SUMMARY
Rad52 is a key factor for homologous recombination (HR) in yeast. Rad52 helps assemble Rad51-ssDNA nucleoprotein filaments that catalyze DNA strand exchange, and it mediates single-strand DNA annealing. We find that Rad52 has an even earlier function in HR in restricting DNA double-stranded break ends resection that generates 3′ single-stranded DNA (ssDNA) tails. In fission yeast, Exo1 is the primary resection nuclease, with the helicase Rqh1 playing a minor role. We demonstrate that the choice of two extensive resection pathways is regulated by Rad52. In rad52 cells, the resection rate increases from ~3–5 kb/h up to ~10–20 kb/h in an Rqh1-dependent manner, while Exo1 becomes dispensable. Budding yeast Rad52 similarly inhibits Sgs1-dependent resection. Single-molecule analysis with purified budding yeast proteins shows that Rad52 competes with Sgs1 for DNA end binding and inhibits Sgs1 translocation along DNA. These results identify a role for Rad52 in limiting ssDNA generated by end resection.
In Brief
The ends of DNA breaks are processed to generate single-stranded DNA (ssDNA) in preparation for break repair. Excessive ssDNA may lead to genome instability. Yan et al. find that the DNA repair protein Rad52 restricts the length of ssDNA by limiting access and translocation of RecQ helicases on DNA.
Graphical Abstract
INTRODUCTION
The initial step of homologous recombination (HR) induced at DNA double-strand breaks (DSBs) is resection of the 5′ strands to produce 3′ single-stranded DNA (ssDNA) tails (reviewed in Symington, 2016). The single-strand binding protein RPA binds the ssDNA tails and recruits DNA damage checkpoint factors, including budding yeast Mec1-Ddc2 and human ATR-ATRIP. However, ssDNA-bound RPA must be replaced by the DNA strand exchange protein Rad51 for HR to proceed. Specific “recombination mediators” assist in RPA displacement from ssDNA and hence are indispensable for the assembly of the Rad51-ssDNA nucleoprotein filament, commonly referred to as the presynaptic filament. The major recombination mediators are Rad52 in yeast and BRCA2 in mammals (Benson et al., 1998; Jensen et al., 2010; Liu et al., 2010; New et al., 1998; Shinohara and Ogawa, 1998; Sung, 1997). Both Rad52 and BRCA2 directly bind Rad51 and ssDNA to promote RPA-Rad51 exchange.
DNA end resection is initiated by the Mre11-Rad50-Xrs2 (MRX) complex in budding yeast and its equivalent MRE11-RAD50-NBS1 (MRN) complex in fission yeast and humans. MRX or MRN acts in conjunction with Sae2 (Ctp1 in fission yeast and CtIP in human) to produce a ssDNA gap that serves as the entry point for the long range resection machinery harboring Exo1, a 5′→3′ exonuclease, or a RecQ helicase (Sgs1 in budding yeast and BLM or WRN in humans) together with the 5′ flap endonuclease and helicase Dna2 (Cejka et al., 2010; Mimitou and Symington, 2008; Nicolette et al., 2010; Niu et al., 2010; Zhu et al., 2008). These latter resection enzymes act in parallel, and only simultaneous elimination of both Sgs1 or Dna2 and Exo1 strongly impair long-range resection. The same short- and long-range end resection pathways exist in other eukaryotes (reviewed in Cejka, 2015; Symington, 2016). Interestingly, in fission yeast, Exo1 predominates over the long-range pathway that is mediated by Sgs1 ortholog Rqh1 (Langerak et al., 2011). Preferential use of one long-range resection pathway over the other has also been described for vertebrate species (Hoa et al., 2015; Liao et al., 2008; Sturzenegger et al., 2014; Tomimatsu et al., 2012). Currently, there is little knowledge regarding how the choice of long-range resection pathways is made at the molecular level.
The same resection pathways that act at chromosomal break ends can also operate at telomeres (de Lange, 2018) or stalled replication forks (Rickman and Smogorzewska, 2019). Multiple mechanisms have evolved to protect telomeres and reversed forks from resection enzymes and DSB ends from hyper-resection. In mammals, the chromatin-binding factor 53BP1 functions together with RIF1 and the tetrameric Shieldin complex to prevent hyper-resection at unprotected telomeres and to help channel DSBs into the non-homologous DNA end joining (NHEJ) pathway (de Lange, 2018) and likely to protect stalled replication forks (Her et al., 2018). It is possible that Shieldin restricts resection by recruiting DNA polymerase α to resynthesize DNA lost during resection, with the 3′ ssDNA tail serving as the template (Mirman et al., 2018). The budding yeast ortholog of 53BP1 called Rad9 also protects DSB ends and uncapped telomeres from hyper-resection and stalled replication forks from degradation (Lazzaro et al., 2008; Villa et al., 2018), although it plays only a minor role in NHEJ promotion (Ferrari et al., 2015). Other negative regulators of resection have been identified in humans, including DYNLL1 (He et al., 2018) and the HELB helicase (Tkáĉ et al., 2016). DYNLL1 appears to stabilize the multimeric form of 53BP1 (Becker et al., 2018), while the mechanism of HELB remains to be defined.
Recently, multiple essential HR proteins, including BRCA2, BRCA1, and RAD51 and many others, were shown to protect stalled replication forks from degradation (Rickman and Smogorzewska, 2019; Schlacher et al., 2011). It is not known whether these proteins also control resection at DSBs, and it is difficult to test their participation considering the other essential roles they play in DSB repair. Here, we examined the role of key HR proteins in resection at DSBs using yeast model organisms. We used an experimental system where a nonrepairable DSB is induced (Klein et al., 2019), and therefore, we could distinguish the functions of key HR proteins in repair from their possible functions in resection. We have found that Rad52, but not other Rad51 mediators or Rad51 itself, negatively regulates resection in fission yeast (S. pombe). Rad52 controls Rqh1-mediated resection, with little or no effect on the Exo1 pathway. We further show that budding yeast (S. cerevisiae) Rad52 also inhibits Sgs1-mediated resection both in vivo and in vitro. By single-molecule biophysical analysis, we demonstrate that Rad52 controls resection likely by blocking resection helicase access to DSB ends and inhibiting its translocation along DNA. Together, our results help define an unexpected early role of Rad52 in long-range resection control during HR.
RESULTS
Assays for Analysis of Resection in Fission Yeast
The kinetics of DNA end resection can be easily followed in budding yeast owing to the synchrony of DSB induction with the HO or I-SceI endonuclease (Klein et al., 2019). Here, we followed resection in the distantly related fission yeast using a robust and quantitative assay that employs the I-PpoI endonuclease (TET-I-PpoI) to synchronously induce a single DSB per genome (Sunder et al., 2012). The I-PpoI recognition sequence was inserted at one of two loci, either at lys1 on chromosome I or at arg1 on chromosome III (Figures S1A and S1B). I-PpoI was induced by adding anhydrotetracycline (ahTET) to the media, and a DSB was formed efficiently at both loci as followed by Southern blotting with probes specific for lys1 and arg1. Interestingly, the I-PpoI generated cleavage band disappeared more rapidly (30–60 min) than DSBs generated by the HO endonuclease in the budding yeast (120–240 min) (Zhu et al., 2008), suggesting that initial DSB resection is faster in the fission yeast. The rate of long-range resection was determined using probes specific for sequences at increasing distances from the break. The resection rate in wild-type cells, measured as resection completed in 50% of the cells specified by the probes, ranged from ~3.5 kb/h closer to the DSB to ~5.5 kb/h farther from the DSB. Thus, in the fission yeast, resection at a nonrepairable break slightly increases over time and/or distance from DSB ends.
