Abstract
Autophagy plays crucial roles in the recycling of metabolites, and is involved in many developmental processes. Rice mutants defective in autophagy are male sterile due to immature pollens, indicating its critical role in pollen development. However, physiological roles of autophagy during seed maturation had remained unknown. We here found that seeds of the rice autophagy-deficient mutant Osatg7-1, that produces seeds at a very low frequency in paddy fields, are smaller and show chalky appearance and lower starch content in the endosperm at the mature stage under normal growth condition. We comprehensively analyzed the effects of disruption of autophagy on biochemical properties, proteome and seed quality, and found an abnormal activation of starch degradation pathways including accumulation of α-amylases in the endosperm during seed maturation in Osatg7-1. These results indicate critical involvement of autophagy in metabolic regulation in the endosperm of rice, and provide insights into novel autophagy-mediated regulation of starch metabolism during seed maturation.
Subject terms: Seed development, Abiotic
Introduction
Rice (Oryza sativa L.) is one of the most important crops in the world not only in agriculture but as a model monocot in scientific research. Elucidating the molecular mechanism for the development of its seed/starch endosperm is of particular significance for food production. Endosperm development in rice consists of four stages: coenocytic nuclear division, cellularization, differentiation of an outer aleurone layer and an inner starchy endosperm, and accumulation of starch, proteins and lipids etc.1–6. Starchy endosperm cells occupy most of the space in the caryopsis and provide the main energy source of rice seeds4,6,7. Starch granules are tightly packed in the endosperm to form translucent grains. However, rice grains exposed to excess environmental stresses during ripening as well as flowering stages exhibit a whitish chalky appearance, which causes serious damage to grain quality8 and economical loss because they impair the commercial value9.
Autophagy is an intracellular destructive mechanism evolutionarily conserved among eukaryotes, degrading intracellular proteins, metabolites, and organelles for recycling and quality control10. Isolation membranes emerge in the cytosol to develop a double membrane structure called the autophagosomes to enclose intracellular components, which fuses with the lytic compartments such as the vacuole and lysosomes11. Many of the more than 30 autophagy-related genes (ATG) identified in yeast, are conserved in most eukaryotes, including animals and plants12.
Autophagy is essential in growth, development, and cell survival in many eukaryotes13,14. In plants, autophagy has been suggested to play roles in the recycling of proteins and metabolites, including lipids, at the whole-plant level, and is involved in many physiological processes such as plant nutrition, senescence, hormonal responses, pathogen resistance, stress protection, seed germination, photomorphogenesis, chloroplast maturation, tracheary element formation, and fertile floret development under nutrient-limited conditions15–20. Moreover, autophagy has recently been shown to be induced in the tapetum in anthers at the uninucleate stage and is required for postmeiotic anther development including the programmed cell death (PCD)-mediated degradation of the tapetum and pollen maturation in rice21,22. However, physiological roles of autophagy during seed maturation and its relationship with metabolic regulation in endosperm development had remained poorly understood in plants.
We herein discovered that autophagy deficiency in rice leads to chalky appearance of grains even under normal growth condition. Multidisciplinary analyses including microscopic, biochemical, genetic and proteomic analyses revealed that this is due to misregulation of sugar and starch metabolism mediated at least in part by abnormal upregulation of α-amylase during seed maturation. These results indicate critical roles of autophagy in seed maturation, grain quality control during reproductive development in rice, and suggest the impact of controlling autophagy in agriculture.
Results and Discussion
Chalky appearance of grains of the Osatg7-1 mutant under normal growth conditions
Rice autophagy-deficient mutants (Osatg7-1, Osatg7-2 and Osatg9) show severe male sterility, fail to accumulate lipidic and starch components in pollen grains at the flowering stage, exhibit reduced pollen germination activity, and have limited anther dehiscence, indicating that autophagy is involved in pollen maturation in rice21–23. The Osatg7-1 mutant showed complete sporophytic male sterility in regular growth chambers and greenhouses or under pot cultivation outdoors21. On the other hand, we have found that the Osatg7-1 mutant produce seeds at a very low frequency in a specific greenhouse (Fig. 1a, 2016A) or paddy fields (Fig. 1a, 2017B and 2018B). The fertility rate of the Osatg7-1 mutant (1.3 ± 1.3 {n = 30}, 2016A; 4.3 ± 4.1{n = 61}, 2017B; 1.8 ± 1.7{n = 40}, 2018B) was much lower than those of the wild type (WT; 91.6 ± 8.6{n = 12}, 2016A; 94.8 ± 2.9 {n = 24}, 2017B; 84.7 ± 4.7 {n = 20}, 2018B).
Figure 1.
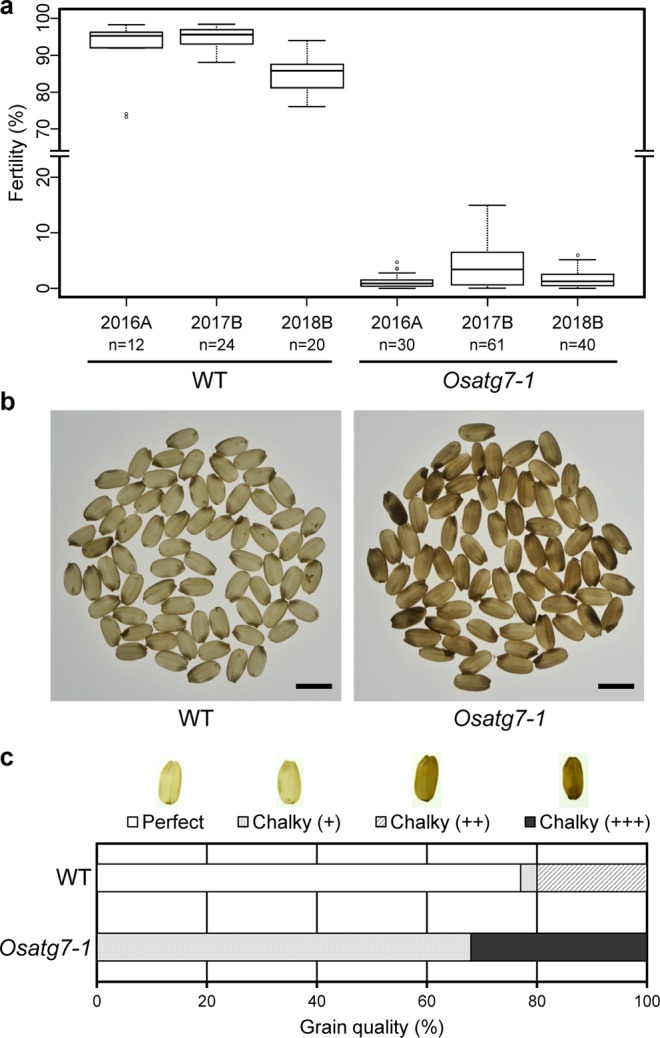
Effects of autophagy disruption on rice reproductive development grown in the paddy field. (a) The Osatg7-1 mutant exhibited a severe sterile phenotype under both greenhouse (2016A) and paddy field conditions (2017B and 2018B). The Box limits represent the upper and lower quartiles, the bold line represents the median, and the whiskers the minimum and maximum within 1.5 IQR of the lower or the upper quartile values. The unfilled circles indicate to suspected outliers. n = 12–61 independent samples. (b) The Osatg7-1 mutant had grains with a chalky appearance. Photographs were taken under back lighting. Scale bar: 5 mm. (c) Viable chalky grains shown in (b) were quantified and categorized. Data are the mean of 100 seeds of rice.
