Summary
No genetic modifiers of MS severity have been independently validated, leading to a lack of insight into genetic determinants of the rate of disability progression. We investigated genetic modifiers of MS severity in prospectively-acquired training (N=205) and validation (N=94) cohorts, using the following advances: 1. We focused on 113 genetic variants previously identified as related to MS severity; 2. We used a novel, sensitive outcome: MS Disease Severity Scale (MS-DSS); 3. Instead of validating individual alleles, we used a machine learning technique (Random Forest) that captures linear and complex non-linear effects between alleles to derive a single Genetic Model of MS Severity (GeM-MSS).
The GeM-MSS consists of 19 variants located in vicinity of 12 genes implicated in regulating cytotoxicity of immune cells, complement activation, neuronal functions and fibrosis. GeM-MSS correlates with MS-DSS (r=0.214; p = 0.043) in a validation cohort that was not used in the modeling steps. The recognized biology identifies novel therapeutic targets for inhibiting MS disability progression.
Keywords: Multiple Sclerosis, severity, single nucleotide polymorphism, machine learning
Introduction
The International Multiple Sclerosis (MS) Genetic Consortium (IMSGC) identified and validated over 200 MS susceptibility genes (International Multiple Sclerosis Genetics Consortium et al., 2017; Sawcer et al., 2011), but thus far has failed to validate genetic variants associated with MS severity (George et al., 2016; Sawcer et al., 2011). These negative results suggest that mechanisms that predispose subjects to develop MS largely differ from those that mediate central nervous system (CNS) destruction (i.e. disease severity) and that the latter remain undefined. The current study is based on the hypothesis that while there are genetic influences on the rate of CNS destruction (likely on the side of effector immune responses, susceptibility of CNS tissue to injury and its ability to repair), the effect sizes of common genetic variants identified in genome-wide association studies are too small to be reliably detectable using insensitive clinical outcomes. Additionally, distinct mechanisms may drive destruction of CNS tissue in different MS patients, as suggested by pathological heterogeneity of acute MS lesions (Lucchinetti et al., 2000). Thus, aggregating several candidate common genetic variants in models that allow complex, including non-linear, interactions between alleles and accommodate heterogeneity in disease mechanisms has a greater potential for success.
MS severity can be defined as the speed of accumulation of neurological disability and is traditionally measured by two outcomes based on the broadly-available Expanded Disability Status Scale (EDSS; (Kurtzke, 1983)), but differ in the measurement of the time-aspect of MS severity: the older MS Severity Score (MSSS; (Roxburgh et al., 2005) uses MS disease duration as a measurement of time, whereas the newer age-related MS severity score (ARMSS; (Manouchehrinia et al., 2017)) uses age. Unfortunately, multiple investigators have observed that MSSS and ARMSS do not predict future disability progression rates in moderately-sized MS cohorts (Confavreux & Vukusic, 2006; Weideman, Barbour, et al., 2017), likely because EDSS is a discrete scale ranging from 0 to 10, which cannot reliably measure individualized disability progression rates in intervals shorter than 10 years. Using machine learning, we developed a MS severity outcome called MS Disease Severity Scale (MS-DSS)(Weideman, Barbour, et al., 2017) based on the data-optimized, continuous disability scale Combinatorial Weight-adjusted disability scale (CombiWISE; (Kosa et al., 2016), ranging from 0–100) that also adjusts disability progression slopes for therapeutic effect of applied treatments. By explaining a much larger proportion of variance of future disability progression rates, MS-DSS has more sensitivity in detecting biological modifiers of MS severity in comparison to MSSS or ARMSS. Consequently, we asked whether MS-DSS can be used to develop and validate a single nucleotide polymorphism (SNP)-based genetic model of MS severity (GeM-MSS) in a small, prospectively-acquired and densely-phenotyped longitudinal cohort of MS patients.
Because this cohort is underpowered for discovery research, we used a candidate gene approach with genes previously linked to MS severity, with poor reproducibility of individual variants in independent validation cohorts (Isobe et al., 2016; Jokubaitis & Butzkueven, 2016; Muhlau, Andlauer, & Hemmer, 2016; Sadovnick et al., 2017).
As a machine-learning algorithm of choice, we selected the Random Forest (RF), which captures complex interactions between alleles and disease heterogeneity, to model a total of 113 candidate genetic variants against MS-DSS in the training cohort. We validated the optimized model in a validation cohort that was not used in any aspect of modeling. While the validated GeM-MSS explained only a small proportion of MS severity variance, the described approach is applicable to larger cohorts that will allow for screening of significantly larger numbers of genetic variants.
Subjects and Methods
Study population
In a blinded fashion, we genotyped 426 prospectively-acquired subjects evaluated under natural history protocol “Comprehensive multimodal analysis of neuroimmunological diseases of the CNS” (ClinicalTrials.gov ). The study was approved by the Combined Neuroscience Institutional Review Board of the NIH, and all patients signed written informed consent. Upon unblinding diagnostic categories, 299 genotyped patients had confirmed diagnosis of MS based on 2010 revisions of McDonald’s MS diagnostic criteria (Polman et al., 2011). Based on quality control filters described below, the final MS cohort was randomly split into training (N=205) and validation (N=94) sub-cohorts balanced for race, age, gender and family history of MS. The demographic and clinical data of these sub-cohorts was compared between diagnostic groups using an analysis of variance followed by Tukey’s test with a Holm adjustment for multiple comparisons (Table 1).
Table 1.
Demographic data of training and validation cohorts
| RR-MSa | SP-MSb | PP-MSc | |
|---|---|---|---|
| Training (n = 205) | |||
| n | 86 | 42 | 77 |
| Females/Males | 54/32 | 21/21 | 39/38 |
| Age, years | 43.99 (18.01–76.43) | 54.28 (31.24–73.82)e | 57.94 (27.61–74.66)e |
| Disease Duration, years | 13.91 (4–44) | 26.52 (9–49)e | 17.38 (6–45)d,f |
| MS-DSS | 1.44 (0.56–3.42) | 2.43 (0.87–5.19)e | 2.30 (0.34–4.90)e |
| MSSS | 2.83 (0.16–9.57) | 6.21 (1.43–9.82)e | 6.64 (0.64–9.85)e |
| Validation (n = 94) | |||
| n | 47 | 28 | 19 |
| Females/Males | 30/17 | 10/18 | 11/8 |
| Age, years | 43.39 (24.95–65.44) | 53.44 (22.02–68.04)d | 59.19 (34.90–69.68)e |
| Disease Duration, years | 14.51 (3–33) | 23.32 (8–33)d | 19.39 (8–46)e |
| MS-DSS | 1.61 (0.51–2.31) | 2.49 (0.81–5.33)d | 2.22 (0.81–4.43)d |
| MSSS | 2.98 (0.26–8.75) | 6.98 (1.7–9.56)e | 6.71 (2.55–9.56)e |
Continuous data are shown as the mean of all measurements and the minimum and maximum values are in parentheses.
Relapsing-remitting MS
Secondary progressive MS
Primary progressive MS
p < 0.05 when compared to RR-MS using one-way ANOVA with Holm adjustment for multiple comparisons
p < 0.0001 when compared to RR-MS using one-way ANOVA with Holm adjustment for multiple comparisons
p < 0.0001 when compared to SP-MS using one-way ANOVA with Holm adjustment for multiple comparisons
Genotyping and quality control
SNP genotyping was performed using Illumina Human Omni Express v1.0 NeuroX array on EBV-transformed peripheral blood mononuclear cells (PBMCs) and whole-blood extracted DNA samples using the standard protocol recommended by the manufacturer (Illumina, Inc. San Diego). The OmniExpress NeuroX array is an Illumina Infinium iSelect HD Custom Genotyping array containing >710,000 markers and an additional 24,706 custom variants designed for neurological disease studies. Of the custom variants, approximately 12,000 are designed to study Parkinson’s disease and are applicable to both large population studies of risk factors and to investigations of familial diseases and known mutations (Nalls et al., 2015).
