Abstract
Objective:
To develop an approach for a retrospective analysis of post-exposure serum samples using diverse molecular profiles.
Methods:
The 236 molecular profiles from 800 de-identified human serum samples from the Department of Defense Serum Repository were classified as smokers or non-smokers based on direct measurement of serum cotinine levels. A machine-learning pipeline was used to classify smokers and non-smokers from their molecular profiles.
Results:
The refined supervised support vector machines with recursive feature elimination predicted smokers and non-smokers with 78% on the independent held-out set. Several of the identified classifiers of smoking status have previously been reported and four additional miRNAs were validated with experimental tobacco smoke exposure in mice, supporting the computational approach.
Conclusions:
We developed and validated a pipeline that shows retrospective analysis of post-exposure serum samples can identify exposures to environmental exposures.
Keywords: support vector machines, tobacco use, molecular profiling
Introduction
A consistent problem in environmental medicine is the lack of individualized real-time measurements of environmental exposures. Environmental exposure events, such as industrial accidents, exposures to polluted environments, or intentional contamination, rarely occur in the presence of adequate environmental monitoring, and individualized exposure monitoring almost never occurs. The long-term goal of our research is to determine whether post-exposure surveillance using biomarkers can be used to assess individual environmental exposures and predict future health risks.
Our study originated with United States Department of Defense (DoD) interest in understanding the health risks posed to service personnel deployed to areas that used open burn-pits to dispose of waste and trash [1]. Burn pits have been used to dispose of all solid waste materials, including medical wastes, plastics, metals, rubber, paints, solvents, munitions, and wood. Open burn pits have been gradually phased out in favor of controlled incineration but are still used today in some countries where service personnel are deployed. Burn pits create downwind exposures to airborne pollutants including particulate matter, polyaromatic hydrocarbons (PAHs) such as naphthalene and anthracene, and polychlorinated dibenzo-p-dioxins and polychlorinated furans [2]. These exposures vary with wind direction, season, and waste segregation policies [3–5]. Of necessity, environmental monitoring is a much lower priority than operational readiness and force protection. Static area monitors may not be established in the best locations or in adequate numbers. Personal breathing zone monitoring is limited or non-existent, due to problems such as concerns about service member mobility and agility with the added weight and bulk of the monitoring equipment; the difficulty of keeping batteries charged and sample filters changed in the field, and the logistical problem of collecting and analyzing thousands of daily samples [6]. Nevertheless, the DoD has a strong interest in maintaining the health and readiness of service personnel and in understanding how exposures to hazardous environments may impact the long-term health of veterans.
Some recent epidemiological studies have investigated the potential contribution of deployment related environmental exposures to post deployment chronic illnesses, including chronic respiratory conditions, among service members and veterans [7–11]. These studies compared deployed with non-deployed personnel, or between personnel at different locations, and they produced findings ranging from no association between deployment and disease to evidence of increased symptoms and specific lung conditions. A recent study reported that deployment to Afghanistan or Iraq was associated with an elevated risk of post deployment respiratory symptoms and new onset of asthma [12]. However, because of the previously described problems with environmental monitoring, acquisition of disease symptoms after deployment cannot be associated with any particular environmental exposure.
The same concerns apply to anyone who receives a poorly-defined occupational or non-occupational exposure to a potential hazard. Large scale industrial contamination events, such as Love Canal, and smaller-scale releases such as rail car derailments or industrial accidents, can expose dozens to thousands of people to potential hazards. Epidemiological studies can assess broad population-based risks [13], but have little to no ability to determine individual exposures or estimate future health risks currently exists.
Here, we describe a robust computational model for distinguishing exposed and unexposed individuals when exposure history is unknown and environmental samples have not been collected. The computational model takes the measurements of analytes from serum sample that can identify individuals as exposed to air pollution, and here we develop the model for a well-characterized inhaled toxicant, tobacco smoke. This is a proof of concept that we can take serum analytes when environmental exposures are unknown and predict what the exposures were, using the serum biomarkers. Further, we can prioritize the biomarkers contributing to the exposure mediated health status. Chronic tobacco smoke is associated with biomarkers of exposure and effect related to cancers; cardiovascular disease; lung disease including chronic obstructive pulmonary disease, emphysema, and chronic bronchitis; peripheral vascular disease; and many other chronic conditions [14, 15]. Several prior studies have identified serum biomarkers of smoking, primarily inflammatory cytokines and chemokines [16–18], and also metabolic deficits [19–21]. Tobacco use can be accurately assessed in de-identified serum samples by measuring cotinine levels, a nicotine metabolite with a longer half-life than nicotine [22, 23].
We applied our computational model to discriminate a broad range of panel consisting of 236 serum analytes between smokers and non-smokers using direct measurement of serum cotinine when smoking history is unknown. We identified a biological signature of tobacco use within the biomarker panel. The biomarkers with highest classification weights corresponded to cytokines and miRNAs that are previously known, or newly identified here, to be associated with smoking. These results suggest that our model may also be able to classify subjects as exposed or not exposed to other environmental hazards, and that the significant features used for classification may represent novel biomarkers of exposure to those environmental hazards.
Materials and Methods
Ethical Considerations
This study used serum samples that were de-identified and approved for use by Institutional Review Boards at the Uniformed Services University and the University of Rochester.
Study Design
The overall study population and design has been described in detail elsewhere [1, 24, 25]. Briefly, we obtained serum samples from the DoD Serum Repository for 400 active duty service personnel originally drawn between 2006 and 2012. The 800 serum sample size was determined in the original study using a power of 0.8 and alpha of 0.1. Two serum samples were obtained for each subject, drawn about one year apart (median, 422 days; IQR, 369–480 days). For this study investigating biomarkers related to tobacco use, the samples were considered as individual data points. From 400 donors and 800 serum samples, 25 samples were excluded from analysis for missing values for at least one data-type. We also obtained basic demographic information (age and sex) for each donor.
Biomarker determination
The serum samples were processed for several classes of biomarkers. RNA was extracted, and quantitative PCR was used to determine the levels of 144 human miRNAs most commonly found in serum. The methods and initial results are fully described elsewhere [25]. Metabolomics analysis was used to determine the levels of 59 metabolites, mainly related to nutritional and health status, and this has also been reported previously [26]. Benzo(a)pyrene diol epoxide (BPDE)-protein adducts were measured by ELISA (OxiSelect™ BPDE Protein Adduct ELISA Kit, Cell Biolabs Inc., San Diego, CA). Serum levels of 25 polychlorinated dibenzo-p-dioxins and dibenzofurans were quantified [1, 2, 24]. Several of the dioxins and furans had low measurement. Hence, we performed Principal Co-ordinate Analysis (PCoA), Nonlinear Multi-dimensional Scaling (NMDS) and principal component analysis (PCA) to obtain a metric combining critical dioxin and furans. These methods produced similar results and hence the first principal component (PC1) was used in subsequent analyses. Specifically, PC1 explained over 90% of the total variance in dioxins and furans and was used in regression and SVM modeling. Serum IgE was measured by ELISA (Bethyl Laboratories, Montgomery, TX) according to the manufacturer’s directions. A panel of 21 cytokines and chemokines associated with inflammation and B-and T-cell activation were measured by multiple assay (Luminex, Austin, TX) as previously described [25]. A panel of nine acute phase proteins and cytokines related to cardiovascular risk (α2 macroglobulin, C-reactive protein, Fetuin A, L-selectin, Serum amyloid A, Platelet factor 4, Adipsin, vW Factor, and Haptoglobin) were also measured by multiplex assay. As per the Center of Disease Control (CDC) guidelines of active smokers, cotinine was measured with a commercial EIA kit (Calbiotech, Spring Valley, CA). Cotinine values ≥10 ng/ml were defined as smokers and values <1 ng/ml were defined as non-smokers. Values between 1 and 10 ng/ml are consistent with exposure to secondhand smoke (N=8 of 775 samples) and were grouped with the nonsmokers for this analysis. The final analysis included 236 individual biomarkers, plus the PC1 for dioxins and furans. See Supplementary Table 1 for a complete list of the biomarkers included in this analysis.
