathways. These include di-hydroxyhexadecanoic acid (m/z 287.2248, 29 s) and hydroxypalmitate (m/z 271.2297, 116 s) (Fig 4C); a list of annotated features is given in Supplemental Table 6.
Figure 4. Stratification and identification of features correlating with BaP concentration.
A. The mean concentration of BaP was 3.34 ng/mL for the serum samples (n=30). Concentration is plotted against frequency among the samples with one sample have a concentration of 37.3 ng/mL. B. Unsupervised 2-way way hierarchal clustering heat-map. 400 features were found to be correlated with BaP concentration (shown in gray bars as ng/mL). Of these 400 features, 44 were negatively associated with BaP, while 356 features were positively associated with BaP. C. Type I Manhattan plot of m/z features plotted against the −log P. Shown in gray are the 5723 non-significant features identified after filtering and normalization. 400 features were found to associate with BaP concentration at FDR < 0.2, indicated by the blue dotted line. Features that were positively associated with BaP concentrations (356/400) are shown in red. These included accurate mass matches to dihydroxyhexadecanoic acid (m/z 287.2248, 29 s) and 16-hydroxy-palmitate (m/z 271.2297, 116 s). A relatively small number of features (44/400) was negatively associated with BaP concentration (blue circles). D. Type II Manhattan plot (−log P as a function of retention time on the C18 column) showed that features associated with BaP included both polar species eluting early and relatively non-polar species eluting later.
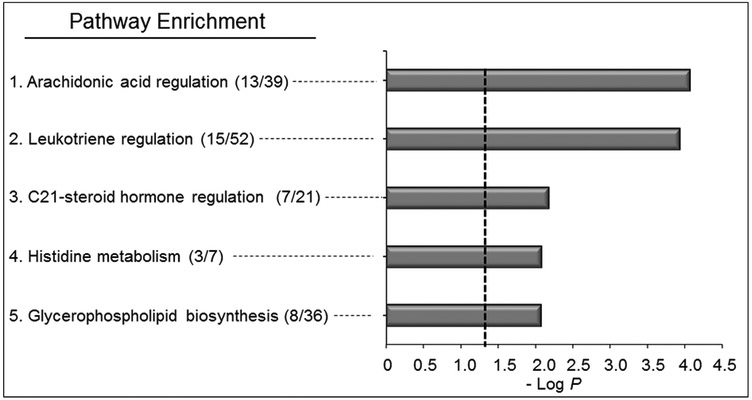
Pathway enrichment analysis of the 400 features using mummichog showed five metabolic pathways varying with BaP (P < 0.05, Supplemental Table 7), including 1) arachidonic acid regulation (P = 2.52 × 10−4), 2) leukotriene metabolism (P = 3.36 × 10−4), 3) C21-steroid hormone regulation (P = 0.01), 4) histidine metabolism (P = 0.01), and 5) glycerophospholipid metabolism (P = 0.01) (Fig 5). Four of these pathways are associated with fatty acids and lipid metabolism while the other is associated with amino acid metabolism. Metabolite matches within each these pathways are shown in Supplemental Table 7. KEGG pathway analysis also showed the serum features to map to energy metabolism, fatty acid and lipid metabolism, and biosynthesis of amino acids, consistent with results from mummichog analysis.
Figure 5. Pathway enrichment analysis of A549 cells after exposure to BaP.
Pathway analysis of BaP exposed features from human serum with different levels of BaP. A total of (5/119) enriched pathways were determined (Filled gray bars indicate significance less than P < 0.05, and the cutoff is indicated by the dotted line). These pathways include arachidonic acid regulation (13/39), leukotriene regulation (15/52), C21-steroid hormone regulation (7/21), histidine metabolism (3/7), and glycerophospholipid biosynthesis (8/36). Numbers in the parentheses indicate the overlap of the metabolites as matched through KEGG and the Edinburgh human metabolic network databases matching using m/z and RT.