We used the Genotyping Analysis Module within Genome Studio version 1.9.4 to perform sample and variant quality control (QC) checks. The threshold call rate for sample inclusion was 95%. QC of sample handling was determined by comparing the reported sex with genotypic sex estimated from X chromosome heterogeneity. X chromosome heterogeneity calculations were based on common SNPs from the International HapMap Project (The International HapMap Consortium et al., 2007). Samples considered heterozygosity outliers (>±3 standard deviations [SD] from the sample mean) or with discrepancies between reported sex and genotypic estimated sex were excluded. All 299 samples used in the analysis passed QC.
Variant quality control checks were performed in PLINK v1.9. We excluded variants with a minor allele frequency (MAF) ≤5%, missingness ≥5%, and genotypes that deviated from Hardy-Weinberg equilibrium (p<1×10−5). Of the 720,107 genotyped autosomal variants, 595,556 variants passed all genotyping quality control filters.
Genotype Imputation
Data was prepared for imputation using HRC-1000G-check-bim.pl v4 (http://www.well.ox.ac.uk/~wrayner/tools/#Checking) and PLINK v2. Imputation was done on the Michigan imputation server, using the Haplotype Reference Consortium (HRC) reference panel version r1.1,2016 (http://www.haplotype-reference-consortium.org/) on unphased data. Importantly, given that our cohort includes samples of African American ancestry, this panel has been shown to estimate genotypes for common variants in African American participants with high imputation quality (Vergara et al., 2018). Molecular genotyping of HLA-DRB1 alleles was provided by the NIH blood bank as a clinically-approved test and reported to investigators via medical records. We removed variants with an imputation quality score <0.5 or a MAF <0.05 as additional quality control measures.
Strategy for identification of candidate SNPs
We used the following publicly available databases to identify literature that reported variants associated with MS severity: Ensembl (Zerbino et al., 2018), Phenotype-Genotype Integrator (Ramos et al., 2013), GWAS Catalog (MacArthur et al., 2017), and PubMed. Studies were selected based on the criteria that supplementary material was publicly available and that the objective of the study was discovery rather than replication. We used “multiple sclerosis severity”, “multiple sclerosis age of onset”, and “genome-wide association study” to identify literature sources that reported genetic associations with MS severity. Previously published studies used three general strategies to measure MS severity: 1. MSSS, 2. age of disease onset, or 3. destructiveness of CNS tissue measured by MRI. We included studies that used any of these three strategies. We excluded copy number variants, insertions, and deletions, and we required that the reported association p-value be <10−5 for this analysis.
Modeling outcome MS-DSS and additional assessment outcomes: MSSS and ARMSS
MS-DSS (Weideman, Barbour, et al., 2017) is assigned by a statistical model using gradient boosting machines. MS-DSS includes disability measured by a highly sensitive Combinatorial Weight-Adjusted Disability Scale (CombiWISE)(Kosa et al., 2016), mathematically-adjusted for the efficacy of administered treatments using a published formula (Weideman, Tapia-Maltos, Johnson, Greenwood, & Bielekova, 2017), the amount of CNS-tissue destruction measured by Combinatorial MRI scale of CNS tissue destruction (COMRIS-CTD)(Kosa et al., 2015), and additional features of lower variable importance, including demographic data. The model utilizes the following cross-sectional data, listed in order of statistical importance: 1. Therapy-adjusted CombiWISE divided by patient age (CombiWISE/Age), 2. CombiWISE; 3. COMRIS-CTD; 4. Time to first therapy, which measures the delay (in years) from disease onset to initiation of treatment; 5. Difference in therapy-adjusted and measured CombiWISE, that reflects the variant of the disease that is treatable by current immunomodulatory treatments, 6. Age, and 7. Family history of MS. MS-DSS, the modeling outcome in current study, is automatically-calculated from user-inputted raw data via a web-interface: https://bielekovalab.shinyapps.io/msdss/.
Even though we previously determined that MSSS (Roxburgh et al., 2005) and its later modification, ARMSS (Manouchehrinia et al., 2017), are too insensitive to predict future rates of accumulation of disability in moderately-sized MS cohorts (Weideman, Barbour, et al., 2017), we assessed the correlation between GeM-MSS and these widely-used MS severity scales as sensitivity analyses.
Random Forest (RF) based Genetic Model of MS Severity (GeM-MSS)
The RF algorithm (Breiman, 2001 and Hastie, Tibshirani, and Friedman 2009) is a highly-successful ensemble learning method suited for high-dimensional data (such as genomics) that aggregates many individual decision trees. A decision tree is a modeling approach used in classification and regression problems that utilizes several features (e.g., laboratory tests) to classify an outcome (e.g., presence or absence of a disease or level of disease severity) by finding the optimal split (e.g., a concentration of an analyte) for each “branch” of the decision tree. The main problem of tree-based classifiers is an “overfit” of the data, making predictions from the classifiers unstable. RF partially mitigates this problem by averaging together results from multiple decision trees (often thousands) that are constructed using bootstrapped samples of the training data, with observations not used to build the tree forming an “out-of-bag” (OOB) group. RF further alleviates the “overfit” problem by introducing an element of random selection in features considered when performing splits (i.e., the algorithm only considers a random subset of features for every split in the tree-building process). The main tuning parameters in a RF are the number of trees to grow (ntree) and the number of variables to sample for each node split (mtry). In the current study, trees were grown until the OOB error stabilized and the default mtry was used (approximately where p is the total number of available features). Briefly, the OOB error is a measure of the RF model accuracy that is derived from the OOB samples, and the final OOB error estimate for the model is the mean prediction error on each sample using only the predictions from trees where the sample was in the OOB group. Since the RF models have still the potential to overfit the training data, we used iterative process adapted from previous studies (Calle, Urrea, Boulesteix, & Malats, 2011; Gregorutti, Michel, & Sainte-Pierre, 2017) where the least important variables ranked by permutation variable importance (Breiman, 2001) were removed and the RF was rebuilt until the OOB error increased by more than 1% (Figure 1). To ensure the stability of the variable importance results, 30 individual RF models using different random seeds were constructed and were averaged together at each iteration. The final set of predictive variants were identified as the variants remaining in the model before the OOB error increased. The predictions from the OOB error stabilized model, referred henceforth as GeM-MSS, were assessed in an validation cohort that was not used in the model building. In all analyses, we used the implementation of the RF algorithm from the ranger package (Wright & Ziegler, 2017) in R version 3.4.4 (R Core Team, 2018) that has been adapted for high-dimensional datasets.