Function of the MRN Complex, Ctp1, Exo1, Rqh1, and Fft3 in Resection
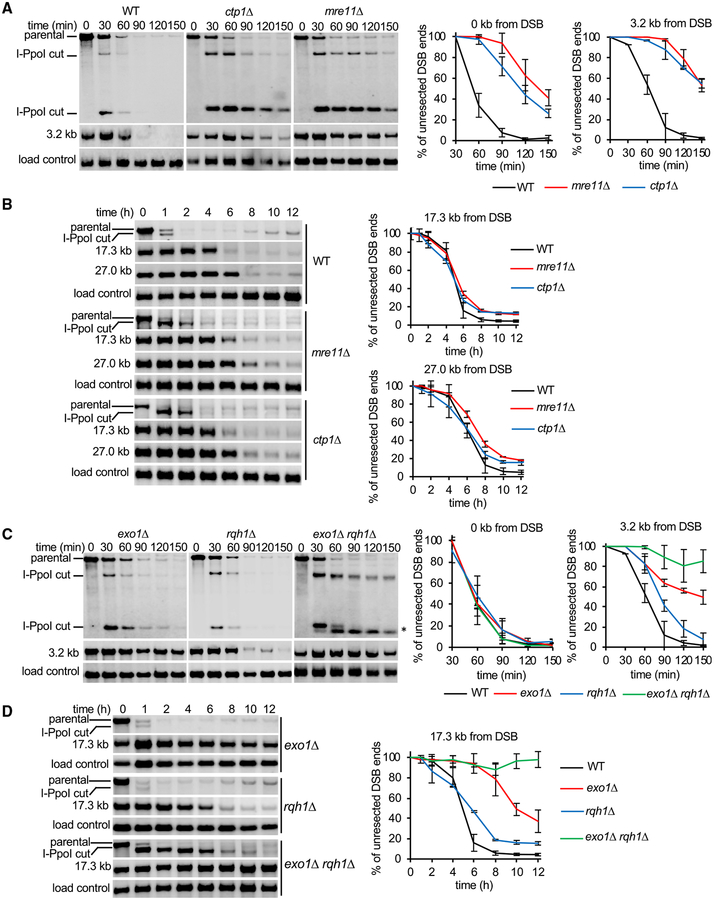
To ascertain the role of previously identified resection factors (Symington, 2016), we disrupted exo1, rqh1, mre11, ctp1, rsc1, and fft3 genes. Initial (0.5–2.5 h after DSB induction) and long-range resection (1–12 h after DSB induction) was measured. As reported by others (Langerak et al., 2011; Limbo et al., 2007), Mre11 and Ctp1 were important for initial resection, with a clear resection delay (~60–90 min) in mutant compared to wild-type cells (Figure 1A). However, most mutant cells eventually initiate resection, with the rate of long-range resection (measured at 17 or 27 kb away from the DSB) being comparable to that in wild-type cells (Figure 1B). These results confirm that initial resection in fission yeast is similar to humans, where CtIP plays an important role (Sartori et al., 2007), but is different from the budding yeast, where Sae2 has a less pronounced role in resection at “clean” DSBs (Shim et al., 2010). The delay in ctp1Δ cells is partially suppressed by deleting yku70, supporting the idea that the MRN complex with Ctp1 can overcome the inhibitory effect of the Ku heterodimer on resection (Figure S2A) (Langerak et al., 2011; Mimitou and Symington, 2010; Shim et al., 2010). The RSC complex, which is important for initial resection in the budding yeast (Chen et al., 2012; Shim et al., 2007), does not play any role in resection in the fission yeast, as revealed in the analysis of rsc1Δ mutant cells (Figure S2B). Furthermore, Exo1 is essential for extensive resection, whereas deletion of rqh1 results in only a modest decrease in resection, as measured at 3.2 or 17.3 kb from the DSB (Figures 1C and 1D). These results suggest that the Rqh1-dependent pathway is either not proficient or is negatively regulated in the fission yeast. Simultaneous deletion of exo1 and rqh1 imparts the same severe resection phenotype as the budding yeast exo1Δ sgs1Δ double mutant, with only short ssDNA (~100 nt) that is generated by MRN-Ctp1 (Figure 1C). Finally, we examined the role of Fft3 and found that similar to its budding yeast and human counterparts (ScFun30 and hSMARCAD1, respectively) (Chen et al., 2012, 2016; Costelloe et al., 2012), it plays an important role in long-range resection (Figure S2B).
Figure 1. Role of MRN, Ctp1, Exo1, and Rqh1 in Initial and Extensive Resection.
(A and B) Southern blot analysis of initial resection (0–150 min) (A) and extensive resection (0–12 h) (B) at lys1 locus in wild-type, mre11, and ctp1 cells.(C and D) Southern blot analysis of initial resection (0–150 min) (C) and extensive resection (0–12 h) (D) at lys1 locus in wild-type and exo1, rqh1, or exo1 rqh1 mutant cells. Restriction-enzyme-digested DNA was separated on 0.8% agarose gels. Primers used to prepare DNA probes for Southern blotting and restriction enzymes used to digest genomic DNA are presented in Table S3. Plots show kinetics of resection and error bars denote SD (n = 3). An asterisk in (C) denotes unresected DSB end.
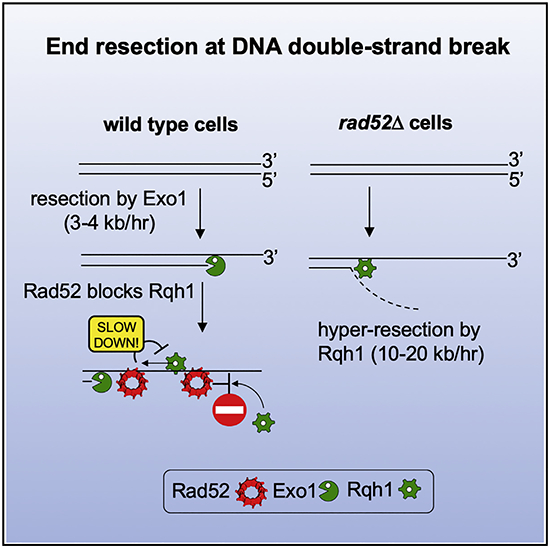
Rad52 Restrains DSB Ends Resection
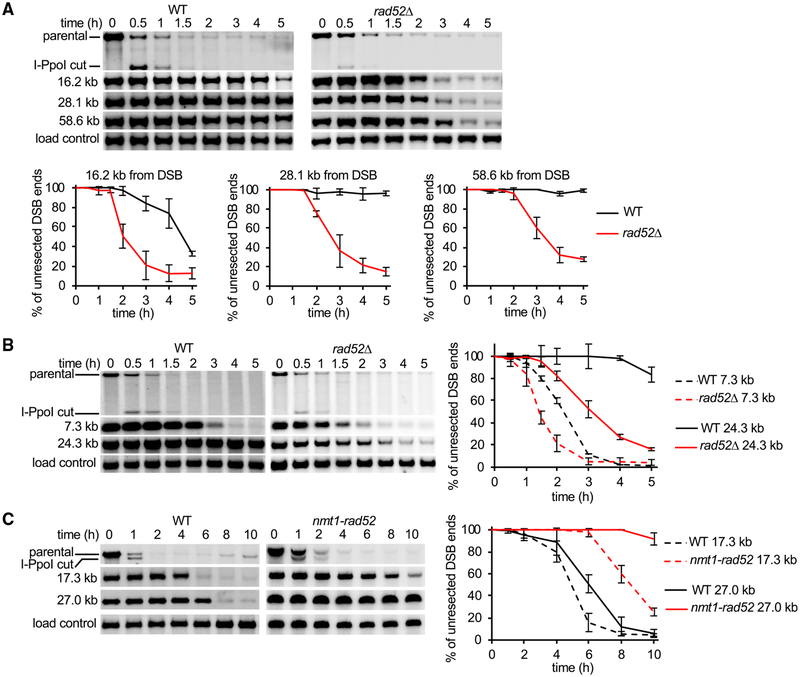
Rad52 is an essential protein for HR in yeast, where it mediates loading of Rad51 onto RPA-coated ssDNA to nucleate assembly of the presynaptic filament (Sung, 1997). Unexpectedly, we found that rad52Δ mutant cells exhibit a highly elevated rate of resection at the lys1 locus, as sequences nearly 60 kb away from the DSB became resected within 3 h (rate of up to 20 kb/h) (Figure 2A). Enhanced resection was also observed at the arg1 locus, demonstrating that the inhibitory activity of Rad52 in resection is not locus specific (Figure 2B). To examine whether other proteins that function in Rad51 presynaptic filament assembly and Rad51 itself are also involved in the negative control of resection, we examined rad51Δ, rhp55Δ, and swi5Δ mutants. The rate of long-range resection in these mutants was either comparable to wild-type (swi5Δ) or marginally faster than wild-type (rad51Δ and rhp55Δ), indicating that the resection regulatory function of Rad52 is unique among these proteins (Figure S2C). We next tested whether overexpression of Rad52 would slow the resection rate in wild-type cells. As expected, overexpression of Rad52 using the nmt1 promoter had a very strong negative effect on resection, with a rate of ~2 kb/h measured at 17 kb away from the DSB and no detectable resection at 27 kb from the DSB by 10 h after DSB induction (Figure 2C). Together, deletion of rad52 gene speeds up DSB ends resection, while rad52 overexpression slows it down.
Figure 2. Rad52 Negatively Controls Extensive Resection in Fission Yeast.