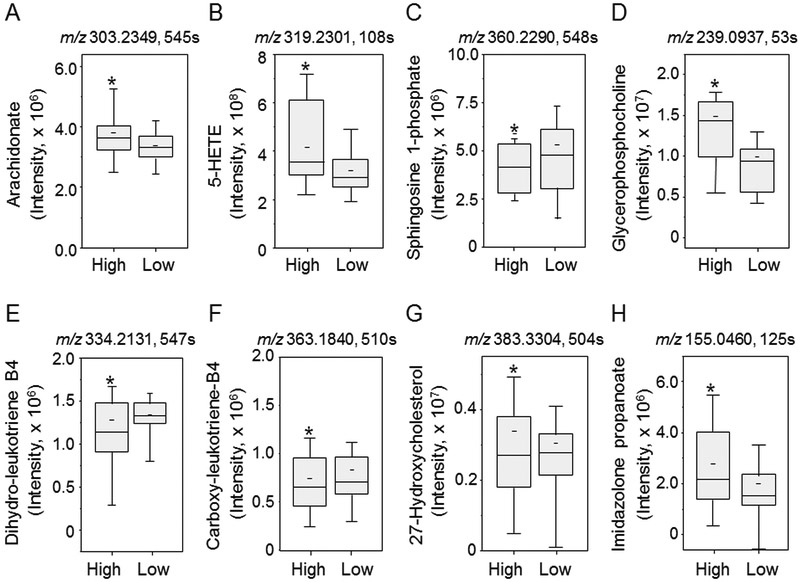
Box plots for representative metabolites in these pathways are shown in Fig 6. In arachidonic acid regulation, arachidonate (6A) and 5-HETE (6B) were increased in samples with high BaP. In glycerophospholipid metabolism, sphingosine 1-phosphate (6C) was decreased with BaP and glycerophosphocholine (6D) was increased with BaP. In leukotriene regulation, both 20-OH-10,11-dihydroleukotriene B4 (6E) and 20-carboxyleuktriene B4 (6F) were decreased with higher BaP. For C21 steroid hormone regulation, 27-hydroxycholesterol (6G) was decreased in association high BaP. For histidine metabolism, 4-imidazolone-5-propanoate (6H) was increased with high BaP. These results provide additional evidence that BaP has a broad range of metabolic effect that include lipid and energy metabolism as well as steroid, xenobiotic and amino acid metabolism.
Figure 6. Comparison of selected metabolites between groups of human sera.
Human sera samples were divided into two categories: high and low groups relative to the median BaP concentration. Selected metabolites from each metabolic pathway were annotated through mummichog pathway analysis and were compared using raw non-normalized spectral intensity between BaP high and BaP low groups. Arachidonic acid regulation, A. arachidonate (m/z 303.2349, 545 s) and B. 5-HETE (m/z 319.2301, 108 s). Glycerophospholipids: C. sphingosine 1-phosphate (m/z 360.2290 548 s) D. glycerophosphocholine (m/z 239.0937, 53 s), leukotriene regulation: E. 20-OH-10,11,dihydro-leukotriene B4(m/z 334.2131, 547 s) F. 20-carboxyleukotriene B4 (m/z 363.1840, 510 s), C21-steroid hormone regulation: G. 27-hydroxy cholesterol (m/z 383.3304, 504 s) and histidine metabolism: H. 4-Imidazolone-5-propanoate (m/z 155.0460, 125 s).
DISCUSSION
BaP is a polycyclic aromatic hydrocarbon rated 8th on the ATSDR’s substance priority list (5). BaP is bio-activated by members of the cytochrome P450 family to form carcinogenic intermediates which can bind to DNA and ultimately lead to cancer initiation. In addition, BaP is associated with cardiovascular disease, asthma, and other diseases (36). The present research adds to the growing evidence that the diverse disease effects of BaP are due to broad metabolic effects, perhaps interacting with diet and other factors impacting disease outcome. The results emphasize a need to gain additional understanding of non-mutagenic mechanisms of BaP to mitigate against adverse effects.