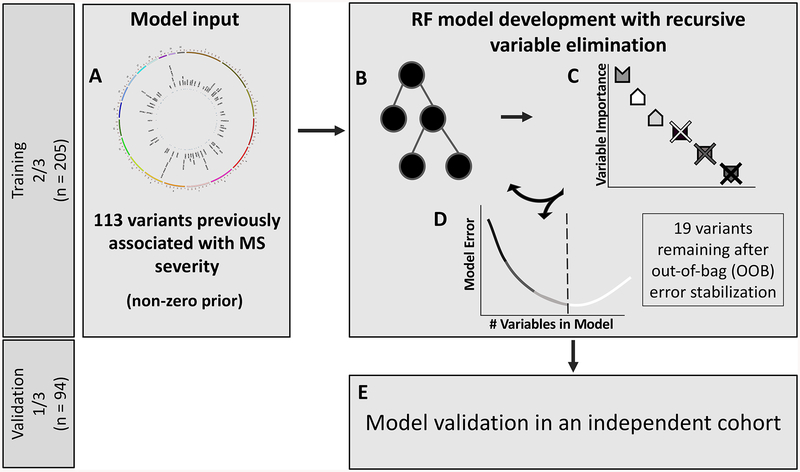
Figure 1. Schematic diagram of the random forest (RF) and variable elimination pipeline.
(A) Each SNP identified in public databases and literature search comprised the feature space available for the RF modeling step. (B) In a RF, features are assembled into several decision trees that differentiate the observations. The relative importance of each feature can be calculated by assessing the change in out-of-bag (OOB) error of the RF when the feature values are permuted. (C) Features can be sorted by order of importance, allowing for removal of the least important feature. The pipeline continues to remove the least important variant from the RF model and rebuilding the forest until the (D) change in OOB error of the subsequent model exceeds 1%. (E) Such a stabilized model is then tested in the independent validation cohort.
Data and Code Availability
All custom code developed and used in this manuscript are available in the supplementary information files. Genetic and phenotype data are available in the database of Genotypes and Phenotypes (dbGaP).
Results
Identification of candidate SNPs
The information about the genotyped SNPs and their published sources are summarized in Supplementary Information Table 1. In total, 116 SNPs were identified for association with MS severity. After QC, 113 variants located in the vicinity of 95 unique genes remained for analysis.
Development of RF genetic model (GeM-MSS) and its optimization based on variable-importance data
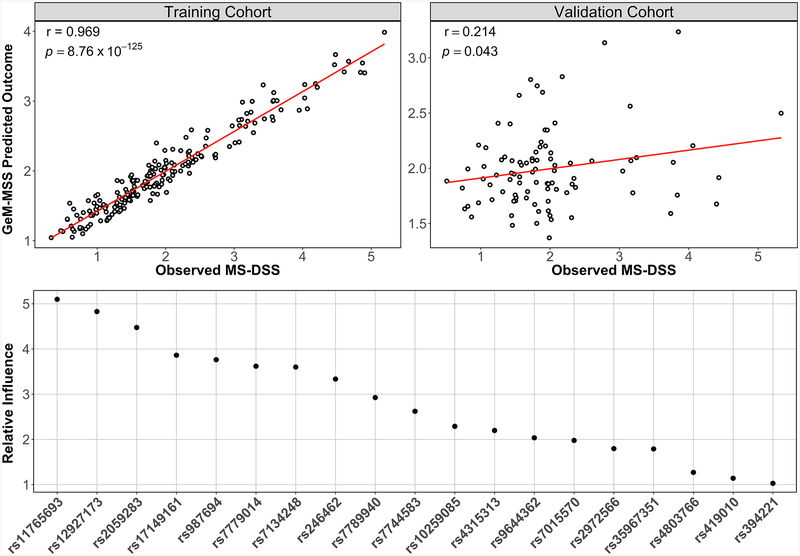
We applied the RF technique to model MS-DSS based on 113 SNPs previously reported for association with MS severity (Figure 1). The model was built and optimized in the training cohort, while its general validity was tested in the validation cohort balanced for race, age, gender and family history of MS (Table 1). Because of the high likelihood that some (perhaps majority) of the tested 113 SNPs represent “noise”, we adopted an iterative process of discarding the least important SNP in each subsequent iteration of the RF model and evaluated the performance of this simplified iteration using the OOB error of the RF. This iterative process of model optimization stopped after observing at least a 1% increase in the error of predicting the OOB samples. This indicates that all remaining genetic variants are important in predicting MS severity of all subjects in the training cohort. The resulting GeM-MSS had 19 remaining variants. Compared to the model with all 113 variants, the OOB error of the GeM-MSS decreased by 14.4% (initial OOB error = 1.059 vs GeM-MSS OOB error = 0.907), even though the root mean squared error (RMSE) in both models was similar (initial RMSE = 0.444 vs GeM-MSS RMSE = 0.464). We observed a strong and statistically-significant correlation between the model-predicted and measured MS-DSS in the training cohort (Figure 2A; r = 0.969; p = 8.76 × 10−125).
Figure 2. The GeM-MSS model predicts future accumulation of disease progression in training and independent validation cohorts.
(A) GeM-MSS correlates strongly with the observed MS-DSS in the training cohort. (B) We observed a weak, but significant correlation between GeM-MSS and the observed MS-DSS in the validation cohort. MS-DSS was calculated according to a published formula (Weideman, Barbour, et al., 2017) using clinical data from the last visit. (C) Relative influence of the 19 remaining variants in GeM-MSS. The random forest (RF) assigned the greatest variable importance to variants in the YWHAG gene are assigned the greatest importance to the predicted outcome. Subsequent variants represent the following genes in order of importance: XYLT1, CAMK2D, KDM2B, MAGI2, ARID1B, C1GALT1, CHD13, CSMD1, PSD3, PVRL2, SLAMF7
Because machine learning techniques have the potential to overfit the training data, we tested the validity of the model in a cohort that was not used in any step of the modeling. In this validation cohort, we observed a statistically-significant correlation, although of much lower strength, between the model-predicted and observed MS-DSS (Figure 2B; r = 0.214; p = 0.043).
Assessment of GeM-MSS with MSSS and ARMSS
Since MS-DSS is a complex model that uses data that may not be available for all MS cohorts, we evaluated the ability of GeM-MSS to predict MS severity outcomes that were not used for its development but are available for genotyped MS cohorts that belong to other investigators.
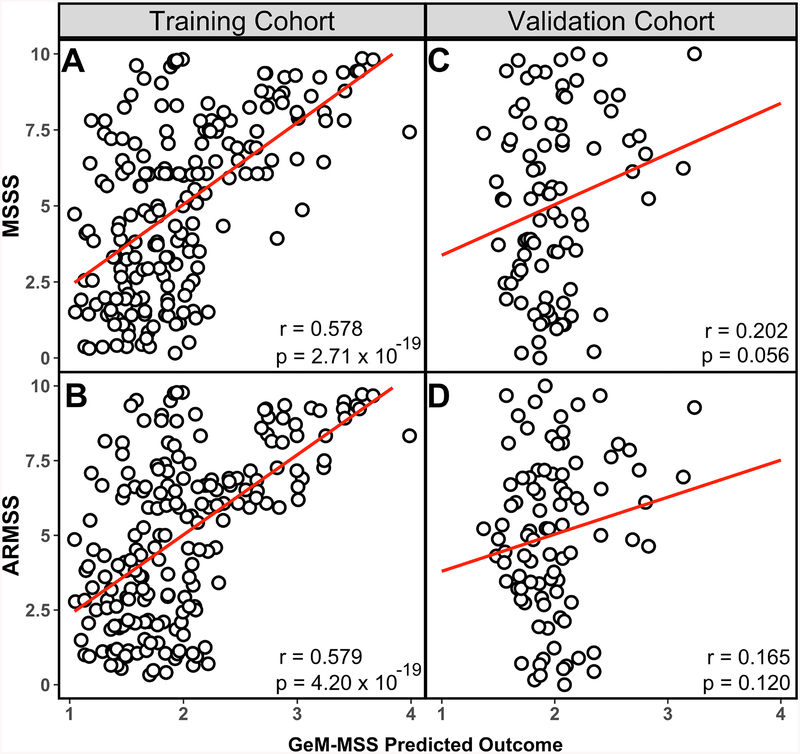
Thus, we assessed the correlation between GeM-MSS and MSSS and ARMSS in the training and validation cohorts. We observed a moderate correlation when comparing GeM-MSS to MSSS (Figure 3A; r = 0.578, p = 2.71 × 10−19) and ARMSS (Figure 3B; r = 0.579, p = 4.20 × 10−19) in the training data. In the smaller validation cohort, we observed positive, although non-significant associations between GeM-MSS and EDSS-based MS severity scores (Figure 3D; r = 0.202; p = 0.056 for MSSS and Figure 3E; r = 0.165; p = 0.120 for ARMSS).