(A and B) Southern blot analysis of resection in wild-type and rad52Δ strains at the (A) lys1 locus or (B) arg1 locus. DNA was separated on 0.8% agarose gels. (C) Southern blot analysis of resection in cells with overexpressed Rad52. Plots show kinetics of resection, and error bars denote SD (n = 3).
The C-Terminal Region of Rad52 Is Important for Resection Control
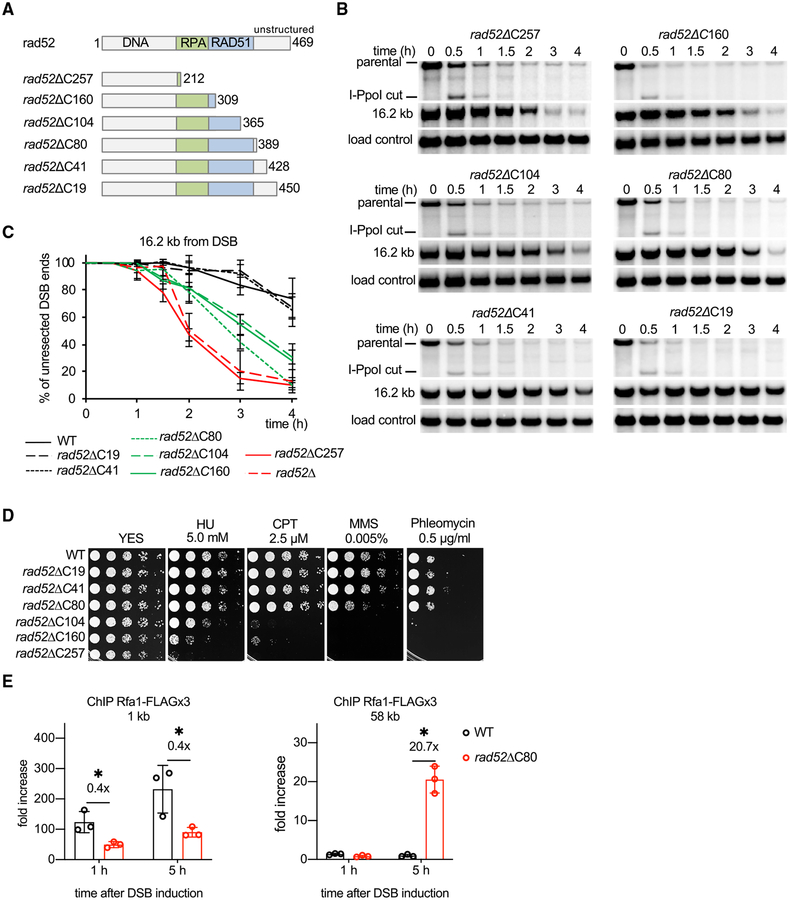
Rad52 harbors conserved domains that mediate its interactions with DNA, Rad51, and RPA, whereas the C-terminal 80 amino acids are less conserved. To identify the region(s) required for the role of fission yeast Rad52 as an anti-resection factor, we inserted a stop codon at several positions in the rad52 coding frame and analyzed resection kinetics in the resulting mutants (Figure 3A). Three classes of mutants were isolated: those that have a resection rate similar to the rad52 null mutant, an intermediate rate of resection, or a wild-type resection rate (Figures 3B and 3C). Based on these results, we conclude that (1) Rad52 variants that harbor DNA and RPA interaction activities are able to partially inhibit resection; (2) Rad51 interaction is dispensable for blocking resection, which is consistent with the nearly normal resection rate in rad51Δ cells; and (3) the C-terminal region of Rad52 is also important for blocking resection. We compared the DNA damage sensitivity of these mutants and found, as expected, mutants that lack the DNA-, RPA-, or Rad51-binding attribute are extremely sensitive to methanesulfonate (MMS), hydroxyurea (HU), camptothecin (CPT), or phleomycin. In contrast, rad52ΔC80 mutant cells, which exhibit an increased rate of resection, are only sensitive to high doses of MMS at 30°C, even though sensitivity to a broad range of DNA-damaging agents (MMS, CPT, and phleomycin) was seen at 37°C (Figures 3D, S3A, and S3B). Thus, rad52ΔC80 is a separation-of-function mutant, being partially deficient in blocking resection but largely proficient in DNA repair at 30°C. Nuclear localization of the rad52ΔC80 protein, formation of DNA nuclear foci upon DSB induction, and stability of the protein are comparable to the wild-type counterpart (Figures S4A and S4B). These observations suggest that the Rad52 C terminus may be directly involved in resection control.
Figure 3. Role of the C-Terminal Region of Rad52 in Extensive Resection.
(A) Schematic showing major Rad52 domains and truncation mutants analyzed.
(B) Southern blot analysis of resection in indicated rad52 truncation mutants.
(C) Plots showing kinetics of resection in rad52 mutants. Error bars denote SD (n = 3).
(D) Analysis of sensitivity to DNA damage of wild-type and rad52 mutant strains. 5-fold serial dilutions were made, and 2 μL was spotted onto yeast extract with supplements (YES) or YES with hydroxyurea (HU), camptothecin (CPT), methyl methanesulfonate (MMS), or phleomycin.
(E) ChIP-qPCR analysis of recruitment of RPA (Rfa1 subunit, called Ssb1 in fission yeast) in wild-type and rad52ΔC80 cells at 1 and 58 kb from the DSB end at lys1 locus. Error bars denote SD (n = 3). One-tailed p values were shown (*p < 0.05).
As an independent measure of ssDNA formation, we monitored loading of the ssDNA-binding protein RPA in wild-type and rad52ΔC80 cells by chromatin immunoprecipitation (ChIP) and qPCR using primers specific for a sequence either 1 or 58 kb away from the DSB end. By 5 h after DSB induction, RPA loading was detectable at 58 kb from DSB only in rad52ΔC80 cells (Figure 3E). Interestingly, we detected a lower level of RPA loading close to the DSB in rad52ΔC80 cells, which could reflect a previously reported role of Rad52 in stabilizing RPA on ssDNA (Gibb et al., 2014).
DNA-Binding and Strand Annealing Activity of Rad52 Plays a Role in Resection Control
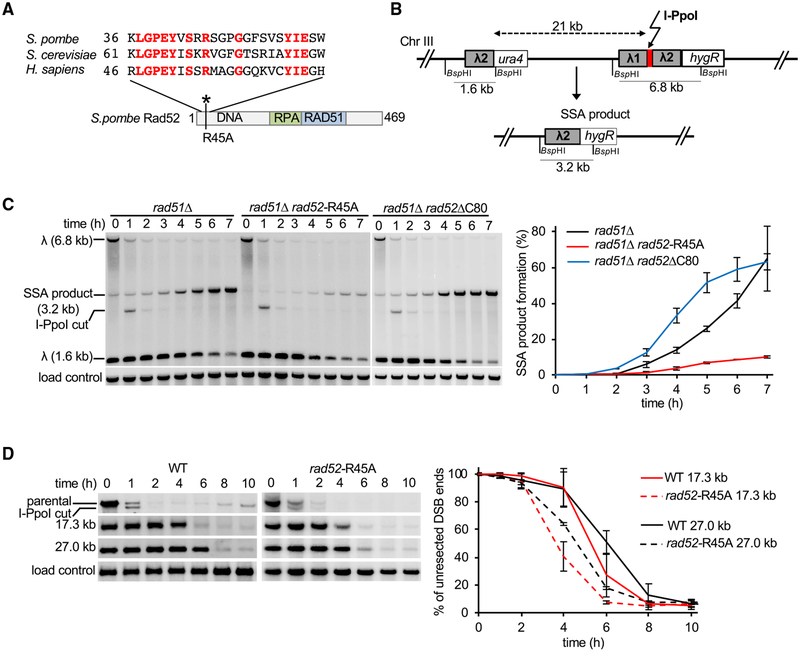
An evolutionarily conserved arginine-70 (R70) of budding yeast Rad52, equivalent to R45 in the fission yeast ortholog (Figure 4A), is needed for its DNA-binding and annealing activities but dispensable for Rad51 loading (Bai et al., 1999; Shi et al., 2009). To test the relevance of DNA binding in resection control, we constructed the fission yeast rad52-R45A mutant and first examined it for the ability to mediate single-strand annealing (SSA), a type of DSB repair that entails the annealing of complementary strands derived from two direct DNA repeats and the deletion of the intervening sequence. The SSA reporter harbors direct repeats of a 755-bp sequence from the bacteriophage lambda inserted 21 kb apart on chromosome III (Figure 4B). I-PpoI expression is slightly leaky and could lead to SSA product formation even before tetracycline is added. To eliminate these cells SSA was designed to include essential genes between the direct repeats. The analysis was done in the rad51Δ mutant background to exclude alternative repair pathways. 50% of rad51Δ cells completed repair via SSA by 6–7 h, whereas only 10% of rad51Δ rad52-R45A cells completed repair at that time. Thus, as expected, rad52-R45A cells are severely impaired for SSA (Figure 4C). We confirmed that the rad52-R45A mutant protein is properly localized to the nucleus, forms DNA damage foci upon DSB induction, and mutant rad52-R45A cells are much more resistant to DNA damage when compared to rad52Δ cells (Figures S4A and S4C). Importantly, rad52-R45 mutant cells resect DSB ends faster than wild type, albeit not as fast as rad52Δ mutant cells (Figure 4D). These results thus show that the DNA-binding and annealing activity of Rad52 is needed for its role as an anti-resection factor. Finally, the rad52-R45A;ΔC80 double mutant behaves like the rad52Δ mutant in exhibiting a greatly enhanced resection rate, suggesting that DNA-binding and annealing and C-terminal domains are two Rad52 regions needed for resection control (Figure S4D).