Data processing:
Correcting for Confounding Factors with Multiple Regression:
Age and sex are known confounding variables that could affect the development of the classifier [27–30]. We used linear regression with ordinary least squares regression models [31, 32] to correct for the effects of confounding variables. Briefly, the ordinary least squares regression method estimates the unknown parameters in a linear regression model by minimizing the sum of the squares of the differences between the observed responses in the given dataset and those predicted by a linear function of a set of explanatory variables [31, 32]. Specifically, we developed ordinary least squares regression models using 775 samples for each of the 236 molecular profiles using age and sex as covariates. For each model, we computed regression coefficient, standard error, t-statistics, and p-values for both age and sex along with ordinary and adjusted R square, mean square error, F-statistics, residuals, and the significance of the overall model. The residuals of the models were used for SVM classification (Supplementary table 2). Correcting for age and sex quantifies their effects on all molecular profiles that can possibly distort the association between molecular profiles and cotinine levels.
Machine Learning Models for Classification:
We used 236 molecular profiles from the 775 samples as an input to the machine learning models. All the features were standardized (zero mean and unit variance) before inputting to the model. Class labels were assigned to each subject based on their cotinine levels (cotinine ≥10 ng/mL = smoker, cotinine <10 ng/mL = non-smoker). A comparative analysis of several statistical and machine learning models was performed. Specifically, 12 different statistical and machine learning methods were used to classify smokers and non-smokers based on the molecular profiles. These models include random forest, k-nearest neighbors, artificial neural networks, linear discriminant analysis, multilayer perceptron, decision trees, logistic regression, naïve Bayes, linear and non-linear support vector machines, and their modifications (see Supplementary Table 3 for details). 80% data was used for training and validation, and remaining 20% for the independent testing of the models. Additionally, for training set, we used repeated 10-fold cross validation to evaluate the model performance. The training samples were randomly partitioned into 10 equal size subsamples. Of the 10 subsamples, a single subsample was retained as the validation data for testing the model, and the remaining 9 subsamples were used as training set for the models. This cross-validation process was then repeated 10 times, with each subsample used exactly once as the validation dataset. The results from the 10-fold validation subsamples were then averaged to produce a single estimation for overall validation-set accuracy of the models. The models were further evaluated on the remaining 20% independent held out set to produce an estimation of test-set performance. The performance of these 12 models were compared using several performance indicators such as accuracy, area under the curve, kappa statistics, sensitivity, specificity, and the generalization ability. The computational analyses were conducted in RStudio version 1.0.143 and Matlab 2016a environment.
Improving the SVM Classifier with Recursive Feature Elimination:
To minimize over fitting the training data and to increase the ability to generalize to unseen data, selecting the most informative features is critical [33, 34]. We used the wrapper method to identify the most informative features by removing the redundant features and less informative features [34, 35]. Specifically, the recursive feature elimination (RFE)[36] technique was used with support vector machine as a classifier. The SVM with RFE approach has been extensively and successfully used for the biomarker selection lately [37–42].
Briefly, at the initial stage, the SVM was trained using all the features. The predictive features were ranked based on the absolute weight assigned to them. At each subsequent stage, the feature with least absolute weight was removed. This process was repeated in a recursive manner, and in each iteration, the efficiency of the classifier was measured by the 10-fold cross validation (explained above). SVM recursive feature elimination identified a small sub-set of optimal features that had better predictive power than using all of the features.
Animal model of cigarette smoke exposure
We used an animal model of cigarette smoke exposure to validate some of the novel miRNA biomarkers identified in the human serum samples. Animal experiments were approved by the University of Rochester University Committee on Animal Research (UCAR) under protocol number 2007–127. Groups of C57BL/6 mice were exposed to air or mainstream tobacco smoke for one hour, twice per day, with a one hour break between exposures, five days per week for five weeks, as previously described [43, 44]. At the end of the exposure protocol, the mice were euthanized with pentobarbital and the lungs were removed for processing. Total RNA was prepared from the right lung of each mouse by the Trizol method as previously described [45]. RNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, Delaware). MiRNA cDNA was generated from 100 ng of total RNA per sample using the TaqMan microRNA reverse transcription kit (Thermo Scientific) following the manufacturer’s instructions. MiRNA qPCR was run using the TaqMan Universal PCR master mix (Thermo Scientific). The specific miRNAs analyzed with corresponding TaqMan Primers sets (Applied Biosystems) were let-7a (4427975 “000377”), let-7g (4427975 “002282”), miR-93–5p (4427975 “001090”), miR-29a (4427975 “002112”) and U6 (4395470 “001973). To analyze changes in miRNA expression, miRNA expression was normalized to expression of U6 snRNA.
Results
Data summary
Of the initial 800 samples with 236 diverse molecular profiles, 25 samples with missing values were removed and the remaining 775 samples were used in subsequent analyses. There were 631 (81%) male and 144 (19%) female subjects. The minimum age in the study population was 18 years, the maximum age was 53 years, and the median age was 26 years (Table 1). The computational model included 21 cytokines, nine cardiovascular markers, 144 miRNAs, 59 metabolites, along with IgE, BPDE-protein adducts, and 25 dioxins (see Materials and Methods for details). The computational pipeline included a linear support vector machine classifier with a recursive feature elimination heuristic (Figure 1). Principal component was performed on the dioxins and furans measurements and retained only the first principal component (PC1) to use in our subsequent analysis. PC1 explained over 90% of the total variance, suggesting that it is a good representation of the variation in dioxin and furan concentrations (Figure 2).
Table 1. Demographic Characteristics of the Study Cohort.
| Counts (Percentage) | |
|---|---|
| Age | |
| 17–19 | 5 (1) |
| 20–29 | 243 (61) |
| 30–39 | 104 (26) |
| 40+ | 48 (12) |
| Gender | |
| Male | 325 (81) |
| Female | 75 (19) |
| Race/Ethnicity | |
| White | 254 (63) |
| Black | 89 (23) |
| Other | 57 (14) |
| Rank | |
| E1-E4 | 154 (39) |
| E5-E9 | 189 (47) |
| O1-O3 | 40 (10) |
| O4-O9 | 17 (4) |
| Service | |
| Army | 226 (57) |
| Coast Guard | 1 (0) |
| Air Force | 68 (17) |
| Marine Corps | 24 (6) |
| Navy | 81 (20) |
Figure 1.
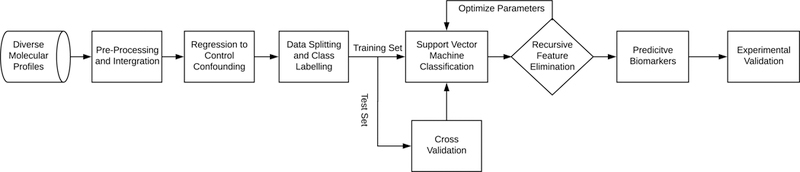
Flow diagram of the steps involved in development of our computational model.
Figure 2.
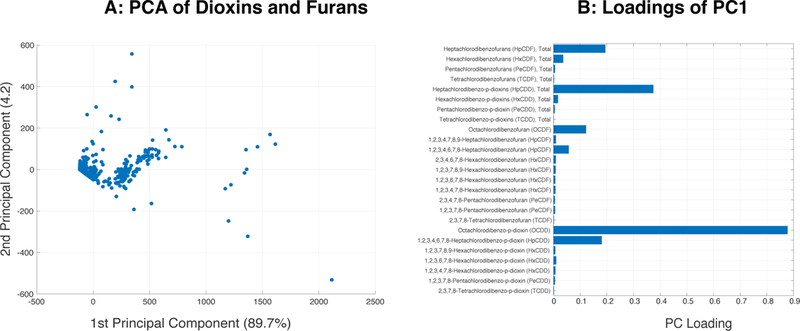
Principal component analysis of the polychlorinated dibenzo-p-dioxins and dibenzofuran data. (A) Graph of the first two principle components. (B) Loading values for individual dioxins and furans in principal component 1. The most heavily weighted dioxins and furans (total heptachlorodibenzofurans, total heptachlorodibenzo-p-dioxins, and octachlorodibenzo-p-dioxin), correspond to the most frequently detected species as previously reported [1, 25].