Previous research showed that the mean concentration of BaP in the serum of individuals in the current study was 3.37 ng/mL after deployment (37). This value is in line with previously reported values which are within the range of 0.005–3.7 ng/mL for a variety of different exposure studies (38-40). The highest observed value for BaP detected for the individuals in the current study was 177.5 ng/mL (0.7 μM). Due to the relatively rapid elimination of BaP, we selected a somewhat higher value (3 μM) for the in vitro studies to model immediate exposure within the lung.
High resolution metabolomics (HRM) uses ultra-high-resolution mass spectrometry with advanced data extraction methods to obtain global analysis of metabolic effects on most human metabolic pathways (41, 42). Computational tools such as mummichog for pathway enrichment analysis (33) and xMSannotator for metabolite annotation (43) use probability-based approaches to complement targeted analyses of specific metabolites. Probability-based identification of CoA-biosynthesis as a top pathway supports the utility of the approach (Fig 2). Similarly, detection of BaP metabolites, hydroxy-BaP, BaP-epoxide, and BaP-glucuronide (Fig 1C; 1D) as top features in cell culture analyses confirms the utility of this top-down metabolomics strategy (22, 24). Many BaP metabolites have specific activities based on regio- and stereo-selectivity and these are important in carcinogenic mechanisms. Some of the metabolites detected in the cell studies, such as epoxides, were not detected in the human serum samples, perhaps due to limited stability. Additionally, a limitation of an untargeted metabolomics approach, as used in the current study, is that many isomeric forms are not distinguished without detailed studies using authentic standards and MS/MS validation. Thus, future studies will need to be designed to bridge this important gap.
The approach is particularly useful for integration of knowledge from in vitro and in vivo studies because of the focus on pathway level effects; specifically, effects on a metabolic pathway can be detected in both cells and plasma even though transport and distribution may result in detection of different metabolites in the pathways in the cells and plasma.
Previous research showed that BaP is an agonist of aryl hydrocarbon receptor (AHR), a system that suppresses fatty acid oxidation and synthesis (44-46). In both cell studies and human sera, we found BaP associated with several lipid and fatty acid pathways. In both, we found changes in sphingolipids, sphinganine 1-phosphate in cells (Fig 3C) and sphingosine 1-phosphate in serum (Fig 6C), that function in signaling and cellular regulation (47). In cells, glycerophospholipid and fatty acid pathways were altered in response to BaP (Fig 2; 3), including pantothenate, a precursor to CoA which was increased, and CoA, which was decreased in association with BaP. The results suggest that BaP inhibits one or more enzymes in the conversion pathway, which could cause downstream disruption in fatty acid oxidation as well as the Krebs cycle metabolic pathways. Analyses of human sera further show association of multiple oxidized fatty acids and the sterol, 27-hydroxycholesterol, with BaP. Collectively, the results show that BaP causes widespread effect on lipid and fatty acid metabolism, perhaps reflecting multiple molecular targets in addition to AhR and PLA2 in the lungs and the liver of individuals exposed.
Effects on diverse pathways of amino acid metabolism are also indicated by the available research. We previously found that sulfur amino acid metabolism, a pathway of particular importance for glutathione-dependent detoxification systems, varied with serum BaP concentration (22). In the present studies, BaP treatment of A549 cells caused changes in proline metabolism (Fig 3C, D), and histidine metabolism was found to vary with BaP concentration in serum (Fig 6H). These multiple amino acid pathway effects could reflect downstream effects of xenobiotic metabolism or toxicity, i.e., glutathione functions in BaP metabolism, proline and hydroxyproline from collagen turnover, histidine metabolism from immune signaling. These findings are also similar to previously published effects by others who observed that PAH’s correlated with fatty acid, amino acid, and carnitine pathways; however, whether these perturbations are adaptive or causative remains unclear and remain the focus for future studies (48). Although speculative, altered amino acid pathways could impact cardiovascular diseases dependent upon redox control (49), lung diseases dependent upon extracellular matrix turnover (50), and asthma and other allergic responses dependent upon histamine metabolism (51).