Figure 4. Analysis of DNA-Binding Domain of Rad52 in Resection Control.
(A) Sequence comparison of yeasts and human Rad52.
(B) Schematic of new SSA assay between two ~0.75-kb λ2 repeats inserted at arg1 locus and 21 kb upstream of arg1.
(C) Southern blot analysis of SSA in indicated rad52 mutants deficient in DNA binding (rad52-R45A) or C-terminal domain (rad52ΔC80). Plot shows kinetics of SSA product formation. Error bars denote SD (n = 3).
(D) Comparison of extensive resection kinetics in wild-type and rad52-R45A mutant cells. Plots show kinetics of resection, and error bars denote SD (n = 3).
Faster Kinetics of SSA in rad52ΔC80
In SSA, faster resection would facilitate the exposure of homologous sequences to stimulate product formation. Consistent with this premise, in the rad51Δ rad52ΔC80 mutant, 50% of cells complete DSB repair by SSA in less than 5 h, nearly 2 h faster when compared to rad51Δ cells (Figure 4C). This result also provides clear evidence that even though both the rad52-R45A and rad52ΔC80 mutant proteins are impaired for resection control, rad52ΔC80, unlike rad52-R45A (Figure 4C), is proficient in supporting SSA.
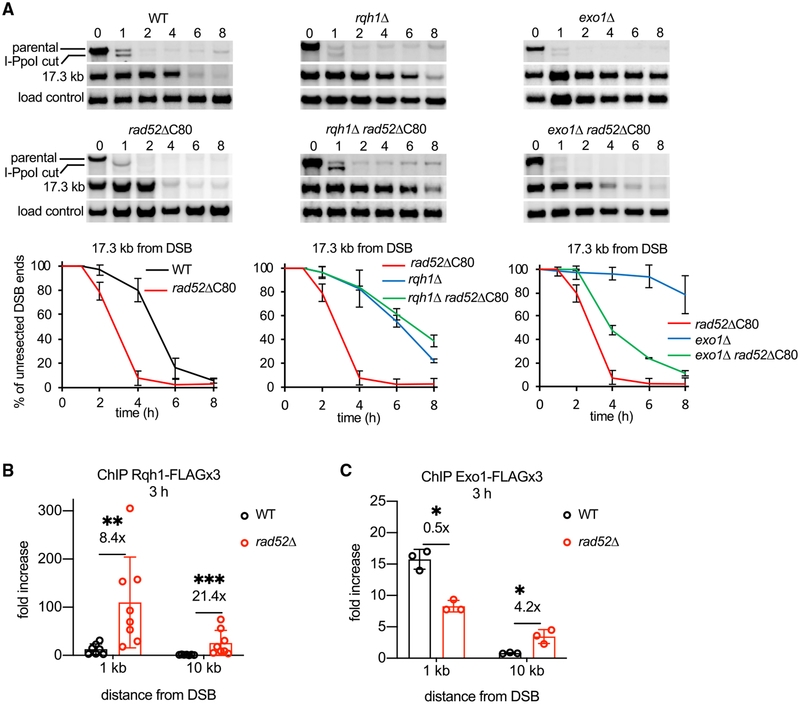
Rad52 Inhibits Rqh1-Mediated Resection
Rad52 might interfere with resection by blocking both the Exo1 and Rqh1 pathways or just one of them. To distinguish between these possibilities, we analyzed resection in rad52ΔC80 exo1Δ and rad52ΔC80 rqh1Δ double mutants. We used rad52ΔC80 instead of rad52Δ, because rad52Δ rqh1Δ is inviable (Wilson et al., 1999). Resection in the rad52ΔC80 rqh1Δ double mutant is much slower than in rad52ΔC80 and comparable to the rqh1Δ mutant, meaning that Rqh1 is required for the faster resection observed in rad52ΔC80 (Figure 5A). In contrast, in rad52ΔC80 exo1Δ cells, resection is much faster than in exo1Δ, thus revealing that Exo1 becomes dispensable for resection in the rad52ΔC80 mutant background (Figure 5A). Similarly, Exo1 becomes dispensable for resection in DNA-binding mutant rad52-R45A (Figure S5A). We note, however, that even though Rad52 acts predominantly on Rqh1-mediated resection, resection was markedly slower than in wild-type cells upon overexpression of Rad52, suggesting that Rad52 at high levels could affect the Exo1 pathway as well (Figure 2C).
Figure 5. Rad52 Controls Rqh1-Mediated Resection and Rqh1 Recruitment.
(A) Epistasis analysis between Rad52 and enzymes mediating two extensive resection pathways, Exo1 and Rqh1. Southern blot analysis of resection in indicated mutants. Plots show kinetics of resection, and error bars denote SD (n = 3).
(B and C) ChIP-qPCR analysis of Rqh1 and Exo1 loading in wild-type and rad52 cells at a DSB at lys1 locus at 1 and 10 kb from DSB ends. Error bars denote SD (Rqh1-FLAG: wild type [WT], n = 7, rad52Δ, n = 8. Exo1-FLAG: n = 3). (B) One-tailed p values were shown (**p < 0.01, ***p < 0.001). (C) One-tailed p values were shown (*p < 0.05).
Rad52 Restricts Resection Independently of Crb2
We considered several potential mechanisms for how Rad52 interferes with Rqh1-mediated resection. Rad52 could act with Crb2, a fission yeast ortholog of human 53BP1 and budding yeast Rad9, which have both been shown to negatively control resection (Bunting et al., 2010; Lazzaro et al., 2008). Human 53BP1 acts together with many other proteins, including RIF1 and the multi-subunit Shieldin complex, to block resection (Greenberg, 2018; Setiaputra and Durocher, 2019), but no Rad9 interactors that can block resection have been identified in yeast. As expected, Crb2 negatively regulates resection, although Rif1 and Rev7 in fission yeast have no such effect (Figures S5B and S5C). The resection increase observed in crb2Δ is not as dramatic as in rad52Δ mutant cells (Figure S5B). We also compared resection in the crb2Δ, rad52ΔC80 and crb2Δ rad52ΔC80 mutants in order to further define the relationship between Crb2 and Rad52. We note that the crb2Δ rad52Δ mutant could not be tested because it is not viable (Wilson et al., 1999). The crb2Δ rad52ΔC80 cells exhibit an increased rate of resection when compared to both single mutants, indicating that these proteins control resection via different mechanisms. Interestingly, we found that Crb2, similar to Rad52, also acts predominantly on Rqh1-mediated resection (Figure S5D). Therefore, while both Crb2 and Rad52 suppress Rqh1-mediated resection, they operate independently of each other.
Enhanced DSB Recruitment of Rqh1 in rad52Δ Cells
We next addressed the possibility that Rad52 interferes with the recruitment of resection factors to DSBs. To test this, we compared recruitment of Rqh1 and Exo1 in wild-type and rad52Δ cells. For this, we attached a FLAG-tag to these proteins and examined their recruitment by ChIP and qPCR. We found that Rqh1 recruitment is markedly (8- to 21-fold) increased in rad52Δ cells 1 and 10 kb away from the DSB ends. In comparison, Exo1 recruitment was not increased or increased just slightly (Figures 5B and 5C). These results provide evidence that Rad52 interferes with the DSB recruitment of Rqh1.