Biomarkers associated with age and sex
We performed regression analysis for age and sex to determine the impact of these known confounding variables on the levels of molecular profiles. Age was significantly associated with 66 features, of which 46 (70%) were miRNAs (Supplementary Table 2). This result is in broad agreement with other studies showing significant differences in serum miRNAs with age. Let-7a-5p had the strongest association with age in our population (p=). Several miRNAs including let-7a-5p, miR-126–3p, miR-142–3p, miR-30b-5-and miR-30c-5p, have been previously reported to be associated with age [46, 47]. Eotaxin was the only cytokine associated with age among the 22 cytokines and chemokines tested (p=0.0002) and has been independently reported to be associated with aging [48].
Sex was significantly associated with 48 features, with the largest categories being metabolites (24, 50%) and miRNAs (16, 33%) (Supplementary Table 2). Relatively few miRNAs associated with sex in this study overlapped those associated in a previous survey [47], although miR-145–5p, miR-146b-5p and miR-19a-3p were in common.
Machine leaning model for predicting smokers and non-smokers from molecular profiles
Each of the 775 samples was assigned a class label (smoker or non-smoker) based on serum cotinine < 10 ng/ml (non-smoker) or ≥ 10 ng/ml (smoker). Based on this criterion, there were 296 smokers and 479 non-smokers. Principal component analysis (PCA) was performed to investigate grouping among smokers and non-smokers based on their miRNA, cytokine, CVD markers and metabolites separately. None of the PCAs showed obvious separation between smokers and non-smokers (Supplementary Figure 1). To classify smokers and non-smokers we evaluated 12 different statistical and machine learning methods including random forest, k-nearest neighbors, artificial neural networks, linear discriminant analysis, multilayer perceptron, decision trees, logistic regression, naïve Bayes, linear and non-linear support vector machines were used. We preferred the above models over simpler (conventional statistical) models because of the complex nature of association between the biomarkers. There existed multicollinearity among the predictor variables (such as most of 144 miRNAs are correlated). Many statistical tests assume that observations are independent leading biased estimates. Lately, a large number of published studies provide significant evidence that the machine learning models are better capturing such complex relationships compared to the simpler models. The model comparison was performed using 80% data for training the models and remaining 20% for independent testing of the models. For evaluating the robustness of the trained models, 10-fold cross validation and held-out accuracies were analyzed (additional details are in methods sections). For all 12 models the repeated 10-fold cross validation performance between 59–67% on 80% training set and between 57%−70% on 20% independent test set (Supplementary Table 3) was observed. The model comparison revealed following methods: SVM with class weights, logistic regression, neural networks, naïve Bayes and multilayer perceptron, had a balance between sensitivity and specificity. We selected linear SVM with class weights which made the feature selection considerable more feasible compared to the other methods [49] for further studies to classify smokers and non-smokers. Briefly, Linear SVM model optimizes a hyperplane that separates the smoker and non-smoker classes as best as possible. The hyperplane is represented by the coordinates of a vector (feature weights) which is orthogonal to the hyperplane. For any data point, if the dot product with the vector is positive, it belongs to the positive class, if it is negative it belongs to the negative class. Moreover, the absolute value of the feature weights relative to the other ones gives an indication of how important the feature was for the separation compared to all other features [50].
As 94 of 236 features had significant associations with age, sex, or both, we used regression models to obtain age-and sex-corrected molecular profiles as inputs to the SVM classifier. The 10-fold cross validation accuracy with adjusted molecular profiles on the training set was 62.65% (ROC-AUC 0.62) and the accuracy on independent held out set was 59.1% with a sensitivity of 52.24% and a specificity of 62.11% (see Supplemental Table 4). We also obtained the relative absolute weights of the features from the trained SVM model. The top five biomarkers with the highest absolute weights assigned by the model were indoleacrylic acid, tryptophan, oxoproline, IL-1β, and creatinine (see Supplementary Table 4 for complete details). These weights provide a relative indication of their importance in the decision making process of the classifier.
Improving the model with recursive feature elimination
In order to improve predictive accuracy and find a smaller set of robust biomarkers that can be tested experimentally, we employed recursive feature elimination by selecting a subset of input features that maximized the performance and generalizability of the model. In scenarios where using all features in the model lead to a biased (overfitted) model, the SVM-RFE approach has a potential to pick a subset of relevant features that can produce a generalized and robust model [38, 41]. Briefly, at the initial stage, the SVM was trained using all the 236 features. Predictive features were ranked based on the absolute weight assigned to them. At each subsequent stage, the feature with least weight was removed. This process was performed in recursive manner, and in each iteration, the efficiency of the classifier was measured by 10-fold cross validation. Specifically, we ran SVM-RFE analysis using all the available data and measured the model performance through 10-fold cross validation (as explained in Methods). The accuracy of the SVM model improved with recursive feature elimination (Figure 3). The highest achieved 10-fold cross validation accuracy was 78% with ROC-AUC of 0.80 and with 87 features (roughly one third of the original features). The smallest feature set that achieved ≥75% training and cross-validation accuracy had 45 features (Figure 4A and Supplementary table 5). For this feature set, the training accuracy was 78% (ROC-AUC=0.82) and the 10-fold cross validation accuracy was 75% (ROC-AUC=0.78). In order to make sure the results from the subset of 45 features are generalized and robust we performed additional experiments. We split the data into 80% as training set and 20% as independent held out set. We performed the repeated 10-fold cross validation training set as discussed in the earlier section but this time with only 45 features. The mean 10-fold cross-validation performance on the training set was 71.2% (ROC-AUC=0.75). The SVM performance on the independent held-out set was 78.6% (with a sensitivity of 72.9% and a specificity of 82.1%). (Supplementary Table S6). The performance of linear SVM model with 45 features was superior to all other 11 models used for the comparative analysis in this study. Random forest was the second best model with an accuracy of 73.38% (CI: 65.7%, 80.2%). The ability of the 45 feature set to separate smokers and non-smokers using weighted PCA is shown in Figure 4B. These 45 features represent a biological signature of tobacco use in our study cohort. This feature set includes 3 members of the tryptophan biosynthesis pathway, 16 other metabolites, 7 inflammatory cytokines, 2 cardiovascular markers, and 17 miRNAs. The pattern suggests a general disruption in the regulation of inflammation and energy metabolism consistent with other studies of tobacco smokers. Specific results are discussed in detail below.
Figure 3.
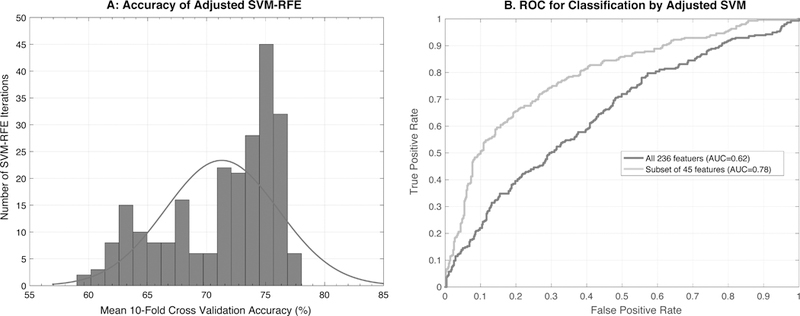
Performance of SVM with recursive feature elimination. (A) Frequency of recursive feature eliminations (y-axis) resulting into mean 10-fold cross-validation accuracy (x-axis). (B) ROC curve (receiver operating characteristic curve) showing performance of a classification with 45 selected features compared to all 236 features shows improvement in the AUC (area under the curve).
Figure 4.
