Abstract
The bacterial, acidobacterial, and fungal communities in wetlands can undergo perturbations by various human activities, such as disturbances caused by cultivation and during the process of system restoration. In this study, we investigated the relationships between the composition of the soil bacterial, acidobacterial, and fungal communities and the transformation of wetlands by human activities in the Sanjiang Plain. Soil microbial communities were assessed in wetland soils collected from pristine marsh, neighboring cropland (wetland turned into arable land), and land that had been reforested with Larix gmelinii. The alpha-diversities of bacteria, Acidobacteria, and fungi were affected by land-use change and were highest in the arable land and lowest in the wetland soils. The soil microbial community structures were also altered with changing land-use. Canonical correlation analyses showed that beta-diversity was significantly affected by soil pH, available phosphorus, soil nitrogen, and total organic carbon. Overall, our results showed that the agricultural cultivation of wetlands changes the available soil carbon, nitrogen, and phosphorus pools, thereby influencing the bacterial, acidobacterial, and fungal diversity and community structure. Once the soil microbial community has been altered by human activity, it might be difficult to restore it to its original state. These findings highlight the importance of effectively maintaining the diversity of soil bacterial, Acidobacterial, and fungal communities despite land use change in order to sustain a microbial community diversity and ecosystem function.
Subject terms: Microbial ecology, Wetlands ecology
Introduction
Globally, marsh wetlands are currently threatened by human activities such as land reclamation and forestation1–3. These activities can affect water cycle patterns, with downstream effects on deposition and the removal of organic and inorganic matter that result in a reduction of soil organic matter, which can ultimately produce barren areas with low groundwater tables. In recent years, the impacts that cultivation4, pollution5, and wetland soil microbial community dynamics have on wetland carbon and nitrogen cycles (e.g., greenhouse gas fluxes to the atmosphere)6 have attracted increasing attention. It has been demonstrated that soil microbes are crucial players in ecosystem functioning7, but there are many microbial taxa in soils. Different microbial groups can differentially impact soil organic matter decomposition, carbon and nitrogen cycling, and the overall stability of the ecosystem8. Microbial community structures, with their direct effects on soil functions, reflect changes in the soil nutrient status, and to some extent they are sentinels of the impact of environmental changes9. Therefore, understanding the taxonomical and functional dynamics of the soil microbiome will improve our knowledge of the processes of organic matter decomposition, nitrogen fixation, plant–soil interactions, and degradation of organic pollutants10–12.
The Sanjiang Plain is the largest wetland in China. It is one of the key regions for global wetland and biodiversity conservation, and the region is also an important grain production and reserve base in China. In recent years, human activities such as cultivation and over-utilization have caused a dramatic decline in the wetland area and have weakened the ecological functions of the remaining wetland. In addition, changes in the soil microbial community structure have been observed13–15. The black soil of the Sanjiang marsh wetland in northeastern China has a high humus content and unusual microbial community structures2 due to very cold climatic conditions in the winter. Acidobacteria is a new phylum of bacteria that play an important role in soil ecological processes; this phylum accounts for 20–50% of all bacteria, making it one of the most widely distributed and diverse bacterial phyla in natural environments16–19. In a previous study, we determined that Acidobacteria was the most abundant bacterial phylum in the soils of the Sanjiang Plain11. Therefore, studying the effects of land use on wetland soil microbial communities is very important for better understanding the future changes in wetland ecosystem function and services under global environmental change. Such knowledge will help us to create better policies to effectively protect the Sanjiang wetland.
There is some research on the effects of land- use change on soil physicochemical and biogeochemical characteristics in wetlands and other ecosystems20–22. Land-use change can have significant and long-lasting effects on soil carbon and nutrient content, soil texture, and soil pH and that these effects largely arise from changes in the above-ground vegetation and associated management practices across land-use types20–22. However, less is known about how wetland change may affect the soil microbiome because of the multiple soil physicochemical factors that are influenced by land-use change. Specifically, many soil variables, such as soil organic matter, texture, salinity and pH, have a significant impact on the soil microbiome, and all of these soil factors may be altered by changes in land-use type23–26. Some studies have found that the microbial structure of pristine soil microbes are similar after land use patterns change, indicating that land use patterns do not affect the structure and function of soil microbes27,28. However, some studies have found that once land use patterns change, microbial communities cannot recover their natural state even after years of recovery29. Peralta et al. have shown that wetland soil microbes can only recover from microbial functions in wetland soils only the microbe overcomes the limitation of soil and physicochemical factors30. Xu et al. found that the soil microbial community structure could not be restored to the original state after the change of land use pattern, but the microbial function was restored indicating functional microbial redundancy31. Similarly, our previous study showed that bacterial community metabolic functions changed when wetlands were changed into arable land in the Sanjiang Plain15. Overall, the few and sometimes contradictionary studies on the changes in microbial community structure and their drivers (e.g. soil physicochemical properties) limit our understanding of soil biogeochemical processes after the reclamation of marshes in farmland and plantation.
To gain deeper insight into how soil biogeochemical processes are affected by wetland changes, we used a typical degraded wetland in Sanjiang plain, northeastern China and selected two dominant typical habitats by human activity disturbance: a restored forest (planted on degraded wetlands in 2000) and a managed arable land (soybean planted in 2007), which is a typical example of an inland freshwater marsh that is dramatically changing. Using Illumina MiSeq sequencing technology we examined changes in soil bacterial, acidobacterial and fungal communities, and their relationships with soil physiochemical properties in three different land use types originating from marsh wetland cultivation. Our specific objectives were (i) to elucidate the changes in soil microbial communities in marshes that have experienced long periods of cultivation and forest plantation, (ii) to investigate the relationship between microbial communities and soil properties, and (iii) to gain insight into soil biogeochemical processes after marsh changes. The data presented here will help quantify the influence of land-use pattern changes on wetland soil microbes in northeastern China. This research is of great practical significance for improving environment management and utilization of microbial resources, and provides scientific guidance for wetland protection and management in northeastern China.
Results
Soil bacterial, Acidobacterial, and fungal community composition
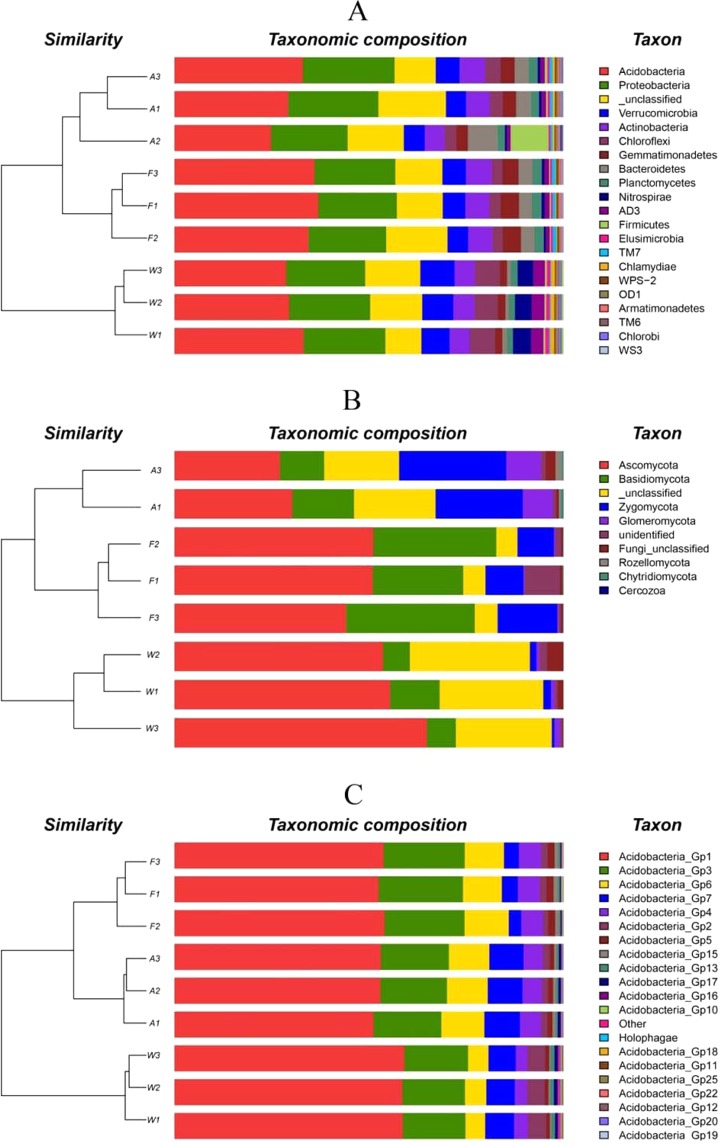
A total of 3917 bacterial OTUs included here belonged to 40 taxonomic groups. In decreasing order of abundance, the six most abundant phyla were as follows: Acidobacteria (31.8%) > Proteobacteria (21.1%) > Verrucomicrobia (6.4) > Actinobacteria (4.1%) > Chlorobi (3.2%) > Gemmatimonadetes (3.2%) (Fig. 1A, Table 1). These were considered to constitute the main bacterial phyla and, together with a significant fraction of unclassified OTUs (9%), they represented approximately 80% of the data from bacterial phyla. There were 30 phyla with a relative abundance <1%. The similarity of soil bacterial community structure was divided into two parts (Fig. 1A): wetlands (W1–W3) had similar bacterial community structures, while arable lands (A1–A3) and forests (F1–F3) had similar bacterial community structures.
