SUMMARY
Follicular regulatory T (TFR) cells are a specialized suppressive subset that controls the germinal center (GC) response and maintains humoral self-tolerance. The mechanisms that maintain TFR lineage identity and suppressive activity remain largely unknown. Here, we show that expression of Blimp1 by FoxP3+ TFR cells is essential for TFR lineage stability, entry into the GC, and expression of regulatory activity. Deletion of Blimp1 in TFR cells reduced FoxP3 and CTLA-4 expression and increased pro-inflammatory cytokines and spontaneous production of autoantibodies, including elevated IgE. Maintenance of TFR stability reflected Blimp1-dependent repression of the IL-23R-STAT3 axis and activation of the CD25-STAT5 pathway, while silenced IL-23R-STAT3 or increased STAT5 activation rescued the Blimp1-deficient TFR phenotype. Blimp1-dependent control of CXCR5/CCR7 expression also regulated TFR homing into the GC. These findings uncover a Blimp1-dependent TFR checkpoint that enforces suppressive activity and acts as a gatekeeper of GC entry.
In Brief
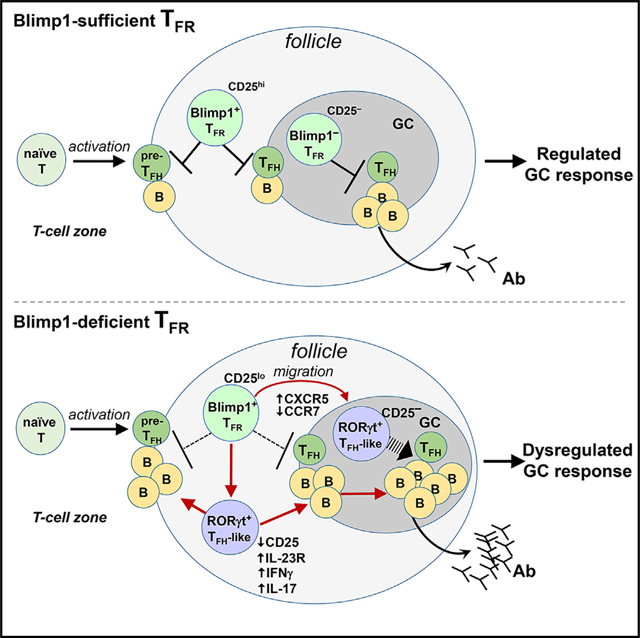
Wang et al. identify Blimp1 as a critical transcription factor for the proper positioning and stable expression of the suppressive activity of TFR cells that control GC responses. In the absence of Blimp1, unstable TFR cells prematurely migrate into the GC and differentiate into TFH-like cells to promote dysregulated GC responses.
Graphical Abstract
INTRODUCTION
Germinal centers (GCs) are specialized dynamic structures that provide a unique niche for B cells to generate high-affinity antibody (Ab) responses to microbial pathogens after infection or vaccination. The GC response takes place in the context of substantial cell death and apoptosis, which provides a potential arsenal of self-antigens that may activate autoreactive Ab responses. Under these circumstances, the induction of cognate GC B cells by follicular helper T cells (TFH) may result in excessive Ab responses that include autoantibodies to self-tissues (Crotty, 2011, 2014). Since dysregulated GC responses may be at the root of an array of systemic autoimmune diseases (Crotty, 2011, 2014; Leavenworth et al., 2013, 2015), insight into mechanisms that control these responses is essential.
There is abundant evidence that immune responses and self-tolerance are stringently controlled by FoxP3+ regulatory T cells (Treg). FoxP3+ Treg are composed of a central Treg (cTreg) component and several tissue-specific sublineages of effector Treg (eTreg), including the recently defined subset of follicular regulatory T cells (TFR) that regulate GC responses through interactions with activated TFH and GC B cells (Chung et al., 2011; Leavenworth et al., 2015; Linterman et al., 2011; Sage and Sharpe, 2015; Smigiel et al., 2014). TFR cells share several features with TFH cells, including the expression of ICOS, PD-1, and CXCR5 receptors that contribute to TFR differentiation and follicular localization (Chung et al., 2011; Linterman et al., 2011; Wing et al., 2017). TFR cells also co-opt the expression of Bcl6, the cardinal transcription factor (TF) that guides follicular CD4+ T cell differentiation (Chung et al., 2011; Leavenworth et al., 2015; Linterman et al., 2011). The differentiation of Treg precursors into TFR cells is associated with signs of cellular activation and the upregulation of genes expressed by eTreg, including GITR, CTLA-4, ICOS, KLRG1, and the Blimp1 TF (Linterman et al., 2011). Although it is likely that strong T cell receptor (TCR) signals favor TFR cell differentiation (Kallies et al., 2006; Linterman et al., 2011), the mechanisms that ensure the maintenance of lineage identity and expression of regulatory activity by TFR are not well defined.
TFR cells, like other eTreg, express the Blimp1 TF (Cretney et al., 2011; Linterman et al., 2011; Vasanthakumar et al., 2015). Recent analyses suggest that Blimp1 may not make a significant contribution to TFR differentiation and may even have a negative impact on the TFR response. This view is supported by findings that Blimp1 expression may reduce TFR expansion and development (Botta et al., 2017; Linterman et al., 2011), and that the downregulation of Blimp1 expression is associated with the acquisition of TFR effector activity and navigation into the GC (Wing et al., 2017).
Here, we report that Blimp1 expression is essential to maintain TFR lineage stability, appropriate positioning in the GC, and effective regulatory activity. Blimp1 regulates CTLA-4 expression and signals transmitted by interleukin (IL)-23R and CD25 to maintain the TFR phenotype. The upregulation of IL-23R by Blimp1-deficient TFR resulted in enhanced STAT3 signaling, diminished FoxP3 expression, and impaired regulatory activity. Blimp1-deficient TFR cells displayed reduced CTLA-4 expression and acquired an effector T cell phenotype and expression of IL-4, which was accompanied by high levels of immunoglobulin E (IgE) and serum autoantibodies. Blimp1-dependent control of the CXCR5-CCR7 axis was also essential for the correct positioning of TFR within the GC. These findings suggest that the expression of Blimp1 in TFR is essential for differentiation into functional TFR with a stable phenotype.
RESULTS
FoxP3-Specific Deletion of Blimp1 Leads to Dysregulated GC Responses
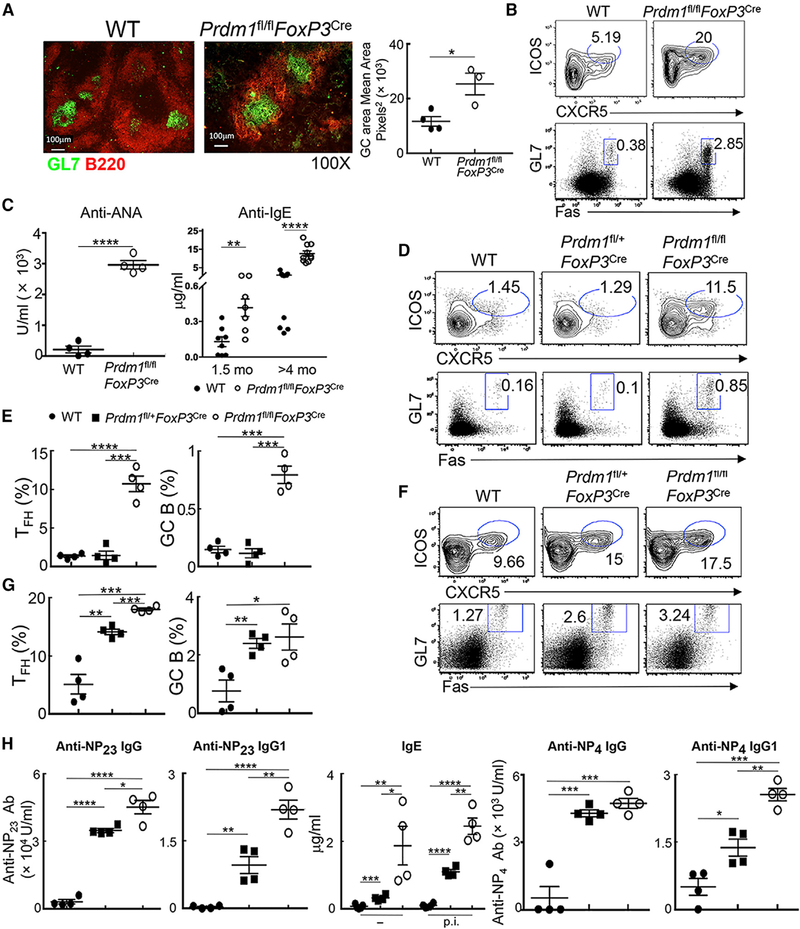
To investigate the contribution of Blimp1 to the differentiation and regulatory function of FoxP3+ TFR, we generated mice in which Prdm1 alleles were deleted in FoxP3+ T cells (Prdm1fl/flFoxP3Cre mice) and confirmed Blimp1 deficiency in TFR cells (Figures S1A and S1B). We noted a 3-fold increase in the GC area at 4–5 months of age, a 6- to 8-fold increase in the frequency of TFH cells (CD4+CD3+ICOShiCXCR5+FoxP3−), and a 5- to 10-fold increase in B220+GL-7+Fas+GC B cells compared with FoxP3Cre (wild-type [WT]) controls (Figures 1A and 1B). This TFH-GC expansion was associated with high titers of anti-nuclear autoantibodies (ANAs) and IgE levels beginning at 1.5 months and increasing with age (Figure 1C). We also observed an increased frequency and number of TFH and GC B cells and (CD4+FoxP3−CD44hiCD62Llo) effector T cells in Prdm1fl/flFoxP3Cre (knockout [KO]) mice compared with WT controls in the steady state at an early age (6 weeks old) (Figures 1D, 1E, and S1C).
Figure 1. FoxP3-Specific Deletion of Blimp1 Leads to Dysregulated GC Responses.
(A) Histology of splenic GCs, and quantification of GL7+ GC areas from 4- to 5-month-old FoxP3Cre (WT) and Prdm1fl/flFoxP3Cre (KO) mice (n = 3–4/group).
(B) Flow cytometry of splenic TFH (CD4+CD3+ICOShiCXCR5+FoxP3−) and GC B (B220+GL7+Fas+) from 4- to 5-month-old mice.
(C) (Left) Serum ANA levels from 4- to 5-month-old mice (n = 4/group). (Right) Serum IgE levels from 1.5- and >4-month-old WT and KO mice (n = 7–11/group).
(D–G) Flow cytometry and frequency of splenic TFH and GC B from 6-week-old mice before (D and E) or after (F and G) immunization with NP-KLH in complete Freund’s adjuvant (CFA) (n = 4/group).