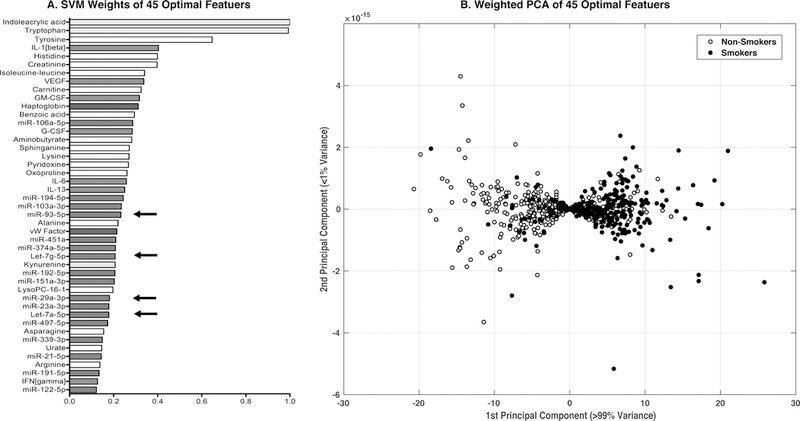
Features identified by SVM with recursive feature elimination to classify subjects into smokers and non-smokers. Serum biomarkers were analyzed by SVM as described. (A) The smallest feature set to achieve ≥75% training and cross-validation accuracy is shown, with the relative predictive weights. Features of interest are highlighted in red (miRNAs), green (cytokines), blue (cardiovascular markers) and yellow (the tryptophan pathway). Arrows indicate miRNAs selected for further validation. (B) Weighted principal component analysis of the 775 subjects using the 45 feature set. Blue dots represent non-smokers and red dots represent smokers. Using the SVM weights, the PCA is able to classify the subjects by smoking status.
Comparison between adjusted and un-adjusted models
To evaluate the impact of the age and sex adjustments on the outcome, we also trained an SVM classifier using unadjusted data and compared it with the age-and sex-adjusted SVM model (described above). The accuracies of both the models were comparable when using the full 236 feature set (Supplementary Tables 7). The 10-fold cross validation accuracy of an unadjusted model was 60% (ROC-AUC=0.61) whereas that of the age-sex adjusted model was 63% (ROC-AUC=0.62) respectively. Using SVM-RFE approach, we noticed the improvement in the model performance. The smallest feature set in the unadjusted model that exceeded 75% training and cross validation accuracy consisted of 50 features, compared to 45 features with the age-sex adjusted model (Supplementary Tables 5 and 8). Among these two optimal feature sets, 36 were common to both models, suggesting that the unadjusted and age-sex adjusted models were both able to identify the most significant predictive features of tobacco use, although the weights assigned to these features were somewhat different.
Biological validation of miRNA results.
Many of the cytokine and metabolic markers identified by the SVM as classifying smokers from non-smokers have previously been reported to have associations with cigarette smoking, as discussed below. MicroRNAs represent a relatively novel class of biomarker and much fewer data have been published on the relationships between specific miRNAs and environmental exposures. Since miRNAs are also highly conserved among mammalian species [51], we next queried a mouse model of cigarette smoke exposure to determine if the miRNAs that were classifiers of smoking in our study population were also targeted by cigarette smoke in the mouse model. We selected 4 miRNAs that contributed to classifying tobacco exposure in the SVM results based on absolute weights, and their relevance in literature in deployed military service personnel [24]. The expression of the four miRNAs, miR-29a, miR-93, let-7a, and let-7g, was measured in mouse lung tissue from mice that had been exposed sub-chronically to mainstream cigarette smoke. Expression of miR-93 decreased with smoke exposure while let-7g increased, but these results were not significant. Excitingly, however, cigarette smoke significantly altered expression of both miR-29a and let-7a (Figure 5), consistent with their contribution to the classification of smokers among service personnel. SVM weights do not provide insights into the direction of change. The fact that the selected 4 miRNAs showed differences in mouse models upon smoke exposure encourages further evaluation of these markers in human and mice serum with larger sample sizes. Other miRNAs from the 45 features will also be investigated in future.
Figure 5.
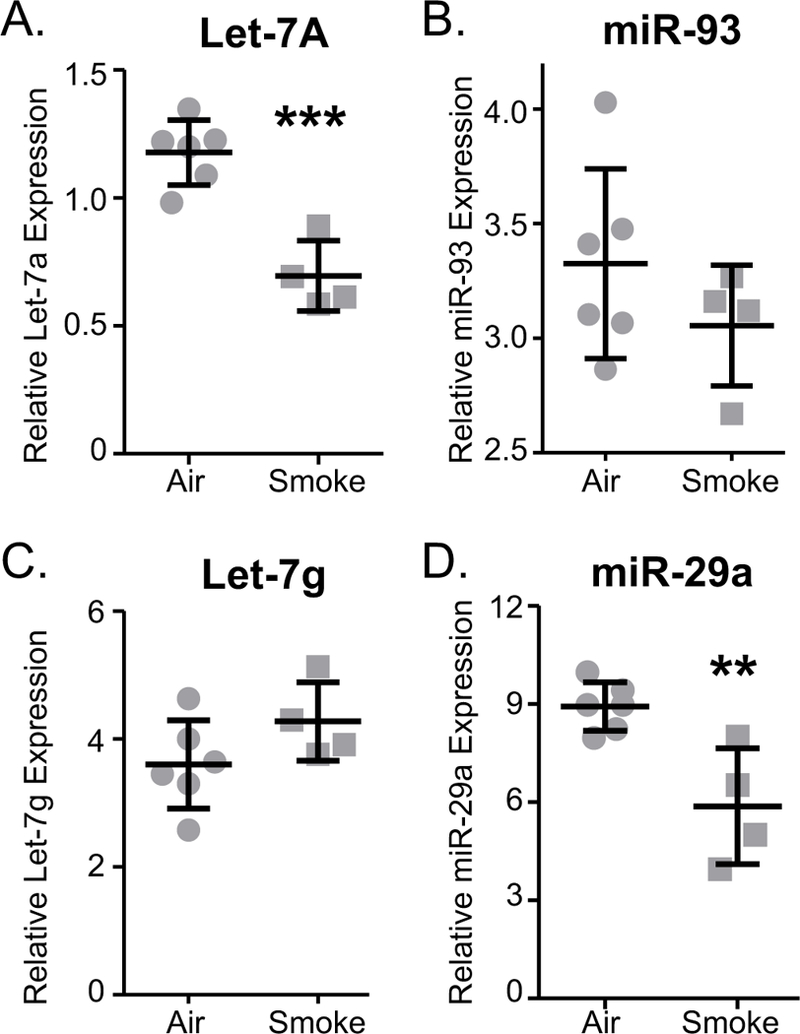
Tobacco smoke exposure alters miRNA levels in mouse lungs. Groups of mice were exposed to tobacco smoke as described in the Methods. Total RNA was prepared from lung tissue and levels of miRNAs (A) let-7a, (B) miR-93, (C) let-7g, and (D) miR-29a were determined by RT-PCR. N=4–6 samples per group. **P≤0.01, ***P≤0.0001 by t-test.
Discussion
Military service personnel deployed to operations in Iraq and Afghanistan between 2006 and 2012 were exposed to multiple poorly-defined environmental hazards including sand and dust storms, endemic organisms, air pollution from aviation and motor pool operations, and most notably, air pollution from open burn pits (summarized in [1]). Past experience with Agent Orange and Gulf War Illness has sensitized the DoD to the need to be able to determine individual exposures, with the aim of understanding risks to future health based on those exposures. The present study is part of an ongoing project to investigate whether it is possible to identify exposure to burn pits and other deployment hazards using intensive biomarker measurements in pre-and post-deployment serum samples. The study design is reviewed in depth elsewhere [1]. Here, our goal was to develop a machine learning method that could use serum biomarkers to classify subjects by exposure groups and identify a biological signature of exposure to an environmental hazard. The biomarkers that distinguish exposed from non-exposed subjects represent biological processes that have been impacted by the environmental exposure and may be useful in understanding and predicting immediate and future health impacts.
To validate this model, we took advantage of the fact that information on subjects’ tobacco use can be obtained independently from the serum samples by measuring cotinine, a metabolite of nicotine with a longer serum half-life than nicotine [22, 23]. Cotinine was used as a discrete variable since CDC has clear definition of active smoker (cotinine >=10 ng/ml). If the model classified subjects by smoking status with high confidence, we felt that the model would also be able to classify subjects by exposure to other environmental toxins, such as those encountered during deployment to areas using open burn pits. Using 236 biomarkers, the SVM-RFE model achieved a peak cross-validation accuracy of 78% with 87 biomarkers. For a more manageable number of features, we found that the smallest feature set to achieve both ≥75% cross-validation and independent testing accuracy used only 45 biomarkers.