Figure 1.
Histogram and cluster analysis of soil bacterial (panel A), fungal (B) and acidobacterial (C) communities in different land use types, for three forests samples (F1–F3), three wetland samples (W1–W3) and three arable land samples (A1–A3).
Table 1.
Differences in relative abundances of all detected phyla in the different land use habitat types in the Sanjiang Wetland, northeastern China.
| Taxonomy | Relative abundance (mean ± sd; n = 3) | One-way ANOVA | |||
|---|---|---|---|---|---|
| Domain | Phylum | Wetland | Forest | Arable land | |
| Bacteria | Acidobacteria | 30.5 ± 1.38 | 35.8 ± 0.74 | 29.0 ± 2.36 | 0.058 ns |
| Bacteria | Proteobacteria | 20.7 ± 0.21 | 20.3 ± 0.24 | 22.2 ± 1.24 | 0.259 ns |
| Bacteria | unclassified | 12.3 ± 1.53 | 13.2 ± 1.27 | 14.1 ± 1.96 | 0.732 ns |
| Bacteria | Verrucomicrobia | 8.0 ± 0.4 | 5.7 ± 0.22 | 5.5 ± 0.30 | 0.003** |
| Bacteria | Actinobacteria | 5.3 ± 0.09 | 6.4 ± 0.15 | 6.0 ± 0.42 | 0.063 ns |
| Bacteria | Chloroflexi | 6.3 ± 0.09 | 2.7 ± 0.15 | 3.4 ± 0.424 | 0.000** |
| Bacteria | Gemmatimonadetes | 1.9 ± 0.01 | 4.5 ± 0.21 | 3.3 ± 0.17 | 0.000** |
| Bacteria | Bacteroidetes | 1.0 ± 0.06 | 3.5 ± 0.11 | 5.0 ± 1.31 | 0.027* |
| Bacteria | Planctomycetes | 1.6 ± 0.06 | 2.4 ± 0.04 | 2.1 ± 0.14 | 0.002** |
| Bacteria | Nitrospirae | 4.3 ± 0.17 | 0.6 ± 0.08 | 0.6 ± 0.04 | 0.000** |
| Bacteria | AD3 | 3.2 ± 0.10 | 1.2 ± 0.08 | 1.1 ± 0.07 | 0.000** |
| Bacteria | Firmicutes | 0.5 ± 0.00 | 0.3 ± 0.03 | 3.6 ± 3.07 | 0.389 |
| Bacteria | Chlamydiae | 1.0 ± 0.03 | 0.2 ± 0.01 | 0.5 ± 0.02 | 0.000** |
| Bacteria | Others | 3.5 ± 0.08 | 3.1 ± 0.08 | 3.5 ± 0.08 | 0.018* |
| Fungi | Ascomycota | 57.99 ± 6.06 | 48.74 ± 3.98 | 28.71 ± 2.26 | 0.217 ns |
| Fungi | Zygomycota | 1.41 ± 0.68 | 11.48 ± 3.33 | 24.96 ± 3.63 | 0.032* |
| Fungi | unclassified | 27.40 ± 3.15 | 5.69 ± 0.23 | 20.12 ± 1.22 | 0.078 ns |
| Fungi | Basidiomycota | 9.01 ± 3.21 | 29.31 ± 5.27 | 13.62 ± 3.16 | 0.337 ns |
| Fungi | Glomeromycota | 1.11 ± 0.38 | 0.32 ± 0.04 | 8.36 ± 0.82 | 0.007** |
| Fungi | unidentified | 1.08 ± 0.84 | 3.75 ± 4.66 | 0.98 ± 0.24 | 0.024* |
| Fungi | Others | 0.03 ± 0.01 | 0.19 ± 0.06 | 0.55 ± 0.25 | 0** |
| Fungi | Fungi_unclassified | 1.98 ± 1.97 | 0.51 ± 0.08 | 1.60 ± 1.31 | 0.067 |
| Fungi | Rozellomycota | 0.00 ± 0.00 | 0.02 ± 0.01 | 1.10 ± 0.88 | 0.000** |
| Acidobacteria | Acidobacteria_Gp1 | 28.95 ± 0.24 | 28.62 ± 0.95 | 31.08 ± 0.22 | 0.035* |
| Acidobacteria | Acidobacteria_Gp10 | 0.03 ± 0.00 | 0.10 ± 0.01 | 0.20 ± 0.05 | 0.015* |
| Acidobacteria | Acidobacteria_Gp11 | 0.08 ± 0.06 | 0.01 ± 0.02 | 0.00 ± 0.00 | 0.162 ns |
| Acidobacteria | Acidobacteria_Gp12 | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.084 ns |
| Acidobacteria | Acidobacteria_Gp13 | 0.49 ± 0.00 | 0.30 ± 0.02 | 0.28 ± 0.05 | 0.040* |
| Acidobacteria | Acidobacteria_Gp15 | 0.18 ± 0.01 | 0.43 ± 0.04 | 0.60 ± 0.06 | 0.084 ns |
| Acidobacteria | Acidobacteria_Gp16 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.09 ± 0.00 | 0.400 ns |
| Acidobacteria | Acidobacteria_Gp17 | 0.36 ± 0.05 | 0.30 ± 0.03 | 0.12 ± 0.00 | 0.092 ns |
| Acidobacteria | Acidobacteria_Gp18 | 0.10 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.053 ns |
| Acidobacteria | Acidobacteria_Gp19 | 0.02 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.018* |
| Acidobacteria | Acidobacteria_Gp2 | 2.26 ± 0.08 | 0.92 ± 0.05 | 0.91 ± 0.12 | 0.244 ns |
| Acidobacteria | Acidobacteria_Gp20 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.521 ns |
| Acidobacteria | Acidobacteria_Gp22 | 0.02 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.224 ns |
| Acidobacteria | Acidobacteria_Gp23 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | ns |
| Acidobacteria | Acidobacteria_Gp25 | 0.05 ± 0.01 | 0.03 ± 0.00 | 0.01 ± 0.00 | 0.071 ns |
| Acidobacteria | Acidobacteria_Gp3 | 7.97 ± 0.07 | 9.47 ± 0.09 | 12.21 ± 0.44 | 0.018* |
| Acidobacteria | Acidobacteria_Gp4 | 1.61 ± 0.15 | 2.81 ± 0.11 | 3.31 ± 0.04 | 0.200 ns |
| Acidobacteria | Acidobacteria_Gp5 | 0.47 ± 0.03 | 0.65 ± 0.05 | 1.03 ± 0.02 | 0.168 ns |
| Acidobacteria | Acidobacteria_Gp6 | 2.60 ± 0.09 | 5.81 ± 0.14 | 6.11 ± 0.42 | 0.031* |
| Acidobacteria | Acidobacteria_Gp7 | 3.56 ± 0.14 | 4.88 ± 0.08 | 2.15 ± 0.28 | 0.200 ns |
| Acidobacteria | Holophagae | 0.11 ± 0.01 | 0.07 ± 0.01 | 0.01 ± 0.00 | 0.272 ns |
| Acidobacteria | Other | 0.22 ± 0.02 | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.112 ns |
Note: The level of significance determined by one way ANOVA is listed (**P < 0.01, *P < 0.05, ns: not significant).
A total of 1818 fungal OTUs included here belonged to 9 phyla groups. In decreasing order of abundance, the four most abundant fungal phyla were Ascomycota (47.2%) > Basidiomycota (17.8%) > Zygomycota (11.1%) > Glomeromycota (2.6%). These were considered to constitute the main fungal phyla and, together with a significant fraction of unclassified OTUs (17.4%), they represented approximately 90% of the data from fungal phyla. There were three phyla with a relative abundance < 1% (Fig. 1B, Table 1). Meanwhile, there was a high abundance of unclassified phyla in the wetland soils (Fig. 1B, Table 1). The Zygomycota was the main phylum in arable land soils, while Basidiomycota was most abundant in forest soils (Fig. 1B, Table 1). The similarity of these three land use types in soil fungal community structure was consistent with that of bacteria (Fig. 1B). The fungal community structures of wetland soils (W1 to W3) were different from those of arable land (A1 to A3) and forest (F1 to F3), which had similar fungal community structures.