Rad52 Inhibits Resection in Budding Yeast
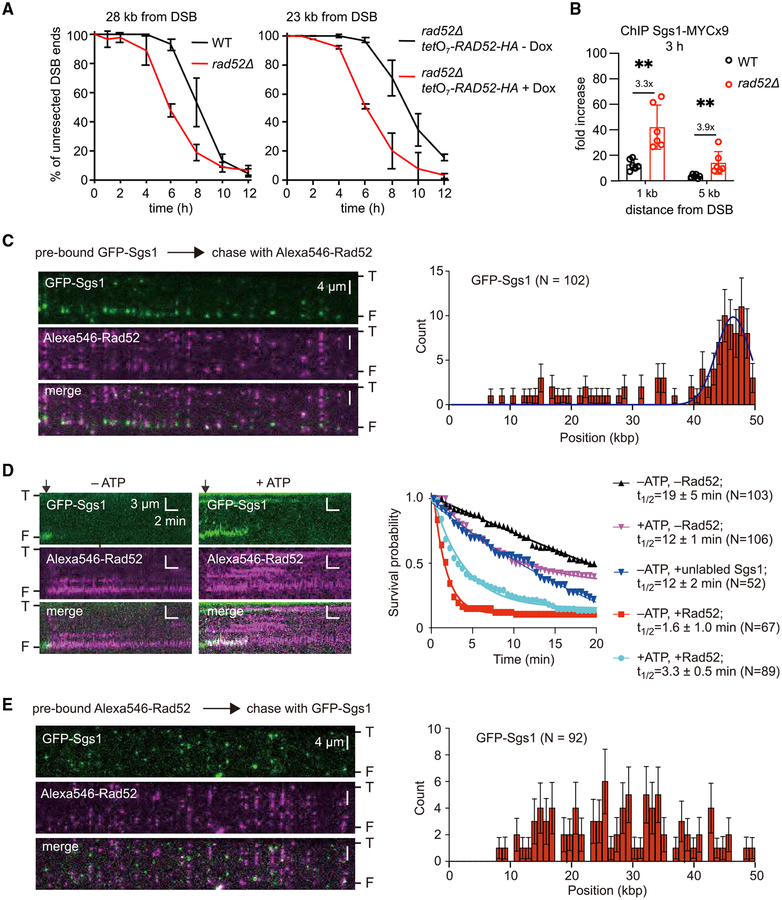
To test whether the impact of Rad52 on DSB end resection is conserved among species, we tested Rad52 from budding yeast, which is an evolutionarily very distant organism from fission yeast (Sipiczki, 2000). We followed resection at an HO-endonuclease-induced DSB at the MAT locus in a yeast haploid strain in which the HML and HMR homologous sequences were deleted to prevent repair by HR (Zhu et al., 2008). We found that the absence of Rad52 also speeds up resection in budding yeast cells, albeit not to the same degree as observed in fission yeast (Figures 6A and S6A). Next, we used the doxycycline-regulated tetO7 promoter to examine the effect of Rad52 overexpression on resection in budding yeast cells. Rad52 is highly expressed in the absence of doxycycline, whereas its expression is extinguished upon adding doxycycline. Overexpression of Rad52 is inhibitory for resection (Figures 6A and S6B–S6D). As observed in fission yeast, also in budding yeast, Sgs1, but not Exo1, was essential to observe faster resection in rad52 mutants (Figures S6F–S6H). Furthermore, we found that the DSB recruitment of Sgs1, but not Exo1, is also significantly enhanced in rad52Δ cells (Figures 6B and S6E). These results suggest that similar to fission yeast, budding yeast Rad52 also interferes with DSB end resection, likely by restricting the recruitment of Sgs1.
Figure 6. Rad52 Can Block DNA End Binding by Sgs1.
(A) Analysis of resection kinetics at DSB at MAT locus in budding yeast wild-type cells and cells that either lack or overexpress Rad52. Genomic DNA was digested with EcoRI; primers used to prepare DNA probes for Southern blotting are presented in Table S4. Corresponding Southern blots are shown in Figures S6A and 6B. Dox, doxycycline.
(B) ChIP-qPCR analysis of Sgs1 loading at MAT locus 3 h after DSB induction in wild-type and rad52Δ cells. Error bars denote SD (n = 6). One-tailed p values were shown (**p < 0.01).
(C) (Left) Wide-field images of GFP-Sgs1 (green) pre-bound to dsDNA ends after addition of Alexa546-Rad52 (magenta); “T” and “F” refer to the tethered and free DNA ends. (Right) Initial distribution of pre-bound GFP-Sgs1 (N = 102) that was chased by addition of Alexa546-Rad52; error bars represent 95% confidence intervals (CIs), and the blue line represents a Gaussian fit to the data.
(D) (Left) Kymograph showing the dissociation of end-bound GFP-Sgs1 (green) after addition of Alexa546-Rad52 (magenta). Arrowheads denote the time point of the Alexa546-Rad52 injection. (Right) Survival probability for end-bound GFP-Sgs1 (±ATP) chased with Rad52 or unlabeled Sgs1, as indicted. Note, that the minus Rad52 data curves are reproduced from Xue et al. (2019) for comparison.
(E) (Left) Wide-field images showing dsDNA pre-bound by Alexa546-Rad52 (magenta) and then chased with GFP-Sgs1 (green). (Right) The corresponding GFPSgs1 binding distribution. Error bars represent 95% CIs.
DNA Curtain Analysis of Rad52 for Interference with Sgs1 Activity
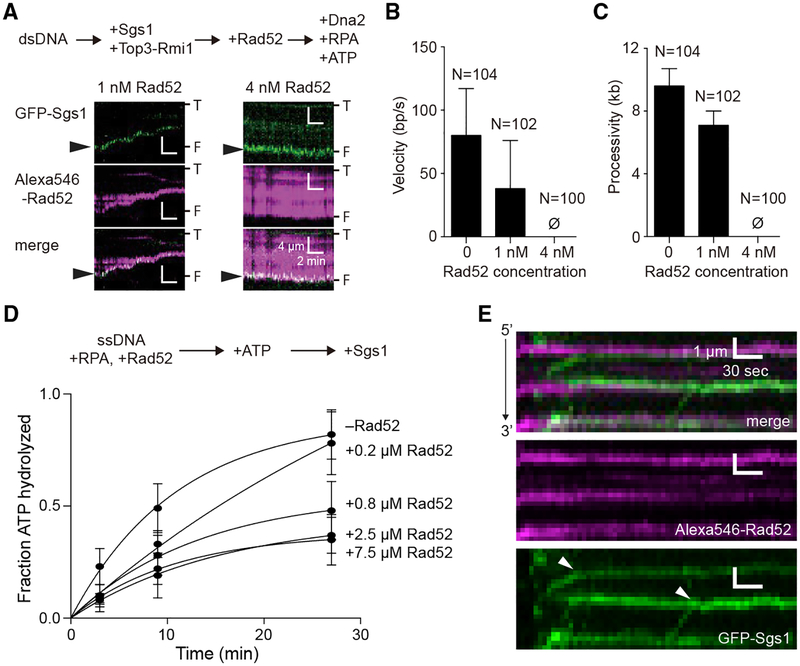
Our genetic studies provide evidence that Rad52 acts as a negative regulator of DNA end resection. To further assess the potential mechanisms by which Rad52 might inhibit the Sgs1-mediated DNA end resection pathway, we examined how Rad52 affected the behavior of Sgs1 in single-molecule assays. We chose budding yeast resection proteins, because these proteins have been well characterized (Cejka et al., 2010; Niu et al., 2010). We have previously developed DNA curtain assays for visualizing protein-DNA interactions in real time with total internal reflection fluorescence microscopy (TIRFM) (reviewed in Bell and Kowalczykowski, 2016). Using these assays, we have recently examined the recruitment of S. cerevisiae GFP-tagged Sgs1 to DNA ends and Sgs1-mediated end resection at the single-molecule level (Xue et al., 2019). These studies revealed that GFP-Sgs1 is preferentially recruited to short (≥12 nt) 3′ or 5′ ssDNA overhangs (Xue et al., 2019). We asked whether Rad52 would alter the pattern of GFP-Sgs1 recruitment to dsDNA ends bearing a 30-nt 3′ ssDNA overhang. As we have previously shown, GFP-Sgs1 was readily targeted to these DNA ends (Figure 6C) and exhibited a lifetime of ~12 to ~19 min, depending upon whether or not ATP was present (Xue et al., 2019) (Figure 6D). However, when Alexa546-Rad52 was added after Sgs1, it reduced the lifetime of end-bound GFP-Sgs1 by ~4-fold to ~12-fold, again depending upon whether or not ATP was present (Figures 6C and 6D). Notably, the half-life of GFP-Sgs1 is 12 ± 1.0 min when chased with dark Sgs1 (in the absence of ATP, to prevent translocation), compared to a half-life of 3.3 ± 0.5 min when chased with Rad52; the 4-fold difference in dissociation rates emphasizes the potential role of Rad52 in dislodging Sgs1 from DNA ends (Figure 6D). These findings indicate that once Sgs1 is bound to the DNA ends, it can be removed by Rad52. Moreover, when Rad52 was added to the DNA before Sgs1, there was a drastic decrease in the recruitment of GFP-Sgs1 to the DNA ends (Figure 6E). These results support a model in which Rad52 directly competes with Sgs1 for initial binding to DNA ends and Rad52 can also displace Sgs1 that is already bound at DNA ends.