Figure 3. Correlation of GeM-MSS with MS severity scales for training and validation cohorts.
(A) GeM-MSS correlates with the MS severity scale (MSSS) and its complement, (B) the age-related MS severity score (ARMSS) in the training cohort. (C) In the validation cohort, GeM-MSS correlates with MSSS, and shows a similar, but non-significant trend when compared to (D) ARMSS
Biological interpretation of validated GeM-MSS
To obtain biological interpretation of GeM-MSS, the variable importance ranking, MAF, and nearest gene of the 19 remaining variants are given in Table 2. In total, the GeM-MSS outcome predicted by the 19 variants explains approximately 4.4% of the variance in MS-DSS. Each of the variants remaining in GeM-MSS is relatively common and had a MAF greater than 0.10. These variants are within the vicinity of 12 genes, including YWHAG, XYLT1, CAMK2D, and KDM2B. Three genes (YWHAG, XYLT1, PVRL2) were represented by several SNPs and represent regions of high linkage disequilibrium (LD) within the genes. The most important variant in the model (rs11765693) is located in the YWHAG gene and is succeeded by two variants in the XYLT1 gene (rs12927173 and rs2059283) (Figure 2C). Interestingly, the signals in the XYLT1 and YWHAG genes are amongst the most significant associations to MS severity in each of the discovery cohorts that initially identified these associations (Supplemental Information References 23 and 24, respectively). This result provided evidence that efforts to replicate the top genetic associations to MS severity may be improved, in part, by using MS-DSS as an outcome.
Table 2.
Summary of variants in GeM-MSS
| Chromosome | Nearest Gene | Variant | Type | Alleles | MAF | Model Rank |
|---|---|---|---|---|---|---|
| 1 | SLAMF7 | rs35967351 | Intronic | A/T | 0.25 | 16 |
| 4 | CAMK2D | rs987694 | Intronic | G/A | 0.34 | 5 |
| 6 | ARID1B | rs7744583 | Intronic | G/A | 0.33 | 10 |
| 7 | C1GALT1 | rs10259085 | Intronic | C/T | 0.50 | 11 |
| MAGI2 | rs246462 | Intronic | A/G | 0.34 | 8 | |
| YWHAG | rs11765693 | Intronic | A/G | 0.29 | 1 | |
| rs17149161 | Intronic | C/A | 0.29 | 4 | ||
| rs7779014 | Intronic | C/T | 0.29 | 6 | ||
| rs7789940 | Intergenic | A/G | 0.29 | 9 | ||
| 8 | CSMD1 | rs9644362 | Intronic | G/C | 0.18 | 13 |
| PSD3 | rs7015570 | Intronic | A/G | 0.21 | 14 | |
| 12 | KDM2B | rs7134248 | Intronic | T/C | 0.48 | 7 |
| 16 | CDH13 | rs4315313 | Intronic | T/C | 0.43 | 12 |
| XYLT1 | rs2059283 | Intronic | T/G | 0.48 | 3 | |
| rs12927173 | Intronic | T/C | 0.48 | 2 | ||
| 19 | PVRL2 | rs4803766 | Intronic | G/A | 0.48 | 17 |
| rs2972566 | Intronic | G/C | 0.48 | 15 | ||
| rs419010 | Intronic | C/T | 0.48 | 18 | ||
| rs394221 | Intronic | C/T | 0.48 | 19 |
Several selected variants are predominantly expressed in the CNS (especially in neurons), such as tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (YWHAG), membrane associated guanylate kinase, WW and PDZ domain containing 2 (MAGI2), CUB and Sushi multiple domains 1 (CSMD1). CSMD1 and MAGI2 variants have been associated with cognitive dysfunction; whereas, YWHAG, which belongs to the 14-3-3 family of proteins, participates in glutamate-induced cell death. These neuronal variants had high variable importance in the GeM-MSS model. In addition to its neuronal role, CSMD1 also inhibits formation of the complement membrane attack complex in the CNS, and, therefore, may prevent CNS tissue destruction (Kraus et al., 2006).
Two variants in the vicinity of Xylosyltransferase I (XYLT1) had the second and third highest variable importance. XYLT1 catalyzes the first step in biosynthesis of glycosaminoglycans, including chondroitin sulfate proteoglycans, which have been shown to impede remyelination (Lau et al., 2012). Finally, several variants with high variable importance (i.e., MAGI2, YWHAG, XYLT1) participate in, or regulate the process of epithelial-mesenchymal trans-differentiation, which is linked to fibrosis. Therefore, restructuring the extracellular matrix into gliotic scar may be a crucial limiting step for CNS repair in MS.
On the side of the immune system, the selected variants highlight the importance of cytotoxic cells (T cells and NK cells) and their interaction with B cells and plasma cells. The cross-linking of the signaling lymphocytic activation molecule family 7 (SLAMF7) enhances interferon- γ production and cytotoxicity (Comte et al., 2017) of NK cells and T cells. SLAMF7 is also expressed on plasma blasts, plasma cells and activated B cells (Llinas et al., 2011), possibly mediating co-stimulatory functions of these cells of humoral immunity for CD8+ T cells and NK cells via homotypic interactions. Finally, SLAMF7 is also highly expressed on macrophages, where it mediates phagocytosis (Chen et al., 2017) and thus may contribute to myelin stripping in MS. On the other hand, poliovirus receptor-related 2 (PVRL2, also called herpesvirus entry mediator B and NECTIN2 or CD112) binds to a pair of negative (i.e., T cell immunoglobulin and ITIM domain [TIGIT]) and positive (i.e., CD226 [DNAM-1]) regulators of NK and cytotoxic T cell functions (Stein, Tsukerman, & Mandelboim, 2017). These immune-related variants had lower variable importance in comparison to the CNS-enriched variants.
Discussion
Genetic modifiers of the MS disease course remain elusive. In fact, it is hard to even estimate how much of the MS severity variance is genetically determined. Intuitively, a reasonable estimate of the genome-wide genetic contribution to MS severity might be up to 50%, considering the effect of treatments, environment, and stochastic processes. The previous attempts to link MS susceptibility variants to MS severity were not successful (Isobe et al., 2016; Jokubaitis & Butzkueven, 2016; Muhlau et al., 2016; Sadovnick et al., 2017). If these results were true negatives, then genetic variants that predispose patients to acquiring MS under favorable environmental conditions did not influence the rate of CNS tissue destruction or its recovery. This conclusion is counterintuitive because therapeutic success of FDA-approved immunomodulatory treatments on accumulation of MS disability (Weideman, Tapia-Maltos, et al., 2017) leaves no doubt that the immune system partakes in CNS tissue destruction; at least some of the MS susceptibility variants linked to dysregulated immune responses would be expected to also influence destruction of CNS tissue. Therefore, we considered it likely that previous negative results were, to a certain extent, due to Type II errors. This conclusion is strongly supported by GeM-MSS, which contains SNPs linked to effector immune mechanisms known to be associated with destruction of varied human tissues, including CNS, such as complement-mediated cell-lysis (i.e., formation of the terminal membrane attack complex) and cellular cytotoxicity.