(H) Serum anti-NP23 Ig, anti-NP4 Ig, IgG1, and IgE levels from 6-week-old mice after immunization with NP-KLH in CFA (n = 4/group).
p.i., post immunization; −, before immunization. For (B)–(H), the data represent one of four independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S1.
Blimp1 is expressed by eTreg subsets located in both lymphoid and non-lymphoid tissues (e.g., intestine, skin, fat, possibly lung), and may be essential for IL-10 production (Cretney et al., 2013). However, examination of non-lymphoid tissues (skin, adipose tissue, lung) from 4– 5-month-old Prdm1fl/fl FoxP3Cre mice failed to detect histological abnormalities, with the exception of mild to moderate inflammation in the colon (Figure S1D), which is consistent with diminished eTreg IL-10 production, as noted by others (Cretney et al., 2011; Kallies et al., 2006; Martins et al., 2006). In view of a recent study that IL-10 production by TFR cells promotes the GC response (Laidlaw et al., 2017), the above findings, along with the observations of increased numbers and sizes of GCs, suggested that the primary contribution of Blimp1 expression by Treg may entail an IL-10-independent regulation of humoral self-tolerance, perhaps by FoxP3+ TFR cells.
Prdm1fl/flFoxP3Cre Mice Develop Dysregulated GC Responses after Antigen Challenge
To understand the contribution of Blimp1 to Treg-mediated control of Ab responses, we analyzed antigen-specific GC responses after immunizing 6-week-old mice with the hapten NP coupled to KLH (NP-KLH). One week after immunization, Prdm1fl/flFoxP3Cre mice displayed increased TFH and GC B cells along with CD4+ effector T cells (Figures 1F, 1G, and S1C) and markedly increased (>20-fold) anti-NP IgG titers (Figure 1H) compared to WT mice. The partial loss of Blimp1 expression in heterozygous Prdm1fl/+FoxP3Cre mice also resulted in increased numbers of TFH and GC B cells (Figures 1F and 1G) and elevated anti-NP titers after immunization (Figure 1H). The increased frequency of TFH and GC B cells that is apparent at 3 weeks after the primary immune response persisted 7 days after the secondary challenge (Figure S1E). Given the critical role of IL-21 and IL-4 cytokine-producing TFH in inducing IgE and IgG1 responses (Harada et al., 2012), we determined the numbers of IL-21/IL-4-producing TFH after immunization (Weinstein et al., 2016) and observed a significant increase in both the numbers of TFH and cytokine-producing TFH cells in KO mice at day 7 after immunization (Figure S1F). Thus, expression of Blimp1 by FoxP3+ T cells may be required to suppress the GC response, including “natural” IgE levels and Abs produced after immunization with foreign antigen.
Abnormal Treg and TFR Cell Homeostasis in Prdm1fl/flFoxP3Cre Mice
In view of the robust GC responses of Prdm1fl/flFoxP3Cre mice, we were surprised to note the increased numbers of FoxP3+ Treg in the steady state and after immunization (Figures 2A, 2B, and S2A), as well as in the 4- to 5-month-old mice (Figures S2B and S2C). Analysis of mixed bone marrow chimeras, generated by the reconstitution of Rag2−/− hosts with CD45.2+Prdm1fl/flFoxP3Cre and B6.CD45.1+ bone marrow cells, and as a control, a mixture of WT and B6.CD45.1+ cells, revealed that Blimp1 deficiency within the FoxP3+ lineage mainly affected eTreg but not cTreg (Figure S2D). We then analyzed eTreg subsets, TFR (CD4+CD3+PD-1+CXCR5+FoxP3+) and non-TFR (CD4+CD3+CXCR5−FoxP3+) cells. For unimmunized 7- to 9-week-old mice, although Blimp1 deficiency resulted in increased FoxP3+ TFR and non-TFR cells, the 14.5-fold TFR increase compared to that in WT mice was substantially greater than the 1.2-fold increase noted for total Treg and non-TFR Treg (1.8-fold) (Figure S2A). An analysis of 4- to 5-month-old unimmunized mice also showed that the numbers of Blimp1 KO TFR were dramatically increased compared to WT TFR (~8- to 10-fold in the spleen and mesenteric lymph node [mLN]), while the numbers of total Blimp1-deficient Treg were only slightly increased compared to WT Treg, with the exception of a significant increase in Blimp1-deficient non-TFR in mLNs (~4-fold) (Figures S2B and S2C). Immunization with 4-hydroxy-3-nitrophenylacetyl-ovalbumin (NP-OVA) also resulted in the substantial expansion of TFR cells from Prdm1fl/flFoxP3Cre mice compared to WT mice, while the expansion of non-TFR was modest in comparison (Figures 2A and 2B). Although FoxP3+ Treg and TFR cells from Prdm1fl/flFoxP3Cre mice expressed similar levels of Bcl2 (Figure S2E), they displayed increased Ki67 expression and diminished levels of annexin V, indicating a relatively high rate of proliferation and reduced apoptosis (Figure 2C). The heightened TFR proliferative response was apparently uncoupled to TFR activation, as judged by reduced expression of the CD69 marker of TFR (but not CXCR5− Treg) (Figure 2C). These results suggested that Blimp1 may normally limit the survival and expansion of FoxP3+ eTreg cells in vivo.
Figure 2. Expanded TFR Cells Express an Abnormal Phenotype and Altered GC Distribution after Immunization of Prdm1fl/flFoxP3Cre Mice.
(A and B) Flow cytometry (left) and numbers (right) of splenic (A) or mLN (B) Treg (FoxP3+CD4+) (upper), TFR (CD4+CD3+PD-1+CXCR5+FoxP3+), and non-TFR (CD4+CD3+CXCR5−FoxP3+) (bottom) from WT and KO mice (7–9 weeks old) 10 days post-immunization with NP-KLH in CFA.
(C) Histogram of Ki67, annexin V, and CD69 expression in mLN TFR cells in (B) (upper) and quantification of mean fluorescence intensity (MFI) in mLN TFR and CXCR5− Treg cells (bottom).
(D) Expression of TFR-associated gene products by splenic TFR from (A) and quantification of MFI (right).
(E) IL-17A, IFNγ, and IL-4 expression (left) and frequency (right) by splenic TFR (CD4+CD3+PD-1+BTLA+FoxP3+) cells from WT or KO mice (6–8 weeks old) at day 7 post-immunization with NP-OVA in CFA.
(F) Frequency of CD25lo (left), CD25int (center), and CD25hi (right) splenic TFR from WT and KO mice (6–8 weeks old) 10 days post-immunization with NP-OVA in CFA. (Bottom) FoxP3 and CTLA-4 expression in the CD25-expressing TFR subsets.
(G) MFI of FoxP3, CTLA-4, GITR, and RORγt expression and frequencies of IL-17A+ cells in YFP+ Blimp1-deficient (KO) compared to YFP− Blimp1-sufficient (WT) TFR cells from the spleens of female Prdm1fl/flFoxP3Cre/+ mice (6–8 weeks old) 10 days post-immunization with NP-OVA in CFA.
(H and I) CXCR5 (H) and CCR7 (I) expression in the splenic TFR cells in (A) and mLN TFR cells in (B).
(J) Histology of 7-week-old WT and KO mice 14 days post-immunization with NP-OVA in CFA. TFR cells (CD3+FoxP3+, white arrows) and TFH cells (CD3+FoxP3−) in the GC area (Ki67+) within the B cell follicle (IgD+). Insets indicate the B cell follicles. (Bottom) Numbers of TFR and TFH cells in the GC.
In (A)–(I), the data are representative of four independent experiments (A–F, n = 4/group; G, n = 5/group; H and I, n = 4–5/group). In (J), the data are representative of two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S2.
Impaired Suppressive Phenotype of Treg and TFR Cells in Prdm1fl/flFoxP3Cre Mice
Examination of expanded FoxP3+ TFR and non-TFR conventional Treg in the spleen of unimmunized and immunized young Prdm1fl/flFoxP3Cre mice revealed reduced expression of FoxP3 and other Treg-associated molecules, including CTLA-4, compared to cells from WT mice (Figures 2D and S2F), suggesting a functionally impaired phenotype (Wing et al., 2014). We also noted that TFR cells but not non-TFR cells from unimmunized Prdm1fl/flFoxP3Cre mice produced substantially higher levels of interferon γ (IFNγ) compared to WT cells (Figure S2G). Moreover, TFR cells from immunized Prdm1fl/flFoxP3Cre mice produced increased IL-17A, IFNγ, and IL-4 pro-inflammatory cytokines (Figure 2E). These results suggested that Blimp1 deficiency resulted in the expansion of FoxP3lo Treg, particularly FoxP3lo TFR, that expressed reduced levels of Treg-associated receptors and increased levels of effector cytokines.
The expression of CD25 on TFR cells is downregulated as they mature and migrate into the GC, and these highly differentiated CD25lo/− TFR cells express low but significant levels of Blimp1 compared to their naive Treg precursors (Wing et al., 2017). Our finding that TFR cells from Prdm1fl/flFoxP3Cre mice displayed reduced CD25 expression (Figure 2D) prompted us to fully define the impact of Blimp1 deficiency on TFR cells that expressed progressively diminishing levels of CD25 (Figure S2H). Although all Blimp1-deficient CD25-expressing TFR expressed reduced levels of FoxP3 and CTLA-4, the highest level of FoxP3 was expressed by the CD25hi TFR subset (Figure 2F), suggesting that Blimp1 deficiency resulted in the expansion of TFR cells with an impaired suppressive phenotype, including a mature “GC” CD25lo TFR subset.
To examine the phenotype of Blimp1-deficient TFR cells under more physiological conditions, we analyzed female heterozygous Prdm1fl/flFoxP3Cre/+ mice. Due to the X-linked nature of the FoxP3Cre knockin transgene, these mice have both YFP+ Blimp1-deficient Treg and YFP− Blimp1-sufficient Treg. A comparison of the phenotype of YFP+ and YFP− TFR cells revealed that YFP+ Blimp1-deficient TFR cells express reduced levels of FoxP3, CTLA-4, and GITR, as well as increased levels of RORγt and IL-17A at day 10 after NP-OVA immunization (Figure 2G). These findings indicate that Blimp1-deficient TFR display an impaired suppressive phenotype and upregulated inflammatory cytokine production in a relatively non-inflammatory setting.