Sgs1 possesses robust ATP-dependent DNA helicase activity (Cejka et al., 2010; Cejka and Kowalczykowski, 2010; Niu et al., 2010), and Sgs1 also translocates rapidly along ssDNA (Crickard et al., 2019). Therefore, we asked whether Rad52 might also affect these DNA motor activities of Sgs1. Surprisingly, DNA end resection assays containing GFP-Sgs1, the Top3-Rmi1 complex (an accessory factor of Sgs1), Dna2, RPA, and ATP revealed marked inhibition of end resection when Rad52 was bound to the DNA (Figures 7A–7C). We were unable to find any evidence of end resection in the presence of 4 nM Alexa546-Rad52 (Figures 7A–7C). In addition, bulk biochemical assays revealed that Rad52 exerts a marked inhibitory effect on Sgs1 ATP hydrolysis activity (Figure 7D). Moreover, ssDNA curtain assays showed that Rad52 also blocks Sgs1 translocation on ssDNA (Figure 7E). Indeed, although Sgs1 readily bound to and translocated along RPA-ssDNA, 78% (N = 100/128) of the observed encounters between Sgs1 and Rad52 on RPA-ssDNA resulted in Sgs1 completely stopping; the remaining fraction either bypass Rad52 (16.4%; N = 21/128), dissociate from the ssDNA (3.9%; N = 5/128), or evict Rad52 from the ssDNA (1.6%; N = 2/128). Taken together, our findings reveal three distinct Rad52 inhibitory effects on Sgs1 by outcompeting Sgs1 for binding to short ssDNA overhangs at DNA ends, displacing end-bound Sgs1, and attenuating the ATP hydrolysis and DNA translocase activities of Sgs1.
Figure 7. Rad52 Inhibits the Motor Activities of Sgs1.
(A) Kymographs depicting GFP-Sgs1 (green) during dsDNA end resection assays on DNA pre-bound by Alexa546-Rad52 (magenta). Arrowheads highlight the position of GFP-Sgs1 at the DNA ends at the beginning of the measurement. “T” and “F” refer to the tethered and free ends of the DNA.
(B) Velocity of the dsDNA resection machinery (GFP-Sgs1, Top3-Rmi1, Dna2, and RPA) in assays containing either no Rad52, 1 nM or 4 nM Alexa546-Rad52, as indicated. Error bars represent SD obtained from Gaussian fits to the data.
(C) Processivity of the dsDNA resection machinery (GFP-Sgs1, Top3-Rmi1, Dna2, and RPA) in assays containing no Rad52 or 1 nM or 4 nM Alexa546-Rad52, as indicated. Error bars represent SD obtained from Gaussian fits to the data.
(D) Sgs1 ATP hydrolysis activity in the presence of Rad52. Error bars represent the SD of three independent experiments.
(E) Kymographs depicting the translocation activity of GFP-Sgs1 (green) on RPA-ssDNA (unlabeled) in the presence of Alexa546-Rad52 (magenta). White arrowheads in the GFP-Sgs1 image illustrate the positions at which translocating molecules of GFP-Sgs1 terminate translocation upon encountering Alexa546-Rad52. Note that the minus Rad52 data columns in (B) and (C) are reproduced from Xue et al. (2019) for comparison.
We next asked whether Rad52 might similarly affect end resection by exonuclease 1 (Exo1). We do not have an active fluorescently tagged version of Exo1, so we are unable to conduct a comparable in-depth analysis of Exo1 end-binding activity. However, we can observe DNA end resection in reactions with unlabeled Exo1 and YOYO1-stained DNA (Figure S7A), revealing a resection velocity and processivity of 20.0 ± 8.2 bp/s and 11.1 ± 5.7 kb, respectively (Figures S7B–S7D). As shown above, the presence of 1 nM Rad52 caused a 54% decrease in the velocity of Sgs1-mediated end resection and a 21% decrease processivity. However, for reactions involving Exo1, there was no statistically significant effect of Rad52 on the velocity of Exo1-mediated end resection, although there is a 19% decrease in Exo1 processivity (Figure S7). Thus, the biggest effect of Rad52 observed in these assays comparing the Exo1 and Sgs1 datasets is the >50% reduction in velocity of the Sgs1-mediated end resection pathway.
DISCUSSION
Studies in the budding and fission yeasts, frogs, and humans have identified two long-range resection pathways, one being reliant on Exo1 and the other on a helicase from RecQ family together with the nuclease/helicase Dna2 (Symington, 2016). How cells regulate the usage of these two resection pathways has remained unknown, although it has been determined that one pathway is often dominant over the other (Hoa et al., 2015; Liao et al., 2008; Sturzenegger et al., 2014; Tomimatsu et al., 2012). Proper control of initial and long-range DNA end resection is important for the efficiency and fidelity of DSB repair, as decreased or elevated resection both lead to genomic instability. Longer ssDNA may expose homology to repetitive sequences on a potential donor DNA molecule, which would lead to ectopic recombination events to compromise genomic stability (Ochs et al., 2016). Repair-specific DNA synthesis during HR and alternative end joining (Alt-EJ) associated with long ssDNA tails are more mutation prone, thus tempering the extent of long-range resection would be expected to minimize mutagenic events during repair (Harris et al., 1994; Ponder et al., 2005; Sinha et al., 2017; Strathern et al., 1995). Finally, damage to ssDNA appears to give rise to clustered mutations (Burch et al., 2011; Chan and Gordenin, 2015; Chan et al., 2012; Yang et al., 2008, 2010). Thus, the level of ssDNA generated during DNA damage repair needs to be tightly controlled.
We have devised several new assays to examine the kinetics of DSB end resection and SSA in the fission yeast. Our study reveals that Rad52, in addition to being a recombination mediator of Rad51 presynaptic filament assembly, also negatively regulates Rqh1-mediated resection in fission yeast and to a lesser degree Sgs1 mediated resection in budding yeast. In fission yeast, this regulatory function of Rad52 is independent of Rad51 and Crb2, a known negative regulator of resection. We considered the possibility that Rad52 could anneal DNA strands generated as a result of strand separation by Rqh1. Rad52 could anneal two strands separated by helicase only if the 5′ strand is not immediately degraded by Dna2, which would require some minimal uncoupling of strand unwinding and 5′ strand degradation. Analysis of DNA fragments size released during resection by budding yeast Sgs1 and Dna2 indicates that longer fragments of ~100 nt are generated, providing the potential opportunity for Rad52 to reanneal the strands behind Sgs1 (Levikova et al., 2017; Miller et al., 2017). Annealing activity of Rad52 likely reduces amount of long 5′ flap substrates for Dna2 produced during replication (Lee et al., 2014). Similarly, Rad52 could anneal strands separated by Sgs1. However, the rad52-R45A, which is severely impaired for annealing, shows only a moderate increase in resection rate (Figure 4D), suggesting that DNA annealing is not the only Rad52 activity controlling resection. The other possibility is that Rad52 could simply compete with Rqh1 or Sgs1 for binding to partially unwound DSB ends. Indeed, Rqh1 and Sgs1 loading at DSB ends is increased in the absence of Rad52 in cells (Figures 5B and 6B). Moreover, by single-molecule analysis, we have found that Rad52 binding to DNA ends suppresses Sgs1 recruitment and Rad52 can even dislodge Sgs1 bound to short ssDNA overhangs present at DNA ends (Figures 6C–6E). Finally, our data also demonstrate that Rad52 interferes with ATP hydrolysis by Sgs1 and in doing so blocks the ability of Sgs1 to translocate along DNA (Figures 7A–7E). Together, these findings suggest that Rad52 acts as a negative regulator of Sgs1 activities in the early stages of HR.