Our primary model was age-and sex-adjusted, after we found multiple strong associations of selected biomarkers with age and sex (Supplementary Table 3) consistent with prior reports [46–48]. However, it is interesting that the results of an unadjusted model were qualitatively similar. The most highly weighted features in each model were similar, although the absolute weights differed. The smallest feature set using the unadjusted model with ≥75% training and cross-validation accuracy contained 50 features (Supplementary Table 7), comparable to the corresponding set from the adjusted model (45 features, Supplementary Table 5), and 36 of the features were common to both models. This finding implies that tobacco smoking, as an example of toxic environmental exposure, has overall greater biological impact on the biomarkers we measured than either age or sex. Nevertheless, we believe that age and sex adjustments will be important for future studies, especially if the biological effects of the injury or exposure under study are subtler.
The choice of using machine learning technique was done to limit bias due to multicollinearity among the predictor variables (such as most of 144 miRNAs are correlated). Many statistical tests assume that observations are independent. The application of these tests to correlated observations could lead to the over or under estimation of P-values in certain cases that may produce biased estimates. We believe machine learning algorithm are better at unraveling the complex hidden relationships in the feature space. Comparison with different statistical machine learning techniques reveals that the simpler linear SVM model provided comparable accuracy and a better sensitivity. Moreover, unlike some of the other methods, feature selection with SVM was feasible. Additionally, weighted SVM allowed us to address slight imbalance in the class with 296 smokers and 479 non-smokers. Particularly, class weights penalize the misclassification cost for the minority class (smokers in our case) instead of giving equal weight to the both classes. Since the prior probability of being non-smoker was 0.62 and that of being smoker was 0.38, we designed the cost matrix based on these prior class probabilities. The model, therefore, avoided being skewed towards the majority class and produced generalized results. We demonstrated a 20.2% improvement (from 58.4% to 78.6%) in the model performance by identifying and training machine learning model on a subset of more relevant and informative features. However, though SVM can reveal a set of features with potentially complex relationships, biological validation of this multivariate association is a multistep process. Here we begin to investigate selected features one by one and in future they will be studied in depth.
To understand if the model identified classifiers with real biological importance, we turned to the substantial body of literature on the impact of tobacco smoking on human health. We expected that the model would identify cytokines and other biomarkers that have been previously shown to be associated with smoking using independent subject cohorts, and this result is indeed what we found. In the cardiovascular panel, haptoglobin and vWF were predictive factors for tobacco use, consistent with previous reports [46, 52–54]. Similarly, creatinine has a reported association with smoking [55, 56]. Among cytokines, smoking has previously been reported to elevate serum levels of IL-6, IL-1β, VEGF and IL-13 [57–62]. Two miRNAs that were recently reported to be associated with smoking, miR-106a-5p and miR-374a-5p, [63–67] are also highly weighted classifiers in our model.
Since comparatively little information exists on serum miRNAs relative to other biomarkers, we selected four miRNAs for further validation. These miRNAs, miR-29a, miR-93, let-7a, and let-7g, were highly weighted classifiers of smoking in both the age-and sex-adjusted model and the unadjusted model. They were also associated with exposure to polyaromatic hydrocarbons or polychlorinated dibenzo-p-dioxins and dibenzofurans, which are toxic products of burn pit combustion [24, 25]. Since many miRNAs are highly conserved across mammalian species [51], we quantified these miRNAs in lung tissue of mice exposed to mainstream tobacco smoke for 5 weeks. The exposure is roughly equivalent to a 1–1/2 to 2 pack per day habit in humans. It gives good acute, sub-chronic, and chronic results, including inflammation and emphysematous changes, and has been used for pre-clinical animal testing of several potential anti-inflammatory therapies to reduce acute inflammation and chronic airspace changes. Furthermore, the described experiments from lung samples is a first step in a multistep follow-up study. Lungs are the most likely source of miRNAs since they are directly affected by smoking hence lung RNA was reasonable site to evaluate for mechanistic understanding. Among the four miRNAs selected by SVM classifier model, two miRNAs (let-7g and miR-93) were affected by cigarette smoke exposure, although not significantly. However, the other two (let-7a and miR-29a) were significantly reduced with cigarette smoke exposure (Figure 5). Taken together with other published miRNA and cytokine data, this result provides reasonable confirmation that the SVM model has identified classifiers that have significant and biologically relevant associations with tobacco use. Further experiments are required to completely understand the mechanism of miRNA regulation.
In addition to classifying current smoking status, the SVM model identified features that may indicate links between exposures and future health risk. MiRNAs can regulate hundreds of genes and impact multiple disease pathways, can be regulated epigenetically, and can exhibit dysregulation long after the original toxic exposure. Additionally, miRNAs can persist in serum because their short length and sequestration in protein complexes, lipocomplexes, exosomes, or microparticles making them resistant to ribonucleases [68, 69]. An important characteristic of the smoking classifiers identified by our model is that many of them have been implicated in promoting inflammation and disease. Strong evidence indicates that smokers exhibit chronic low-level inflammation that is a significant risk factor for multiple diseases associated with smoking including cardiovascular and cerebrovascular disease, diabetes, and cancer [70–72]. For example, mir-106a has recently been shown to be involved in TH17 cell differentiation [73]. Expression of miR-106a decreased production of IL-17a, which led to a loss of differentiation and function of T helper cells. TH17 cell mediated inflammation is also associated with lung diseases such as COPD and asthma. Let-7a and miR-29a, both important classifiers identified here and independently validated in our mouse model of smoking, regulate inflammatory pathways in the cell. Here, we show lower expression of let-7a and miR-29a in our smoking cohorts, and this outcome is also in line with previous reports showing lower levels of let-7a and miR-29a in lung cancer [74, 75]. Let-7a, part of the larger let-7 family of miRNAs, regulates proliferation, glucose metabolism, and cell differentiation and is often thought of as a tumor suppressor [76]. One key target of let-7a is HMGA1 (High-mobility group protein HMG-I/HMG-Y), a chromatin binding protein that plays a role in numerous cancers [77]. Interestingly, elevated levels of HMGA1 (that would be consistent with lower levels of the targeting miRNA, let-7a) are observed in lung cancer [78]. Mir-29a, like let-7a, is a tumor suppressor miRNA and targets the mRNA for CDC42, a key mediator cell cycle progression [79]. Thus, many of the cytokines and miRNAs that are altered in subjects with elevated cotinine, and therefore act as classifiers of smoking, have associations with known diseases caused by smoking, including inflammation and cancer.
The overall goal of this study was to develop a machine learning model that is generalizable to any environmental exposure, such as occupational exposures, industrial accidents, and military exposures to burn pits during deployment; tobacco smoking is simply a well-defined environmental insult we could use to confirm that the model works. Exposure hazards may not be recognized until well after the exposure (e.g. Love Canal and similar contamination events, the use of Agent Orange, exposure to burn pits in forward operating areas), and the exact nature of the hazard and traditional exposure assessment methods may be inadequate to identify people at risk of future harm. The ability to classify individuals by exposure will allow identification of persons who might benefit from enhanced medical surveillance, while the biomarkers that differentiate exposed from non-exposed subjects identify affected biological pathways that may be associated with future disease risk. Of course, that ability will be dependent on the strength of the exposure and the degree of biological impact. It will likely be more difficult to detect signatures of more subtle exposures. The effect of smoking itself as a co-factor in future studies also must be considered. As this study demonstrates, tobacco use has a strong biological signature independent of age or sex. Although tobacco use rates are declining in the United States, approximately 15% of Americans over age 18 smoke cigarettes [80]. The rate is higher among the military (38% in our cohort, sampled between 2006 and 2012) and over 40% in some regions of the world [81]. Self-reported smoking status tracks lower than independent measures such as serum cotinine [23]. As a result, future studies using serum biomarkers to evaluate environmental exposures should correct not only for age and sex, but for tobacco exposure as determined by an unbiased quantitative method.