Altered GC Distribution of TFR Cells in Prdm1fl/flFoxP3Cre Mice
Further analysis of TFR-specific gene expression revealed that TFR cells from Prdm1fl/flFoxP3Cre mice expressed elevated levels of Bcl6 and CXCR5 (Figures 2D and 2H) (Linterman et al., 2011). The upregulation of CXCR5 along with diminished CCR7 expression in follicular T cells may be essential for navigation from the T cell zone into the GC (Crotty, 2011; Wing et al., 2017). Prdm1fl/flFoxP3Cre mice had fewer CCR7+ TFR and, in general, lower CCR7 levels compared to WT TFR (Figures 2I and S2I). Confocal analysis revealed a higher number of TFR and TFH cells within the GC of Prdm1fl/flFoxP3Cre mice compared to WT mice 2 weeks after NP-OVA immunization (Figure 2J). These results suggested that FoxP3-specific deletion of Blimp1 resulted in an enrichment of TFR cells in the GC that express a functionally impaired phenotype.
FoxP3-Specific Ablation of Blimp1 Impairs TFR Suppressive Activity after Adoptive Transfer
To determine whether Blimp1-deficient conventional (non-TFR) Treg may contribute to dysregulated GC responses, we compared Bcl6fl/flPrdm1fl/flFoxP3Cre to Bcl6fl/flFoxP3Cre mice that do not contain TFR cells (Figures 3A and 3B). While the latter strain contains an intact (Blimp1-sufficient) conventional Treg population, the former strain harbors only Blimp1-deficient Treg (Figures 3A and 3B). The frequency of TFH, GC B cells, and serum Ab titers were substantially reduced in Bcl6fl/flPrdm1fl/flFoxP3Cre mice (to levels similar to Bcl6fl/fl FoxP3Cre and WT mice) compared with Prdm1fl/flFoxP3Cre mice (Figures 3A and 3B), indicating that Blimp1-deficient conventional Treg do not contribute significantly to the increased frequency of TFH and GC B cells or the dysregulated Ab responses observed in Prdm1fl/flFoxP3Cre mice.
Figure 3. Blimp1 Deficiency Impairs TFR Suppressive Activity and Alters Gene Expression by TFR Cells.
(A and B) Splenic TFR, TFH, GC B, and expression of IL-17A in Treg cells from the indicated mouse strains (6–8 weeks old) 10 days post-immunization with NP-OVA in CFA (A). Frequencies of Treg, TFR, TFH, GC B cells, and IL-17A+ Treg cells, FoxP3 MFI, and total and high-affinity anti-NP IgG titers (B).
(C–G) (C) Schematic presentation of experimental protocol. CD45.2+ WT, KO, and CD45.1+ mice were immunized with NP-OVA in alum. Seven days later, sorted TFR (CD4+CD3+PD-1+CXCR5+YFP+) along with CD45.1+ TFH (CD4+CD3+PD-1+CXCR5+GITR−) and GL7− B cells (B220+GL7−) were transferred into Rag2−/− hosts followed by immunization with NP-OVA in alum before analysis (day 7).
(D) Fluorescence-activated cell sorting (FACS) profile (upper) and numbers (bottom) of TFR (CD45.2+CD4+CD3+PD-1+CXCR5+FoxP3+) and ex-TFR (CD45.2+CD4+CD3+CXCR5−) cells.
(E) Intracellular IL-17A and IFNγ expression by donor TFR cells.
(F) Histogram (upper) and MFI (bottom) of the indicated markers by donor CD45.2+ WT TFR cells (red) and CD45.2+ KO TFR cells (blue).
(G) Flow cytometry of donor CD45.1+ TFH (CD45.2−CD4+CD3+PD-1+CXCR5+) and GC B cells (CD19+GL7+) 7 days post-immunization.
(H) Tcra−/− mice were transferred with sorted TFR (WT versus KO) along with CD45.1+ TFH cells, followed by immunization with NP-OVA in CFA. Serum ANA, anti-NP23 IgG, IgG1, and IgE levels were analyzed 10 days post-immunization.
In (A) and (B), the data are pooled from two independent experiments (n = 5/group). In (C)–(H), the data are representative of three independent experiments (D and F, n = 4/group; H, n = 3–4/group). N.S., no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S2.
Analysis of purified TFR cells from Prdm1fl/flFoxP3Cre mice indicated that they failed to inhibit in vitro IgG production by mixtures of TFH and B cells compared to WT counterparts (Figures S2J and S2K). We then transferred purified TFR from CD45.2+ Blimp1-deficient or WT donors along with CD45.1+ TFH and B cells from NP-OVA-immunized mice into Rag2−/− hosts and challenged with NP-OVA (Figures 3C and S2L). After the transfer, TFR cells included a subpopulation of ex-TFR cells that were PD-1−CXCR5−, reflecting the dynamic state of TFR differentiation during the GC response (Wing et al., 2017) (Figure 3D). Transferred Blimp1-deficient TFR expressed increased levels of IFN-γ and IL-17A (Figure 3E), reduced levels of FoxP3, CD25, CTLA-4, and GITR (Figure 3F), and demonstrated impaired regulatory activity, as judged by increased numbers of TFH and GC B cells (Figure 3G), heightened IgG, IgG1, and IgE anti-NP responses, and elevated ANAs (Figure 3H). These changes resulted solely from the differences in Blimp1 expression in TFR cells, indicating that Blimp1 expression is essential for the suppressive activity expressed by isolated TFR.
Tamoxifen-Induced Blimp1 Deletion Impairs Lineage Stability and Functional TFR Differentiation
To define the precise stage at which Blimp1 affects TFR differentiation, we used an inducible Blimp1 deletion system to circumvent potential developmental defects secondary to inflammatory or other changes in the environment. We generated Prdm1fl/fliCre+ or Prdm1fl/fliCre− mice after crossing Prdm1fl/fl mice with Rosa26-Cre-ERT2 (referred to as iCre) transgenic mice to allow the conditional deletion of Blimp1 after the administration of tamoxifen. We treated Prdm1fl/fliCre+ mice and Prdm1fl/fliCre− control mice with tamoxifen 1 day before isolation of CD25hi Treg and co-transferred these cells along with naive CD4+ T cells into Tcra−/− hosts, followed by immunization with NP-OVA and injection of tamoxifen into these hosts for 3 more days (Figure S3A). Acute reduction of Blimp1 in Treg immediately before immunization resulted in increased numbers of TFR and CD138+ plasma cells in adoptive hosts (Figures S3B and S3C).
To determine whether continued Blimp1 expression was required for suppressive activity in differentiated TFR in the absence of other FoxP3+ eTreg, we isolated CD4+PD-1+CXCR5+GITR+ TFR cells (CD45.2+) from Prdm1fl/fliCre+ mice or Prdm1fl/flCre− control mice 6 days after NP-OVA immunization and 1 day after tamoxifen administration. We transferred these cells along with TFH cells from immunized CD45.1+ mice into Tcra−/− hosts before challenge with NP-OVA and injection of tamoxifen for 3 more days (Figure 4A). We observed substantially reduced Blimp1 and increased Bcl6 expression by TFR cells from Prdm1fl/fliCre+ mice (Figure 4B), along with an increased frequency of TFR, TFH, and GC B cells (Figure 4C). Defective suppressive activity of FoxP3lo TFR after acute Blimp1 depletion was accompanied by increased production of pro-inflammatory IL-17A and IFNγ cytokines (Figures 4D and 4E), suggesting that acute deletion of Blimp1 led to the differentiation of TFR into TH effector-like cells. TH1/TH17-like conversion was not apparent in Blimp1-deleted ex-TFR cells (PD1−CXCR5−) (Figures 4D and 4E). These results suggested that transferred Blimp1-deleted and control TFR cells both contained “ex-TFR” cells that had lost the TFR phenotype, but only the Blimp1-deleted TFR population displayed functional instability that may provide the de novo helper function for B cells that promote dysregulated GC and Ab responses.
Figure 4. Blimp1 Deletion in TFR Cells after Immunization Impairs TFR Suppressive Activity.
(A) Schematic diagram of experimental protocol. Donor CD45.2+ Prdm1fl/fliCre− (WT), Prdm1fl/fliCre+ (Blimp1-deleted; del), and CD45.1 + mice were immunized with NP-OVA in CFA for 7 days; tamoxifen was administered to WT and Del mice on day 6. On day 7, donors were euthanized and sorted CD45.2+ TFR (CD4+CD3+PD-1+CXCR5+GITR+) along with CD45.1+ TFH cells (CD4+CD3+PD-1+CXCR5+GITR−) were transferred into Tcra−/− hosts followed by immunization with NP-OVA in CFA and tamoxifen administration daily from days 7 to 9. Spleens from euthanized hosts were analyzed on day 18.
(B) Histogram (upper) and MFI (bottom) of Blimp1 and Bcl6 expression in donor CD45.2+ TFR cells after tamoxifen injection. Ctrl, TFR cells from Prdm1fl/flFoxP3Cre (left), or Bcl6fl/flFoxP3Cre mice (right).
(C) Frequency of CD45.2+ TFR (CD45.2+CD4+PD-1+CXCR5+FoxP3+), CD45.1+ TFH (CD45.2−CD4+Bcl6+CXCR5+), and GC B cells (B220+CD19+GL7+Fas+).
(D and E) Expression (D) and quantification (E) of IL-17A, IFNγ, and IL-10 by donor CD45.2+ TFR and ex-TFR cells.
(F) WT and KO mice were immunized with NP-OVA in CFA. Seven days later, TFR (WT versus KO: CD4+CD3+PD-1+CXCR5+YFP+) and non-TFR (WT versus KO: CD4+CD3+CXCR5+YFP+) were sorted and transferred (105/mouse) into Tcra−/− hosts followed by immunization with NP-OVA in CFA; hosts were further challenged with NP-OVA in IFA at day 13. Spleens or mLNs were analyzed on day 20 (n = 3/group). Splenic (upper) or mLN (center) TFR, TFH, intracellular expression of IL-17A by donor TFR and non-TFR cells. At right, serum anti-NP30 IgG, and anti-NP4 IgG levels.
In (A)–(E), the data are representative of two independent experiments (B–E: n = 3–4/group). N.S., no significance, *p < 0.05, **p < 0.01, and ***p < 0.001 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S3.
Blimp1-Deficient TFR but Not Conventional Treg Display Lineage Instability and Conversion into TFH-like Cells
To further test the above proposition, we transferred TFR or non-TFR cells from Prdm1fl/flFoxP3Cre mice or WT mice separately into Tcra−/− hosts followed by analysis of TFR and TFH cells and the Ab response after NP-OVA immunization (Figure 4F). The results showed that mice transferred with Blimp1-deficient TFR cells had the highest Ab titers associated with the highest frequencies of TFH compared to the other groups that had similar levels of TFH cells Ab titers. Although both Blimp1-deficient TFR and non-TFR cells expressed the pro-inflammatory cytokine IL-17A, the former had the largest portion that converted into TH17-like cells (Figure 4F). These findings indicate that Blimp1-deficient TFR but not conventional Treg acquired TH activity, showed impaired suppressive activity and contributed to the dysregulated GC responses, and that continued Blimp1 expression by TFR is required for the maintenance of TFR functional stability.