CONCLUSIONS
This study showed that BaP caused metabolic changes in lipid and fatty acid metabolic pathways in a lung cell line that are similar to results reported by Walker et al. (22). The data further indicated that changes occur in mitochondria fatty acid and energy metabolism. Additional effects on amino acid metabolism were also noted, suggesting that BaP has broader effects on metabolism, which could contribute to multiple disease processes associated with BaP exposure.
Supplementary Material
Acknowledgements
The authors would like to acknowledge ViLinh Tran and Ken Liu for their technical expertise with the mass spectrometer. This public health surveillance project was supported by funding from the Department of Defense award (306889–1.00–64239), and National Institute of Health (award R01 ES023485, P30 ES019776 and S10 OD 018006).
Footnotes
Disclaimer
The opinions expressed are those of the authors and do not necessarily reflect the official positions of the Uniformed Services University, the U.S. Departments of Defense, the Army and the Air Force, the U.S. Army Public Health Center (Provisional) or Emory University.
Conflicts of interest: None to declare
References
- 1.Cavalieri E, Rogan E. Role of radical cations in aromatic hydrocarbon carcinogenesis. Environ Health Perspect. 1985;64:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalieri EL, Rogan EG. The approach to understanding aromatic hydrocarbon carcinogenesis. The central role of radical cations in metabolic activation. Pharmacol Ther. 1992;55:183–199. [DOI] [PubMed] [Google Scholar]
- 3.Devanesan PD, RamaKrishna NV, Todorovic R, et al. Identification and quantitation of benzo[a]pyrene-DNA adducts formed by rat liver microsomes in vitro. Chem Res Toxicol. 1992;5:302–309. [DOI] [PubMed] [Google Scholar]
- 4.RamaKrishna NV, Gao F, Padmavathi NS, et al. Model adducts of benzo[a]pyrene and nucleosides formed from its radical cation and diol epoxide. Chem Res Toxicol. 1992;5:293–302. [DOI] [PubMed] [Google Scholar]
- 5.Mumtaz MaG J. Toxicological profile for Polycyclic Aromatic Hydrocarbons (PAHs) In: (ATSDR) AfTSaDR, ed. Atlanta, GA: U.S. Department of Health and Human Services, Public Health Service; 1995. [Google Scholar]
- 6.Butler JP, Post GB, Lioy PJ, Waldman JM, Greenberg A. Assessment of carcinogenic risk from personal exposure to benzo(a)pyrene in the Total Human Environmental Exposure Study (THEES). Air Waste. 1993;43:970–977. [DOI] [PubMed] [Google Scholar]
- 7.Hattemer-Frey HA, Travis CC. Benzo-a-pyrene: environmental partitioning and human exposure. Toxicol Ind Health. 1991;7:141–157. [DOI] [PubMed] [Google Scholar]
- 8.Stavric B, Klassen R. Dietary effects on the uptake of benzo[a]pyrene. Food Chem Toxicol. 1994;32:727–734. [DOI] [PubMed] [Google Scholar]
- 9.Hall M, Grover PL. Polycyclic Aromatic Hydrocarbons: Metabolism, Activation and Tumour Initiation In: Cooper CS, Grover PL, eds. Chemical Carcinogenesis and Mutagenesis I. Berlin, Heidelberg: Springer Berlin Heidelberg; 1990:327–372. [Google Scholar]
- 10.Hoffmann D, Hoffmann I, El-Bayoumy K. The Less Harmful Cigarette: A Controversial Issue. A Tribute to Ernst L. Wynder. Chemical Research in Toxicology. 2001;14:767–790. [DOI] [PubMed] [Google Scholar]
- 11.Kasala ER, Bodduluru LN, Barua CC, Sriram CS, Gogoi R. Benzo(a)pyrene induced lung cancer: Role of dietary phytochemicals in chemoprevention. Pharmacol Rep. 2015;67:996–1009. [DOI] [PubMed] [Google Scholar]
- 12.Gentner NJ, Weber LP. Intranasal benzo[a]pyrene alters circadian blood pressure patterns and causes lung inflammation in rats. Arch Toxicol. 2011;85:337–346. [DOI] [PubMed] [Google Scholar]