The method is generalizable to the study of other environmental exposures where post-exposure serum samples may be obtained in lieu of personal monitoring. Identifying those individuals in an at-risk population who were actually exposed would allow for more cost-effective medical surveillance focusing on those exposed individuals. Future studies will examine other exposure scenarios including dioxins and polyaromatic hydrocarbons, as part of the larger overall study plan. Since there were also strong effects of age, sex and smoking status, corrections for these factors should be included in future models of environmental exposures.
Supplementary Material
Principal component analysis (PCA) to investigate grouping based on smoking status in the (A) miRNA, (B) cytokine, (C) cardiovascular markers and (D) metabolites. None of the groups showed obvious separation between smokers (red dots) and non-smokers (blue dots).
Acknowledgments:
This work was supported in part by The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. grant number HT9404-13-1-0030, the National Institute of Environmental Health Sciences Grant # P30-ES01247 and NIH training grant # T32HL066988.
Footnotes
Conflict of Interest: none declared.
Ethics statement
This study used serum samples that were de-identified at various levels and was approved by Institutional Review Boards at the Uniformed Services University and the University of Rochester. Animal experiments were approved by the University of Rochester University Committee on Animal Research (UCAR) under protocol number 2007-127.
Data, code and materials
Code developed for this manuscript is available at https://github.com/Thakar-Lab/ML_Exposure_Prediction. All primary data analysis and the results can be found in Supplementary tables S1–S8.
References
- [1].Mallon CT, Rohrbeck MP, Haines MK, Jones DP, Utell M, Hopke PK, Phipps RP, Walker DI, Thatcher T, Woeller CF, et al. 2016. Introduction to Department of Defense Research on Burn Pits, Biomarkers, and Health Outcomes Related to Deployment in Iraq and Afghanistan. J Occup Environ Med 58, S3–S11. (doi: 10.1097/JOM.0000000000000775). [DOI] [PubMed] [Google Scholar]
- [2].Xia X, Carroll-Haddad A, Brown N, Utell MJ, Mallon CT & Hopke PK 2016. Polycyclic Aromatic Hydrocarbons and Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans in Microliter Samples of Human Serum as Exposure Indicators. J Occup Environ Med 58, S72–79. (doi: 10.1097/JOM.0000000000000743). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Masiol M, Mallon CT, Haines KM Jr., Utell MJ & Hopke PK 2016. Airborne Dioxins, Furans, and Polycyclic Aromatic Hydrocarbons Exposure to Military Personnel in Iraq. J Occup Environ Med 58, S22–30. (doi: 10.1097/JOM.0000000000000771). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Masiol M, Mallon CT, Haines KM Jr., Utell MJ & Hopke PK 2016. Source Apportionment of Airborne Dioxins, Furans, and Polycyclic Aromatic Hydrocarbons at a United States Forward Operating Air Base During the Iraq War. J Occup Environ Med 58, S31–37. (doi: 10.1097/JOM.0000000000000759). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Blasch KW, Kolivosky JE & Heller JM 2016. Environmental Air Sampling Near Burn Pit and Incinerator Operations at Bagram Airfield, Afghanistan. J Occup Environ Med 58, S38–43. (doi: 10.1097/JOM.0000000000000792). [DOI] [PubMed] [Google Scholar]
- [6].Report C f.R.o.t.D.s.E.P.M.S.P. 2010. Review of the Department of Defense Enhanced Particulate Matter Surveillance Program Report. Washinton, DC, National Academies Press. [PubMed] [Google Scholar]
- [7].Falvo MJ, Osinubi OY, Sotolongo AM & Helmer DA 2015. Airborne hazards exposure and respiratory health of Iraq and Afghanistan veterans. Epidemiol Rev 37, 116–130. (doi: 10.1093/epirev/mxu009). [DOI] [PubMed] [Google Scholar]
- [8].Liu J, Lezama N, Gasper J, Kawata J, Morley S, Helmer D & Ciminera P 2016. Burn Pit Emissions Exposure and Respiratory and Cardiovascular Conditions Among Airborne Hazards and Open Burn Pit Registry Participants. J Occup Environ Med 58, e249–255. (doi: 10.1097/JOM.0000000000000776). [DOI] [PubMed] [Google Scholar]
- [9].Rohrbeck P, Hu Z & Mallon CT 2016. Assessing Health Outcomes After Environmental Exposures Associated With Open Pit Burning in Deployed US Service Members. J Occup Environ Med 58, S104–110. (doi: 10.1097/JOM.0000000000000802). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. 2017In Assessment of the Department of Veterans Affairs Airborne Hazards and Open Burn Pit Registry (eds. D.A. Butler, A.N. Styka & D.A. Savitz. Washington (DC). [PubMed] [Google Scholar]
- [11].Smith B, Wong CA, Boyko EJ, Phillips CJ, Gackstetter GD, Ryan MA, Smith TC & Millennium Cohort Study T 2012. The effects of exposure to documented open-air burn pits on respiratory health among deployers of the Millennium Cohort Study. J Occup Environ Med 54, 708–716. (doi: 10.1097/JOM.0b013e31825107f9). [DOI] [PubMed] [Google Scholar]
- [12].Falvo MJ, Abraham JH, Osinubi OY, Klein JC, Sotolongo AM, Ndirangu D, Patrick-DeLuca LA & Helmer DA 2016. Bronchodilator Responsiveness and Airflow Limitation Are Associated With Deployment Length in Iraq and Afghanistan Veterans. J Occup Environ Med 58, 325–328. (doi: 10.1097/JOM.0000000000000675). [DOI] [PubMed] [Google Scholar]
- [13].Coughlin SS, Kang HK & Mahan CM 2011. Selected Health Conditions Among Overweight, Obese, and Non-Obese Veterans of the 1991 Gulf War: Results from a Survey Conducted in 2003–2005. Open Epidemiol J 4, 140–146. (doi: 10.2174/1874297101104010140). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yanbaeva DG, Dentener MA, Creutzberg EC, Wesseling G & Wouters EF 2007. Systemic effects of smoking. Chest 131, 1557–1566. (doi: 10.1378/chest.06-2179). [DOI] [PubMed] [Google Scholar]
- [15].Services., U.S.D.o.H.a.H. 2014. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. . [Google Scholar]
- [16].Cockayne DA, Cheng DT, Waschki B, Sridhar S, Ravindran P, Hilton H, Kourteva G, Bitter H, Pillai SG, Visvanathan S, et al. 2012. Systemic biomarkers of neutrophilic inflammation, tissue injury and repair in COPD patients with differing levels of disease severity. PloS one 7, e38629. (doi: 10.1371/journal.pone.0038629). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bade G, Khan MA, Srivastava AK, Khare P, Solaiappan KK, Guleria R, Palaniyar N & Talwar A 2014. Serum cytokine profiling and enrichment analysis reveal the involvement of immunological and inflammatory pathways in stable patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 9, 759–773. (doi: 10.2147/COPD.S61347). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Golpe R, Martin-Robles I, Sanjuan-Lopez P, Perez-de-Llano L, Gonzalez-Juanatey C, Lopez-Campos JL & Arellano-Orden E 2017. Differences in systemic inflammation between cigarette and biomass smoke-induced COPD. Int J Chron Obstruct Pulmon Dis 12, 2639–2646. (doi: 10.2147/COPD.S141068). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jones DP, Walker DI, Uppal K, Rohrbeck P, Mallon CT & Go YM 2016. Metabolic Pathways and Networks Associated With Tobacco Use in Military Personnel. J Occup Environ Med 58, S111–116. (doi: 10.1097/JOM.0000000000000763). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang Y, Hui Q, Walker DI, Uppal K, Goldberg J, Jones DP, Vaccarino V & Sun YV 2018. Untargeted metabolomics reveals multiple metabolites influencing smoking-related DNA methylation. Epigenomics 10, 379–393. (doi: 10.2217/epi-2017-0101). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gu F, Derkach A, Freedman ND, Landi MT, Albanes D, Weinstein SJ, Mondul AM, Matthews CE, Guertin KA, Xiao Q, et al. 2016. Cigarette smoking behaviour and blood metabolomics. Int J Epidemiol 45, 1421–1432. (doi: 10.1093/ije/dyv330). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benowitz NL, Bernert JT, Caraballo RS, Holiday DB & Wang J 2009. Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am J Epidemiol 169, 236–248. (doi: 10.1093/aje/kwn301). [DOI] [PubMed] [Google Scholar]