We investigated these seeds in order to understand if autophagy plays any roles in seed developmental processes in rice, and found that the majority of the Osatg7-1 had grains with whitish chalky appearance under normal growth condition in the paddy field (Fig. 1b). Under this condition (2017B), 77% of WT seeds were perfect grains, while 0% in Osatg7-1 and all grains showed chalky appearance (Fig. 1c). The Osatg7-1 grains were smaller than WT, and the weight of grains of the Osatg7-1 mutant (20.10 ± 2.09** [mg]; n = 120, **P value < 0.01) were slightly smaller than those of WT grains (20.79 ± 1.05 [mg]; n = 120). These phenotypes of the autophagy-deficient mutant were quite similar to those observed in the WT seeds suffering from heat stress during seed maturation24. These results indicate that autophagy plays critical roles in seed development and maturation in rice.
Abnormal morphology of starch granules in the endosperm of the Osatg7-1 mutant
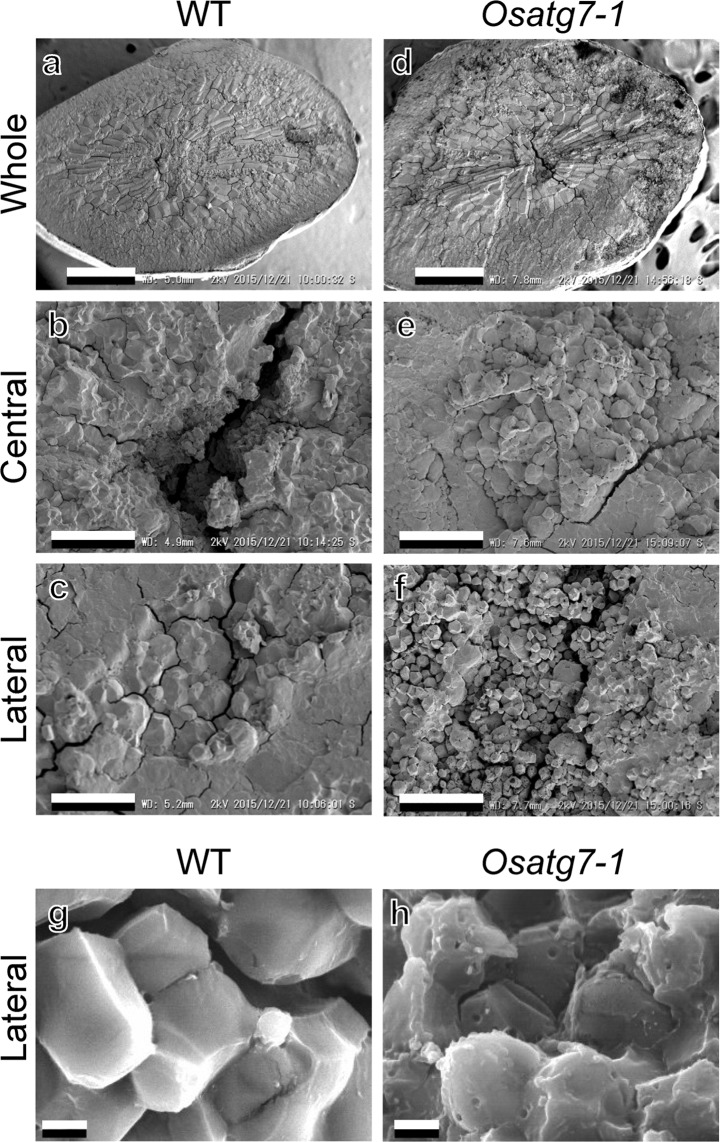
Chalkiness is shown to be derived from the opaque portion in the rice endosperm due to the loose packing of starch granules and protein bodies25,26. We performed a scanning electron microscopic (SEM) analysis to reveal the starch granule morphology of the Osatg7-1 mutant. As shown in Fig. 2, WT starch granules were tightly packed in the whole endosperm, and their shape was polygonal with sharp edges (Fig. 2a–c). On the contrary, in the chalky grains of the Osatg7-1 mutant, the starch granules of the central region had a similar shape and tight packing to that of WT (Fig. 2e), whereas those of the mutant in the lateral part were loosely packed and several small pits were often observed on the surface of the starch granules (Fig. 2f). These small pits of the mutant were also confirmed by electron probe micro analysis (EPMA) (Fig. 2h). The size of starch granules in the lateral region of the endosperm of WT and Osatg7-1 were 4.67 ± 1.33 µm and 2.09 ± 0.53 µm, respectively, indicating that starch granules in the lateral region of the endosperm were smaller and sparser in the Osatg7-1 mutant than in WT. These phenotypes of the autophagy-deficient mutant appear to share some similarities with those of the wild type seeds suffering from heat stress during ripening24 (Fig. 2f).
Figure 2.
Abnormal morphology of starch granules in the endosperm of the Osatg7-1 mutant. (a–f) The ultrastructure of mature seeds in WT (left) and the Osatg7-1 mutant (right). Close up of central (b,e) and lateral (c,f) regions of whole images (a,d). Images were taken by SEM. Scale bars: 500 μm (a,d) and 30 μm (b,c,e,f). Starch granules at the lateral region of the endosperm were smaller and sparser in the Osatg7-1 mutant than in WT. (g–h) Close up of starch granules at the lateral region. Images were taken by EPMA. Scale bars: 2 μm. The starch granules of the Osatg7-1 mutant had small pits. Similar results were obtained from 5 independent samples.
A transmission electron microscopic (TEM) analysis was performed to elucidate the ultrastructure of protein bodies (PBs) in the endosperm of mature rice grains. In the mature endosperm, subcellular structures (PB-I, PB-II, and starch granules) in sectioned rice grains were perfectly preserved in WT and the Osatg7-1 mutant. PB-I was observed in the endosperm as large, spherical structures with diameters of 1–3 μm, whereas PB-II was electron-dense, irregularly-shaped structures with diameters of 2–4 μm (Supplementary Fig. 1). Therefore, no significant differences were noted in the size or morphology of PBs in the starch endosperm between the Osatg7-1 mutant and WT (Supplementary Fig. 1). These results indicate that the main cause of the chalky grains of the Osatg7-1 mutant is the abnormal (small and sparse) morphology of starch granules.