Validation of GeM-MSS also support our hypothesis that common genetic variants are unlikely to exert a strong negative influence on CNS tissue destruction or its recovery. Rather, complex and often non-linear relationships between immune-related MS susceptibility genes and genes expressed predominantly in CNS tissue likely mediate susceptibility versus resistance of CNS tissue to injury or affect recovery mechanisms such as remyelination or synaptogenesis. The variable importance metrics provide strong support to CNS-driven mechanisms such as glutamate-induced cell death and neurodegenerative processes previously linked with cognitive dysfunction, which may affect neurogenesis and CNS repair in general. Another pathogenic process that GeM-MSS identified is the restructuring of extracellular matrix in a form of fibrotic scar that may prevent remyelination and possibly also new synaptogenesis. Because in other organs, fibrosis is often consequence of chronic inflammation, we consider it likely that this process is immune-related in MS as well.
Finally, we would like to discuss technical aspects of our and other published studies: aggregating effects of multiple variants into a single genetic model with greater predictive power is intuitive and at least partially supported by published literature (Pan et al., 2016). Modeling 116 validated MS susceptibility variants in a cohort of 125 early MS cases followed for 5 years, Pan et al (Pan et al., 2016) developed an additive model (Cumulative Genetic Risk Score; [CGRS]) consisting of 7 MS susceptibility SNPs. If the patient had more than 2 of the 7 risk genotype variants, CGRS showed a dose-response relationship with MS severity measured by annualized change in EDSS; their model explained 32.7% of variance in disability progression. However, the publication of CGRS did not include validation in an independent cohort. GeM-MSS selected only one the 7 variants from CGRS (rs35967351; SLAMF7). When we attempted to validate the CGRS dose response relationship in our combined training and validation cohorts that is almost 3 times larger than the training cohort used for modeling CGRS, we observed no evidence of a linear relationship (Supplementary Information). Instead, we saw slight, non-significant increase in the measured MS severity in subjects with at least 3 risk alleles when compared to two or less risk alleles using several MS severity outcomes, including the sensitive MS-DSS.
It is unrealistic for 7 frequent genetic risk alleles to explain 32.7% of the variance in disability progression. This would imply unusually high effect sizes, which should have been easily identified/validated in previous and much larger studies (George et al., 2016; Sawcer et al., 2011). Thus, the performance of CGRS, derived from a small, unvalidated cohort represents an overfit (Ioannidis, 2008). The strong model performance in the training cohort should not be perceived as an automatic guarantee of the model’s clinical utility. Instead, an independent validation cohort, that did not contribute in any way to feature selection or model development, is an absolute requirement for assessing the true value of any model. The correlation coefficient of 0.969 in the training cohort indicates that even the final GeM-MSS model grossly overestimated the true relationship between the 19 risk alleles and MS severity in the training cohort. The performance of GeM-MSS in the validation cohort, demonstrating mild statistical significance and explaining less than 10% of MS severity variance, is much more credible. Yet, the obtained p-value of 0.04 indicates 4% chance of validating these findings in a similar cohort if no relationship is present. Therefore, we would welcome an independent validation of GeM-MSS from investigators with larger genomic/clinical datasets. We consider the probability that GeM-MSS can be validated by independent investigators that own large cohorts of genotyped MS patients with linked EDSS, high. We base this belief on the fact that GeM-MSS correlates significantly with EDSS-based MS severity scores that did not contribute to model development in the training cohort, and that even in the small validation cohort we observed a positive association between GeM-MSS and MSSS and ARMSS.
In conclusion, this study provides a genetic model of MS severity that aggregates several previously-identified common genetic variants and provides important genetic insight into MS disability progression. The introduced technical advances (MS-DSS, combining SNPs into a single model that captures non-linear effects and disease heterogeneity) can be used to further improve GeM-MSS through multicenter collaborations. Only such a collaborative assembly of densely genotyped and phenotyped data can perform genome-wide search of additional variants contributing to MS severity.
Supplementary Material
Acknowledgments and Funding
The study was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID) and the Clinical Center/US National Institutes of Health (NIH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest Statement
The authors declare no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Bibliography
- Breiman Leo. (2001). Random Forests. Machine Learning, 45(1), 5–32. doi: 10.1023/a:1010933404324 [DOI] [Google Scholar]
- Calle ML, Urrea V, Boulesteix AL, & Malats N (2011). AUC-RF: A New Strategy for Genomic Profiling with Random Forest. Human Heredity, 72(2), 121–132. doi: 10.1159/000330778 [DOI] [PubMed] [Google Scholar]
- Chen J, Zhong MC, Guo H, Davidson D, Mishel S, Lu Y, Rhee I, Perez-Quintero LA, Zhang S, Cruz-Munoz ME, Wu N, Vinh DC, Sinha M, Calderon V, Lowell CA, Danska JS, & Veillette A (2017). SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature, 544(7651), 493–497. doi: 10.1038/nature22076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte D, Karampetsou MP, Yoshida N, Kis-Toth K, Kyttaris VC, & Tsokos GC (2017). Signaling Lymphocytic Activation Molecule Family Member 7 Engagement Restores Defective Effector CD8+ T Cell Function in Systemic Lupus Erythematosus. Arthritis Rheumatol, 69(5), 1035–1044. doi: 10.1002/art.40038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confavreux C, & Vukusic S (2006). Natural history of multiple sclerosis: a unifying concept. Brain, 129(Pt 3), 606–616. [DOI] [PubMed] [Google Scholar]