- [23].Max W, Sung HY & Shi Y 2009. Who is exposed to secondhand smoke? Self-reported and serum cotinine measured exposure in the U.S., 1999–2006. Int J Environ Res Public Health 6, 1633–1648. (doi: 10.3390/ijerph6051633). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Woeller CF, Thatcher TH, Van Twisk D, Pollock SJ, Croasdell A, Hopke PK, Xia X, Thakar J, Sime PJ, Mallon TM, et al. 2016. MicroRNAs as Novel Biomarkers of Deployment Status and Exposure to Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans. J Occup Environ Med 58, S89–96. (doi: 10.1097/JOM.0000000000000769). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Woeller CF, Thatcher TH, Van Twisk D, Pollock SJ, Croasdell A, Kim N, Hopke PK, Xia X, Thakar J, Mallon CT, et al. 2016. Detection of Serum microRNAs From Department of Defense Serum Repository: Correlation With Cotinine, Cytokine, and Polycyclic Aromatic Hydrocarbon Levels. J Occup Environ Med 58, S62–71. (doi: 10.1097/JOM.0000000000000742). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Accardi CJ, Walker DI, Uppal K, Quyyumi AA, Rohrbeck P, Pennell KD, Mallon CT & Jones DP 2016. High-Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. J Occup Environ Med 58, S80–88. (doi: 10.1097/JOM.0000000000000770). [DOI] [PubMed] [Google Scholar]
- [27].Brookhart MA, Sturmer T, Glynn RJ, Rassen J & Schneeweiss S 2010. Confounding control in healthcare database research: challenges and potential approaches. Med Care 48, S114–120. (doi: 10.1097/MLR.0b013e3181dbebe3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Greenland S & Morgenstern H 2001. Confounding in health research. Annu Rev Public Health 22, 189–212. (doi: 10.1146/annurev.publhealth.22.1.189). [DOI] [PubMed] [Google Scholar]
- [29].Blair A, Stewart P, Lubin JH & Forastiere F 2007. Methodological issues regarding confounding and exposure misclassification in epidemiological studies of occupational exposures. Am J Ind Med 50, 199–207. (doi: 10.1002/ajim.20281). [DOI] [PubMed] [Google Scholar]
- [30].Skelly AC, Dettori JR & Brodt ED 2012. Assessing bias: the importance of considering confounding. Evid Based Spine Care J 3, 9–12. (doi: 10.1055/s-0031-1298595). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Craven BD & Islam SMN 2011. Ordinary least-squares regression. SAGE Dict Quant Manag Res, 224–228. [Google Scholar]
- [32].Kutner MH, Nachtsheim CJ, Neter J & Li W 1996. Applied Linear Statistical Models. . 5. [Google Scholar]
- [33].Kojadinovic I, Wottka T, Remia I & Réunion UD 2000. Comparison between a filter and a wrapper approach to variable subset selection in regression problems. Proc ESIT, 14–15. [Google Scholar]
- [34].Guyon I & Elisseeff A 2003. An Introduction to Variable and Feature Selection. .J Mach Learn Research 3(3), 1157–1182. [Google Scholar]
- [35].Kohavi R & John GH 1997. Wrappers for feature subset selection. . Artif Intell. 97(1–2), 273–324. [Google Scholar]
- [36].Guyon I, Weston J, Barnhill S & Vapnik V 2002. Gene selection for cancer classification using support vector machines. Mach Learn. 46(1–3), 389–422. [Google Scholar]
- [37].Guyon I, Weston J, Barnhill S & Vapnik V 2002. Gene selection for cancer classification using support vector machines. Mach Learn 46, 389–422. (doi:Doi 10.1023/A:1012487302797). [DOI] [Google Scholar]
- [38].Duan KB, Rajapakse JC, Wang HY & Azuaje F 2005. Multiple SVM-RFE for gene selection in cancer classification with expression data. Ieee T Nanobiosci 4, 228–234. (doi: 10.1109/Tnb.2005.853657). [DOI] [PubMed] [Google Scholar]
- [39].Maldonado S & Weber R 2009. A wrapper method for feature selection using Support Vector Machines. Inform Sciences 179, 2208–2217. (doi: 10.1016/j.ins.2009.02.014). [DOI] [Google Scholar]
- [40].Zhang X, Lu X, Shi Q, Xu XQ, Leung HC, Harris LN, Iglehart JD, Miron A, Liu JS & Wong WH 2006. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC bioinformatics 7, 197. (doi: 10.1186/1471-2105-7-197). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou X & Tuck DP 2007. MSVM-RFE: extensions of SVM-RFE for multiclass gene selection on DNA microarray data Bioinformatics (Oxford, England: ) 23, 1106–1114. (doi: 10.1093/bioinformatics/btm036). [DOI] [PubMed] [Google Scholar]
- [42].Guan W, Zhou M, Hampton CY, Benigno BB, Walker LD, Gray A, McDonald JF & Fernandez FM 2009. Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC bioinformatics 10, 259. (doi: 10.1186/1471-2105-10-259). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hsiao HM, Thatcher TH, Colas RA, Serhan CN, Phipps RP & Sime PJ 2015. Resolvin D1 Reduces Emphysema and Chronic Inflammation. The American journal of pathology 185, 3189–3201. (doi: 10.1016/j.ajpath.2015.08.008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thatcher TH, McHugh NA, Egan RW, Chapman RW, Hey JA, Turner CK, Redonnet MR, Seweryniak KE, Sime PJ & Phipps RP 2005. Role of CXCR2 in cigarette smoke-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol 289, L322–328. (doi: 10.1152/ajplung.00039.2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hsiao HM, Sapinoro RE, Thatcher TH, Croasdell A, Levy EP, Fulton RA, Olsen KC, Pollock SJ, Serhan CN, Phipps RP, et al. 2013. A novel anti-inflammatory and pro-resolving role for resolvin D1 in acute cigarette smoke-induced lung inflammation. PloS one 8, e58258. (doi: 10.1371/journal.pone.0058258). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Burgess KS, Philips S, Benson EA, Desta Z, Gaedigk A, Gaedigk R, Segar MW, Liu Y & Skaar TC 2015. Age-Related Changes in MicroRNA Expression and Pharmacogenes in Human Liver. Clin Pharmacol Ther 98, 205–215. (doi: 10.1002/cpt.145). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ameling S, Kacprowski T, Chilukoti RK, Malsch C, Liebscher V, Suhre K, Pietzner M, Friedrich N, Homuth G, Hammer E, et al. 2015. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC medical genomics 8, 61. (doi: 10.1186/s12920-015-0136-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, et al. 2011. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94. (doi: 10.1038/nature10357). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cortes C & Vapnik V 1995. Support-Vector Networks. . Mach Learn. 20 (3), 273–297. [Google Scholar]
- [50].Burges CJC 1998. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min Knowl Discov. 2, 121–167. (doi:doi: 10.1023/A:1009715923555). [DOI] [Google Scholar]
- [51].Agarwal V, Bell GW, Nam JW & Bartel DP 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. (doi: 10.7554/eLife.05005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bartholo TP, Costa CH & Rufino R 2014. Evaluation of von Willebrand factor in COPD patients. J Bras Pneumol 40, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lavie L & Lavie P 2008. Smoking interacts with sleep apnea to increase cardiovascular risk. Sleep Med 9, 247–253. (doi: 10.1016/j.sleep.2007.03.018). [DOI] [PubMed] [Google Scholar]