- 13.Szema AM, Salihi W, Savary K, Chen JJ. Respiratory symptoms necessitating spirometry among soldiers with Iraq/Afghanistan war lung injury. J Occup Environ Med. 2011;53:961–965. [DOI] [PubMed] [Google Scholar]
- 14.Szema AM. Climate change, allergies, and asthma. J Occup Environ Med. 2011;53:1353–1354. [DOI] [PubMed] [Google Scholar]
- 15.Barhoumi R, Mouneimne Y, Chapkin RS, Burghardt RC. Effects of fatty acids on benzo[a]pyrene uptake and metabolism in human lung adenocarcinoma A549 cells. PLoS One. 2014;9:e90908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhai W, Wang H, Xia X, Zhang C. Benzo(a)pyrene promotes A549 cell migration and invasion through up-regulating Twist. Arch Toxicol. 2015;89:451–458. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Peng H, Xiao J, et al. Benzo(a)pyrene enhances the EMT-associated migration of lung adenocarcinoma A549 cells by upregulating Twist1. Oncol Rep. 2017;38:2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bak Y, Jang HJ, Seo JH, et al. Benzo[a]pyrene Alters the Expression of Genes in A549 Lung Cancer Cells and Cancer Stem Cells. J Microbiol Biotechnol. 2018;28:425–431. [DOI] [PubMed] [Google Scholar]
- 19.Wen X, Walle T. Preferential induction of CYP1B1 by benzo[a]pyrene in human oral epithelial cells: impact on DNA adduct formation and prevention by polyphenols. Carcinogenesis. 2005;26:1774–1781. [DOI] [PubMed] [Google Scholar]
- 20.Hukkanen J, Lassila A, Paivarinta K, et al. Induction and regulation of xenobiotic-metabolizing cytochrome P450s in the human A549 lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 2000;22:360–366. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SY, Shao D, Liu H, et al. Metabolomics analysis reveals that benzo[a]pyrene, a component of PM2.5, promotes pulmonary injury by modifying lipid metabolism in a phospholipase A2-dependent manner in vivo and in vitro. Redox Biol. 2017;13:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker DI, Pennell KD, Uppal K, et al. Pilot Metabolome-Wide Association Study of Benzo(a)pyrene in Serum From Military Personnel. J Occup Environ Med. 2016;58:S44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woeller CF, Thatcher TH, Van Twisk D, et al. Detection of Serum microRNAs From Department of Defense Serum Repository: Correlation With Cotinine, Cytokine, and Polycyclic Aromatic Hydrocarbon Levels. J Occup Environ Med. 2016;58:S62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker DI, Mallon CT, Hopke PK, et al. Deployment-Associated Exposure Surveillance With High-Resolution Metabolomics. J Occup Environ Med. 2016;58:S12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jungnickel H, Potratz S, Baumann S, Tarnow P, von Bergen M, Luch A. Identification of lipidomic biomarkers for coexposure to subtoxic doses of benzo[a]pyrene and cadmium: the toxicological cascade biomarker approach. Environ Sci Technol. 2014;48:10423–10431. [DOI] [PubMed] [Google Scholar]
- 26.Go YM, Kim CW, Walker DI, et al. Disturbed flow induces systemic changes in metabolites in mouse plasma: a metabolomics study using ApoE(−)/(−) mice with partial carotid ligation. Am J Physiol Regul Integr Comp Physiol. 2015;308:R62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Go YM, Walker DI, Liang Y, et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci. 2015;148:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uppal K, Soltow QA, Strobel FH, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel RM, Roback JD, Uppal K, Yu T, Jones DP, Josephson CD. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion. 2015;55:544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of teh Royal Statistical Society Seriese B. 1995;57:289–300. [Google Scholar]