Involvement of autophagy in the regulation of starch and sugar metabolism during rice seed maturation
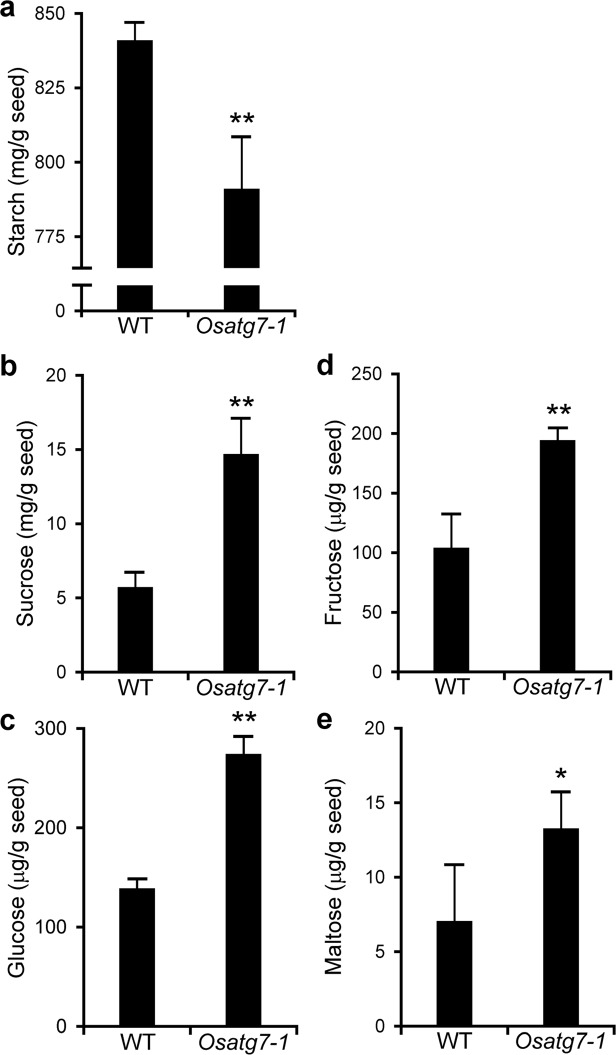
Quality of rice is essentially influenced by the chemical composition of the storage of starch and protein derived from nitrogen (N) and carbon (C) metabolism during grain filling27. Autophagy contributes to efficient N and C remobilization at the whole-plant level and plays an important role in the biomass production of rice28. To investigate the effects of disruption of autophagy on the sugar and starch contents of mature seeds, we conducted comprehensive analyses on the soluble sugar and starch levels of grains at the mature stage in the Osatg7-1 mutant and WT. Endogenous level of starch in grains were lower in the Osatg7-1 mutant than in WT (Fig. 3a), while the levels of a number of soluble sugars were higher in Osatg7-1 (Fig. 3b–e). Maltose, a typical degradation product of starch in the endosperm, also accumulated in the chalky grains of the Osatg7-1 mutant (Fig. 3e), suggesting that the abnormal activation of starch degradation occurs in the endosperm of the autophagy-deficient mutant, Osatg7-1.
Figure 3.
Effects of autophagy disruption on starch and sugar contents in mature grains of rice. Compositions of starch and soluble sugar contents in the grains of WT and the Osatg7-1 mutant at the mature stage. The amounts of starch (a), sucrose (b), glucose (c), fructose (d), and maltose (e) were measured. Data are means ± SD; n = 4 independent samples. *P < 0.05, **P < 0.01; significantly different from the controls.
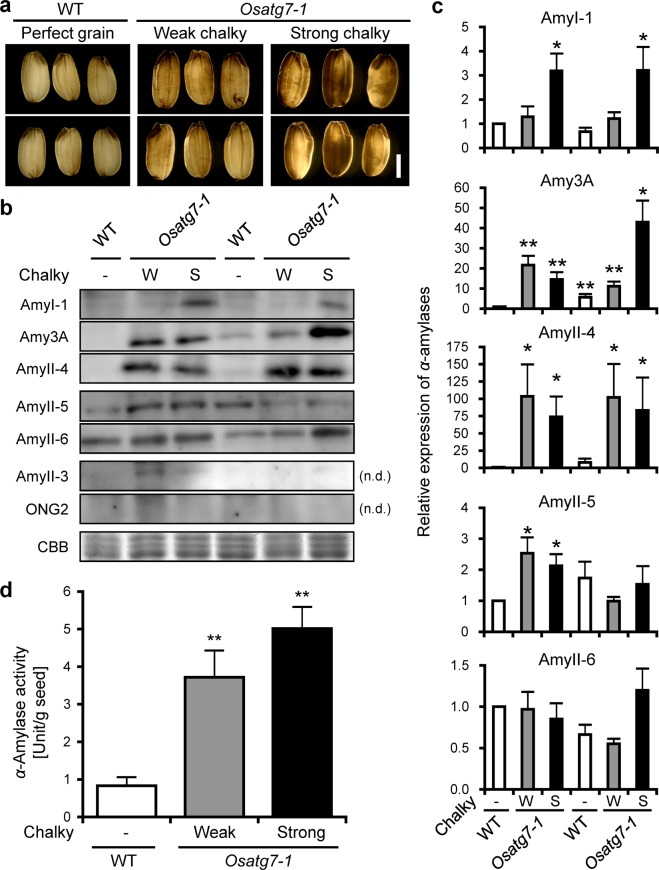
Rice has at least eight members of α-amylase, which are classified into three subfamilies: Amy1, Amy2 and Amy3 and play a role in the first committed step for the degradation of reserve starch29,30. We then characterized the amounts of α-amylase isoforms in the chalky grains of the Osatg7-1 mutant at a mature stage by immunoblot analysis (Fig. 4a–c). The results obtained showed that AmyI-1 (Amy1A) and AmyII-4 (Amy3D) were more strongly expressed in the chalky grains of the Osatg7-1 mutant than in WT (Fig. 4b,c). Moreover, the expression of α-amylase 3 A, which is one of the enzymes potentially involved in this chalky appearance through the degradation of starch accumulating in the developing grain, was also enhanced in Osatg7-1 grains, while no significant difference was observed for the other amylases (AmyII-3, II-5, and II-6) or α-glucosidase (ONG2) between WT and the Osatg7-1 mutant (Fig. 4b,c). As expected, the enzymatic activity of α-amylases was also higher in Osatg7-1 grains than in WT at a mature stage (Fig. 4d). These data were consistent with a previous study showing that the ectopic overexpression of α-amylases, such as Amy1A and Amy3D, resulted in chalky grains even under normal growth conditions31. Furthermore, Amy1A and Amy3D were shown to be mainly localized in the outer layers of rice grains, while α-glucosidase and AmyII-3 were mainly localized in the inner layers24. Since the Osatg7-1 mutant has starch granules with an abnormal morphology at the lateral region of the endosperm, the abnormal expression and activation of α-amylases may occur at the lateral region of the Osatg7-1 endosperm during rice seed maturation.
Figure 4.
Effects of autophagy disruption on the expression and activity of α-amylases in mature grains of rice. (a) A photograph of the chalky grains of the Osatg7-1 mutant at a mature stage grown in the paddy field. Pictures were taken under backlighting. (b) Immunoblotting images. Whole chalky grains at a mature stage were subjected to protein analysis, followed by SDS-PAGE and immunoblotting with specific antibodies28. Left and right parts indicate the sample of 2016 and 2017, respectively. The perfect proteins of Amy-II-3 and ONG2 were not detected (n.d.). CBB, Coomassie Brilliant Blue. W and S indicate a weak and strong chalky grain phenotype, respectively. The full-length blots are presented in Supplementary Figures 2 and 3. (c) Quantitation of α-amylase expression. The amount of each α-amylase isoform in WT grains was normalized to 1. Data are means ± SD; n = 4 independent samples. *P < 0.05; significantly different from the controls. (d) Quantitation of α-amylase activity in WT and Osatg7-1 mutant grains at a mature stage. Enzyme activity was corrected by the protein content of the respective extracts. Data are means ± SD; n = 4 independent samples. **P < 0.01; significantly different from the controls.
Effects of autophagy disruption on starch quality in the endosperm during rice seed maturation
Starch quality is an important parameter influencing the quality of rice grains. Previous studies indicated that environmental stresses, including temperature, at the grain filling stage alter the starch composition of rice grains32. Furthermore, the contents and fine structures of amylose and amylopectin in starch were shown to affect the physicochemical characteristics and textural properties of rice grains33. Therefore, we characterized the starch composition of the chalky grains of the Osatg7-1 mutant using the gel-filtration chromatography34. The results obtained showed no significant differences in the contents and structure of amylose and amylopectin in starchy endosperms between the Osatg7-1 mutant and WT grains (Supplementary Table 1), suggesting that the reasons for the loosely-packed starch granules of the Osatg7-1 mutant were factors other than the chain-length distribution of amylopectin.