Mechanism of Blimp1-Dependent Regulation of the TFR Suppressive Phenotype and Lineage Stability
Comparison of the transcriptional profiles of WT and Prdm1fl/flFoxP3Cre TFR cells (Figure 5A) revealed that ~460 genes were upregulated and 300 genes were downregulated in Blimp1-deficient TFR cells. Ingenuity pathway analysis of differentially (1.5-fold cutoff) expressed genes showed that Blimp1 deficiency affected pathways associated with cytokine signaling and TH cell differentiation (Figure S4A). Genes that regulate the differentiation of TH2 cells (Il4), TH17 cells (Il23r), and TFH cells (Cxcr5, Bcl6, Il21) were strongly upregulated, while genes associated with suppressive activity (Il10, Il2ra, GzmB) were downregulated (Figure 5B), suggesting that diminished suppressive activity of Blimp1-deficient TFR cells may be associated with conversion to T effector cells.
Figure 5. Mechanism of Blimp1-Dependent Control of TFR Differentiation: Contribution of the IL-23R-STAT3 Axis and CXCR5-CCR7 Expression.
(A) WT and KO mice (6 weeks old) were immunized with NP-OVA in CFA. Seven days later, TFR cells (CD4+CD3+PD-1+CXCR5+YFP+) were sorted for microarray analysis. Differential gene expression in TFR from WT and KO mice (>1.5-fold) is shown.
(B) Pathway analysis revealed control of genes associated with TH cell differentiation by Blimp1.
(C) Genes related to cytokine-cytokine receptor interaction by DAVID.
(D) IL-23R expression by donor TFR cells in Tcra−/− hosts after tamoxifen-induced deletion of Blimp1, as in Figure 4.
(E) IL-23R expression in the CD25-expressing TFR subsets from WT and KO mice (6–8 weeks old) 10 days post-immunization.
(F) FoxP3-GFP reporter mice were immunized with NP-OVA in CFA. Seven days later, Treg (CD4+CD3+GFP+CD25+) and CD4+ naive T cells (CD4+CD3+GFP−CD44−) were sorted, chromatin prepared, and ChIP-PCR analyses performed for Blimp1, acetylated H3 (AcH3), and H3K27me3 at Blimp1-binding sites at the 3rd intron of Il23r, the 1st intron of Cxcr5, the 5′ distal element of CTLA-4, the 3rd intron of Ccr7, the FoxP3 CNS2, the 1st intron of Il2ra, and a non-specific region (C, control) of Il2ra. Data are shown as the percentage of input. Naive cells were used as controls for the anti-Blimp1 assay. Rabbit IgG isotype (R-iso) and mouse IgG isotype (M-iso) served as controls for the anti-AcH3 assay and anti-H3K27 assay, respectively.
(G) Prdm1fl/fliCre− (WT) and Prdm1fl/fliCre+ (Del) mice (8 weeks old) were treated with tamoxifen at day 0, followed by immunization with NP-OVA in CFA at day 1 and injection of tamoxifen daily for 4 days. Expression of pSTAT3 and pSTAT5 in TFR cells at day 7 post-immunization. At right, pSTAT3:pSTAT5 ratios in TFR cells are shown (n = 4/group).
In (D), (E), and (G), the data represent one of two experiments. In (F), the data represent one of three independent experiments. **p < 0.01 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S4.
Blimp1 Represses IL-23R-STAT3 Signaling and CXCR5 Expression but Retains CD25-STAT5 Activation in Differentiating TFR Cells
Gene Ontology (GO) analysis indicated that the gene groups upregulated in Blimp1-deficient TFR included genes that regulated cytokine-cytokine receptor interactions (Figures 5C, S4B, and S4C), and that the Il23r gene was one of the most upregulated genes in Blimp1-deficient TFR (Figures 5B and 5C). TFR cells after tamoxifen-induced Blimp1 depletion confirmed the increased expression of IL-23R by Blimp1-deleted TFR cells (Figure 5D). TFR cells from Prdm1fl/flFoxP3Cre mice, including the mature “GC” CD25lQ subset, expressed higher levels of IL-23R than TFR cells from WT mice (Figure 5E). Consistent with expression at the protein levels (Figures 2D and 2H), Blimp1-deficient TFR also expressed elevated Cxcr5 and reduced Il2ra and Ccr7 at the RNA levels (Figures 5B and 5C). These results suggested that Blimp1 may downregulate IL-23R and CXCR5 expression but positively regulate CD25 and CCR7 expression by TFR cells.
To define Blimp1-dependent regulation of the Il23r, Il2ra, Cxcr5, and Ccr7 genes, we analyzed the interaction of Blimp1 with these gene loci according to chromatin immunoprecipitation (ChIP)-PCR. Blimp1 bound to the 3rd intron of the Il23r gene and the 1st intron of the Cxcr5 gene in Treg, but not naive CD4+ T cells isolated from immunized mice (Figure 5F). These interactions were associated with the presence of repressive chromatin H3K27me3 but not acetylated histone H3 (AcH3) marks (Figure 5F). Blimp1 also bound to the 1st intron of Il2ra and the 3rd intron of Ccr7 as well as the 5′ distal element of the CTLA-4 gene, which contained AcH3 activation marks (but not H3K27me3 repressive marks) (Figure 5F). Blimp1 did not bind to the FoxP3 gene, suggesting that Blimp1 does not directly regulate FoxP3 expression (Figure 5F) (Garg et al., 2019). Thus, Blimp1 may repress the transcription of the Il23r and Cxcr5 genes but activate the transcription of the Il2ra, Ccr7, and CTLA-4 genes in Treg, which is consistent with the view that Blimp1 can function as both a transcriptional activator and a repressor (Minnich et al., 2016).
The ability of FoxP3+ Treg to maintain high levels of FoxP3 expression and lineage stability in the face of intense inflammatory responses depends in part on robust CD25-STAT5 activation and binding of activated STAT5 to the FoxP3 CNS2 intronic element (Feng et al., 2014; Kim et al., 2015). In contrast, the engagement of IL-23R promotes STAT3 activation to promote Treg conversion into TH17-like cells (Laurence et al., 2012). We tested the premise that Blimp1 may modulate the balance between IL-23R-STAT3 and CD25-STAT5 signals in TFR in favor of the latter. Analysis of tamoxifen-induced Blimp1 deletion in differentiating TFR revealed increased phosphorylation of STAT3 (pSTAT3) and decreased phosphorylation of STAT5 (pSTAT5) in all CD25-expressing TFR subsets without further cytokine-mediated activation (Figure 5G), suggesting that Blimp1 may repress IL-23R-STAT3 signaling while retaining the CD25-STAT5 pathway in TFR cells.
Silencing IL-23R-STAT3 Signaling Rescues the Blimp1-Deficient TFR Phenotype
We then asked whether forced reduction of IL-23R-STAT3 activation in Blimp1-deficient TFR cells could remedy TFR instability. We used lentiviral vectors that expressed the Thy1.1 reporter and small hairpin RNA (shRNA) targeting the Il23r gene to knock down IL-23R expression in Blimp1-deficient Treg before transfer of sorted Thy1.1+ Treg and naive CD4+ T cells (CD45.1+) into Tcra−/− hosts and immunization with NP-OVA (Figure 6A). Ten days later, we noted that transferred IL-23R-shRNA (Thy1.1+) FoxP3+ Treg expressed diminished levels of IL-23R and Bcl6 (Figure S5A), while the frequency of TFH and GC B cells and anti-NP Ab titers were reduced (Figures 6B, 6C, and S5B). The expression of CD25, Helios, and granzyme B (i.e., genes associated with suppressive activity) was markedly increased (Figures S5A and S5C), while the expression of RORγt, IL-17A, and IFNγ was substantially reduced in Blimp1-deficient TFR cells following IL-23R knockdown (Figures 6B, 6C, S5B, and S5C). These findings indicated that silencing IL-23R could rescue suppressive activity and restore the phenotype of Blimp1-deficient TFR cells.
Figure 6. Silencing the IL-23R-STAT3 Axis Rescues the Blimp1-Deficient TFr Phenotype.
(A) Schematic presentation of experimental protocol. KO Treg cells were sorted and cultured withanti-CD3andanti-CD28 plusIL-2for3days. Cells were infected with lentivirus expressing the Thy1.1 reporter plus IL-23R-shRNA or Ctrl-shRNA. On day 4, sorted Thy1.1+ Treg along with CD45.1+ naive CD4+ T cells were transferred into Tcra−/− hosts followed by immunization with NP-OVA in CFA on day 11. Splenocytes were analyzed on day 20.
(B) Thy1.1+ TFR, CD45.2− Thy1.1− TFH and GL7+ B cells, IL-17A, and IFNγ production by Thy1.1+ TFR.
(C) Frequency of CD45.2− Thy1.1− TFH, Bcl6+ IL-17A+, and Bcl6+ IFNγ+ in donor Thy1.1+ TFR cells and in anti-NP IgG titers.
(D–F) WT, KO, Stat3fl/+Prdm1fl/flFoxP3Cre (Stat3fl/+ + KO), and Stat3fl/flPrdm1fl/flFoxP3Cre (Stat3fl/fl + KO) mice were analyzed 10 days post-immunization with NP-OVA in CFA.
(D) Splenic TFR, TFH, GC B cells, and Bcl6+IL-17A+ in Treg.
(E) Frequency of TFR and Bcl6+IL-17A+ in Treg and in serum anti-NP IgG titers.
(F) FoxP3 expression in TFR by the indicated mouse strains.
In (A)–(C), the data represent one of two experiments (C: n = 4/group). In (D)–(F), the results are pooled from two independent experiments (E, n = 4–9/group). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (unpaired two-tailed Student’s t test). Error bars indicate means ± SEMs.
See also Figure S5.
We also analyzed Stat3fl/flPrdm1fl/flFoxP3Cre mice, which deleted both Blimp1 and STAT3 in FoxP3+ T cells (Figure S5D), to determine whether impaired STAT3 activity in Blimp1-deficient TFR cells could also remedy TFR instability. These mice had decreased TFR, TFH, and GC B cell frequencies, reduced anti-NP Ab titers, minimal ectopic IL-17A, and increased FoxP3 expression by TFR cells compared to Prdm1fl/flFoxP3Cre mice after immunization (Figures 6D–6F, S5E, and S5F). Although the ratios of TFH:TFR or GC B:TFR have been positively correlated with the strength of immune responses (Sage et al., 2013; Sage and Sharpe, 2015), our results did not reveal significant differences in the TFH:TFR ratios for each group (Figure S5G). The ratios of GC B:TFR cells were negatively correlated with the Ab response, most likely reflecting the robust expansion of dysfunctional TFR. These data suggested that the genetic status of TFR cells should be considered when evaluating the relation between TFH:TFR ratios and immune response outcomes. These results indicated that reduction of the IL-23R-STAT3 axis could rescue the Blimp1-deficient TFR phenotype and restore suppressive activity.