A total of 1120 Acidobacterial OTUs included here belonged to 25 subgroups. In decreasing order of abundance, the six most abundant phyla were: Acidobacteria_Gp1 (29.6%) > Acidobacteria_Gp3 (9.9%) > Acidobacteria_Gp6 (4.8%) > Acidobacteria_Gp7 (3.5%) > Acidobacteria_Gp4 (2.6%) > Acidobacteria_Gp2 (1.4%) (Fig. 1C, Table 1). These phyla were considered to constitute the main Acidobacterial community and, together with a significant fraction of unclassified OTUs (0.1%), they represented approximately 51.9% of the data from total sequences. In contrast, 49.1% of the total sequences were not of an Acidobacterial origin and were removed prior to subsequent analysis. There were 15 phyla with a relative abundance <1%. Acidobacteria_Gp13, Acidobacteria_Gp18, Acidobacteria_Gp19, and Acidobacteria_G22 were more abundant in wetlands compared with arable lands and forests. Meanwhile, Acidobacteria_Gp6, Acidobacteria_Gp15, and Holophagae were low abundance in the wetlands compared with arable lands and forests (Fig. 1C, Table 1). The soil Acidobacterial community structure similarity of the three land-use types was consistent with those of bacteria and fungi. Acidobacterial community structures in wetlands were different from those in arable lands and forests, which had similar Acidobacterial community structures (Fig. 1C).
Alpha-diversity
The alpha-diversities of the three different land-use patterns are shown in Table 2. For bacteria, the Shannon-Wiener, Chao, and Simpson’s indices differed significantly (P < 0.01) between wetlands, forests and arable lands (Table 2). For Acidobacteria, the Shannon-Wiener and Chao indices were significantly different (P < 0.01) between wetlands and the other two land use patterns. For fungi, the Shannon-Wiener index and the Simpson’s index did not significantly differ (P > 0.05) between the wetlands, forests, and arable lands (Table 2). However, the Chao index differed significantly among the three different land use patterns (P < 0.01) (Table 2).
Table 2.
The α-diversity indices (mean ± sd; n = 3) of bacterial, acidobacterial and fungal OTUs obtained from the soils of three different land use types in the Sanjiang Wetland, northeastern China.
| Organisms | Type | Chao | Shannon | Simpson |
|---|---|---|---|---|
| Bacteria | Wetland | 2894.4 ± 13.8b | 5.9 ± 0.02c | 0.0078 ± 0.00012a |
| Forest | 2748.2 ± 29.0c | 6.1 ± 0.03b | 0.0063 ± 0.00012b | |
| Arable land | 3187.8 ± 55.7a | 6.3 ± 0.02a | 0.0054 ± 0.00018c | |
| Acidobacteria | Wetland | 916.5 ± 8.3b | 4.9 ± 0.00a | 0.0177 ± 0.0009a |
| Forest | 848.7 ± 9.2a | 5.2 ± 0.03b | 0.0203 ± 0.0011b | |
| Arable land | 925.0 ± 5.7b | 5.2 ± 0.02b | 0.0189 ± 0.0005ab | |
| Fungi | Wetland | 405.2 ± 74.3b | 3.3 ± 0.46a | 0.0970 ± 0.0361a |
| Forest | 666.5 ± 37.5a | 3.9 ± 0.12a | 0.0447 ± 0.0051a | |
| Arable land | 696.5 ± 110.5a | 4.2 ± 0.29a | 0.0650 ± 0.0207a |
Note: Different letters denote significant differences between habitats at P < 0.05. α-diversity indexes were calculated at the OTU level.
Beta-diversity
In the analysis of beta-diversity, wetlands were separated from the other two land-use types, suggesting a clear distinction in the bacterial community structures between wetlands and forests and arable land (Fig. 2A). This finding was supported by the results of a principal coordinates analysis (PCoA) and a PERMANOVA (Fig. 2A and Table 3). In general, the PCoA explained 90.71% of the variation in the bacterial communities.
Figure 2.
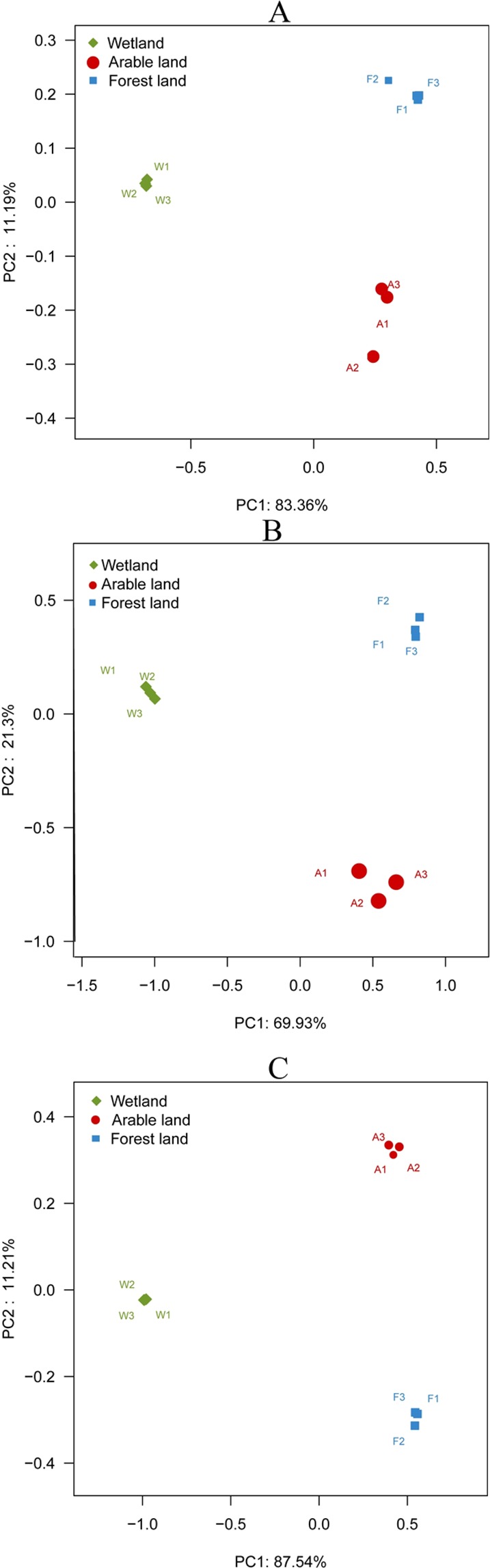
PCoA diagram of bacterial (A), fungal (B) and acidobacterial (C) communities identified from soils in wetland (green), forest (blue), and arable land (red).
Table 3.
PERMANOVA analysis of bacteria, acidobacteria and fungi between three different land use types in Sanjiang plain, northeast of China.
| F | R2 | p | |
|---|---|---|---|
| Bacteria | 25.6** | 0.90 | 0.003 |
| Acidobacteria | 89.1** | 0.97 | 0.004 |
| Fungi | 12.2 | 0.80 | 0.080 |
The PCoA revealed the beta-diversity of the Acidobacterial communities in the wetland, forest, and arable soils (Fig. 2B and Table 3). Overall, the PCoA explained 96.80% of the variation for the Acidobacterial communities. Wetland samples were clustered together and were well separated from the samples from the other habitats (forest and arable land). These results suggest that land use change significantly impacted the soil Acidobacterial communities. Fungal community structures in the wetland samples were also clearly different from the arable land and forest samples (Fig. 2C and Table 3). Overall, the two PCoAs explained 85.83% of the variation in the fungal communities.
Bacterial, Acidobacterial, and fungal beta-diversity in relation to soil physical and chemical properties
Canonical correlation analysis (CCA) of the bacterial data revealed that soil physicochemical properties explained most of the variance in the CCA, for example, the first two axes explained 72.13% (Fig. 3A). The forest and arable land bacterial communities were positively correlated with soil available phosphorus (AP) and soil pH, while the wetlands were positively correlated with total phosphorus (TP), total carbon (TC) total nitrogen (TN), and available nitrogen (AN).
Figure 3.