Effects of autophagy disruption on the proteome of the endosperm in rice grains
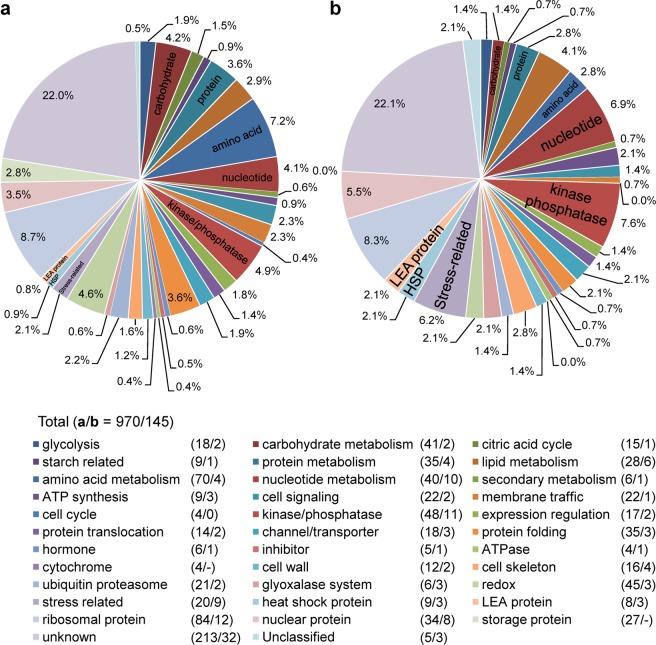
We performed a quantitative shotgun proteomic analysis of starchy endosperms prepared from Osatg7-1 chalky grains and WT grains. The proteins extracted were trypsin-digested and labeled using iTRAQ (isobaric tag for relative and absolute quantitation), followed by a tandem mass spectrometry (MS/MS) analysis24. We identified 970 proteins extracted from Osatg7-1 and WT grains. Among them, 145 proteins, 14.9% of all identified proteins, were deregulated (more than a 1.5-fold difference from WT normal grains; P value < 0.05) in Osatg7-1 chalky grains (Fig. 5 and Supplementary Table 2), suggesting possible involvement of autophagy in the expression of these proteins. Among this population, 130 proteins were up-regulated and 15 were down-regulated (Supplementary Table 2).
Figure 5.
Effects of autophagy disruption on the proteome of the mature grains of rice. Classification of proteins whose expression differed by more than 1.5-fold in mature grains between WT and the Osatg7-1 mutant. Whole seeds were subjected to protein extraction, followed by shotgun proteomic analysis with iTRAQ labeling. Proteins were categorized using UniProtKB database search. (a/b): (a) Total identified proteins (970); (b) >1.5-fold up- or down-regulated proteins in the Osatg7-1 mutant grains (145).
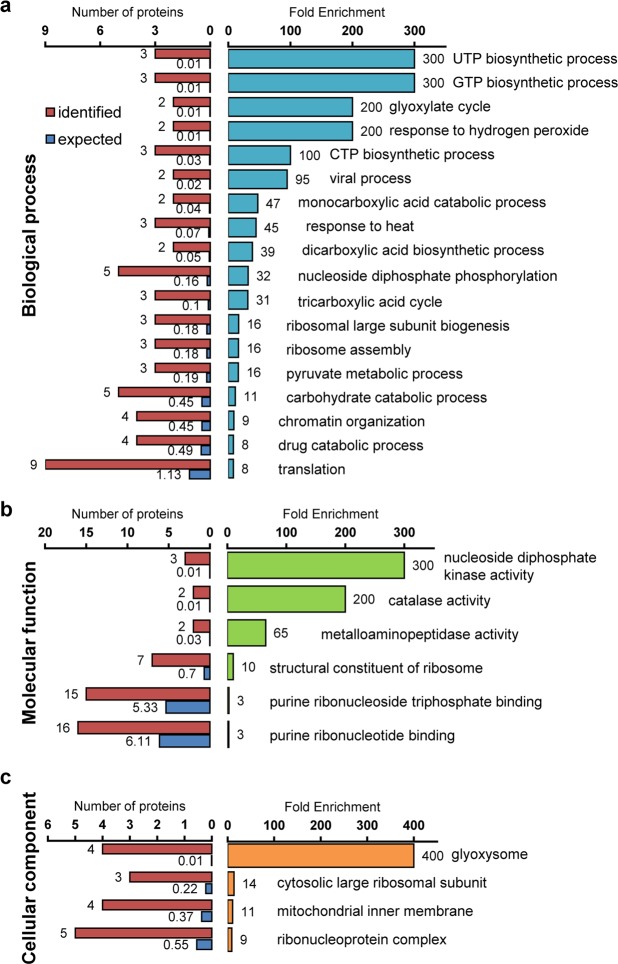
To investigate the biological processes affected by the disruption of autophagy in rice, an analysis of proteins using the InterProScan search was performed. The annotation of protein sequences using InterProScan permitted them to fall into 33 categories including energy and metabolic pathways and the molecular functional group (Fig. 5 and Supplemental Table 2). Our results showed that autophagy affects the carbohydrate, protein, amino acid and nucleotide metabolism. Since autophagy is an intracellular destructive mechanism that degrades intracellular proteins and metabolites for recycling, the source of amino acids may be depleted in the chalky grains of Osatg7-1 mutant. In the molecular functional group, stress-related proteins, HSPs (heat shock proteins) and LEA (late embryogenesis abundant) proteins were ranked at the top of the category, suggesting that the Osatg7-1 mutant gets stressed even under normal growth conditions.
To analyze the effects of autophagy disruption in rice seeds, gene ontology (GO) enrichment analysis using the PANTHER Classification System was performed. The annotation of protein UniProt IDs using the PANTHER website permitted them to fall into 198 GO terms (FDR P < 0.05) including biological process, molecular function, and cellular component (Fig. 6, display only the highest hierarchy, and Supplementary Table 3). Eighty-five of all 145 proteins differentially expressed between WT and the Osatg7-1 grains were mapped to 44321 reference genes contained in the whole rice genome, while the others (60 proteins) were unmapped. Among them, in the biological process group, response to hydrogen peroxide and response to heat were ranked at the top of the category (Fig. 6a). Moreover, in the molecular function and cellular component groups, both catalase activity and peroxisome-enzymatic pathways were ranked at the top of the category occupancy (Fig. 6b,c).
Figure 6.
Effects of autophagy disruption on the biological process, molecular function and cellular component of endosperm cells in rice. GO enrichment analysis using the PANTHER Classification System permitted them to fall into 198 GO terms including biological process (a), molecular function (b), and cellular component (c). Proteins were categorized using the PANTHER databases. The top hierarchies are shown in each process. The significance of each GO term is calculated by comparing GO terms of input proteins (identified; red) with those in the whole transcriptome (expected; blue).