Increased STAT5 Activation in Differentiating Blimp1-Deficient TFR Cells Restores Lineage Stability
Finally, we asked whether increased STAT5 activation in differentiating Blimp1-deficient TFR cells could also mitigate TFR instability. We transduced Blimp1-deficient Treg with a retroviral vector expressing GFP alone (control) or GFP plus constitutively active STAT5 (STAT5ca) before the transfer of GFP+ Treg with CD45.1+ naive CD4+T cells into Tcra−/− hosts, followed by immunization with NP-OVA (Figure S6A). Before transfer or 10 days post-immunization, we observed higher pSTAT5 levels in Treg expressing STAT5CA compared with cells transduced with a control vector (Figures S6D). Expression of STAT5ca in Blimp1-deficient Treg was associated with increased FoxP3 and CD25 expression, reduced TFH and GC B cells (albeit no significance was detected), and diminished production of IL-17A and IFNγ by CD45.2+ (STAT5CA-transduced) TFR compared with TFR cells transduced with control vector (Figures S6B–S6D), suggesting that the forced activation of STAT5 during TFR differentiation can at least partially rescue Blimp1-deficient TFR instability.
These results suggest that the essential contribution of Blimp1 to TFR lineage stability and suppressive activity may reflect repression of the IL-23R-STAT3 axis and maintenance of the CD25-STAT5 pathway.
DISCUSSION
By virtue of their GC-specific localization, TFR cells represent a phenotypically and functionally specialized Treg population that controls the GC and Ab response. Although TFR cells share several features with TFH and Treg, our understanding of the molecular and genetic elements that direct the differentiation of this specialized subset has been incomplete. Here, we have uncovered an essential contribution of the Blimp1 TF to TFR cell differentiation, lineage stability, proper GC localization, and suppressive activity.
The Blimp1 gene is strongly expressed by almost all eTreg (Cretney et al., 2013), including FoxP3+ TFR cells in B cell follicles (Linterman et al., 2011). Early studies of the contribution of Blimp1 to CD4+ T cells that analyzed mice containing a systemic or T cell-specific deletion of Blimp1 have identified the contribution of Blimp1 to T cell expression of IL-10 (Cretney et al., 2011; Kallies et al., 2006; Martins et al., 2006). Histological analysis of non-lymphoid tissues, including lung, skin, and adipose tissues, in mice that carry a FoxP3-specific deletion of Blimp1 failed to reveal obvious abnormalities, with the exception of a mild hyperproliferative colitis apparent at 5 months of age. In contrast, the major phenotype of Prdm1fl/flFoxP3Cre mice was a markedly dysregulated Ab response, which included elevated IgE and autoantibody levels and the expansion of TFH and GC B cells. We used multiple experimental settings for this analysis, including direct examination of Prdm1fl/flFoxP3Cre and heterozygous female Prdm1fl/flFoxP3YFP-Cre+/− mice, adoptive transfer of TFR, inducible deletion of Blimp1 in TFR, and a comparison of the TFH-GC response of Bcl6fl/flPrdm1fl/flFoxP3Cre mice with Prdm1fl/flFoxP3Cre, Bcl6fl/flFoxP3Cre, and FoxP3Cre (WT) mice. We found that Blimp1 was essential for the maintenance of a stable TFR phenotype, suppressive control of Ab responses, and avoidance of atopic IgE and autoantibody responses. Reduced expression of FoxP3 and diminished levels of receptors that normally contribute to TFR inhibitory activity, including CTLA-4, were associated with increased numbers of TFH and GC B cells in the steady state and after immunization. Our results also suggest that Blimp1-deficient TFR rather than Blimp1-deficient non-TFR Treg account for the abnormal immune phenotype of Prdm1fl/flFoxP3Cre mice.
Mutations resulting in diminished FOXP3 expression that result in immune dysregulation (an X-linked [IPEX] syndrome) and genetic deletion of CTLA-4 in Treg are also marked by the loss of humoral tolerance and high serum IgE levels (Bennett et al., 2001; Wildin et al., 2001; Wing et al., 2014). We suggest that reduced FoxP3 expression by Blimp1-deficient TFR secondary to an imbalance in STAT5/STAT3-based signaling may account in part for this abnormal phenotype. The expansion of Blimp1-deficient TFR cells was marked by increased TFR proliferation and reduced apoptosis. Depletion of STAT3 in Blimp1-deficient TFR decreased TFR numbers to levels similar to those of WT TFR, which is consistent with the ability of activated STAT3 to promote T cell proliferation and impede apoptosis (Akira, 2000). The expanded Blimp1-deficient TFR population did not mediate significant suppressive activity and displayed a distorted cellular phenotype.
Diminished FoxP3 expression was accompanied by reduced levels of critical Treg gene products, including CTLA-4, CD25, and GITR, and increased production of the pro-inflammatory cytokines IFNγ and IL-17A. Previous studies have noted that even slight reductions in FoxP3 expression can profoundly affect Treg lineage stability and immunologic function (Di Pilato et al., 2019; Wan and Flavell, 2007). We and others have previously noted that the IL-2-STAT5 axis is essential to this process, reflecting in part an interaction between STAT5 and the FoxP3 CNS2 region (Feng et al., 2014; Kim et al., 2015). However, the contribution of the IL-2-STAT5 axis to TFR differentiation is more complex, reflecting its role in the early (CD25+) and later GC-localized CD25lo/− TFR cells. The latter TFR effector subpopulation expresses low but significant levels of Blimp1 compared to their naive Treg precursors (Wing et al., 2017). We suggest that dynamic changes in Blimp1 expression, partly in response to environmental IL-2, accompany functional TFR differentiation and appropriate navigation from the IL-2-rich T cell zone into IL-2-poor B cell follicles and final localization into the GC (Smigiel et al., 2014). In contrast, upregulation of CXCR5 and strong downregulation of CCR7 may increase the lineage instability of TFR cells devoid of Blimp1 after genetic deletion. Production of pro-inflammatory cytokines by unstable Blimp1-deficient TFR cells may reflect the upregulation of the IL-23R-STAT3 axis, expression of RORγt and IL-17A, and conversion into TH17-like cells. Unstable Blimp1-deficient TFR may also acquire the characteristics of TFH cells, including the expression of IL-4, Bcl6, CXCR5, and IL-21.
In summary, we find that coordinate regulation of the IL-23R-STAT3 and CD-25-STAT5 axes by Blimp1 is essential for the maintenance of TFR stability and suppressive activity. Identification of the molecular factors and signaling pathways that modulate Blimp1 expression in TFR may allow the development of agents that modulate Ab responses in the context of vaccines and autoimmune disease.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Harvey Cantor (Harvey_Cantor@dfci.harvard.edu). Unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Microarray data (GEO: GSE101611) have been deposited in the NCBI GEO.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J (B6), Prdm1fl/fl FoxP3YFP-Cre (FoxP3Cre), Rosa26Cre-ERT2 (iCre+), Bol6fl/fl, Stat3fl/fl, Tcrα−/−(Jackson Labs), Rag2−/−, B6SJL (CD45.1) (Taconic Farms), and B6.FoxP3-GFP reporter mice were housed in pathogen-free conditions. Prdm1fl/fl mice were bred onto FoxP3Cre or Rosa26Cre-ERT2 (iCre+) mice to generate Prdm1fl/flFoxP3Cre, Prdm1fl/+FoxP3Cre, or Prdm1fl/flRosa26Cre-ERT2 (Prdm1fl/fliCre+) mice, respectively. Bol6fl/fl mice were bred onto FoxP3Cre to generate Bol6fl/flFoxP3Cre mice that were further crossed onto Prdm1fl/flFoxP3Cre to yield Bol6fl/flPrdm1fl/flFoxP3Cre mice. Sfaf3fl/fl mice were crossed onto Prdm1fl/flFoxP3Cre to yield Stat3fl/+Prdm1fl/flFoxP3Cre or Stat3fl/flPrdm1fl/flFoxP3Cre mice. All mice were used at the age of 6 to 9 weeks unless otherwise specified. Both sexes (males or females) were randomly included for all experiments in an unblinded fashion. Generally between 3 to 7 mice were used per group, as indicated in each experiment. All experiments were performed in compliance with federal laws and institutional guidelines as approved by DFCI’s and UAB’s Animal Care and Use Committee.
Cell Lines
293 T cells (CRL-3216, ATCC) were grown in DMEM supplemented with 10% FBS at 5% CO2 at 37°C.
METHOD DETAILS
Flow Cytometry and Sorting
Fluorescence dye labeled Abs specific for mouse CD4 (GK1.5, RM4–5), TCRβ (H57–597), CD3 (145–2C11), CD25 (PC61), CD69 (H1.2F3), GITR (DTA-1), CTLA4 (UC10–4B9), KLRG1 (2F1/KLRG1), Blimp1 (5E7), TIGIT (1G9), RORγt (B2D), CCR6 (29–2L17), ST2 (RMST2–2, DIH9), Granzyme B (NGZB, GB11), IL-23R (12B2B64), Bcl2 (3F11), Helios (22F6), Ki-67 (SolA15), CD138 (281–2), CD45.1 (A20), CD45.2 (104), CD19 (1D3), B220 (RA3–6B2), CD44 (IM7), CD62L (MEL-14), Fas (15A7), IgM (II/41), T- and B cell activation antigen (GL7), ICOS (C398.4A), PD-1 (J43, 29F.1A12), CD90.1 (OX-7), IFN-γ (XMG1.2), IL-10 (JES5–16E3), IL-4 (11B11), IL-17A (eBio17B7), Bcl6 (K112–91), FoxP3 (FJK-16 s) and Bim (C34C5) were purchased from BD Biosciences, eBioscience, Biolegend and Cell Signaling Technology. Analysis of CXCR5 expression was performed using a biotinylated anti-CXCR5 (2G8, BD) Ab followed by incubation with APC or APC.Cy7 labeled streptavidin (Biolegend) as previously described (Leavenworth et al., 2015). A short incubation of cells with anti-mouse CD16/CD32 Fc block (BD) was performed prior to surface staining. Intracellular staining for Bcl6, FoxP3 and cytokines was performed using the FoxP3 staining buffer set (eBioscience). To assess pSTAT5 and pSTAT3 levels directly ex vivo, spleens and mLNs were immediately disrupted using glass slides into Cytofix/Cytoperm buffer (BD) for incubation of 30min at room temperature. The cells were washed and resuspended in 90% methanol and incubated on ice for 30min. After additional wash, the cells were stained for surface and intracellular antigens, including pSTAT5 (47/Stat5, pY694) and pSTAT3 (4/P-STAT3, pY705) for 45 min at room temperature (Smigiel et al., 2014). To assess apoptosis, cells were surface stained as described above, washed, and stained with Annexin-V (BD Biosciences) in 1 × Annexin Binding Buffer (BD Biosciences) for 15 min at room temperature, and immediately analyzed by FACS. For IL-21, intracellular staining was performed as previously described (Kashiwakuma et al., 2010). Cells were acquired on a Fortessa X20 using FACSDiva software (BD Biosciences) and analyzed with FlowJo software (Treestar). For cell sorting, single-cell suspension underwent positive enrichment for CD4+T cells or B cells through the use of CD4or CD19 microbeads (Miltenyi Biotec). Enriched cells were labeled with various fluorescent antibodies, as indicated in Figure S2, followed by sorting on a FACSAria II using FACSDiva software (BD Biosciences).