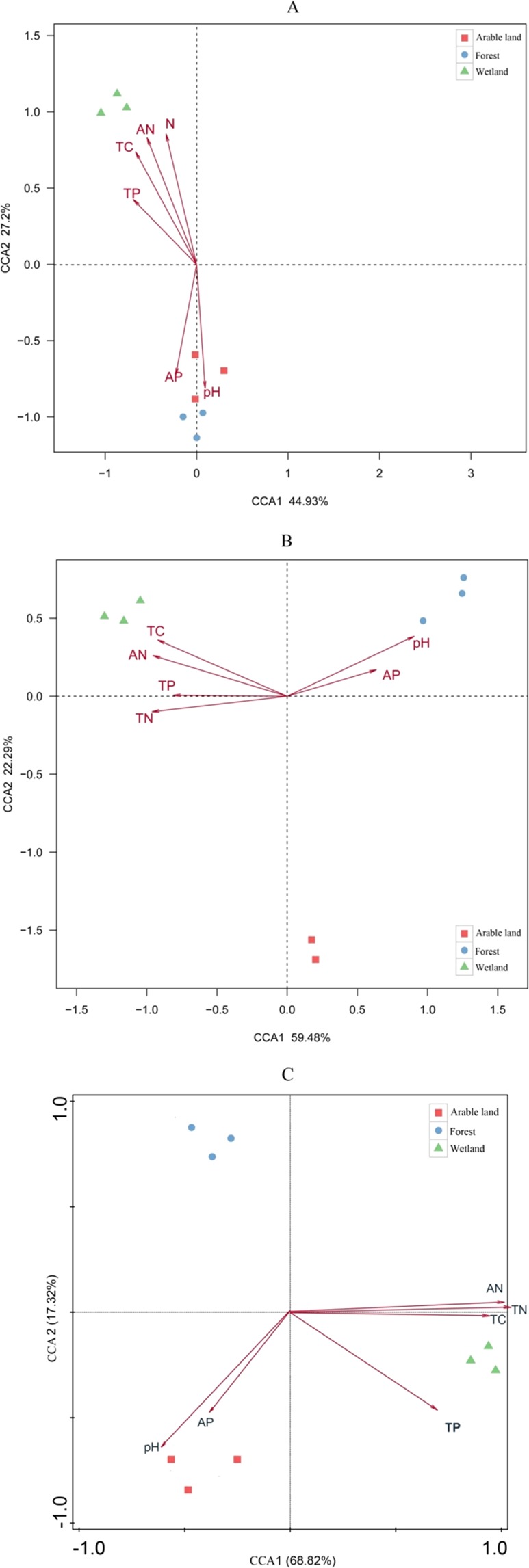
CCA of soil bacterial (A), fungal (B) and acidobacterial (C) community and environmental factors. Samples from wetland (W1–W3), forest (F1–F3) and arable land (A1–A3) are shown. TC: total organic carbon; TN: total nitrogen; AN: available nitrogen; AP: available phosphorus; TP: total phosphorus.
CCA of the fungal data showed that the first two axes explained 81.77% of the total variance (Fig. 3B). The forest and arable land were positively correlated with available phosphorus (AP) and the soil pH, while wetlands were positively correlated with TP, TC, AN, and TN.
CCA of the Acidobacterial data showed that the first two axes explained 86.14% of the variance (Fig. 3C). The wetlands were positively correlated with TC, TN, TP, and AN, while arable land and forest were positively correlated with soil pH and AP.
Discussion
Bacterial diversity, community composition, and the soil environment
In this study, we used Illumina MiSeq sequencing technology to determine the soil microbial community composition in three different land use types. In total, we resolved 40 phyla from soils collected from the Sanjiang Plain, with noticeable differences between wetland, arable land and forest soils. There were significant differences in the microbial communities between the different land-use types (Fig. 1A). By cluster analysis, we established that the communities thriving in arable land soils were relatively similar to those in forest soils—despite the obviously different vegetation in these land use types—indicating that vegetation type is not a good indicator of soil bacterial community structures. The observed bacterial community composition of the wetlands was markedly different from that of the other two soil types. This suggests that changing wetlands into forests or arable land altered the soil properties considerably, with changes in the soil microbial population structure occurring as a consequence32. The variation in bacterial composition indicated that the land-use change disturbed the ecological stability of the original (wetland) microbial community and changed the habitat conditions, which resulted in shifts in the dominant microbial phyla and community diversity33.
We found that major microbial phyla were overrepresented in wetland soils, with a high abundance relative to that in the other land-use types (Fig. 1A). For example, there were significantly higher relative abundances of Chloroflexi and Chlamydiae in the wetland soils compared to the forest and arable land soils. In contrast, the relative abundance of Bacteroidetes and Tenericutes was significantly higher in forest soils, and Planctomycetes and Gemmatimonadetes were higher in arable land soils (Table 1). Our findings are consistent with the literature, indicating that different habitats contain specific bacterial taxa34,35. Wetland soils are characterized by high soil moisture and low pH; this habitat may be suitable for some bacterial phyla such as Chloroflexi and Chlamydiae. For example, Zhang studied the rhizosphere bacterial diversity of three Phragmites australis ecotypes in the Hexi Corridor, China, and found that members of the Chlamydiae phylum only existed in the swamp reed wetland36. Han investigated the microbial community composition in Ebinur Lake, China, and found that Chlamydiae were the main phylum correlating with soil moisture and pH37. In our research, the soil moisture of the wetland was significant higher (p < 0.05) than that of the arable land and forest soils, and the high soil moisture might be a habitat more suitable for Chlamydiae. Hence, the abundance of Chlamydiae in the wetland soils was higher than in the other habitats. The soil microbial community structures changed from wetlands to arable land and forest because of the changes of soil hydrothermal and nutrient conditions38. Changes in the soil microbial community structure are a direct manifestation of the adaptation of that microbiota to changed conditions (due to human activities) that accelerate the decomposition of soil organic carbon and promote the utilization of soil nutrients by plants39.
Diversity indices quantify the diversity of soil microbial communities. Our results indicated that the Shannon-Wiener index of bacteria from the wetland soil was lower than from the other two soil types, suggesting that land-use changes from wetland to forest or arable land will increase the soil bacterial diversity. This result is consistent with Yu et al., who found that soil bacterial diversity was higher in agricultural land (Shannon-Wiener index 3.49–3.69) than in natural restoration land (3.34–3.44)40. The latitude of the area they analyzed is similar to that of the Sanjiang Plain, and the soil conditions are also similar, so the results of their study are comparable to ours. Xu et al. also reported that cultivated land had higher bacterial diversity than pristine marshland on the Sanjiang Plain41.
Moreover, Hartman et al. found that the diversity of the soil bacterial community was lower in natural wetlands than in arable land on the North Carolina coastal plain, finding that fertilization practices increased the soil pH, which resulted in an increase in the bacterial diversity in arable land42. There are many studies that report soil pH is highly positively correlated with soil bacterial diversity43,44. However, Ausec et al. found that the soil bacterial diversity was higher in bog soils with low pH values but high organic carbon content, and they concluded that the soil organic carbon was the key factor determining the soil bacterial diversity45. The importance of soil organic carbon45 is not supported by the present study, because the soil pH and total organic carbon content were 5.8 and 25.6 for the arable land (high diversity), and 5.5 and 52.7 for the wetland (low diversity), respectively (Table 4). Our study suggests that soil pH rather than soil organic carbon content plays a more important role in affecting the soil bacterial diversity.
Table 4.
Physicochemical properties of soils from three different land use habitat types in the Sanjiang Wetland, northeastern China.
| Land use type | pH | Total organic carbon (g/kg) | Total nitrogen (g/kg) | Available nitrogen (mg/kg) | Total phosphorus (mg/kg) | Available phosphorus (mg/kg) | Soil gravimetric moisture (%) |
|---|---|---|---|---|---|---|---|
| Wetland | 5.5 ± 0.0c | 52.7 ± 1.3a | 4.3 ± 0.8a | 455.3 ± 29.6a | 6.4 ± 1.2a | 26.3 ± 1.8a | 52.8 ± 1.0a |
| Arable land | 5.8 ± 0.1b | 25.6 ± 1.4b | 3.1 ± 0.6b | 197.6 ± 7.5b | 3.7 ± 0.7b | 26.7 ± 3.6a | 32.9 ± 3.1c |
| Forest | 7.4 ± 0.1a | 25.1 ± 1.5b | 1.8 ± 0.3c | 143.8 ± 7.2c | 3.2 ± 0.5b | 32.7 ± 4.5a | 47.0 ± 1.2b |
Note: Statistically significant differences (P < 0.05; n = 3) between habitats is indicated by different superscript letters a–c in the same column.
In our study, changes in land use patterns resulted in changes in soil physicochemical properties (P < 0.05) (Table 1). Soil samples from the wetlands had a higher mean moisture content and a lower mean pH than those from the arable land or forest (Table 1). Soil bacterial diversity was affected by soil pH and soil moisture conditions in our study. This result seems counterintuitive because the concentrations of TC, TN, AN, and TP decreased significantly from wetland to forest. This phenomenon may be due to perennial accumulation of organic matter in the swamp meadow wetland soils. Thus, the soil bacteria cannot use the nutrient deposits despite their high abundance in the wetlands. Under new land type regimes, however, the original form of these soil nutrients changed, which provided favorable (more oxic) conditions for the utilization of C and N by soil microbes. The limiting factor of soil microbial diversity in the wetland soils is anaerobic conditions rather than the C and N content.