- George MF, Briggs FB, Shao X, Gianfrancesco MA, Kockum I, Harbo HF, Celius EG, Bos SD, Hedstrom A, Shen L, Bernstein A, Alfredsson L, Hillert J, Olsson T, Patsopoulos NA, De Jager PL, Oturai AB, Sondergaard HB, Sellebjerg F, Sorensen PS, Gomez R, Caillier SJ, Cree BA, Oksenberg JR, Hauser SL, D’Alfonso S, Leone MA, Martinelli Boneschi F, Sorosina M, van der Mei I, Taylor BV, Zhou Y, Schaefer C, & Barcellos LF (2016). Multiple sclerosis risk loci and disease severity in 7,125 individuals from 10 studies. Neurol Genet, 2(4), e87. doi: 10.1212/NXG.0000000000000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorutti B, Michel B, & Sainte-Pierre P (2017). Correlation and variable importance in random forests. Statistics and Computing, 27(3), 659–678. doi: 10.1007/s11222-016-9646-1 [DOI] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Patsopoulos Nikolaos, Baranzini Sergio E., Santaniello Adam, Shoostari Parisa, Cotsapas Chris, Wong Garrett, Beecham Ashley H., James Tojo, Replogle Joseph, Vlachos Ioannis, McCabe Cristin, Pers Tune, Brandes Aaron, White Charles, Keenan Brendan, Cimpean Maria, Winn Phoebe, Panteliadis Ioannis-Pavlos, Robbins Allison, Andlauer Till F. M., Zarzycki Onigiusz, Dubois Benedicte, Goris An, Bach Sondergaard Helle, Sellebjerg Finn, Soelberg Sorensen Per, Ullum Henrik, Wegner Thoerner Lise, Saarela Janna, Cournu-Rebeix Isabelle, Damotte Vincent, Fontaine Bertrand, Guillot-Noel Lena, Lathrop Mark, Vukusik Sandra, Berthele Achim, Biberacher Viola, Buck Dorothea, Gasperi Christiane, Graetz Christiane, Grummel Verena, Hemmer Bernhard, Hoshi Muni, Knier Benjamin, Korn Thomas, Lill Christina M., Luessi Felix, Muhlau Mark, Zipp Frauke, Dardiotis Efthimios, Agliardi Cristina, Amoroso Antonio, Barizzone Nadia, Benedetti Maria Donata, Bernardinelli Luisa, Cavalla Paola, Clarelli Ferdinando, Comi Giancarlo, Cusi Daniele, Esposito Federica, Ferre Laura, Galimberti Daniela, Guaschino Clara, Leone Maurizio A., Martinelli Vittorio, Moiola Lucia, Salvetti Marco, Sorosina Melissa, Vecchio Domizia, Zauli Andrea, Santoro Silvia, Zuccala Miriam, Mescheriakova Julia, van Duijn Cornelia, Bos Steffan D., Celius Elisabeth G., Spurkland Anne, Comabella Manuel, Montalban Xavier, Alfredsson Lars, Bomfim Izaura L., Gomez-Cabrero David, Hillert Jan, Jagodic Maja, Linden Magdalena, Piehl Fredrik, Jelcic Ilijas, Martin Roland, Sospedra Mireia, Baker Amie, Ban Maria, Hawkins Clive, Hysi Pirro, Kalra Seema, Karpe Fredrik, Khadake Jyoti, Lachance Genevieve, Molyneux Paul, Neville Matthew, Thorpe John, Bradshaw Elizabeth, Caillier Stacy J., Calabresi Peter, Cree Bruce A. C., Cross Anne, Davis Mary F., de Bakker Paul, Delgado Silvia, Dembele Marieme, Edwards Keith, Fitzgerald Kate, Frohlich Irene Y., Gourraud Pierre-Antoine, Haines Jonathan L., Hakonarson Hakon, Kimbrough Dorlan, Isobe Noriko, Konidari Ioanna, Lathi Ellen, Lee Michelle H., Li Taibo, An David, Zimmer Andrew Lo Albert, Madireddy Lohith, Manrique Clara P., Mitrovic Mitja, Olah Marta, Patrick Ellis, Pericak-Vance Margaret A., Piccio Laura, Schaefer Cathy, Weiner Howard, Lage Kasper, Compston Alastair, Hafler David, Harbo Hanne F., Hauser Stephen L., Stewart Graeme, D’Alfonso Sandra, Hadjigeorgiou Georgios, Taylor Bruce, Barcellos Lisa F., Booth David, Hintzen Rogier, Kockum Ingrid, Martinelli-Boneschi Filippo, McCauley Jacob L., Oksenberg Jorge R., Oturai Annette, Sawcer Stephen, Ivinson Adrian J., Olsson Tomas, & De Jager, Philip L (2017). The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. doi: 10.1101/143933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP (2008). Why most discovered true associations are inflated. Epidemiology, 19(5), 640–648. doi: 10.1097/EDE.0b013e31818131e7 [DOI] [PubMed] [Google Scholar]
- Isobe N, Keshavan A, Gourraud PA, Zhu AH, Datta E, Schlaeger R, Caillier SJ, Santaniello A, Lizee A, Himmelstein DS, Baranzini SE, Hollenbach J, Cree BA, Hauser SL, Oksenberg JR, & Henry RG (2016). Association of HLA Genetic Risk Burden With Disease Phenotypes in Multiple Sclerosis. JAMA Neurol, 73(7), 795–802. doi: 10.1001/jamaneurol.2016.0980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokubaitis VG, & Butzkueven H (2016). A genetic basis for multiple sclerosis severity: Red herring or real? Mol Cell Probes, 30(6), 357–365. doi: 10.1016/j.mcp.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Kosa P, Ghazali D, Tanigawa M, Barbour C, Cortese I, Kelley W, Snyder B, Ohayon J, Fenton K, Lehky T, Wu T, Greenwood M, Nair G, & Bielekova B (2016). Development of a Sensitive Outcome for Economical Drug Screening for Progressive Multiple Sclerosis Treatment. Front Neurol, 7, 131. doi: 10.3389/fneur.2016.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosa P, Komori M, Waters R, Wu T, Cortese I, Ohayon J, Fenton K, Cherup J, Gedeon T, & Bielekova B (2015). Novel composite MRI scale correlates highly with disability in multiple sclerosis patients. Mult Scler Relat Disord, 4(6), 526–535. doi: 10.1016/j.msard.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, & Holers VM (2006). CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. Journal of Immunology, 176(7), 4419–4430. [DOI] [PubMed] [Google Scholar]
- Kurtzke JF (1983). Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology, 33(11), 1444–1452. [DOI] [PubMed] [Google Scholar]
- Lau LW, Keough MB, Haylock-Jacobs S, Cua R, Doring A, Sloka S, Stirling DP, Rivest S, & Yong VW (2012). Chondroitin sulfate proteoglycans in demyelinated lesions impair remyelination. Ann Neurol, 72(3), 419–432. doi: 10.1002/ana.23599 [DOI] [PubMed] [Google Scholar]
- Llinas L, Lazaro A, de Salort J, Matesanz-Isabel J, Sintes J, & Engel P (2011). Expression profiles of novel cell surface molecules on B-cell subsets and plasma cells as analyzed by flow cytometry. Immunol Lett, 134(2), 113–121. doi: 10.1016/j.imlet.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Lucchinetti C, Bruck W, Parisi J, Scheithauer B, Rodriguez M, & Lassmann H (2000). Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann Neurol, 47(6), 707–717. [DOI] [PubMed] [Google Scholar]