Adoptive Transfer and Immunization
As detailed in individual figure legends, FACS-sorted CD4+ T cell subsets and/ or GL7− B cells were transferred into the indicated hosts before immunization with protein antigens (NP-OVA) emulsified in CFA or alum for an additional 7 days before euthanisia and ex vivo analysis. In some experiments, the hosts were further challenged with protein antigens in IFA at the indicated times. All immunizations were conducted by intraperitoneal injection. Serum was prepared at the indicated time for measurement of primary and secondary antibody titers.
Tamoxifen Treatment
For in vivo experiments involving Cre strains, mice were injected with 1 mg/25g body weight tamoxifen (Sigma) emulsified in sunflower oil (Sigma) intraperitoneally (vol: 100–150 ul) once every 24 hours for 3–4 consecutive days unless otherwise specified.
Enzyme-Linked Immunosorbent Assay (ELISA)
Detection of NP-specific antibodies was performed as described (Kim et al., 2010). Anti-nuclear Antibodies (ANA) in mouse sera were determined by ELISA (Alpha Diagnostic International). IgE was measured using the OptEIA ELISA kit (BD Biosciences).
Generation of Retrovirus and Lentivirus
The retroviral vector RV-pMIG expressing STAT5ca was a gift from Shane Crotty (Johnston et al., 2012). The production of retroviruses was performed as previously described (Leavenworth et al., 2015). The short hairpin RNA targeting the Il23r gene was cloned into the pLKO.3 Thy1.1 lentiviral vector (Addgene 14749). Lentiviral stocks were generated by transfection of 293T cells with this plasmid along with the lentiviral packaging vectors, PsPAX2 (Addgene 12260) and pCMV-VSV-G (Addgene 8454) (Stewart et al., 2003), through the use of TransIT-LT1 transfection reagent (Mirus). Viral supernatants were collected 72 h later before infection of Treg cells as described below.
Infection by Retrovirus and Lentivirus
Retroviral or lentiviral infection of Treg cells was adapted, as previously described (Leavenworth et al., 2015). Briefly, purified CD25+CD4+ T cells were stimulated with plate-bound anti-CD3 (5 μg ml−1) and anti-CD28 (5 mg ml−1) in the presence of 50 U ml−1 recombinant human IL-2 (rhIL-2, Peprotech). Three days post-stimulation, cells were infected with retrovirus expressing GFP alone or GFP plus STAT5ca or lentivirus expressing Thy1.1 or Thy1.1 plus IL-23R-shRNA, and sorted GFP+ or Thy1.1+ Treg along with CD45.1+ naive CD4+ T cells were transferred into Tcra−/− hosts followed by immunization.
ChIP-qPCR
B6.FoxP3-GFP mice were immunized with NP-OVA in CFA for 7 days. GFP+CD25hiCD4+ Treg cells were sorted and fixed for 10 min at 37°C with 1% formaldehyde. Cells were washed twice in ice-cold PBS and cell pellets were stored at −80°C. Chromatin was isolated and immunoprecipitated with antibody to Blimp1 (Santa Cruz, sc-66015), acetylated H3 (AcH3) (Millipore, 06–599) or H3K27me3 (Abcam, ab-6002) and protein G Dynabeads (Thermo Fisher), followed by reverse crosslinking and DNA purification. Quantitative real-time PCR assays were performed for Blimp1, AcH3 or H3K27me3 on Blimp1-binding sites at the indicated gene loci.
Generation of Bone Marrow Chimeras
Rag2−/− mice received a sublethal dose of radiation (600 rads) one day before BM cell transfer. BM cells from donor mice were harvested and depleted of NK1.1+, CD4+, CD8+ and B220+ cells using biotinylated antibodies targeting each subset and anti-biotin microbeads (Miltenyi Biotech). Enriched 5 × 106−107 cells were intravenously injected into Rag2−/− mice. BM cells from Prdm1fl/fl and CD45.1 mice, or Prdm1fl/flFoxP3Cre and CD45.1 mice (5 × 106 cells per strain) were transferred, respectively.
Gene Expression Profiling
TFR cells (PD-1+CXCR5+YFP+CD4+CD3+) were sorted from FoxP3Cre or Prdm1fl/flFoxP3Cre mice 7 d post-immunization with NP-OVA in CFA. RNA was prepared with the RNeasy plus micro kit, according to the manufacturer’s instructions (QIAGEN). RNA amplification, labeling, and hybridization to Mouse Gene 2.0 ST arrays (Affymetrix) were performed at a Core Facility (Dana Farber Cancer Institute).
In Vitro Suppression Assay of TFR Cells
The suppression assay of TFR cells was performed as previously described (Sage et al., 2013). Briefly, sorted GL7− B cells were cultured alone, or with TFH cells in the presence or absence of TFR cells from FoxP3Creor Prdm1fl/flFoxP3Cre mice plus 2 μg/ml soluble anti-CD3 (2C11, BD) and 5 μg/ml anti-IgM (Jackson Immunoresearch). Five days later, IgG in the supernatants was determined by ELISA (Sage et al., 2013).
Immunoblot
The procedure was performed as described previously (Leavenworth et al., 2015). The following antibodies were used: Blimp1 (Santa Cruz, sc-66015) and Actin (Sigma, A-3584).
Quantitative RT-PCR
RNA was extracted using an RNeasy plus micro kit (QIAGEN). Relative quantification real-time PCR was performed with TaqMan gene expression assays, Bcl6 (Mm00477633_m1), Prdm1 (Mm00476128_m1), Rps18 (Mm02601777_g1) and RNA-to-CT™ 1-Step Kit (Life Technologies). All results were first normalized to those of the Rps18 control and are presented as normalized expression for the sample relative to the appropriate comparison conditions, as indicated in the legends.
Immunohistochemistry
To assess immunopathology in multiple organs, mice were fixed with Bouin’s solution (Sigma), and tissue sections were generated from paraffin-embedded tissues and stained with hematoxylin and eosin. For the identification of germinal centers, 7 μm acetone- fixed frozen sections from spleen were air-dried and labeled with phycoerythrin (PE)-conjugated anti-B220 antibody (BD, RA3–6B2) and FITC-conjugated anti-GL-7 antibody (BD, clone GL7). Quantification of GL7-FITC positively-stained areas using ImageJ software (NIH) was depicted as pixel2/area: two diameters of each germinal center (GL7+) were measured, and were divided into 2 to get R1 and R2. Each GC area was estimated according to the formula S = R1 × R2 × 3.14. All the GC areas calculated in one slide equal the total GC area. The mean GC area was calculated by dividing the total GC area by the GC numbers. For confocal analysis of TFR and TFH distribution in the GC, 7 weeks old mice were immunized with NP-OVA in CFA and spleens were collected 14 days post immunization. The spleens were handled and stained as previously described (Vanderleyden and Linterman, 2017). Briefly, spleens were treated with periodate (0.01 M NaIO4)-lysine (0.075 M L-lysine)-paraformaldehyde (1%) for 3 hours at 4C, before incubating overnight with sucrose 30% at 4C. On the following day, spleens were washed with new sucrose 30% and remaining sucrose was removed by dab-drying with a tissue. The spleens were put into cryomold containing OCT® Cryoprotective embedding medium (Sakura Finetek Usa Inc) and placed on a 2-propanol-dry ice cooling bath. The frozen blocks were stored in −80C until sectioning (10μm), blocking (2% BSAand 10% goat serum in PBS), permeabilization (2% Triton-Xin PBS) and staining with hamster anti-mouse CD3ε (clone eBio500A2, eBioscience), Alexa Fluor 568-conjugated goat anti-hamster IgG (ThermoFisher Scientific); eFluor450-con- jugated rat anti-mouse/rat FoxP3 (clone FJK16S, eBioscience); Alexa Fluor 647 conjugated rat anti-mouse IgD (clone 11–26c.2a, BioLegend); rabbit anti-mouse Ki67 (ThermoFisher Scientific), and Alexa Fluor 488 conjugated goat anti-rabbit (ThermoFisher Scientific). Images were captured with a Leica SP5 confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSES
Statistical analyses were performed using two-tailed, unpaired Student’s t test with GraphPad Prism V6 software. Error bars indicate mean ± SEM. A p value of < 0.05 was considered to be statistically significant (* p < 0.05, ** p < 0.01, *** p < 0.001, ****p <0.0001). No exclusion of data points was used. Sample size was not specifically predetermined, but the number of mice used was consistent with previous experience with similar experiments.