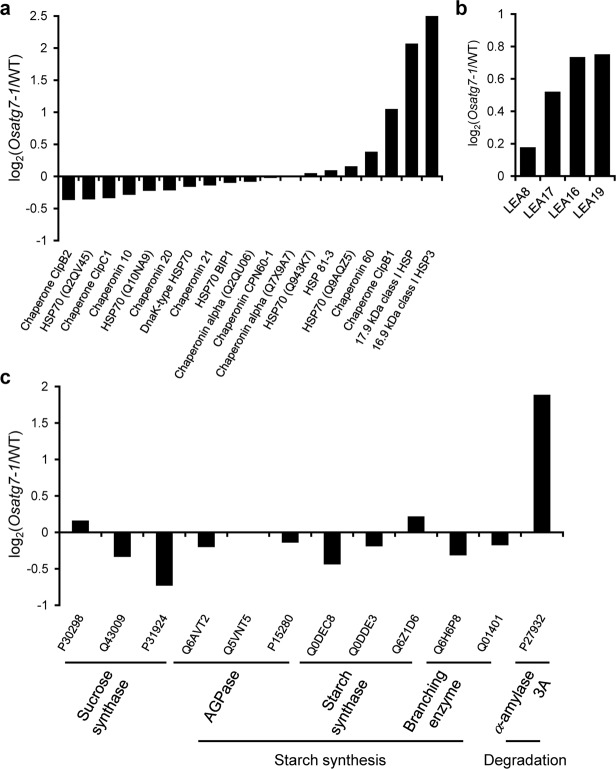
HSPs stabilize protein conformation, prevent aggregation, and, thus, maintain non-native proteins in a competent state for subsequent refolding as molecular chaperones in plants under heat stress35–37. Significant up-regulation of HSPs is a key part of the heat shock response38,39, and the levels of a variety of HSPs as well as chaperones were strongly up-regulated in the chalky grains of the Osatg7-1 mutant (Fig. 7a). In addition, LEA proteins, which are involved in desiccation resistance as well as direct protection of other proteins or membranes, or renaturation of unfolded proteins40,41, highly accumulated in the chalky grains of the Osatg7-1 mutant (Fig. 7b), suggesting that autophagy may be involved in protecting endosperm development from cellular damage caused by environmental stresses, such as heat or water loss, during rice seed maturation.
Figure 7.
Proteomic comparisons of starch metabolism-related enzymes and stress-related proteins in chalky grains of WT and the Osatg7-1 mutant. The comparative expression of heat shock-related proteins (a), LEA proteins (b) and starch metabolism-related enzymes (c) between WT and Osatg7-1 mutant grains.
Autophagy is induced by abiotic stresses42, and autophagy-defective mutants are hypersensitive to environmental stresses in several plant species43–47. It is partly because autophagy plays a critical role in the degradation of potentially toxic misfolded/denatured proteins and their aggregates as a protein quality control mechanism48–50. The present results showed that grains of Osatg7-1 mutant show whity chalky appearance under normal growth conditions, and the SEM analysis revealed smaller and sparser starch granules in the lateral region of the Osatg7-1 endosperm than WT (Figs. 1 and 2). In rice, chalky zone sometimes occurs at the lateral region of the endosperm in developing kernels under high-temperature conditions51, which are called white-back grains. Ring-shaped chalkiness is suggested to be a cell-specific event associated with the disruption of starch accumulation52. In contrast, increase in the frequency of pitting in starch granules observed in the grains ripened under high temperature has been suggested to be due to the premature autolysis of starch by abnormal induction of α-amylases in rice53,54, wheat and maize55, implying that autophagy deficiency may lead to a similar phenotype as heat stress-induced starch autolysis during seed maturation in rice.
However, no significant differences have been observed in the contents and structure of amylose and amylopectin in starchy endosperms between the Osatg7-1 mutant and WT grains (Supplementary Table 1). Slight alternation of amylopectin chain length has been shown to occur in the heat-induced chalky grains and long-length chains seem to increase in chalky grains56, suggesting some difference in the loosely-packed starch granules of the Osatg7-1 mutant and the heat-stressed wild type plants.
In the proteome analysis followed by GO analysis, proteins associated with hydrogen peroxide were ranked at the top of the category occupancy (Fig. 6a). Moreover, the level of catalase, one of the major scavenging enzymes of reactive oxygen species (ROS), was also upregulated in the chalky grains of the Osatg7-1 mutant (Fig. 6b). Autophagy-deficient mutants of Arabidopsis such as atg5 are reported to over-accumulate ROS in leaves18. Autophagy is involved in the turnover of peroxisomes, which play a role in the degradation of hydrogen peroxide57. Moreover, autophagy is involved in the turnover of mitochondria as well as chloroplasts58, which induce over-accumulate ROS in environmental stress conditions, suggesting that the ROS scavenging system may be activated in the anthers of the Osatg7-1 mutant to alleviate such oxidative damages. The relationship among autophagy, redox homeostasis and grain quality (enhanced frequency of chalky grains) as well as environmental stress adaptation would also be an important topic for future research.
Involvement of autophagy in the regulation of starch turnover and α-amylases during rice seed maturation
The present omics analyses revealed that the disruption of autophagy clearly resulted in the differential expression of proteins related to a variety of metabolite and suggested that starch metabolic pathways are strongly affected (Fig. 5). Among them, α-amylase 3 A was more strongly up-regulated in the chalky grains of the Osatg7-1 mutant than in WT grains (Fig. 7c). This result was consistent with those of the immunoblot analysis (Fig. 4b). In contrast, the expression levels of granule-bound starch synthases (GBSS), starch branching enzyme (SBE) BEIIb, ADP-glucose pyrophosphorylases (AGPases), and sucrose synthases (SuSy) were slightly lower in the Osatg7-1 mutant than in WT (Fig. 7c). These results suggest that autophagy participates in the regulation of the starch degradation pathways of the rice endosperm during seed maturation, and the impaired balance between starch biosynthesis and degradation at the seed maturation stage may cause the imperfect filling of endosperm cells with starch granules in the Osatg7-1 mutant.
As osmolytes and signaling molecules, soluble sugars participate in the response and adaptation of plants to environmental stresses59. Under salinity stress, photosynthetic carbon assimilation is inhibited, and starch turnover as well as an accumulation of soluble sugars are induced in rice leaves60. The production of soluble sugars, such as fructose, glucose, maltose, and sucrose, appeared to be at the expense of starch through the overexpression of α-amylase genes31. The accumulation of maltose may be due to starch degradation by the activation of α-amylase, which is a typical endoglucanase and produces oligosaccharides. High temperature stress promotes accumulation of glucose and sucrose in the mature grains of rice24,61,62. Moreover, in the developing seeds of rice, expression of the α-amylase genes Amy1A, Amy1C, Amy3D, and Amy3E were induced under high-temperature stress, and its suppression through an RNA interference (RNAi) strategy mitigated grain damage such as chalkiness8. In the present study, we found significant increase in the content of maltose as well as the activation of α-amylase isoforms in Osatg7-1 grains grown under normal growth conditions (Figs. 3, 4 and 7). These results support de novo starch degradation by the abnormal activation of α-amylase in the chalky grains of the Osatg7-1 mutant and suggest that its chalky-grain phenotype may be at least in part caused by enhanced sensitivity to environmental stresses.
Concluding remarks
Overall, the present study revealed that autophagy plays essential roles in the regulation of starch and sugar metabolism as well as quality control of endosperm cells during seed maturation in rice. These findings shed light on a possible novel strategy for protection of grain quality and crop yield affected by environmental stresses. The relationship among autophagy induction throughout seed development and maturation, quality control in organelles and stress adaptation as well as metabolite regulation during seed maturation, is an important topic for future research.