Acidobacterial diversity, community composition, and the soil environment
We found that the Shannon-Wiener diversity of Acidobacteria from the wetland soils was lower than that from the soils of the other two habitats. This was consistent with the pattern found for total bacteria, demonstrating that land-use change can lead to variations in bacterial and Acidobacterial α-diversity. However, this tendency in total bacterial α-diversity was different with Acibobacterial α-diversity. That is, the Shannon-Wiener diversity of the total bacterial community in wetland soil was significantly different from that in the forest and arable land (Table 2). For Acidobacteria, however, the Shannon-Wiener index for the wetland soils was significantly different from that of the arable land, but not from that of the forest. Our study indicated that soil AP content and pH were the major soil parameters driving bacterial diversity in the arable and forest soils, while TP, TC, TN, and AN were the key soil parameters determining soil Acidobacterial diversity in the wetland soils. Our data suggest that the soil Acidobacterial community was strongly affected by soil pH46,47. Similar to our findings, land use change altered the Acidobacterial community diversity in soybean cropland and adjacent native forests in Amazon forest soils48. Naether et al. analyzed the Acidobacterial community diversity of three forest and grassland areas in Germany; they found significant differences between the grassland and forest, including significant differences between different sites within the same environment49.
In our study, Gp1, Gp3, Gp6, Gp7, Gp4, Gp2, Gp5, and Gp15 (listed in decreasing order from highest to lowest relative abundances >1%) were the main Acidobacterial subgroups in the three land use types investigated. The composition of Acidobacteria in our soil samples was comparable to the relative abundances of dominant Acidobacterial subgroups found in other studies of Acidobacterial communities19,47,49. Generally, Gp1, Gp2, Gp3, Gp4, Gp6, and Gp16 have been found to be the most abundant subgroups in most soils studied, but the relative abundances of those subgroups varied among soil samples50,51. Our study was nearly consistent with these previous results. A notable distinction, however, is that Gp2 has been frequently detected in forest soils in previous studies, accounting for 11.5% of Acidobacteria in Xishuangbanna forest soils52, 29.0% in Changbai mountain soils53, 5.5–33.2% in Shennongjia forest soils54, and it was a main dominant subgroup in Pinus sylvestris forests55. However, in this study, the forest and arable land soils had less Gp2, but the relative abundance in these soils was only approximately 0.98% and 0.91% (Table 2). Eichorst56 et al. also found that there were rarely GP2 or many other subgroups (GP1, GP3, GP4, and GP6) in agricultural soils. Navarrete19 et al. found that the relative abundance of GP2 was higher in Amazon forest soils than agricultural soils, implying that agricultural practices could decrease the soil GP2 abundance. We need to examine the effects of agronomic actives on soil Acidobacterial subgroup GP2 in future studies.
In our study, the community composition of Acidobacteria was affected by the pH, AP, AN, TP, and TC in the soil. Further, the relative abundances of GP1, GP3, GP4, GP5, GP6, and GP15 were positively correlated with soil pH, while the abundance of GP2 and GP7 were negatively correlated with pH (Fig. 3B). Our results are similar with the results of Jones46 and Liu47. In addition, Liu47 et al. demonstrated that soil pH was a key soil physicochemical property that affected soil Acidobacterial subgroup relative abundance. The soil pH adjusted the abundance of Acidobacteria subgroups in the black soils of northeast China. Other than soil pH, previous studies have found that soil environmental factors had a strong relationship with the abundance of soil Acidobacterial subgroups19,56. For example, Navarrete19 found that high aluminum content could limit the relative abundance of the GP6 and GP7 subgroups of Acidobacteria, but high contents of soil Ca, Mg, Mn, and B promoted the relative abundance in the Amazon forest. Eichorst56 et al. found that the relative abundance of Gp 4 was significantly negatively correlated with soil carbon concentration in agricultural and managed grassland soils. Like these studies, our study observed that soil Acidobacterial subgroups showed significant correlations with soil physicochemical properties, with the exception of soil pH. This indicates that soil physicochemical properties, such as pH, TC, TN, TP, and AP could affect the community composition of Acidobacterial communities.
Fungal diversity, community composition, and the soil environment
Fungi play important roles in soil ecosystems, and the community composition of soil fungi is affected by management57 and soil nutrient status58. In this study, we observed that fungal diversity was higher in arable lands and forests than in the wetlands, which is consistent with previous research59. The high soil water content of wetlands results in decreased soil oxygen content. These conditions most likely restrain the survival of fungi, resulting in decreased soil fungal diversity.
The diversity of soil fungi in arable lands and forests was significantly higher than that in the wetlands, presumably because the condition of the soil environment in the arable land was influenced by disturbances such as agricultural tillage, which resulted in a higher soil oxygen content than the wetland soils. Moreover, because of the abundance of litter inputs and the higher soil oxygen content, the diversity of soil fungi in the forest was also higher than that in the wetland60. The high plant diversity in the forest may have also increased the soil fungal biomass and diversity38.
Our results showed that the fungal phyla structures were affected by various soil physicochemical properties in the three studied land-use patterns of the Sanjiang Plain. A CCA showed that soil pH and SOC, TP, AN, and TN had obvious effects on the fungal community structure in the different land-use types (Fig. 3C). The variation in the fungal composition among land use types indicated that soil fungal structure changed when the original wetland was converted into arable land and forest. Land use change resulted in a change in the aboveground vegetation, which caused new habitat conditions and a concomitant increase or decrease of specific fungal taxa (e.g., root-associated), which, in turn, changed the fungal community composition61. Soil organic C and total N are important carbon and nitrogen sources for fungi. Forests and arable lands are rich in plant litter that can promote fungal growth62. In addition, some studies have indicated that fungi might promote their growth by improving their water use efficiency in acidic environments63. Soil P is energy for soil fungal communities, but it is also an important resource for plant growth64. In our study area, shifts in land use changed the soil physiochemical factors—such as pH, TC, TN, and TP—which, in turn, affected the soil fungal community composition.
Vegetation was found to affect the soil microbial community structure13,65. Underground ecosystems with the roots of various plants may have a marked effect on soil microbial structures61. Therefore, the relationships of soil microorganisms, aboveground vegetation, and the soil environment need to be investigated to clarify the interactions between and among them in order to better understand the ecosystem effects on soil microorganisms. Our results showed that there were significant differences in soil fungal structure among the three land use types. This may, at least partly, indicate an important effect of vegetation type associated with land use change on the structure and function of the soil fungal community. Although we widely tested the effects of seven physicochemical properties on soil bacterial, Acidobacterial, and fungal diversity in the present study, other abiotic factors such as soil temperature and soil oxygen content might also be important, and thus future studies should quantify their impacts on soil microorganisms.
Many studies proved that different land use patterns changed vegetation composition, gas permeability, water content, and microclimate characteristics of soil, and these change would affect the microbial community structure and diversity and function66–68. Changes in land use patterns have changed the original niche of soil microbes, therefore, resulting in corresponding changes in the soil microbial community structure. However, these conclusions were only obtained through some statistical tests, and the real situation cannot be unified because of its complexity69. For example, although our study found that soil organic carbon significantly affected soil microbial structure, but the effects of organic carbon interacted with other factors, and other environmental factors also had a significant impact on soil microbes. Therefore, how to distinguish and analyze the microbial community changes that are affected by each single soil factors is very difficult.
We concluded that soil microbial communities were affected by soil physicochemical properties, but the content of soil physicochemical properties did not affect the soil microbial communities and the quality and type of soil physicochemical properties mainly affected the soil microbial communities70,71. In addition, the identity of these soil parameters in the original ecosystem depends on the original nature of each ecosystem in different areas.
Conclusion
We observed differences in the soil properties as well as the characteristics of the soil bacterial, Acidobacterial and fungal communities among the three different land use types in Sanjiang plain. The Shannon diversity index of both soil bacteria and Acidobacteria varied significantly among the three land use types. The bacterial, Acidobacterial and fungal community structures showed similar distinctions between arable land and forest land, but differed with marsh wetland. Our data suggest that soil pH and resource availability (soil organic carbon, total nitrogen, available nitrogen, and total phosphorus concentrations) are the primary drivers of soil bacterial and Acidobacterial and fungal community compositions along the land use changed. This study improved our understanding of soil microbial structure in a typical wetland ecosystem, and the results can be used to predict the impact of land use change from wetland to farmland and forest plantation on the soil microbiome.
Materials and Methods
Site description
The study was performed in the Sanjiang Plain (47°35′N, 133°31′E), northeastern China. The local average monthly temperature ranged from −21.6 °C in January to 21.5 °C in July, with an annual mean temperature of 1.9 °C. The mean annual precipitation was approximately 560 mm, of which 80% occurred from May to October. We selected three land use types (habitats) for this study: wetland, arable land developed from wetlands, and a planted forest on degraded wetlands.