DATA AND CODE AVAILABILITY
The microarray data have been deposited in the NCBI GEO under accession number GEO: GSE101611.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD4 (GK1.5) | BD Biosciences | Cat# 552051; RRID: AB_394331 |
| Anti-mouse CD4 (RM4–5) | Biolegend | Cat# 100531; 100559; RRID: AB_493374; RRID: AB_2562608 |
| Anti-mouse TCRβ (H57–597) | Biolegend | Cat# 109220; RRID: AB_893624 |
| Anti-mouse CD3 (145–2C11) | Biolegend | Cat# 100233; 100306; RRID: AB_2561387; RRID: AB_312671 |
| Anti-mouse CD25 (PC61) | BD Biosciences | Cat# 553866; RRID: AB_395101 |
| Anti-mouse CD25 (PC61) | Biolegend | Cat# 102025; RRID: AB_830744 |
| Anti-mouse CD69 (H1.2F3) | Biolegend | Cat# 104512; RRID: AB_493564 |
| Anti-mouse GITR (DTA-1) | Biolegend | Cat# 126308; 126317; RRID: AB_1089125; RRID: AB_2563385 |
| Anti-mouse CTLA-4 (UC10–4B9) | Biolegend | Cat# 106306; RRID: AB_313255 |
| Anti-mouse KLRG1 (2F1/KLRG1) | Biolegend | Cat# 138410; 138418; RRID: AB_10643582; RRID: AB_2563015 |
| Anti-mouse Blimp1 (5E7) | BD Biosciences | Cat# 563643; RRID: AB_2738342 |
| Anti-mouse TIGIT (1G9) | Biolegend | Cat# 142103; RRID: AB_10895760 |
| Anti-mouse RORγt (B2D) | eBioscience | Cat# 12698182; RRID: AB_10807092 |
| Anti-mouse CCR6 (29–2L17) | Biolegend | Cat# 129807; RRID: AB_1227498 |
| Anti-mouse ST2 (RMST2–2) | eBioscience | Cat# 46933580; RRID: AB_2573882 |
| Anti-mouse ST2 (DIH9) | Biolegend | Cat# 145303; RRID: AB_2561914 |
| Anti-mouse Granzyme B (NGZB) | eBioscience | Cat# 11889882; RRID: AB_10733414 |
| Anti-mouse Granzyme B (GB11) | Biolegend | Cat# 515408; RRID: AB_2562196 |
| Anti-mouse IL-23R (12B2B64) | Biolegend | Cat# 150903; RRID: AB_2572188 |
| Anti-mouse Bcl2 (3F11) | BD Biosciences | Cat# 554221; RRID: AB_395312 |
| Anti-mouse Bim (C34C5) | Cell Signaling Technology | Cat# 12186; RRID: AB_2797842 |
| Anti-mouse Helios (22F6) | Biolegend | Cat# 137229; 137220; RRID: AB_2561639; RRID: AB_10690535 |
| Anti-mouse Ki-67 (SolA15) | eBioscience | Cat# 12569882; RRID: AB_11150954 |
| Anti-mouse CD138 (281–2) | BD Biosciences | Cat# 553713; RRID: AB_394999 |
| Anti-mouse CD45.1 (A20) | Biolegend | Cat# 110728; RRID: AB_893346 |
| Anti-mouse CD45.2 (104) | Biolegend | Cat# 109837; RRID: AB_2561393 |
| Anti-mouse CD19 (1D3) | BD Biosciences | Cat# 551001; RRID: AB_394004 |
| Anti-mouse B220 (RA3–6B2) | BD Biosciences | Cat# 553090; RRID: AB_394620 |
| Anti-mouse B220 (RA3–6B2) | eBioscience | Cat# 25045282; RRID: AB_469627 |
| Anti-mouse CD44 (IM7) | Biolegend | Cat# 103028; RRID: AB_830785 |
| Anti-mouse CD62L (MEL-14) | Biolegend | Cat# 104441; RRID: AB_2561537 |
| Anti-mouse Fas (15A7) | BD Biosciences | Cat# 554258; RRID: AB_395330 |
| Anti-mouse IgM (II/41) | eBioscience | Cat# 11579081; RRID: AB_465244 |
| Anti-mouse T-/ B cell activation Ag (GL7) | Biolegend | Cat# 144606; RRID: AB_2562185 |
| Anti-mouse ICOS (C398.4A) | Biolegend | Cat# 313508; 313518; RRID: AB_416332; RRID: AB_10641280 |
| Anti-mouse PD-1 (J43) | BD Biosciences | Cat# 551892; RRID: AB_394284 |
| Anti-mouse PD-1 (29F.1A12) | Biolegend | Cat# 135216; 135213; RRID: AB_10689635; RRID: AB_10689633 |
| Anti-mouse IFN-γ (XMG1.2) | BD Biosciences | Cat# 554412; RRID: AB_395376 |
| Anti-mouse IL-10 (JES5–16E3) | BD Biosciences | Cat# 554467; RRID: AB_395412 |
| Anti-mouse IL-4 (11B11) | Biolegend | Cat# 504112; RRID: AB_493323 |
| Anti-mouse IL-17A (eBio17B7) | eBioscience | Cat# 50717782; RRID: AB_11220280 |
| Anti-Human/mouse Bcl6 (K112–91) | BD Biosciences | Cat# 561522; RRID: AB_10717126 |
| Anti-mouse FoxP3 (FJK-16 s) | eBioscience | Cat# 25577382; 45577382; RRID: AB_891552; RRID: AB_914351 |
| Anti-mouse CXCR5 (2G8) | BD Biosciences | Cat# 551960; RRID: AB_394301 |
| Streptavidin-APC | Biolegend | Cat# 405207 |
| Streptavidin-APC.Cy7 | Biolegend | Cat# 405208 |
| Anti-mouse CD90.1 (OX-7) | Biolegend | Cat# 202535; RRID: AB_2562643 |
| Anti-mouse NK1.1 Biotin (PK136) | Biolegend | Cat# 108704; RRID: AB_313391 |
| Anti-mouse B220 Biotin (RA3–6B2) | Biolegend | Cat# 103204; RRID: AB_312989 |
| Anti-mouse CD4 Biotin (GK1.5) | Biolegend | Cat# 100404; RRID: AB_312689 |
| Anti-mouse CD8 Biotin (53–6.7) | Biolegend | Cat# 100704; RRID: AB_312743 |
| Anti-Human/mouse pSTAT5 (47/Stat5, pY694) | BD Biosciences | Cat# 612599; RRID: AB_399882 |
| Anti-Human/mouse pSTAT3 (4/P-STAT3, pY705) | BD Biosciences | Cat# 612569; RRID: AB_399860 |
| Annexin-V | BD Biosciences | Cat# 561012; RRID: AB_2034024 |
| Anti-human IgG Fc (HP6017) | Biolegend | Cat# 409319; RRID: AB_2563329 |
| Goat anti-mouse IgG Fc HRP | Invitrogen | Cat# A16084; RRID: AB_2534758 |
| Rat anti-mouse IgG1 (X56) HRP | BD Biosciences | Cat# 559626; RRID: AB_397292 |
| Purified Anti-Mouse CD16/CD32 (Fc Block) | BD Biosciences | Cat# 553142; RRID: AB_394657 |
| Purified NA/LE anti-mouse CD3 (145–2C11) | BD Biosciences | Cat# 553057; RRID: AB_394590 |
| Purified NA/LE anti-mouse CD28 (37.51) | BD Biosciences | Cat# 553294; RRID: AB_394763 |
| AffiniPure F(ab’)2 Fragment Goat anti-IgM | Jackson Immunoresearch | Cat# 115–006–020; RRID: AB_2338469 |
| Purified anti-Blimp1 (3H2E8) | Santa Cruz Biotechnology | Cat# sc-66015; RRID: AB_1119615 |
| Purified anti-acetylated H3 (rabbit polyclonal) | Millipore | Cat# 06–599; RRID: AB_2115283 |
| Rabbit IgG, polyclonal – Isotype control | Abcam | Cat# ab-171870; RRID: AB_2687657 |
| Purified anti-H3K27me3 (mouse monoclonal) | Abcam | Cat# ab-6002; RRID: AB_1977539 |
| Mouse IgG isotype control | Thermo Fisher Scientific | Cat# 10400C; RRID: AB_2532980 |
| Monoclonal anti-β-actin-Peroxidase (AC-15) | Sigma | Cat# A-3584; RRID: AB_2765165 |
| Hamster anti-mouse CD3ε (eBio500A2) | eBioscience | Cat# 14–0033–85; RRID: AB_837129 |
| Alexa Fluor 568 goat anti-hamster IgG | Thermo Fisher Scientific | Cat# A-21112; RRID: AB_2535761 |
| Alexa Fluor 647 rat anti-mouse IgD (11–26c.2a) | BioLegend | Cat# 405708; RRID: AB_893528 |
| Rabbit anti-mouse Ki67 (polyclonal) | Thermo Fisher Scientific | Cat# PA5–19462; RRID: AB_10981523 |
| Alexa Fluor 488 goat anti-rabbit IgG | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_143165 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TransIT-LT1 | Mirus | Cat# MIR2300 |
| Tamoxifen | Sigma | Cat# T5648 |
| Sunflower seed Oil | Sigma | Cat# S5007 |
| Anti-mouse CD4 microbeads | Miltenyi Biotec | Cat# 130–049–201; RRID: AB_2722753 |
| Anti-mouse CD19 microbeads | Miltenyi Biotec | Cat# 130–052–201 |
| Anti-biotin microbeads | Miltenyi Biotec | Cat# 130–090–485; RRID: AB_244365 |
| CFA | Sigma | Cat# F5881 |
| IFA | Sigma | Cat# F5506 |
| Imject Alum | Thermo Fisher Scientific | Cat# 77161 |
| NP16-OVA (16 loading) | Biosearch Technologies | Cat# N-5051–100 |
| NP23-KLH (23 loading) | Biosearch Technologies | Cat# N-5060–25 |
| NP23-BSA (23 loading) | Biosearch Technologies | Cat# N-5050–10 |
| NP4-BSA (4 loading) | Biosearch Technologies | Cat# N-5050–10 |
| rhIL-2 | Peprotech | Cat# 200–02 |
| Mouse IL-21R Fc Chimera Protein | R&D Systems | Cat# 596-MR-100 |
| Leukocyte Activation cocktail | BD Biosciences | Cat# 550583 |
| Formaldehyde (16%, w/v), Methanol-free | Pierce | Cat# 28906 |
| Protein G Dynabeads | Thermo Fisher Scientific | Cat# 10003D |
| Bouin’s solution | Sigma | Cat# HT10132 |
| Critical Commercial Assays | ||
| FoxP3 staining Buffer Set | eBioscience | Cat# 00552300 |
| BD Cytofix/Cytoperm Solution kit | BD Biosciences | Cat# 554714 |
| Anti-nuclear Antibodies (ANA) Elisa kit | Alpha Diagnostic | Cat# 5210 |
| IgE OptEIA ELISA Set | BD Biosciences | Cat# 555248 |
| RNeasy plus micro kit | QIAGEN | Cat# 74034 |
| TaqMan RNA-to-CT™ 1-Step Kit | Thermo Fisher Scientific | Cat# 4392653 |
| ChIP-IT® Express Enzymatic | Active Motif | Cat# 53009 |
| Chromatin IP DNA Purification Kit | Active Motif | Cat# 58002 |
| Deposited Data | ||
| Microarray dataset | This paper | GEO: GSE101611 |
| Experimental Models: Cell Lines | ||
| 293 T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6: C57BL/6J | Jackson Laboratories | Jax:000664; RRID: IMSR_JAX: 000664 |
| Mouse: Prdm1fl/fl: B6.129-Prdm1tm1Clme/J | Jackson Laboratories | Jax:008100; RRID: IMSR_JAX:008100 |
| Mouse: FoxP3Cre: B6.129(Cg)-Foxp3tm4(YFP/cre)Ayr/J | Jackson Laboratories | Jax:016959; RRID: IMSR_JAX:016959 |
| Mouse: iCre+: B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J | Jackson Laboratories | Jax:008463; RRID: IMSR_JAX:008463 |
| Mouse: Bcl6fl/fl: B6.129S(FVB)-Bcl6tm1.1Dent/J | Jackson Laboratories | Jax:023727; RRID: IMSR_JAX:023727 |
| Mouse: Stat3fl/fl: B6.129S1-Stat3tm1Xyfu/J | Jackson Laboratories | Jax:016923; RRID: IMSR_JAX:016923 |
| Mouse: Tcrα−/−: B6.129S2-Tcratm1Mom/J | Jackson Laboratories | Jax:002116; RRID: IMSR_JAX:002116 |
| Mouse: FoxP3-GFP: B6.Cg-Foxp3tm1Mal/J | Jackson Laboratories | Jax:018628; RRID: IMSR_JAX:018628 |
| Mouse: Rag2−/−: B6.129S6-Rag2tm1Fwa N12 | Taconic | Model# RAGN12; RRID: IMSR_TAC:ragn12 |
| Mouse: B6SJL: B6.SJL-Ptprca-BoyAiTac | Taconic | Model# 4007; RRID: IMSR_CMMR:PST4007 |
| Oligonucleotides | ||
| ChIP-PCR Il23r forward | Integrated DNA Technologies | CTTGGCAAACTTCCTTCCTATTAAC |
| ChIP-PCR Il23r reverse | Integrated DNA Technologies | AAACAGTGCTGACTACTT GGCAT |