- 33.Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol. 2016;29:1956–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schymanski EL, Jeon J, Gulde R, et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol. 2014;48:2097–2098. [DOI] [PubMed] [Google Scholar]
- 36.Podechard N, Lecureur V, Le Ferrec E, et al. Interleukin-8 induction by the environmental contaminant benzo(a)pyrene is aryl hydrocarbon receptor-dependent and leads to lung inflammation. Toxicol Lett. 2008;177:130–137. [DOI] [PubMed] [Google Scholar]
- 37.Xia X, Carroll-Haddad A, Brown N, Utell MJ, Mallon CT, Hopke PK. Polycyclic Aromatic Hydrocarbons and Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans in Microliter Samples of Human Serum as Exposure Indicators. J Occup Environ Med. 2016;58:S72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh VK, Patel DK, Ram S, Mathur N, Siddiqui MK, Behari JR. Blood levels of polycyclic aromatic hydrocarbons in children of Lucknow, India. Arch Environ Contam Toxicol. 2008;54:348–354. [DOI] [PubMed] [Google Scholar]
- 39.Pleil JD, Stiegel MA, Sobus JR, Tabucchi S, Ghio AJ, Madden MC. Cumulative exposure assessment for trace-level polycyclic aromatic hydrocarbons (PAHs) using human blood and plasma analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:1753–1760. [DOI] [PubMed] [Google Scholar]
- 40.Al-Daghri NM, Alokail MS, Abd-Alrahman SH, Draz HM. Polycyclic aromatic hydrocarbon distribution in serum of Saudi children using HPLC-FLD: marker elevations in children with asthma. Environ Sci Pollut Res Int. 2014;21:12085–12090. [DOI] [PubMed] [Google Scholar]
- 41.Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker DI, Go Y-M, Liu K, Pennell KD, Jones DP. Chapter 7 - Population Screening for Biological and Environmental Properties of the Human Metabolic Phenotype: Implications for Personalized Medicine In: Holmes E, Nicholson JK, Darzi AW, Lindon JC, eds. Metabolic Phenotyping in Personalized and Public Healthcare. Boston: Academic Press; 2016:167–211. [Google Scholar]
- 43.Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem. 2017;89:1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanos R, Murray IA, Smith PB, Patterson A, Perdew GH. Role of the Ah receptor in homeostatic control of fatty acid synthesis in the liver. Toxicol Sci. 2012;129:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH, Wada T, Febbraio M, et al. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsunawa M, Amano Y, Endo K, et al. The aryl hydrocarbon receptor activator benzo[a]pyrene enhances vitamin D3 catabolism in macrophages. Toxicol Sci. 2009;109:50–58. [DOI] [PubMed] [Google Scholar]
- 47.Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Zheng Y, Zhao B, et al. Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. J Proteome Res. 2015;14:2583–2593. [DOI] [PubMed] [Google Scholar]
- 49.Patel D, Alhawaj R, Kelly MR, et al. Potential role of mitochondrial superoxide decreasing ferrochelatase and heme in coronary artery soluble guanylate cyclase depletion by angiotensin II. Am J Physiol Heart Circ Physiol. 2016;310:H1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson WH, Ritzenthaler JD, Roman J. Lung extracellular matrix and redox regulation. Redox Biol. 2016;8:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comhair SA, Erzurum SC. Redox control of asthma: molecular mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2010;12:93–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.