- MacArthur Jacqueline, Bowler Emily, Cerezo Maria, Gil Laurent, Hall Peggy, Hastings Emma, Junkins Heather, McMahon Aoife, Milano Annalisa, Morales Joannella, Pendlington Zoe May, Welter Danielle, Burdett Tony, Hindorff Lucia, Flicek Paul, Cunningham Fiona, & Parkinson Helen. (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research, 45(D1), D896–D901. doi: 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manouchehrinia A, Westerlind H, Kingwell E, Zhu F, Carruthers R, Ramanujam R, Ban M, Glaser A, Sawcer S, Tremlett H, & Hillert J (2017). Age Related Multiple Sclerosis Severity Score: Disability ranked by age. Mult Scler, 1352458517690618. doi: 10.1177/1352458517690618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlau M, Andlauer TF, & Hemmer B (2016). HLA Genetic Risk Burden in Multiple Sclerosis. JAMA Neurol, 73(12), 1500–1501. doi: 10.1001/jamaneurol.2016.4329 [DOI] [PubMed] [Google Scholar]
- Nalls MA, Bras J, Hernandez DG, Keller MF, Majounie E, Renton AE, Saad M, Jansen I, Guerreiro R, Lubbe S, Plagnol V, Gibbs JR, Schulte C, Pankratz N, Sutherland M, Bertram L, Lill CM, DeStefano AL, Faroud T, Eriksson N, Tung JY, Edsall C, Nichols N, Brooks J, Arepalli S, Pliner H, Letson C, Heutink P, Martinez M, Gasser T, Traynor BJ, Wood N, Hardy J, Singleton AB, International Parkinson’s Disease Genomics, Consortium, & Parkinson’s Disease meta-analysis, consortium. (2015). NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol Aging, 36(3), 1605 e1607–1612. doi: 10.1016/j.neurobiolaging.2014.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Simpson S Jr., van der Mei I, Charlesworth JC, Lucas R, Ponsonby AL, Zhou Y, Wu F, & Taylor BV (2016). Role of genetic susceptibility variants in predicting clinical course in multiple sclerosis: a cohort study. J Neurol Neurosurg Psychiatry, 87(11), 1204–1211. doi: 10.1136/jnnp-2016-313722 [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O’Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, & Wolinsky JS (2011). Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol, 69(2), 292–302. doi: 10.1002/ana.22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2018). R: A Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
- Ramos Erin M., Hoffman Douglas, Junkins Heather A., Maglott Donna, Phan Lon, Sherry Stephen T., Feolo Mike, & Hindorff Lucia A. (2013). Phenotype–Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. European Journal Of Human Genetics, 22, 144. doi:10.1038/ejhg.2013.9610.1038/ejhg.2013.96https://www.nature.com/articles/ejhg201396#supplementary-informationhttps://www.nature.com/articles/ejhg201396#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh RH, Seaman SR, Masterman T, Hensiek AE, Sawcer SJ, Vukusic S, Achiti I, Confavreux C, Coustans M, le Page E, Edan G, McDonnell GV, Hawkins S, Trojano M, Liguori M, Cocco E, Marrosu MG, Tesser F, Leone MA, Weber A, Zipp F, Miterski B, Epplen JT, Oturai A, Sorensen PS, Celius EG, Lara NT, Montalban X, Villoslada P, Silva AM, Marta M, Leite I, Dubois B, Rubio J, Butzkueven H, Kilpatrick T, Mycko MP, Selmaj KW, Rio ME, Sa M, Salemi G, Savettieri G, Hillert J, & Compston DA (2005). Multiple Sclerosis Severity Score: using disability and disease duration to rate disease severity. Neurology, 64(7), 1144–1151. doi: 10.1212/01.WNL.0000156155.19270.F8 [DOI] [PubMed] [Google Scholar]
- Sadovnick AD, Traboulsee AL, Zhao Y, Bernales CQ, Encarnacion M, Ross JP, Yee IM, Criscuoli MG, & Vilarino-Guell C (2017). Genetic modifiers of multiple sclerosis progression, severity and onset. Clin Immunol, 180, 100–105. doi: 10.1016/j.clim.2017.05.009 [DOI] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E, D’Alfonso S, Blackburn H, Boneschi FM, Liddle J, Harbo HF, Perez ML, Spurkland A, Waller MJ, Mycko MP, Ricketts M, Comabella M, Hammond N, Kockum I, McCann OT, Ban M, Whittaker P, Kemppinen A, Weston P, Hawkins C, Widaa S, Zajicek J, Dronov S, Robertson N, Bumpstead SJ, Barcellos LF, Ravindrarajah R, Abraham R, Alfredsson L, Ardlie K, Aubin C, Baker A, Baker K, Baranzini SE, Bergamaschi L, Bergamaschi R, Bernstein A, Berthele A, Boggild M, Bradfield JP, Brassat D, Broadley SA, Buck D, Butzkueven H, Capra R, Carroll WM, Cavalla P, Celius EG, Cepok S, Chiavacci R, Clerget-Darpoux F, Clysters K, Comi G, Cossburn M, Cournu-Rebeix I, Cox MB, Cozen W, Cree BA, Cross AH, Cusi D, Daly MJ, Davis E, de Bakker PI, Debouverie M, D’Hooghe M B, Dixon K, Dobosi R, Dubois B, Ellinghaus D, Elovaara I, Esposito F, Fontenille C, Foote S, Franke A, Galimberti D, Ghezzi A, Glessner J, Gomez R, Gout O, Graham C, Grant SF, Guerini FR, Hakonarson H, Hall P, Hamsten A, Hartung HP, Heard RN, Heath S, Hobart J, Hoshi M, Infante-Duarte C, Ingram G, Ingram W, Islam T, Jagodic M, Kabesch M, Kermode AG, Kilpatrick TJ, Kim C, Klopp N, Koivisto K, Larsson M, Lathrop M, Lechner-Scott JS, Leone MA, Leppa V, Liljedahl U, Bomfim IL, Lincoln RR, Link J, Liu J, Lorentzen AR, Lupoli S, Macciardi F, Mack T, Marriott M, Martinelli V, Mason D, McCauley JL, Mentch F, Mero IL, Mihalova T, Montalban X, Mottershead J, Myhr KM, Naldi P, Ollier W, Page A, Palotie A, Pelletier J, Piccio L, Pickersgill T, Piehl F, Pobywajlo S, Quach HL, Ramsay PP, Reunanen M, Reynolds R, Rioux JD, Rodegher M, Roesner S, Rubio JP, Ruckert IM, Salvetti M, Salvi E, Santaniello A, Schaefer CA, Schreiber S, Schulze C, Scott RJ, Sellebjerg F, Selmaj KW, Sexton D, Shen L, Simms-Acuna B, Skidmore S, Sleiman PM, Smestad C, Sorensen PS, Sondergaard HB, Stankovich J, Strange RC, Sulonen AM, Sundqvist E, Syvanen AC, Taddeo F, Taylor B, Blackwell JM, Tienari P, Bramon E, Tourbah A, Brown MA, Tronczynska E, Casas JP, Tubridy N, Corvin A, Vickery J, Jankowski J, Villoslada P, Markus HS, Wang K, Mathew CG, Wason J, Palmer CN, Wichmann HE, Plomin R, Willoughby E, Rautanen A, Winkelmann J, Wittig M, Trembath RC, Yaouanq J, Viswanathan AC, Zhang H, Wood NW, Zuvich R, Deloukas P, Langford C, Duncanson A, Oksenberg JR, Pericak-Vance MA, Haines JL, Olsson T, Hillert J, Ivinson AJ, De Jager PL, Peltonen L, Stewart GJ, Hafler DA, Hauser SL, McVean G, Donnelly P, & Compston A (2011). Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 476(7359), 214–219. doi:nature10251 [pii] 10.1038/nature10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Tsukerman P, & Mandelboim O (2017). The paired receptors TIGIT and DNAM-1 as targets for therapeutic antibodies. Hum Antibodies, 25(3–4), 111–119. doi: 10.3233/HAB-160307 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium, Frazer Kelly A., Ballinger Dennis G., Cox David R., Hinds David A., Stuve Laura L., Gibbs Richard A., Belmont John W., Boudreau Andrew, Hardenbol Paul, Leal Suzanne M., Pasternak Shiran, Wheeler David A., Willis Thomas D., Yu Fuli, Yang Huanming, Zeng Changqing, Gao Yang, Hu Haoran, Hu Weitao, Li Chaohua, Lin Wei, Liu Siqi, Pan Hao, Tang Xiaoli, Wang Jian, Wang Wei, Yu Jun, Zhang Bo, Zhang Qingrun, Zhao Hongbin, Zhao Hui, Zhou Jun, Gabriel Stacey B., Barry Rachel, Blumenstiel Brendan, Camargo Amy, Defelice Matthew, Faggart Maura, Goyette Mary, Gupta Supriya, Moore Jamie, Nguyen Huy, Onofrio Robert C., Parkin Melissa, Roy Jessica, Stahl Erich, Winchester Ellen, Ziaugra Liuda, Altshuler David, Shen Yan, Yao Zhijian, Huang Wei, Chu Xun, He Yungang, Jin Li, Liu Yangfan, Shen Yayun, Sun Weiwei, Wang Haifeng, Wang Yi, Wang Ying, Xiong Xiaoyan, Xu Liang, Waye Mary M. Y., Tsui Stephen K. W., Xue Hong, Wong J. Tze-Fei, Galver Luana M., Fan Jian-Bing, Gunderson Kevin, Murray Sarah S., Oliphant Arnold R., Chee Mark S., Montpetit Alexandre, Chagnon Fanny, Ferretti Vincent, Leboeuf Martin, Olivier Jean-François, Phillips Michael S., Roumy Stéphanie, Sallée Clémentine, Verner Andrei, Hudson Thomas J., Kwok Pui-Yan, Cai Dongmei, Koboldt Daniel C., Miller Raymond D., Pawlikowska Ludmila, Taillon-Miller Patricia, Xiao Ming, Tsui Lap-Chee, Mak William, Qiang Song You, Tam Paul K. H., Nakamura Yusuke, Kawaguchi Takahisa, Kitamoto Takuya, Morizono Takashi, Nagashima Atsushi, Ohnishi Yozo, Sekine Akihiro, Tanaka Toshihiro, Tsunoda Tatsuhiko, Deloukas Panos, Bird Christine P., Delgado Marcos, Dermitzakis Emmanouil T., Gwilliam Rhian, Hunt Sarah, Morrison Jonathan, Powell Don, Stranger Barbara E., Whittaker Pamela, Bentley David R., Daly Mark J., de Bakker Paul I. W., Barrett Jeff, Chretien Yves R., Maller Julian, McCarroll Steve, Patterson Nick, Pe’er Itsik, Price Alkes, Purcell Shaun, Richter Daniel J., Sabeti Pardis, Saxena Richa, Schaffner Stephen F., Sham Pak C., Varilly Patrick, Altshuler David, Stein Lincoln D., Krishnan Lalitha, Vernon Smith Albert, Tello-Ruiz Marcela K., Thorisson Gudmundur A., Chakravarti Aravinda, Chen Peter E., Cutler David J., Kashuk Carl S., Lin Shin, Abecasis Gonçalo R., Guan Weihua, Li Yun, Munro Heather M., Steve Qin Zhaohui, Thomas Daryl J., McVean Gilean, Auton Adam, Bottolo Leonardo, Cardin Niall, Eyheramendy Susana, Freeman Colin, Marchini Jonathan, Myers Simon, Spencer Chris, Stephens Matthew, Donnelly Peter, Cardon Lon R., Clarke Geraldine, Evans David M., Morris Andrew P., Weir Bruce S., Tsunoda Tatsuhiko, Johnson Todd, Mullikin James C., Sherry Stephen T., Feolo Michael, Skol Andrew, Zhang Houcan, Zeng Changqing, Zhao Hui, Matsuda Ichiro, Fukushima Yoshimitsu, Macer Darryl R., Suda Eiko, Rotimi Charles N., Adebamowo Clement A., Ajayi Ike, Aniagwu Toyin, Marshall Patricia A., Nkwodimmah Chibuzor, Royal Charmaine D. M., Leppert Mark F., Dixon Missy, Peiffer Andy, Qiu Renzong, Kent Alastair, Kato Kazuto, Niikawa Norio, Adewole Isaac F., Knoppers Bartha M., Foster Morris W., Wright Clayton Ellen, Watkin Jessica, Gibbs Richard, Belmont John W., Muzny Donna, Nazareth Lynne, Sodergren Erica, Weinstock George M., Wheeler David A., Yakub Imtaz, Gabriel Stacey B., Onofrio Robert C., Richter Daniel J., Ziaugra Liuda, Birren Bruce W., Daly Mark J., Altshuler David, Wilson Richard K., Fulton Lucinda L., Rogers Jane, Burton John, Carter Nigel P., Clee Christopher M., Griffiths Mark, Jones Matthew C., McLay Kirsten, Plumb Robert W., Ross Mark T., Sims Sarah K., Willey David L., Chen Zhu, Han Hua, Kang Le, Godbout Martin, Wallenburg John C., L’Archevêque Paul, Bellemare Guy, Saeki Koji, Wang Hongguang, An Daochang, Fu Hongbo, Li Qing, Wang Zhen, Wang Renwu, Holden Arthur L., Brooks Lisa D., McEwen Jean E., Guyer Mark S., Ota Wang Vivian, Peterson Jane L., Shi Michael, Spiegel Jack, Sung Lawrence M., Zacharia Lynn F., Collins Francis S., Kennedy Karen, Jamieson Ruth, & Stewart John. (2007). A second generation human haplotype map of over 3.1 million SNPs. Nature, 449, 851. doi:10.1038/nature0625810.1038/nature06258https://www.nature.com/articles/nature06258#supplementary-informationhttps://www.nature.com/articles/nature06258#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara Candelaria, Parker Margaret M., Franco Liliana, Cho Michael H., Valencia-Duarte Ana V., Beaty Terri H., & Duggal Priya. (2018). Genotype imputation performance of three reference panels using African ancestry individuals. Human Genetics, 137(4), 281–292. doi: 10.1007/s00439-018-1881-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weideman AM, Barbour C, Tapia-Maltos MA, Tran T, Jackson K, Kosa P, Komori M, Wichman A, Johnson K, Greenwood M, & Bielekova B (2017). New Multiple Sclerosis Disease Severity Scale Predicts Future Accumulation of Disability. Front Neurol, 8, 598. doi: 10.3389/fneur.2017.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weideman AM, Tapia-Maltos MA, Johnson K, Greenwood M, & Bielekova B (2017). Meta-analysis of the Age-Dependent Efficacy of Multiple Sclerosis Treatments. Front Neurol, 8, 577. doi: 10.3389/fneur.2017.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright Marvin N., & Ziegler Andreas. (2017). ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. 2017, 77(1), 17. doi: 10.18637/jss.v077.i01 [DOI] [Google Scholar]
- Zerbino Daniel R., Achuthan Premanand, Akanni Wasiu, Amode M Ridwan, Barrell Daniel, Bhai Jyothish, Billis Konstantinos, Cummins Carla, Gall Astrid, Girón Carlos García, Gil Laurent, Gordon Leo, Haggerty Leanne, Haskell Erin, Hourlier Thibaut, Izuogu Osagie G., Janacek Sophie H., Juettemann Thomas, To Jimmy Kiang, Laird Matthew R., Lavidas Ilias, Liu Zhicheng, Loveland Jane E., Maurel Thomas, McLaren William, Moore Benjamin, Mudge Jonathan, Murphy Daniel N., Newman Victoria, Nuhn Michael, Ogeh Denye, Ong Chuang Kee, Parker Anne, Patricio Mateus, Riat Harpreet Singh, Schuilenburg Helen, Sheppard Dan, Sparrow Helen, Taylor Kieron, Thormann Anja, Vullo Alessandro, Walts Brandon, Zadissa Amonida, Frankish Adam, Hunt Sarah E., Kostadima Myrto, Langridge Nicholas, Martin Fergal J., Muffato Matthieu, Perry Emily, Ruffier Magali, Staines Dan M., Trevanion Stephen J., Aken Bronwen L., Cunningham Fiona, Yates Andrew, & Flicek Paul. (2018). Ensembl 2018. Nucleic Acids Research, 46(D1), D754–D761. doi: 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All custom code developed and used in this manuscript are available in the supplementary information files. Genetic and phenotype data are available in the database of Genotypes and Phenotypes (dbGaP).