| ChIP-PCR Il2ra forward | Integrated DNA Technologies | TCGGAGAGGGATTCGGTAGCTTGA |
| ChIP-PCR Il2ra reverse | Integrated DNA Technologies | TGATAGCCTGCTGCTCAGAACTGGG |
| ChIP-PCR Il2raCtrl forward | Integrated DNA Technologies | TTACAGCAGTGCCTCCCTTG |
| ChIP-PCR Il2raCtrl reverse | Integrated DNA Technologies | GGGAGTGAGTGGGGTTAGGA |
| ChIP-PCR Cxcr5 forward | Integrated DNA Technologies | GGGCAGGAAGAACAGAGTAAG |
| ChIP-PCR Cxcr5 reverse | Integrated DNA Technologies | CTGCTAACCACAGAGGAAGAC |
| ChIP-PCR Ccr7 forward | Integrated DNA Technologies | CACTCAAGCCAAGACAGCTA |
| ChIP-PCR Ccr7 reverse | Integrated DNA Technologies | GACTACTCAACCAGGGTGTTC |
| ChIP-PCR Ctla4 forward | Integrated DNA Technologies | AATATGTTTCTCTGCGGGCACCA |
| ChIP-PCR Ctla4 reverse | Integrated DNA Technologies | GCCTAAGTAAACCCCAGATCAGC |
| ChIP-PCR FoxP3 forward | Integrated DNA Technologies | CACCCTACCTGGGCCTATCC |
| ChIP-PCR FoxP3 reverse | Integrated DNA Technologies | GCTTCATCGGCAACAAGGAG |
| shRNA targeting sequence: Il23r | Integrated DNA Technologies | CCTACATAGATACCAAGTATA |
| TaqMan probe for Bcl6 | Thermo Fisher Scientific | Mm00477633_m1 |
| TaqMan probe for Prdm1 | Thermo Fisher Scientific | Mm00476128_m1 |
| TaqMan probe for Rps18 | Thermo Fisher Scientific | Mm02601777_g1 |
| Recombinant DNA | ||
| RV-pMIG-STAT5CA | Johnston et al., 2012 | N/A |
| pLKO.3 Thy1.1 | Benoist and Mathis Lab (Harvard Medical School) | RRID: Addgene_14749 |
| PsPAX2 | https://tronolab.epfl.ch | RRID: Addgene_12260 |
| pCMV-VSV-G | Stewart et al., 2003 | RRID: Addgene_8454 |
| Software and Algorithms | ||
| FlowJo | FlowJo, LLC | v. 10; RRID: SCR_008520 |
| FACSDiva | BD Bioscience | RRID: SCR_001456 |
| Prism 6 | GraphPad | v. 6; RRID: SCR_002798 |
| ImageJ | NIH | RRID: SCR_003070 |
| DAVID Bioinformatics Resources | https://david.ncifcrf.gov | v. 6.8; RRID: SCR_001881 |
| Ingenuity Pathway analysis (IPA) | QIAGEN | RRID: SCR_008653 |
Highlights.
FoxP3-specific ablation of Blimp1 results in expansion of dysfunctional TFR cells
Inducible deletion of Blimp1 in TFR cells impairs TFR stability and function
Blimp1 controls CTLA4 expression, IL-23R-CD25 and CXCR5-CCR7 axes in TFR cells
Blimp1 controls appropriate homing and positioning of TFR cells into the GC
ACKNOWLEDGMENTS
We acknowledge C. Benoist and D. Mathis (pLKO.3 Thy1.1 plasmid), D. Trono (psPAX2 plasmid), B. Weinberg (pCMV-VSV-G plasmid), S. Crotty (STAT5ca plasmid), D. Campbell (phospho-STAT staining protocol), H. Nakajima (IL-21 staining protocol), Y. Li and D. Pan (microarray analysis), and A. Angel (manuscript/figure preparation). This study was supported in part by grants from the NIH (AI48125/AI37562, H.C.), the LeRoy Schecter Research Foundation (H.C.), and the National Natural Science Foundation of China (81971482, E.S.); fellowships from Sahlgrenska Academy, the University of Gothenburg, and The Foundation Blanceflor Boncompagni Ludovisi, nee Bildt (H.R.), and the Benacerraf Fellowship in Immunology (L.W.); and start-up funds from the University of Alabama at Birmingham (J.W.L.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.012.
REFERENCES
- Akira S (2000). Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19, 2607–2611. [DOI] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, White-sell L, Kelly TE, Saulsbury FT, Chance PF, and Ochs HD (2001). The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet 27, 20–21. [DOI] [PubMed] [Google Scholar]
- Botta D, Fuller MJ, Marquez-Lago TT, Bachus H, Bradley JE, Weinmann AS, Zajac AJ, Randall TD, Lund FE, Leon B, and Ballesteros-Tato A (2017). Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol 18, 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. (2011). Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med 17, 983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, and Kallies A (2011). The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol 12, 304–311. [DOI] [PubMed] [Google Scholar]
- Cretney E, Kallies A, and Nutt SL (2013). Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol. 34, 74–80. [DOI] [PubMed] [Google Scholar]
- Crotty S (2011). Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol 29, 621–663. [DOI] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pilato M, Kim EY, Cadilha BL, Prüßmann JN, Nasrallah MN, Seruggia D, Usmani SM, Misale S, Zappulli V, Carrizosa E, et al. (2019). Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 570, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, and Rudensky AY (2014). Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G, Muschaweckh A, Moreno H, Vasanthakumar A, Floess S, Lepennetier G, Oellinger R, Zhan Y, Regen T, Hiltensperger M, et al. (2019). Blimp1 Prevents Methylation of Foxp3 and Loss of Regulatory T Cell Identity at Sites of Inflammation. Cell Rep. 26, 1854–1868.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Tanaka S, Motomura Y, Harada Y, Ohno S, Ohno S, Yanagi Y, Inoue H, and Kubo M (2012). The 3’ enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity 86, 188–200. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, and Crotty S (2012). STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med 209, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, and Nutt SL (2006). Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol 7, 466–474. [DOI] [PubMed] [Google Scholar]
- Kashiwakuma D, Suto A, Hiramatsu Y, Ikeda K, Takatori H, Suzuki K, Kagami S, Hirose K, Watanabe N, Iwamoto I, and Nakajima H (2010). B and T lymphocyte attenuator suppresses IL-21 production from follicular Th cells and subsequent humoral immune responses. J. Immunol 185, 2730–2736. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Verbinnen B, Tang X, Lu L, and Cantor H (2010). Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature 467, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TA, Chan S, et al. (2015). Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 350, 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Lu Y, Amezquita RA, Weinstein JS, Vander Heiden JA, Gupta NT, Kleinstein SH, Kaech SM, and Craft J (2017). Interleukin-10 from CD4+ follicular regulatory T cells promotes the germinal center response. Sci. Immunol 2, eaan4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Amarnath S, Mariotti J, Kim YC, Foley J, Eckhaus M, O’Shea JJ, and Fowler DH (2012). STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity 37, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavenworth JW, Tang X, Kim HJ, Wang X, and Cantor H (2013). Amelioration of arthritis through mobilization of peptide-specific CD8+ regulatory T cells. J. Clin. Invest 123, 1382–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavenworth JW, Verbinnen B, Yin J, Huang H, and Cantor H (2015). A p85a-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat. Immunol 16, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. (2011). Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med 17, 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, and Calame K (2006). Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol 7, 457–465. [DOI] [PubMed] [Google Scholar]
- Minnich M, Tagoh H, Bonelt P, Axelsson E, Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M, and Busslinger M (2016). Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol 17, 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, and Sharpe AH (2015). T follicular regulatory cells in the regulation of B cell responses. Trends Immunol. 36, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, Francisco LM, Carman CV, and Sharpe AH (2013). The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat. Immunol 14, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, and Campbell DJ (2014). CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med 211, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderleyden I, and Linterman MA (2017). Identifying Follicular Regulatory T Cells by Confocal Microscopy. Methods Mol. Biol 1623, 87–93. [DOI] [PubMed] [Google Scholar]
- Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S, Fagarasan S, Mielke LA, Afshar-Sterle S, Masters SL, et al. (2015). The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol 16 276–285 [DOI] [PubMed] [Google Scholar]
- Wan YY, and Flavell RA (2007). Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770. [DOI] [PubMed] [Google Scholar]
- Weinstein JS, Herman EI, Lainez B, Licona-Limón P, Esplugues E, Flavell R, and Craft J (2016). TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol 17, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. (2001). X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet 27, 18–20. [DOI] [PubMed] [Google Scholar]
- Wing JB, Ise W, Kurosaki T, and Sakaguchi S (2014). Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41, 1013–1025. [DOI] [PubMed] [Google Scholar]
- Wing JB, Kitagawa Y, Locci M, Hume H, Tay C, Morita T, Kidani Y, Matsuda K, Inoue T, Kurosaki T, et al. (2017). A distinct subpopulation of CD25− T-follicular regulatory cells localizes in the germinal centers. Proc. Natl. Acad. Sci. USA 114, E6400–E6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data have been deposited in the NCBI GEO under accession number GEO: GSE101611.