Methods
Plant materials and growth conditions
Surface-sterilized seeds of wild-type rice Oryza sativa L. cv. Nipponbare (NB) and the Osatg7-1 mutant21 were germinated on MS medium containing 0.8% agar and grown for 10 days in a growth chamber under long-day conditions (16-h light/8-h darkness, 28 °C). Seedlings were transplanted into soil and grown in the greenhouse at NIES in Tsukuba (Japan) or the paddy field at NIG in Mishima (Japan).
Tos17-insertional rice Osatg7-1 mutant (OsATG7−/−), wild-type (OsATG7 +/+), and heterozygous (OsATG7 +/−) plants were selected in seed pools obtained from heterozygous plants by genomic PCR using the following primer mixture: OsATG7 forward primer 5′-CATACTACCACCTCAGCTTGCTAG-3′, Tos-17 forward primer 5′-ACTATTGTTAGGTTGCAAGTTAGTTAAGA-3′, and OsATG7 reverse primer 5′-GCATTCAGGAAAACCTCGTATCG-3′. The original parental cultivars of the Tos17-insertional mutant and NB were also used as control plants for Osatg7-1.
Scanning electron microscopic (SEM) and Electron Probe Micro Analysis (EPMA)
SEM and EPMA analyses of the mature rice endosperm was carried out as described previously24. Rice grains were cracked with a razor blade, and the cracked surfaces were coated with gold for 90 s using a vaporizer (IB-3, EIKO, Tokyo, Japan) and subjected to SEM (JSM6510LA, JEOL, Tokyo, Japan) or EPMA (EPMA-8705, Shimazu, Kyoto, Japan). Observation conditions were as follows: acceleration voltage, 10 kV; magnification, 1,000–10,000. These analyses were carried out using three biological replicates with at least four technical repetitions of rice grains per replication of the mounted specimens.
Transmission electron microscopic (TEM) analysis
TEM analysis of the mature rice endosperm was carried out as described previously63. Samples were fixed with 4% w/v paraformaldehyde in Dullbecco’s PBS (−) (Nacalai Tesque, Kyoto, Japan) for 3 h. Specimens were rinsed in the same buffer and the samples were infiltrated overnight with D-PBS (−). Dehydrated specimens were embedded in LR White resin (London Resin, Hampshire, UK). Resin sections (200 nm) were cut with a diamond knife using Ultracut UCT. Ultrathin sections were stained with 2% w/v uranyl acetate. After staining, samples were examined with TEM (JEM-1220, JEM, Tokyo) at 80 kV.
Measurement of soluble sugars and starch content
Rice grains were pulverized using a Shake Master NEO (Biomedical Science Inc.) with a stainless steel bead (6 mm, Biomedical Science Inc.) and three zirconia beads (3 mm, Nikkato Inc.) for 5 min at 1,750 rpm. Soluble sugars were extracted from 5–10 mg powdered rice grain with 1 mL of 80% EtOH containing 20 μg/mL methyl glucose (mGlc) as internal control for 1 h and the supernatant was recovered as soluble sugar fraction. The supernatant was completely evaporated by centrifugal dryer and then dissolved with ultrapure water. Contaminated protein in dissolved solution was removed with chloroform twice and then aqueous layer was recovered, evaporated, and then dissolved with ultrapure water again. The glucose, fructose, and maltose in dissolved solution were derivatized with p-aminobenzyl ethyl ester (ABEE) and those contents were determined with ABEE-UPLC system as described previously64. Sucrose was determined by the enzymatic method with invertase. Glucose released from sucrose in invertase solution containing 2 unit/mL invertase solution (Wako, Osaka, Japan) and 25 mM citrate at pH4.8 was determined as sucrose content by Glucose test Kit II (Wako, Osaka, Japan). Those detected sugar amounts were normalized with recovery rate of mGlc.
Starch content was determined by slightly modified method of “Resistant Starch Assay” (Megazyme). Insoluble residue above mentioned was rinsed with 80% EtOH again and then dried overnight at 65 °C. Crushed rice grain (2–3 mg) was hydrophilized with 20 μL of 100% EtOH and gelatinized in 180 μL of maleate buffer (pH4.8) for 10 min at 65 °C. Starch in gelatinized suspension was digested in amylase solution containing 100 unit/mL α-amylase (Megazyme) and 5.5 unit/mL amyloglucosidase (Megazyme) in maleate buffer (pH4.8) for 20 h at 37 °C with shaking. The liberated glucose from insoluble residue was determined as starch content by Glucose Test Kit II (Wako, Osaka, Japan).
Preparation of starch granules and amylopectin for gel filtration
The method used to assess the composition of carbohydrate content in endosperm was performed as described previously34. The eluate refractive index was measured with an RI-8020 detector (Tosoh, Japan). Fractions I, II, and III were divided at the minimum values of the refractive index curves. The contents of apparent amylose, true amylose, and extra-long chains in starch were calculated from the ratios of fractions I, II, and III. The contents of each fraction were confirmed by mixing the eluate with iodine and measuring the λmax value with the spectrophotometer as described previously65.
Measurement of α-amylase activity in embryonic rice
α-Amylase activity was assayed by a modification of the method described previously66. Mature rice grains were ground in liquid nitrogen to fine powder and then suspended in a 10-fold volume of extraction buffer (20 mM Tris-HCl, pH 7.5, 0.01% (w/v) Triton X-100) at 4 °C for 10 min. The suspensions were centrifuged at 15,000 × g at 4 °C for 15 min, and the supernatant was used to determine α-amylase activity. One-hundred μL of substrate solution (50 mM acetate buffer, pH 5.3, 0.5 mM CaCl2, 5 mM EDTA, pH7.5 and 0.34% soluble potato starch) and 100 μL of the supernatant was incubated at 37 °C for 5 min and then the reaction was stopped by the addition of 250 μL KI solution (1 mM). After dilution of with D.W. (up to 1 mL), A620 was determined to measure the α-amylase activity. The activity of α-amylase was calculated using the following equation: units of α-amylase activity = [10 − (A620 Sample/ A620 Blank) × 10]. The data were corrected by the protein content of the respective extracts determined by the Pierce 660 nm Protein Assay Kit (Thermo Fisher Scientific) using bovine serum albumin (BSA) as a standard.
Immunoblot analysis
The immunoblot analysis was performed as described previously24. Crude protein samples from mature grains of rice were separated on a 12% SDS-PAGE gel and blotted onto a PVDF membrane. The membrane was blocked in PBST buffer (8.1 mM Na2HPO4, 137 mM NaCl, 1.5 mM KH2PO4, 2.7 mM KCl and 0.1% Tween-20, pH7.5) with 5% fat-free milk 25 °C for 1 h. Blots were incubated with specific antibodies detecting the amylase isoforms30 at 25 °C for 1 h, and then with horseradish peroxidase (HRP)-linked anti-rabbit immunoglobulin G. Bands were quantitatively detected using chemiluminescent HRP substrate ECL Prime (Amersham) and the chemiluminescent analyzer, LAS3000 (GE Healthcare). Signals were normalized by the intensity of Coomassie brilliant blue staining with the corresponding protein.
Proteomic analysis
Two hundred mg of starchy endosperm prepared from the opaque part of chalky grains or the corresponding part of perfect grains were ground in liquid nitrogen to fine powder and then suspended in 7 M urea, 2 M thiourea, 3% (w/v) CHAPS, 1% (v/v) Triton X-100, and 10 mM DTT. The suspensions were centrifuged at 10,000 × g at 4 °C for 5 min. The supernatants were mixed with 1/10 volume of 100% (w/v) trichloroacetic acid, incubated on ice for 15 min, and centrifuged at 10,000 × g at 4 °C for 15 min. The resulting protein precipitates were washed 3 times in ice-cold acetone and resuspended in 8 M urea. Protein concentration was determined by the Pierce 660 nm Protein Assay Kit (Thermo Fisher Scientific).