Rad52 is a multifunctional HR protein (Mortensen et al., 2009) that provides the mediator activity needed for timely assembly of the Rad51 presynaptic filament and DNA strand-annealing activity. Our studies have uncovered an unexpected Rad52 function in downregulating the resection of DSB ends. It will be of interest to determine whether Rad52 in vertebrate organisms similarly protects DNA ends from extensive degradation. Recent study implicates that indeed human RAD52 could protect stalled replication forks from degradation (Malacaria et al., 2019).
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Grzegorz Ira (gira@bcm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
All fission yeast strains are derived from strain 1913 (h-leu1-32) and are listed in Table S1. I-PpoI enzyme under the control of the TET promoter is integrated at the leu1–32 locus, and an I-PpoI cleavage site marked with the hph marker is integrated in either the lys1 or arg1 locus. In all strains that contain the I-PpoI cleavage site the natural I-PpoI cleavage sites within rDNA repeats were eliminated (Sunder et al., 2012). To integrate the I-Ppol cleavage site at the arg1 locus we cloned the 15 bp long I-Ppol cleavage site within ~1.4 kb of λ DNA and flanked by a downstream hph selectable marker. This cassette was cloned between 1 kb sequences upstream and downstream of the arg1 gene in the pBluescriptKS (+) plasmid (Agilent). To release the targeting cassette this plasmid was cut with KpnI and SacI and transformed. Integration of the I-PpoI cleavage site at arg1 generates arginine auxotrophy. To make the DSB at arg1 locus repairable by SSA λ2::ura4 cassette was amplified by PCR and inserted ~21 kb upstream of the I-PpoI cleavage site. All deletion mutants were generated using standard protocols. To generate gene truncations, we inserted three tandem stop codons at desired positions. All truncation mutants were verified by Sanger sequencing. To generate rad52-R45A, the rad52-R45A::KanMX6 cassette was made by PCR and transformed. To generate the Rad52 overexpression strain in fission yeast kanMX6-Pnmt1 was amplified from strain F2669 (Ohle et al., 2016) with primers specific for rad52 promoter and transformed. Budding yeast strains are derivatives of JKM139 (ho MATa hml::ADE1 hmr::ADE1 ade1-100 leu2-3,112 trp1::hisG’ lys5 ura3–52 ade3::GAL::HO) and listed in Table S2 (Lee et al., 1998). To generate Rad52 overexpression in budding yeast we amplified the RAD52 open reading frame and cloned it into a URA3 marked plasmid pCM189 (harbors the tetO7 promoter) (Garí et al., 1997). RAD52 was amplified using primers Fw- ACA TCG ATA GCG GCC GCA TGA ATG AAA TTA TGG ATA TGG ATG AGA AG and Rw- AGG GCC CTA GCG GCC GCT CAA GCA TAA TCA GGG ACA TCA TAA GGA TAA GTA GGC TTG CGT GCA TGC A and cloned into the NotI site of pCM189 using the In-Fusion cloning Kit (TaKaRa). To shut off the expression of RAD52 under tetO7 promoter, doxycycline was added to the culture to a final concentration of 5 μl/ml at 14 hr before HO induction.
METHOD DETAILS
Media, growth and I-PpoI and HO induction
Fission yeasts were grown in Edinburgh minimal medium glutamate (EMMG), supplemented with the appropriate amino acids (Sunder et al., 2012). Cells were grown overnight to a density of 0.2–0.4 × 107 cells ml−1. I-PpoI was induced by addition of ahTET (Acros Organics) to a final concentration of 4 μM. Induction of HO endonuclease was done by adding galactose (final concentration of 2%) to budding yeast cells grown overnight in YEP raffinose medium (1% yeast extract, 2% peptone, 2% raffinose) to a density of 1×107 cells/ml.
Chromatin immunoprecipitation
The cell cultures and DSB induction were done as described above. Formaldehyde (1% final concentration) was used to crosslink DNA-protein complexes. Crosslinking reactions were quenched after 12 min by the addition of glycine (125 mM) and reactions were continued for another 5 min. Cells were collected by centrifugation and washed first with TBS (125 mM Tris-HCl [pH 7.4], 130 mM NaCl, 5 mM KCl, 0.9 mM CaCl2 and 0.5 mM MgCl2) supplemented with glycine (125 mM) and a second time with TBS alone and stored at −70°C. Cells were lysed with glass beads on a bead beater for 6 minutes in lysis buffer (50 mM HEPES [pH 7.5], 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% Sodium deoxycholate, 1 mg/ml bacitracin, 1mM benzamidine, and 1 mM PMSF). Genomic DNA was sheared to an average size of 0.5 kb by sonication (Misonix Sonicator 3000). Cell debris was removed by centrifugation at 14000 RPM and supernatant was incubated overnight with anti-FLAG antibody (Sigma M2) at 4°C, followed by the addition of protein G-Agarose beads (50 ml, Roche) and continued incubation for 4 hr. Beads were then washed twice with lysis buffer, twice with high salt lysis buffer (lysis buffer with 500 mM NaCl), twice with wash buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 250 mM LiCl, 0.5% Sodium Deoxycholate and 0.5% Igepal CA-630) and twice with 1xTE. DNA-protein complexes were eluted by incubating beads with 100 μl of elution buffer (50 mM Tris-HCl [pH8.0], 10 mM EDTA and 1% SDS) for 30 min at 65°C. Reverse crosslinking was carried out overnight at 65°C, followed by ethanol precipitation and analysis by qPCR using the PowerUp SYBR Green Master Mix (Applied Biosystems). The primers used in qPCR are listed in Table S5. Enrichment was normalized to another locus (act1 in fission yeast and ACT1 in budding yeast).
Fluorescence microscopy
Cells carrying Rad52-GFP rad52-R45A-GFP or rad52ΔC80-GFP were grown as described above in filtered EMMG media. I-PpoI was induced by ahTET as described above. Live-cell pictures were captured using an AxioCam MRm camera (Zeiss), Olympus BX51 microscope, 100× UPlanFI objective lens, and an EGFP filter.
Measurement of DSB ends resection
100 mL or 50 mL of cells (fission yeast or budding yeast respectively) were harvested prior to I-PpoI induction (time 0) and at several time intervals after DSB induction as indicated within the figures. Genomic DNA was extracted using the standard glass bead phenol-chloroform method. DNA was digested with restriction enzymes indicated in Table S3, S4, separated on a 0.8% agarose gel and transferred to a positively charged nylon membrane (PerkinElmer). Southern blotting and hybridization with radiolabeled DNA probes was carried out according to the Church and Gilbert method. The rate of resection (defined here as a number of kilobases of DNA degraded within 1 hour) was estimated by dividing the distance of the restriction site from the DSB end by the time needed to resect 50% of DNA at this site. DSB end resection was analyzed in experiments where at least 95% of the cells had experienced an DSB within 1 hr after break induction. Multiple DNA probes used for hybridization to detect 5′ strand resection beyond the restriction enzyme site of choice, and the sequences of DNA primers used to prepare the probes by PCR, are listed in Table S3 and Table S4. Intensities of bands on Southern blots corresponding to probed DNA fragments were analyzed with ImageQuant TL 7.0 (GE Healthcare). Kinetics of resection was measured based of the normalized intensity of the bands corresponding to restriction fragments at different distance from DSB over time. Quantities of DNA loaded on gels for each time point were normalized using a DNA probe specific for sir2 gene on ChrII (for experiments with cleavage site at lys1 and DNA digestion with HpaI/StuI), gta3 gene on ChrIII (for experiments with cleavage site at lys1 and DNA digestion with EcoRI) or SPBC19G7.18c on ChrII (for experiments with cleavage site at arg1) in fission yeast and TRA1 gene in budding yeast.
Analysis of SSA
To follow the repair of DSB by SSA between two ~0.75 kb λ2 repeats we induced a DSB at the λ repeat inserted at arg1 locus (Figure 4B). SSA is a lethal event as two essential genes are deleted during DSB repair. SSA product accumulation over time was monitored by Southern blotting with probe specific for λ2 sequence. DNA was digested with BspHI and separated on 0.8% agarose gel overnight. The SSA kinetics was measured as the intensity of normalized SSA product band (3.2 kb) divided by the normalized band intensity of the upstream λ2 repeat (1.6 kb) at time 0.