The wetland habitat was a pristine marsh meadow wetland that was seasonally flooded from May to August, and was the typical wetland type occurring in the Sanjiang Plain where Deyeuxia angustifolia is the dominant plant species13. The arable land habitat was converted from a pristine marsh meadow wetland into a soybean plantation in 2007. The soybean plantation was fertilized with 370 kg ha−1 y−1 of fertilizer (N:P:K) each year in May. The forest habitat was plantations of Larix gmelinii that was planted on degraded wetlands in 2000. At the time of this study in 2016, the average height of the Larix gmelinii was about 7 m, the diameter at breast height was about 12 cm, and the average density was 1600 stems ha−1. No fertilization or forest management was conducted in this forest plantation. The wetland and arable land habitat types were approximately 400 m apart, and the forest plantation was located between them at a distance of approximately 200 m from each of the other two habitat types. The wetland, forest, and arable land covered a total area of 3000 m2, 2000 m2, and 1000 m2, respectively.
Soil sampling
Soil samples (0–20 cm depth) were taken on 15 October 2016. Three plots (10 m × 10 m) were established in each habitat, and the distance between any two plots within the same habitat was >50 m. We randomly designated five smaller quadrats (1 × 1 m each) within each standard quadrat. We measured three individual replicates per site, whereas in each site, five individual soil samples were taken and pooled. Soil samples were collected from 5 locations inside each plot using a sterile soil drill and then pooled to obtain a mixed soil specimen (approximately 1 kg of fresh soil) for each plot. We stored field soil samples in an ice box with a temperature near 4 °C, which was transported immediately to the lab for DNA extraction as soon as possible to avoid any freeze-thawing that would affect the integrity of the soil DNA. In the laboratory, approximately 10 g of fresh soil from each sample was transferred into a sterile 50 mL Falcon tube and kept at −80 °C for microbial analysis. The remaining soil was air dried for soil physiochemical analyses.
Determination of soil physiochemical properties
The pH was measured using a pH meter after mixing the soil with water (1:5 w/v) for 30 min. The total organic carbon (TC) and total nitrogen (TN) concentrations of the soil samples were determined using an elemental analyzer (VarioEL III, Elementar Analyses System, Hanau, Germany). Total phosphorus (TP), available phosphorus (AP), and available nitrogen (AN) were digested with H2SO4-HClO4, 0.5 M NaHCO3, and 2.0 M KCl in succession and then assayed using a continuous flow analytical system (SKALAR SAN++, the Netherlands). Soil moisture content was measured gravimetrically. The physiochemical properties of the soil samples are summarized in Table 4.
Soil DNA extraction, PCR amplification, and high-throughput sequencing
DNA was removed from a 0.5 g frozen soil specimen with a MOBIO Power Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA) based on the company’s directions. The extracted DNA was diluted in 100 μL TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) and then kept at −20 °C prior to use. The DNA was quality checked and quantified using a Nano Drop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). We extracted the DNA of each soil sample individually and performed PCR in triplicate for each DNA sample. We amplified the V3-V4 region of bacterial 16S rRNA72 as well as the ITS1 region of fungal ITS rRNA73. We selected the primers ACIDO (5′GCTCAGAATSAACGCTGG3′)/342r (5′CTGCTGCSYCCCGTAG3′) to amplify the Acidobacterial specific phyla74.
The bacterial PCR reactions were performed in a 25 uL mixture containing 0.5 uL of each primer (30 μmol L−1), 1.0–1.5 μL of template DNA (10 ng), and 22.5 μL of Platinum PCR SuperMix (Invitrogen, Shanghai, China). The amplification procedure was as follows: 95 °C for 3 min, followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by an extension at 72 °C for 10 min.
The fungal PCR reactions were performed in a 25 μL mixture containing 0.5 μL of each primer (30 μmol L−1), 1.5 μL of template DNA (10 ng), and 22.5 μL of Platinum PCR SuperMix (Invitrogen, Shanghai, China). The amplification procedure was as follows: 95 °C for 5 min, followed by 30 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by an extension at 72 °C for 10 min.
The Acidobacterial PCR reactions were performed in a 25 uL mixture reaction system similar to that used for bacteria. The amplification procedure was as follows: 95 °C for 5 min, followed by 35 cycles at 95 °C for 60 s, annealing at 63 °C for 60 s, and extension at 72 °C for 60 s, with a final extension at 72 °C for 5 min.
Each sample was amplified in triplicate, and amplicons were removed from 2% agarose gels, purified with the Axy Prep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) based on the company’s directions, and then quantified with QuantiFluor-ST (Promega, Madison, WI, USA). The three purified amplicons from one DNA sample were pooled at equimolar concentrations and paired-end sequenced on a MiSeq PE 300 platform (Illumina, San Diego, CA, USA) based on standard protocols at BIONOVA BioPharm Technology Co., Ltd., Beijing, China.
Sequence analysis
Bacterial, Acidobacterial and fungal raw fastq files were de-multiplexed, quality-filtered, and assessed using QIIME (version 1.17). Forward and reverse reads were merged using PEAR software (version v.0.9.5). Low-quality sequences shorter than 200 bp in length and with an average quality score less than 20 were removed prior to any additional analysis. Exact barcode matching was used, which allowed a two-nucleotide mismatch in primer matching. Reads containing ambiguous characters were also removed. The trimmed sequences were chimera checked, and those with chimeras were removed with the Uchime algorithm75. Only sequences that overlapped by more than 10 bp were assembled based on their overlapped sequences; reads that could not be assembled were discarded. The remaining and unique sequences were clustered at a 97% similarity using CD-HIT76 to yield operational taxonomic units (OTUs). To identify soil bacteria, Acidobacteria and fungi, and to estimate community diversity, the OTUs obtained were compared to the SILVA and UNITE databases.
When taxonomies were finished, the sequences that did not belong to Acidobacteria were removed before subsequent sequence analysis. To correct the sampling effort, the lowest sequencing number was randomly selected per sample and used for subsequent community analysis. All the bacterial sequences have been deposited in GenBank short-read archive under the following accession numbers: forest (SRR7287293, SRR7287316, SRR7287315), arable land (SRR7287330, SRR7287323, SRR7287324) and wetland (SRR7287307, SRR7287310, SRR7287309). All the Acidobacterial sequences have been deposited in GenBank short-read archive under the following accession numbers: forest (SRR7285667, SRR7285668, SRR7285669), arable land (SRR7285654, SRR7285657, SRR7285656) and wetland (SRR7285660, SRR7285653, SRR7285652). All fungal sequences have also been deposited in the GenBank short-read archive: forest (SRR7299284, SRR7299321, SRR7299322), arable land (SRR7299279, SRR7299303, SRR7299304) and wetland (SRR7299286, SRR7299328, SRR7299327).
Statistical analysis
Alpha-diversity of the bacterial, Acidobacterial, and fungal communities included OTU richness, Chao1, and the Shannon-Wiener and Simpson’s indices, and the sequencing depth was expressed as the coverage calculated in MOTHUR. Estimates were calculated by employing the tools Aligner, Complete Linkage Clustering, and Rarefaction of the RDP pyrosequencing pipeline.
Beta-diversity was analyzed by PCoA using the Bray Curtis dissimilarity index. The selected OTUs had a similarity level of 0.03. This simple numeric visualization shows the similarity and overlap of the OTUs within all samples. Differences in the community structure (β-diversity) were assessed by conducting a permutational ANOVA (PERMANOVA, number of permutations = 99 999) with the Adonis function in the Vegan R package and displayed with PCoA ordinations. BioEnv and a canonical correspondence analysis (CCA) were performed to identify normalized OTU abundance and environmental variables that were most frequently related to bacterial, Acidobacterial, and fungal community structures in R using the Vegan package. Based on a diversity distance matrix of bacteria, Acidobacteria, and fungi, three dendrograms were constructed using cluster analysis and the unweighted pair-group method with arithmetic means (UPGMA) at the OTU level. All other statistical tests were performed in R software (v.2.8.1).
Soil physicochemical properties and alpha-diversity indices were analyzed using one-way analysis of variance (ANOVA) with a significance limit of P < 0.05. Relative abundances of all detected phyla were analyzed using one-way ANOVA with significance limits of P < 0.05 and P < 0.01. Least significant difference (LSD) procedures were used to determine significant differences between each habitat type. Standard deviation was calculated as the square root of the variance as a measure of the dispersion of a set of data from its mean.
Acknowledgements
We would first like to thank three referees for valuable comments on the manuscript. This work was funded by National Key Research and Developmental Project of China (2016YFC0500405) and National Natural Sciences Foundation of China (31470019, 31670489, 31700424) and Key Project of Heilongjiang Province (GA19C006-6).We really thank Melissa Dawes for language editing for this paper and also thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author contributions
Hongwei Ni designed this experiment. Xin Sui did the experiment and analyzed data and wrote the manuscript. Beat Frey helped to analyze the data and contributed to the writing of the manuscript. Rongtao Zhang and Libin Yang helped to perform the experiment. Mai-He Li helped to write the manuscript and revised this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mai-He Li, Email: maihe.li@wsl.ch.