The procedure of quantitative shotgun proteomic analysis was essentially identical as described previously24. Each protein preparation (50 μg) was digested in 20 μL of endoproteinase Lys-C (1 μg/ μL) at 37 °C for 3 h, then diluted to 10 times volume by ultrapure water (18.2 MΩ cm). The diluted samples were further digested in 200 μL of trypsin (1 μg/ μL) at 37 °C for 12 h. iTRAQ labeling of peptides were carried out with 4-plex iTRAQ tags the manufacturer’s protocol (Sciex), and the resultant 4 iTRAQ-labeled peptide samples were mixed. iTRAQ analysis was performed by using a DiNa-A-LTQOrbitrap-XL system. The iTRAQ labeled peptides were loaded on a trap column (HiQ sil C-18 W-3; 0.5 mm i.d. × 1 mm, 3 μm particle size) with buffer A consisting of 0.1% (v/v) formic acid and 2% (v/v) acetonitrile in water using a DiNa-A system (KYA Tech., Tokyo, Japan). A linear gradient from 0 to 33% buffer B (0.1% formic acid and 80% acetonitrile in water) for 600 min, 33 to 100% B for 10 min and back to 0% B in 15 min was applied, and peptides eluted from the HiQ sil C-18 W-3 column were directly loaded on a separation column (MonoCap C18 High Resolution 2000; 0.1 mm i.d. × 2000 mm, 2 μm pore size). The separated peptides were introduced into an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) with a flow rate of 300 nL min−1 and an ionization voltage 1.7–2.5 kV.
Liquid chromatography-MS/MS (LC-MS/MS) spectrometer was operated using Xcalibur 2.0 software (Thermo Fisher Scientific). The mass range selected for MS scan was set to 350–1,600 m/z and the top three peaks were subjected to MS/MS analysis. Full MS scan was detected in the Orbitrap, and the MS/MS scans were detected in the linear ion trap and Orbitrap. The normalized collision energy for MS/MS was set to 35 eV for collision-induced dissociation (CID) and 45 eV for higher-energy C-trap dissociation (HCD). High resolution of Fourier transform mass spectrometer (FTMS) was maintained at 60,000 resolutions. Divalent or trivalent ions were subjected to MS/MS analysis in dynamic exclusion mode, and proteins were identified with Proteome Discoverer v. 1.4 software and the SEQUEST HT (Thermo Fisher Scientific) and Ms Amanda67 search tool using the UniProt (http://www.uniprot.org/) O. sativa subsp. japonica database (63,535 proteins) with the following parameters: enzyme, trypsin; maximum missed cleavages site, 2; peptide charge, 2 + or 3 + ; MS tolerance, 5 ppm; MS/MS tolerance, ± 0.5 Da; dynamic modification, carboxymethylation (C), oxidation (H, M, W), iTRAQ 4-plex (K, Y, N-terminus). False discovery rates were < 1%.
Gene ontology analysis
Gene Ontology (GO) enrichment analyses of GO terms were conducted on each set of differentially expressed proteins to the reference list including all genes in Oryza sativa genome (released 2018–11–15) by using the PANTHER overrepresentation test. Enriched GO terms were calculated false discovery rate (FDR) settings and statistically analyzed by Fisher’s exact test (P < 0.05).
Statistical analysis
Significance was assessed using an unpaired Student’s t-test at P < 0.05.
Accession codes
Sequence data from this article can be found in the GenBank databases under the following accession numbers: OsATG7 (Os01g0614900), AmyI-1 (Os02g0765600), Amy3A (Os09g0457400), AmyII-3 (Os08g0473600), AmyII-4 (Os08g0473900), AmyII-5 (Os09g0457600), AmyII-6 (Os09g0457800), and ONG2 (Os06g0675700).
Supplementary information
Acknowledgements
We would like to thank Yuko Hosaka, Jumpei Sawada, Togo Fukunaga, Shohei Shiina and Shoko Saji for their general experimental technical support. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology for challenging Exploratory Research (15K14681, 16H01207 and 17K19274) to K.K., for Scientific Research (C) (18K05562) to T.K., by the Asahi Glass Foundation to T.K, and by the NIG-JOINT (84A2018 and 39A2019) to K.K.
Author contributions
T. Kurusu and K. Kuchitsu designed the research; Y.S., S. Hanamata, S.S., S.O., K. Kaneko, Y.M., T. Koyano, N.F., A.S., T. Masumura and H. Saji performed the research; Y.S., S. Hanamata, K. Kaneko, and T. Kurusu analyzed the data; T. Kurusu, Y.S., S.H., K.N., N.M., T. Mitsui, and K. Kuchitsu wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuri Sera and Shigeru Hanamata.
Contributor Information
Takamitsu Kurusu, Email: kurusu@rs.sus.ac.jp.
Kazuyuki Kuchitsu, Email: kuchitsu@rs.tus.ac.jp.
Supplementary information
is available for this paper at 10.1038/s41598-019-54361-1.