- [54].Wolf GT, Chretien PB, Weiss JF, Edwards BK & Spiegel HE 1982. Effects of smoking and age on serum levels of immune reactive proteins. Otolaryngol Head Neck Surg 90, 319–326. [PubMed] [Google Scholar]
- [55].Chuahirun T & Wesson DE 2002. Cigarette smoking predicts faster progression of type 2 established diabetic nephropathy despite ACE inhibition. Am J Kidney Dis 39, 376–382. (doi: 10.1053/ajkd.2002.30559). [DOI] [PubMed] [Google Scholar]
- [56].El Housseini Y, Phan O, Burnier M & Vogt B 2009. [Smoking and the kidney]. Rev Med Suisse 5, 457–458, 460–452. [PubMed] [Google Scholar]
- [57].Santos S, Peinado VI, Ramirez J, Morales-Blanhir J, Bastos R, Roca J, Rodriguez-Roisin R & Barbera JA 2003. Enhanced expression of vascular endothelial growth factor in pulmonary arteries of smokers and patients with moderate chronic obstructive pulmonary disease. Am J Respir Crit Care Med 167, 1250–1256. (doi: 10.1164/rccm.200210-1233OC). [DOI] [PubMed] [Google Scholar]
- [58].Koyama S, Sato E, Haniuda M, Numanami H, Nagai S & Izumi T 2002. Decreased level of vascular endothelial growth factor in bronchoalveolar lavage fluid of normal smokers and patients with pulmonary fibrosis. Am J Respir Crit Care Med 166, 382–385. (doi: 10.1164/rccm.2103112). [DOI] [PubMed] [Google Scholar]
- [59].Suzuki M, Betsuyaku T, Nagai K, Fuke S, Nasuhara Y, Kaga K, Kondo S, Hamamura I, Hata J, Takahashi H, et al. 2008. Decreased airway expression of vascular endothelial growth factor in cigarette smoke-induced emphysema in mice and COPD patients. Inhal Toxicol 20, 349–359. (doi: 10.1080/08958370701866412). [DOI] [PubMed] [Google Scholar]
- [60].Conklin BS, Zhao W, Zhong DS & Chen C 2002. Nicotine and cotinine up-regulate vascular endothelial growth factor expression in endothelial cells. The American journal of pathology 160, 413–418. (doi: 10.1016/S0002-9440(10)64859-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Barbieri SS, Zacchi E, Amadio P, Gianellini S, Mussoni L, Weksler BB & Tremoli E 2011. Cytokines present in smokers’ serum interact with smoke components to enhance endothelial dysfunction. Cardiovasc Res 90, 475–483. (doi: 10.1093/cvr/cvr032). [DOI] [PubMed] [Google Scholar]
- [62].Aldaham S, Foote JA, Chow HH & Hakim IA 2015. Smoking Status Effect on Inflammatory Markers in a Randomized Trial of Current and Former Heavy Smokers. Int J Inflam 2015, 439396. (doi: 10.1155/2015/439396). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mullany LE, Herrick JS, Wolff RK, Stevens JR & Slattery ML 2016. Association of cigarette smoking and microRNA expression in rectal cancer: Insight into tumor phenotype. Cancer Epidemiol 45, 98–107. (doi: 10.1016/j.canep.2016.10.011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Zhang X, Bowers J, et al. 2009. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America 106, 2319–2324. (doi: 10.1073/pnas.0806383106). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Russ R & Slack FJ 2012. Cigarette-Smoke-Induced Dysregulation of MicroRNA Expression and Its Role in Lung Carcinogenesis. Pulm Med 2012, 791234. (doi: 10.1155/2012/791234). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Takahashi K, Yokota S, Tatsumi N, Fukami T, Yokoi T & Nakajima M 2013. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol 272, 154–160. (doi: 10.1016/j.taap.2013.05.018). [DOI] [PubMed] [Google Scholar]
- [67].Graff JW, Powers LS, Dickson AM, Kim J, Reisetter AC, Hassan IH, Kremens K, Gross TJ, Wilson ME & Monick MM 2012. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PloS one 7, e44066. (doi: 10.1371/journal.pone.0044066). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kumar S, Vijayan M, Bhatti JS & Reddy PH 2017. MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases. Prog Mol Biol Transl Sci 146, 47–94. (doi: 10.1016/bs.pmbts.2016.12.013). [DOI] [PubMed] [Google Scholar]
- [69].Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I, Langley SR, et al. 2013. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res 112, 595–600. (doi: 10.1161/CIRCRESAHA.111.300539). [DOI] [PubMed] [Google Scholar]
- [70].Vanderstocken G, Dvorkin-Gheva A, Shen P, Brandsma CA, Obeidat M, Bosse Y, Hassell JA & Stampfli MR 2018. Identification of Drug Candidates to Suppress Cigarette Smoke-induced Inflammation via Connectivity Map Analyses. American journal of respiratory cell and molecular biology 58, 727–735. (doi: 10.1165/rcmb.2017-0202OC). [DOI] [PubMed] [Google Scholar]
- [71].Cosio BG, Jahn A, Iglesias A, Shafiek H, Busquets X & Agusti A 2015. Haemophilus influenzae induces steroid-resistant inflammatory responses in COPD. BMC Pulm Med 15, 157. (doi: 10.1186/s12890-015-0155-3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bhat TA, Kalathil SG, Bogner PN, Miller A, Lehmann PV, Thatcher TH, Phipps RP, Sime PJ & Thanavala Y 2018. Secondhand Smoke Induces Inflammation and Impairs Immunity to Respiratory Infections. J Immunol 200, 2927–2940. (doi: 10.4049/jimmunol.1701417). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kastle M, Bartel S, Geillinger-Kastle K, Irmler M, Beckers J, Ryffel B, Eickelberg O & Krauss-Etschmann S 2017. microRNA cluster 106a~363 is involved in T helper 17 cell differentiation. Immunology 152, 402–413. (doi: 10.1111/imm.12775). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Fassina A, Cappellesso R & Fassan M 2011. Classification of non-small cell lung carcinoma in transthoracic needle specimens using microRNA expression profiling. Chest 140, 1305–1311. (doi: 10.1378/chest.11-0708). [DOI] [PubMed] [Google Scholar]
- [75].Liu X, Lv X, Yang Q, Jin H, Zhou W & Fan Q 2018. MicroRNA-29a Functions as a Tumor Suppressor and Increases Cisplatin Sensitivity by Targeting NRAS in Lung Cancer. Technol Cancer Res Treat 17, 1533033818758905. (doi: 10.1177/1533033818758905). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Triboulet R, Pirouz M & Gregory RI 2015. A Single Let-7 MicroRNA Bypasses LIN28-Mediated Repression. Cell reports 13, 260–266. (doi: 10.1016/j.celrep.2015.08.086). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Andreozzi M, Quintavalle C, Benz D, Quagliata L, Matter M, Calabrese D, Tosti N, Ruiz C, Trapani F, Tornillo L, et al. 2016. HMGA1 Expression in Human Hepatocellular Carcinoma Correlates with Poor Prognosis and Promotes Tumor Growth and Migration in in vitro Models. Neoplasia 18, 724–731. (doi: 10.1016/j.neo.2016.10.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang Z, Wang Q, Chen F & Liu J 2015. Elevated expression of HMGA1 correlates with the malignant status and prognosis of non-small cell lung cancer. Tumour Biol 36, 1213–1219. (doi: 10.1007/s13277-014-2749-4). [DOI] [PubMed] [Google Scholar]
- [79].Li Y, Wang Z, Li Y & Jing R 2017. MicroRNA-29a functions as a potential tumor suppressor through directly targeting CDC42 in non-small cell lung cancer. Oncol Lett 13, 3896–3904. (doi: 10.3892/ol.2017.5888). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jamal A, Phillips E, Gentzke AS, Homa DM, Babb SD, King BA & Neff LJ 2018. Current Cigarette Smoking Among Adults — United States, 2016. CDC Morbidity and Mortality Weekly Report (MMWR). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].WHO. http://www.who.int/tobacco/publications/surveillance/reportontrendstobaccosmoking/en/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal component analysis (PCA) to investigate grouping based on smoking status in the (A) miRNA, (B) cytokine, (C) cardiovascular markers and (D) metabolites. None of the groups showed obvious separation between smokers (red dots) and non-smokers (blue dots).