Hongwei Ni, Email: nihongwei2000@163.com.
References
- 1.Liu Junjie, Sui Yueyu, Yu Zhenhua, Shi Yu, Chu Haiyan, Jin Jian, Liu Xiaobing, Wang Guanghua. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biology and Biochemistry. 2014;70:113–122. doi: 10.1016/j.soilbio.2013.12.014. [DOI] [Google Scholar]
- 2.Liu Junjie, Zheng Chunyu, Song Changchun, Guo Sida, Liu Xiaobing, Wang Guanghua. Conversion from natural wetlands to paddy field alters the composition of soil bacterial communities in Sanjiang Plain, Northeast China. Annals of Microbiology. 2013;64(3):1395–1403. doi: 10.1007/s13213-013-0784-9. [DOI] [Google Scholar]
- 3.Sui, X. et al. Effects of Long-term Elevated CO 2 Fumigation on Microbial Communities in a Wetland Soil. Journal of Residuals Science & Technology12:93–96, 10.12783/issn.1544-8053/12/S1/14 (2015a).
- 4.Jianguo G, Longhao Z, Dan X, Yeqing S. The bacterial community changes after papermaking wastewater treatment with constructed wetlands. Acta Ecologica Sinica. 2014;34:2005–2010. doi: 10.5846/stxb201307011813. [DOI] [Google Scholar]
- 5.Li F, et al. Changes in soil microbial biomass and functional diversity with a nitrogen gradient in soil columns. Applied Soil Ecology. 2013;64:1–6. doi: 10.1016/j.apsoil.2012.10.006. [DOI] [Google Scholar]
- 6.Li, W. Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Springer US (2015). [DOI] [PubMed]
- 7.Sura S, Waiser MJ, Tumber V, Rainafulton R, Cessna AJ. Effects of a herbicide mixture on primary and bacterial productivity in four prairie wetlands with varying salinities: an enclosure approach. Science of the Total Environment. 2015;512–513:526–539. doi: 10.1016/j.scitotenv.2015.01.064. [DOI] [PubMed] [Google Scholar]
- 8.Ansola G, Arroyo P, Miera LESD. Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Science of the Total Environment. 2014;473–474:63–71. doi: 10.1016/j.scitotenv.2013.11.125. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y, Huang ZG, Wu HT, Lv XG. Carbon and nitrogen distributions and microbial characteristics in the soils of four types of wetlands in Sanjiang Plain, Northeast China. Chinese. Journal of Applied Ecology. 2014;25:2847–2854. doi: 10.13287/j.1001-9332.20140731.001. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Lu X, Tong S, Dai G. Influence of environment and substrate quality on the decomposition of wetland plant root in the Sanjiang Plain, Northeast China. Journal of Environmental Science. 2008;20:1445–1452. doi: 10.1016/S1001-0742(08)62547-4. [DOI] [PubMed] [Google Scholar]
- 11.Sui, X., et al. Study on Bacterial Diversity of Deyeuxia angustifolia Wetland by Application of High-throughput Sequencing Technology in Sanjiang Plain. Soils47:919–925. 10.13758/j.cnki.tr.2015.05.017 (2015b).
- 12.Zheng C, et al. Characterization of the major capsid genes (g23) of T4-type bacteriophages in the wetlands of northeast China. Microbial Ecology. 2013;65:616–625. doi: 10.1007/s00248-012-0158-z. [DOI] [PubMed] [Google Scholar]
- 13.Zhou D, Gong H, Wang Y, Khan S, Zhao K. Driving Forces for the Marsh Wetland Degradation in the Honghe National Nature Reserve in Sanjiang Plain, Northeast China. Environmental Modeling & Assessment. 2009;14:101–111. doi: 10.1007/s10666-007-9135-1. [DOI] [Google Scholar]
- 14.Song CC, et al. Impacts of natural wetland degradation on dissolved carbon dynamics in the Sanjiang Plain, Northeastern. Journal of China Hydrology. 2011;398:26–32. doi: 10.1016/j.jhydrol.2010.11.029. [DOI] [Google Scholar]
- 15.Wang JF, et al. Effects of Different Land Use on Soil Bacterial Functional Diversity in Sanjiang Plain, Northeast China. Journal of Residuals Science & Technology. 2017;14:91–98. doi: 10.12783/issn.1544-8053/14/1/12. [DOI] [Google Scholar]
- 16.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. Journal Bacteriology. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tringe SG, et al. Comparative Metagenomics of Microbial Communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 18.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Applied Environmental Microbiology. 2006;72:1719–28. doi: 10.1128/AEM.72.3.1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarrete AA, et al. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbial Ecology. 2013;83:607–621. doi: 10.1111/1574-6941.12018. [DOI] [PubMed] [Google Scholar]
- 20.Calhoun AJK, Arrigoni J, Brooks RP, Hunter ML, Richter SC. Creating successful vernal pools: a literature review and advice for practitioners. Wetlands. 2014;34:1027–1038. doi: 10.1007/s13157-014-0556-8. [DOI] [Google Scholar]
- 21.Gao J, Zhang X, Lei G, Wang G. Soil organic carbon and its fractions in relation to degradation and restoration of wetlands on the Zoige Plateau, China. Wetlands. 2014;34:235–241. doi: 10.1007/s13157-013-0487-9. [DOI] [Google Scholar]
- 22.David MM, Power ME, Comín FA, Yockteng R. Structural and functional loss in restored wetland ecosystems. Plos Boilogy. 2012;10:e1001247. doi: 10.1371/journal.pbio.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu HJ, et al. Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environmental Science & Technology. 2014;48:9391–9399. doi: 10.1021/es5021058. [DOI] [PubMed] [Google Scholar]
- 24.Wu JP, et al. Analysis of bacterial communities in rhizosphere soil of continuously cropped healthy and diseased konjac. World Journal of Microbiology and Biotechnology. 2017;33:134. doi: 10.1007/s11274-017-2287-5. [DOI] [PubMed] [Google Scholar]
- 25.Mc CKS. The diversity-stability debate. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 26.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 27.Andersen R, Wells C, Macrae M, Price J. Nutrient mineralisation and microbial functional diversity in a restored bog approach natural conditions 10 years post restoration. Soil Biology and Biochemistry. 2013;64:37–47. doi: 10.1016/j.soilbio.2013.04.004. [DOI] [Google Scholar]
- 28.Card SM, Quideau SA. Microbial community structure in restored riparian soils of the Canadian prairie pothole region. Soil Biology and Biochemistry. 2010;42:1463–1471. doi: 10.1016/j.soilbio.2010.05.010. [DOI] [Google Scholar]
- 29.Pellegrino E, et al. Agricultural abandonment in Mediterranean reclaimed peaty soils: long-term effects on soil chemical properties, arbuscular mycorrhizas and CO2 flux. Agriculture, Ecosystems & Environment. 2015;199:164–175. doi: 10.1016/j.agee.2014.09.004. [DOI] [Google Scholar]
- 30.Peralta AL, Ludmer S, Matthews JW, Kent AD. Bacterial community response to changes in soil redox potential along a moisture gradient in restored wetlands. Ecological Engineering. 2014;73:246–253. doi: 10.1016/j.ecoleng.2014.09.047. [DOI] [Google Scholar]
- 31.Xu SQ, et al. Effects of marsh cultivation and restoration on soil microbial communities in the Sanjiang Plain, Northeastern China. European Journal of Soil Biology. 2017;82:81–87. doi: 10.1016/j.ejsobi.2017.08.010. [DOI] [Google Scholar]
- 32.Zhang Wei, Wu Xiukun, Liu Guangxiu, Chen Tuo, Zhang Gaosen, Dong Zhibao, Yang Xuan, Hu Ping. Pyrosequencing Reveals Bacterial Diversity in the Rhizosphere of ThreePhragmites australisEcotypes. Geomicrobiology Journal. 2013;30(7):593–599. doi: 10.1080/01490451.2012.740145. [DOI] [Google Scholar]
- 33.Lynn TM, et al. Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch Microbiology. 2017;199:711–721. doi: 10.1007/s00203-017-1347-4. [DOI] [PubMed] [Google Scholar]
- 34.Bossio DA, et al. Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microbial Ecology. 2005;49:50–62. doi: 10.1007/s00248-003-0209-6. [DOI] [PubMed] [Google Scholar]
- 35.Jangid K, et al. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biology and Biochemistry. 2011;43:2184–2193. doi: 10.1016/j.soilbio.2011.06.022. [DOI] [Google Scholar]
- 36.Zhang W, et al. Pyrosequencing reveals bacterial diversity in the rhizosphere of three ecotypes. Geomicrobiology. 2013;30:593–599. doi: 10.1080/01490451.2012.740145. [DOI] [Google Scholar]
- 37.Han J, et al. Bacterial diversity in bole river entrance soil of ebinur lake wetland, xinjiang by 16s rna gene sequence analysis. Microbiology. 2014;41:2244–2253. doi: 10.13344/j.microbiol.china.140130. [DOI] [Google Scholar]