References
- 1.Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown RC, Lemmon BE, Olsen OA. Development of the endosperm in rice (Oryza sativa L): Cellularization. J Plant Res. 1996;109:301–313. doi: 10.1007/BF02344477. [DOI] [Google Scholar]
- 3.Krishnan S, Dayanandan P. Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.) J Biosci. 2003;28:455–469. doi: 10.1007/BF02705120. [DOI] [PubMed] [Google Scholar]
- 4.Olsen OA. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell. 2004;16(Suppl):S214–227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becraft PW, Yi G. Regulation of aleurone development in cereal grains. J Exp Bot. 2011;62:1669–1675. doi: 10.1093/jxb/erq372. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Gu Y, Zheng Y, Wang H. Structure observation of rice endosperm cell development and its mineral element analysis. Chin J Rice Sci. 2012;26:693–705. [Google Scholar]
- 7.Wu X, Liu J, Li D, Liu CM. Rice caryopsis development II: Dynamic changes in the endosperm. J Integr Plant Biol. 2016;58:786–798. doi: 10.1111/jipb.12488. [DOI] [PubMed] [Google Scholar]
- 8.Hakata M, et al. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol J. 2012;10:1110–1117. doi: 10.1111/j.1467-7652.2012.00741.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Fitzgerald M. Climate change: implications for the yield of edible rice. Plos One. 2013;8:e66218. doi: 10.1371/journal.pone.0066218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurusu T, Hanamata S, Kuchitsu K. Quantitative live cell imaging of autophagic flux and roles of autophagy in reproductive development in plants. Bioimages. 2016;24:1–11. [Google Scholar]
- 12.Avin-Wittenberg T, Honig A, Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma. 2012;249:285–299. doi: 10.1007/s00709-011-0296-z. [DOI] [PubMed] [Google Scholar]
- 13.Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimoto K. Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 2012;53:1355–1365. doi: 10.1093/pcp/pcs099. [DOI] [PubMed] [Google Scholar]
- 15.Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149:220–234. doi: 10.1104/pp.108.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida H, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–155. doi: 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiglione HO, et al. Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 2008;55:1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x. [DOI] [PubMed] [Google Scholar]
- 18.Yoshimoto K, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon SI, Cho HJ, Park OK. Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation. Autophagy. 2010;6:1187–1189. doi: 10.4161/auto.6.8.13429. [DOI] [PubMed] [Google Scholar]
- 20.Izumi M, et al. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 2015;167:1307–1320. doi: 10.1104/pp.114.254078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurusu T, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014;10:878–888. doi: 10.4161/auto.28279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanamata S, Kurusu T, Kuchitsu K. Roles of autophagy in male reproductive development in plants. Front Plant Sci. 2014;5:457. doi: 10.3389/fpls.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurusu T, et al. Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal Behav. 2017;12:e1365211. doi: 10.1080/15592324.2017.1365211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko K, et al. Proteomic and glycomic characterization of rice chalky grains produced under moderate and high-temperature conditions in field system. Rice (N Y) 2016;9:26. doi: 10.1186/s12284-016-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi M, et al. Endosperm structure of White-Belly and White-Core rice grains shown by scanning electron microscopy. Plant Prod Sci. 2014;17:285–290. doi: 10.1626/pps.17.285. [DOI] [Google Scholar]
- 26.Sreenivasulu N, et al. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J Exp Bot. 2015;66:1737–1748. doi: 10.1093/jxb/eru544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald MA, McCouch SR, Hall RD. Not just a grain of rice: the quest for quality. Trends Plant Sci. 2009;14:133–139. doi: 10.1016/j.tplants.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Wada S, et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 2015;168:60–73. doi: 10.1104/pp.15.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang N, Koizumi N, Reinl S, Rodriguez RL. Structural organization and differential expression of rice α-amylase genes. Nucleic Acids Res. 1990;18:7007–7014. doi: 10.1093/nar/18.23.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsui T, Yamaguchi J, Akazawa T. Physicochemical and serological characterization of rice α-amylase isoforms and identification of their corresponding genes. Plant Physiol. 1996;110:1395–1404. doi: 10.1104/pp.110.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakata M, et al. High Temperature-induced expression of rice α-amylases in developing endosperm produces chalky grains. Front Plant Sci. 2017;8:2089. doi: 10.3389/fpls.2017.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsui T, Yamakawa H, Kobata T. Molecular physiological aspects of chalking mechanism in rice grains under high-temperature stress. Plant Prod Sci. 2016;19:22–29. doi: 10.1080/1343943X.2015.1128112. [DOI] [Google Scholar]
- 33.Muench DG, et al. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- 34.Toyosawa Y, et al. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016;170:1255–1270. doi: 10.1104/pp.15.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morimoto RI. Dynamic remodeling of transcription complexes by molecular chaperones. Cell. 2002;110:281–284. doi: 10.1016/S0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- 36.Wang WX, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Zi J, et al. Stress responsive proteins are actively regulated during rice (Oryza sativa) embryogenesis as indicated by quantitative proteomics analysis. Plos One. 2013;8:e74229. doi: 10.1371/journal.pone.0074229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scarpeci TE, Zanor MI, Valle EM. Investigating the role of plant heat shock proteins during oxidative stress. Plant Signal Behav. 2008;3:856–857. doi: 10.4161/psb.3.10.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao JL, Zhou HW, Zhang HY, Zhong PA, Huang YJ. Comparative proteomic analysis of differentially expressed proteins in the early milky stage of rice grains during high temperature stress. J Exp Bot. 2014;65:655–671. doi: 10.1093/jxb/ert435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 41.Olvera-Carrillo Y, Luis Reyes J, Covarrubias AA. Late embryogenesis abundant proteins: versatile players in the plant adaptation to water limiting environments. Plant Signal Behav. 2011;6:586–589. doi: 10.4161/psb.6.4.15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot. 2008;59:4029–4043. doi: 10.1093/jxb/ern244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3:257–258. doi: 10.4161/auto.3847. [DOI] [PubMed] [Google Scholar]
- 44.Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu YM, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9:e1003196. doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi: 10.1371/journal.pgen.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 49.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishimaru T, et al. Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under high-temperature stress. J Cereal Sci. 2009;50:166–174. doi: 10.1016/j.jcs.2009.04.011. [DOI] [Google Scholar]
- 52.Wada H, et al. Rice chalky ring formation caused by temporal reduction in starch biosynthesis during osmotic adjustment under foehn-induced dry wind. Plos One. 2014;9:e110374. doi: 10.1371/journal.pone.0110374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tashiro T, Wardlaw IF. The effect of high-temperature on kernel dimensions and the type and occurrence of kernel damage in rice. Aust J Agr Res. 1991;42:485–496. doi: 10.1071/AR9910485. [DOI] [Google Scholar]
- 54.Tashiro T, Wardlaw IF. The response to high-temperature shock and humidity changes prior to and during the early stages of grain development in wheat. Aust J Plant Physiol. 1990;17:551–561. [Google Scholar]
- 55.Commuri PD, Jones RJ. Ultrastructural characterization of maize (Zea mays L.) kernels exposed to high temperature during endosperm cell division. Plant Cell Environ. 1999;22:375–385. doi: 10.1046/j.1365-3040.1999.00424.x. [DOI] [Google Scholar]
- 56.Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshimoto K, et al. Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci. 2014;127:1161–1168. doi: 10.1242/jcs.139709. [DOI] [PubMed] [Google Scholar]
- 58.Izumi M, Ishida H, Nakamura S, Hidema J. Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell. 2017;29:377–394. doi: 10.1105/tpc.16.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S, Sreeharsha RV, Mudalkar S, Sarashetti PM, Reddy AR. Molecular insights into photosynthesis and carbohydrate metabolism in Jatropha curcas grown under elevated CO2 using transcriptome sequencing and assembly. Sci Rep. 2017;7:11066. doi: 10.1038/s41598-017-11312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubey RS, Singh AK. Salinity induces accumulation of soluble sugars and alters the activity of sugar metabolising enzymes in rice plants. Biol Plantarum. 1999;42:233–239. doi: 10.1023/A:1002160618700. [DOI] [Google Scholar]
- 61.Yamakawa H, Hakata M. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010;51:795–809. doi: 10.1093/pcp/pcq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsutsui K, Kaneko K, Hanashiro I, Nishinari K, Toshiaki M. Characteristics of opaque and translucent parts of high temperature stressed grains of rice. J appl glycosci. 2013;60:61–67. doi: 10.5458/jag.jag.JAG-2012_014. [DOI] [Google Scholar]
- 63.Saito Y, et al. Ultrastructure of mature protein body in the starchy endosperm of dry cereal grain. Biosci Biotechnol Biochem. 2010;74:1485–1487. doi: 10.1271/bbb.100147. [DOI] [PubMed] [Google Scholar]
- 64.Sakamoto S, Yoshida K, Sugihara S, Mitsuda N. Development of a new high-throughput method to determine the composition of ten monosaccharides including 4-O-methyl glucuronic acid from plant cell walls using ultra-performance liquid chromatography. Plant Biotechnol (Tokyo) 2015;32:55–63. doi: 10.5511/plantbiotechnology.15.0113a. [DOI] [Google Scholar]
- 65.Horibata T, Nakamoto M, Fuwa H, Inouchi N. Structural and physicochemical characteristics of endosperm starches of rice cultivars recently bred in Japan. J appl glycosci. 2004;51:303–313. doi: 10.5458/jag.51.303. [DOI] [Google Scholar]
- 66.Jones RL, Varner JE. The bioassay of gibberellins. Planta. 1966;72:155–161. doi: 10.1007/BF00387479. [DOI] [PubMed] [Google Scholar]
- 67.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.