- 38.Sheik CS, et al. Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS One. 2012;7:e40059. doi: 10.1371/journal.pone.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Ximei, Chen Quansheng, Han Xingguo. Soil Bacterial Communities Respond to Mowing and Nutrient Addition in a Steppe Ecosystem. PLoS ONE. 2013;8(12):e84210. doi: 10.1371/journal.pone.0084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu ZH, Wanga GH, Liu JJ, Liu X. Soil microbial communities are affected more by land use than seasonal variation in restored grassland and cultivated Mollisols in Northeast China. European Journal of Soil Biology. 2011;47:357–363. doi: 10.1016/j.ejsobi.2011.09.001. [DOI] [Google Scholar]
- 41.Xu F, Cai TJ, Yang X, Ju CY, Tang Q. Effect of cultivation and natural restoration on soil bacterial community diversity in marshland in the Sanjiang Plain. Acta Ecological Sinina. 2016;36:7412–7421. doi: 10.5846/stxb201601040015. [DOI] [Google Scholar]
- 42.Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proceedings of the national academy of sciences. 2008;105:17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu HY, et al. Soil bacterial diversity in the arctic is not fundamentally different from that found in other biomes. Environmental microbiology. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu JJ, Zheng CY, Song CC, Guo SD, Wang GH. Conversion from natural wetlands to paddy field alters the composition of soil bacterial communities in sanjiang plain, northeast china. Annals of Microbiology. 2014;64:1395–1403. doi: 10.1007/s13213-013-0784-9. [DOI] [Google Scholar]
- 45.Ausec L, Kraigher B, Mandic-Mulec I. Differences in the activity and bacterial community structure of drained grasslandand forest peat soils. Soil Biology and Biochemistry. 2009;41:1874–1881. doi: 10.1016/j.soilbio.2009.06.010. [DOI] [Google Scholar]
- 46.Jones RT, et al. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Isme Journal. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JJ, et al. Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biology and Biochemistry. 2016;95:212–222. doi: 10.1016/j.soilbio.2015.12.021. [DOI] [Google Scholar]
- 48.Mendes LW, Brossi MJDL, Kuramae EE, Tsai SM. Land-use system shapes soil bacterial communities in Southeastern Amazon region. Applied Soil Ecology. 2015;95:151–160. doi: 10.1016/j.apsoil.2015.06.005. [DOI] [Google Scholar]
- 49.Naether A, et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Applied Environmental Microbiology. 2012;78:7398–7406. doi: 10.1128/AEM.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navarrete AA, et al. Differential Response of Acidobacteria Subgroups to Forest-to-Pasture Conversion and Their Biogeographic Patterns in the Western Brazilian Amazon. Frontiers in Microbiology. 2015;6:1443. doi: 10.3389/fmicb.2015.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber CF. Reduced vertical stratification of soil bacterial community structure and composition is associated with Bromus tectorum invasion of sagebrush steppe. Journal of Arid Environments. 2015;115:90–99. doi: 10.1016/j.jaridenv.2015.01.012. [DOI] [Google Scholar]
- 52.Wang CX. Distribution and Diversity of Acidobacteria in Tropical Rain Forest Soil of Xishuangbanna. Microbiology China. 2010;31:24–29. doi: 10.13344/j.microbiol.china.2010.01.024. [DOI] [Google Scholar]
- 53.Shen CC, et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biology and Biochemistry. 2013;57:204–211. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 54.Zhang Y, et al. Community structure and elevational diversity patterns of soil Acidobacteria. Journal of Environment Sciences. 2014;26:1717–1724. doi: 10.1016/j.jes.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Hartmann M, et al. A decade of irrigation transforms the soil microbiome of a semi-arid pine forest. Molocular Ecology. 2016;26:1–13. doi: 10.1111/mec.13995. [DOI] [PubMed] [Google Scholar]
- 56.Eichorst SA, Kuske CR, Schmidt TM. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Applied Environmental Microbiology. 2011;77:586–596. doi: 10.1128/AEM.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu JJ, et al. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biology and Biochemistry. 2015;83:29–39. doi: 10.1016/j.soilbio.2015.01.009. [DOI] [Google Scholar]
- 58.Bainard LD, et al. Arbuscular mycorrhizal fungal communities are influenced by agricultural land use and not soil type among the Chernozem great groups of the Canadian Prairies. Plant & Soil. 2015;387:351–362. doi: 10.1007/s11104-014-2288-1. [DOI] [Google Scholar]
- 59.Cookson WR, Murphy DV, Roper MM. Characterizing the relationships between soil organic matter components and microbial function and composition along a tillage disturbance gradient. Soil Biology and Biochemistry. 2008;40:763–777. doi: 10.1016/j.soilbio.2007.10.011. [DOI] [Google Scholar]
- 60.Wei X, Shao M, Gale WJ, Zhang X, Li L. Dynamics of aggregate-associated organic carbon following conversion of forest to cropland. Soil Biology and Biochemistry. 2013;57:876–883. doi: 10.1016/j.soilbio.2012.10.020. [DOI] [Google Scholar]
- 61.Hill PW, Marsden KA, Jones DL. How significant to plant N nutrition is the direct consumption of soil microbes by roots? New Phytologist. 2013;199:948–955. doi: 10.1111/nph.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delgadobaquerizo M, et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nature Communication. 2016;7:10541. doi: 10.1038/ncomms10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang G, Zheng C, Wang Y, Li Y, Xin Y. Soil organic carbon and microbial community structure exhibit different responses to three land use types in the North China Plain. Acta Agricul Turae Scandinavica Section B-Soil and Plant Science. 2015;65:341–349. doi: 10.1080/09064710.2015.1011223. [DOI] [Google Scholar]
- 64.Wang C, et al. Differences in Arbuscular Mycorrhizal Fungal Community Composition in Soils of Three Land Use Types in Subtropical Hilly Area of Southern China. PLoS One. 2015;6:e0130983. doi: 10.1371/journal.pone.0130983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hua C, et al. Influence of invasive plant coreopsis grandiflora on functional diversity of soil microbial communities. Journal of Environment Biology. 2011;32:567–572. [PubMed] [Google Scholar]
- 66.Michelsen A, Lisanework N, Friis I, Holst N. Comparisons of understorey vegetation and soil fertility in plantations and adjacent natural forests in the Ethiopian highlands. Journal of Applied Ecology. 1996;33:627–642. doi: 10.2307/2404991. [DOI] [Google Scholar]
- 67.Aon MA, Sarena DE, Burgos JL, Cortassa S. (micro)biological, chemical and physical properties of soils subjected to conventional or no-till management: an assessment of their quality status. Soil and Tillage Research. 2001;60:173–186. doi: 10.1016/S0167-1987(01)00190-8. [DOI] [Google Scholar]
- 68.Wang GH, Jin J, Xu MN, Liu XB. Effects of plant, soil and soil management on soil microbial community diversity. Chinese Journal of Ecology. 2006;25:550–556. doi: 10.13292/j.1000-4890.2006.0106. [DOI] [Google Scholar]
- 69.Peng M, Zi XX, Shang W, Wang QY. Effect of different land use on soil physicochemical properties, enzyme activity and microorganism quantity in Daqing. Journal of Northeast Forestry University. 2015;43:79–84. [Google Scholar]
- 70.Gömöryová E, Ujházy K, Martinák M, Dušan Gömöry D. Soil microbial community response to variation in vegetation and abiotic environment in a temperate old-growth forest. Applied Soil Ecology. 2013;68:10–19. doi: 10.1016/j.apsoil.2013.03.005. [DOI] [Google Scholar]
- 71.Demoling F, Figueroa D, Bååth E. Comparison of factors limiting bacterial growth in different soils. Soil Biology & Biochemistry. 2007;39:2485–2495. doi: 10.1016/j.soilbio.2007.05.002. [DOI] [Google Scholar]
- 72.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of microbiological methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fouts DE, et al. Next Generation Sequencing to Define Prokaryotic and Fungal Diversity in the Bovine Rumen. PLoS One. 2012;7:e48289. doi: 10.1371/journal.pone.0048289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SH, Cho JC. Group-specific PCR primers for the phylum Acidobacteria designed based on the comparative analysis of 16S rRNA gene sequences. Journal of microbiological methods. 2011;86:195–203. doi: 10.1016/j.mimet.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Edgar RC, Haas BJ, Clemente JC, Christopher Q, Rob K. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X, et al. Abundance and diversity of polycyclic aromatic hydrocarbon degradation bacteria in urban roadside soils in Shanghai. Applied Microbiology Biotechnology. 2015;99:3639–3649. doi: 10.1007/s00253-014-6299-x. [DOI] [PubMed] [Google Scholar]