Western blotting
Yeast cells were processed using the TCA method. ~4×107 cells were centrifuged and suspended in 500 μl water. 75 μl lysis solution (1.85 M NaOH and 7.5% 2-Mercaptoethanol) was added and incubated for 15 min. Then 75 μl TCA solution (55% Trichloroacetic acid) was added and the mix was centrifuged and the pellets were suspended in the HU buffer (8M Urea, 200 mM Tris-HCl [pH6.8], 1 mM EDTA, 5% SDS, 0.1% bromophenol blue and 1.5% dithiothreitol) and denatured at 60°C. Samples were resolved on a 10% SDS-PAGE gel and transferred onto a 0.2 μm PVDF membrane (Whatman) using a semi-dry method. Antibodies used are listed in the key resources table. Blots were developed by Amersham ECL (GE Healthcare). Images were captured by the Azure c400 Imaging System (Azure Biosystems).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG M2-Peroxidase (HRP) antibody produced in mouse | Sigma-Aldrich | A8592–1MG; RRID:AB_439702 |
| Monoclonal Anti-HA antibody produced in mouse | Sigma-Aldrich | H3663–100UL; RRID:AB_262051 |
| Goat Anti-Mouse Goat anti-mouse IgG-HRP Polyclonal, Hrp Conjugated antibody | Santa Cruz Biotechnology | sc-2005; RRID:AB_631736 |
| PGK1 antibody [22C5D8] | abcam | ab113687; RRID:AB_10861977 |
| Monoclonal ANTI-FLAG M2 antibody produced in mouse | Sigma-Aldrich | F3165–1MG; RRID:AB_259529 |
| Monoclonal Anti-c-Myc antibody produced in mouse | Sigma-Aldrich | M4439–100UL; RRID:AB_439694 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Anhydrotetracycline hydrochloride (ahTET) | Acros Organics | AC23313–1000 |
| cOmplete, Mini Protease Inhibitor Cocktail | Roche | 4693124001 |
| Protein G-Agarose | Roche | 11243233001 |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | R6513–250MG |
| Easytides dATP, [α-32P] | PerkinElmer | BLU512H250UC |
| Hydroxyurea (HU) | Sigma-Aldrich | H8627–100G |
| Camptothecin (CPT) | Sigma-Aldrich | C9911–250MG |
| Phleomycin | InvivoGen | ant-ph-5 |
| Methyl methanesulfonate (MMS) | Sigma-Aldrich | M4016–1G |
| Doxycycline hyclate | Sigma-Aldrich | D9891–5G |
| SNAP-Surface Alexa Fluor 546 | NEB | S9129S |
| Chitin resin | NEB | S6651L |
| cOmplete His-tag resin | Roche | 5893682001 |
| SP Sepharose | GE Healthcare | 28950513 |
| anti-FLAG-M2 resin | Sigma | A1205 |
| PD10 desalting column | GE Healthcare | 17085101 |
| DOPC | Avanti Polar Lipids | 850375C |
| biotinylated-PE | Avanti Polar Lipids | 860562C |
| mPEG 2OOO-DOPE | Avanti Polar Lipids | 880130C |
| quartz slides | Finkenbeiner | N/A |
| Polymethylmethacrylate 25 kDa | Polymer Source | P9790-MMA |
| 3% PMMA 495 kDa | MICROCHEM | N/A |
| AquaSave | Mitsubishi Rayon Co., Ltd | N/A |
| λ-dsDNA | NEB | N3011S |
| M13mp18 ssDNA | NEB | N4040S |
| TLC plates | Millipore | HX71732079 |
| Critical Commercial Assays | ||
| amfiSure ONE PCR Master Mix (2×) | GenDEPOT | P7000–010 |
| In-Fusion HD Cloning Plus | TaKaRa Bio | 638909 |
| Ex Taq DNA Polymerase | TaKaRa Bio | RR001A |
| NucleoSpin Gel and PCR clean-up | MACHEREY-NAGEL | 740609.250 |
| RadPrime DNA Labeling System | Invitrogen | 18428–011 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | A25742 |
| Amersham ECL Prime Western Blotting Detection Reagent | GE Healthcare | RPN2232 |
| Deposited Data | ||
| Unprocessed autoradiographs, western blot images and fluorescence microscopy images of this study | This study | https://doi.org/10.17632/g566xfgd96.1 |
| Experimental Models: Organisms/Strains | ||
| Fission and budding yeast strains used in this study are listed in Table S1 and Table S2. | N/A | N/A |
| Rosetta (DE3)pLys | Novagen | 70956–3 |
| yeast strain BJ5464 | Judith Campbell | N/A |
| SF9 insect cells | ThermoFisher | B82501 |
| DH10Bac | Invitrogen | N/A |
| Turbo E. coli | NEB | C2984H |
| BL21DE3 | Novagen | 69449–3 |
| Oligonucleotides | ||
| Oligonucleotides used in this study are listed in Table S1 | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| SNAP-34-Rad52 | Gibbet al., 2014 | Available upon request |
| pLK79 | Niu et al., 2010 | Available upon request |
| pRSF-Duet-Rmi1 | Niu et al., 2010 | Available upon request |
| pGAL10-DNA2 | Niu et al., 2010 | Available upon request |
| pFastBac-HTB-GFP-Sgs1 | Xue et al., 2019 | Available upon request |
| p11d-tscRPA-30MxeHis6 | Gibbet al., 2014 | Available upon request |
| pET21a-Exo1 | This study | Available upon request |
| pCM189-RAD52-HA | This study | Available upon request |
| pBluescriptKS (+) | Agilent | 212205 |
| pBluescriptKS(+)-arg1 -5’-λ1-I-PpolCS-λ2-hph-arg1-3’ | This study | Available upon request |
| Software and Algorithms | ||
| Image Quant TL 7.0 | GE Healthcare | N/A |
| Prism 6 | GraphPad | N/A |
| Prism 8 | GraphPad | N/A |
| NIS-Element software | Nikon | N/A |
| FIJI | ImageJ software | N/A |
| Other | ||
| Edinburgh Minimal Media Glutamate (EMMG) (Powder) | United States Biological | E2205–15 |
| Storage Phosphor Screen | Molecular Dynamics | N/A |
| Typhoon TRIO Variable Mode Imager | GE Healthcare | N/A |
| MiniBeadBeater-16, Model 607 | BioSpec Products | N/A |
| Misonix Sonicator 3000 Ultrasonic Cell Disruptors | Misonix | N/A |
| 7900HT Fast Real-Time PCR system | Applied Biosystems | N/A |
| MicroAmp Fast Optical 96-Well Reaction Plate, 0.1 mL | Applied Biosystems | 4346907 |
| BX51 microscope | Olympus | N/A |
| AxioCam MRm camera | Zeiss | N/A |
| Azure c400 Imaging System | Azure Biosystems | N/A |
| Inverted TIRF Microscope | Nikon | N/A |
| 488 nm laser | Coherent Sapphire | N/A |
| 561 nm laser | Coherent Sapphire | N/A |
| iXon X3 EMCCD cameras | Andor | N/A |
| GeneScreen Plus Hybridization Transfer Membrane | PerkinElmer | NEF988001PK |
| Westran S, 0.2 μm PVDF membrane | Whatman | 10413096 |
Single-molecule assays
SNAP-Rad52 was expressed using construct SNAP-34-Rad52 in Rosetta (DE3) pLys. Briefly, E. coli was grown at 37°C and induced overnight at 18°C with 0.5 mM IPTG when OD600 reached 0.4–0.6. Cells were harvested by centrifugation and lysed by sonication. The extract was purified over a chitin column (NEB) and was further purified using a Ni-NTA column (Roche). SNAP-Rad52 was labeled with SNAP-Surface Alexa Fluor 546 (NEB) overnight at 4°C. Free dyes was removed with a PD10 desalting column (GE Healthcare) (Gibb et al., 2014; Van Komen et al., 2006). Top3-Rmi1 was purified by co-transforming pLK79 and pRSF-Duet-Rmi1 into Rosetta(DE3)pLys. E. coli was grown at 37°C and induced overnight at 16°C with 0.2 mM IPTG when OD600 reached 0.8. Cells were harvested by centrifugation and lysed by sonication. The extract was purified using a 5 mL SP Sepharose column (GE Healthcare) and was further purified through a Ni-NTA column (Roche). GFP-Sgs1 was purified from insect cells. In brief, a bacmid with GFP-Sgs1 tagged with a HA, a FLAG, and a (His)6 was generated in the E. coli strain DH10Bac (Invitrogen). Cells were disrupted by sonication and the lysate was clarified by centrifugation. The extract was purified with anti-Flag M2 resin (Sigma) and was further purified through a Ni-NTA column. Dna2 was purified in the protease deficient yeast strain BJ5464 using construct pGAL10-DNA2. Cells were cultured at 30°C and induced with 2% galactose when OD600 reached 0.8. Cells were harvested by centrifugation at 4,000 rpm for 10 min at 4°C and lysed by grinding with dry ice. The lysate was clarified by centrifugation using a Beckman 45Ti rotor at 40,000 g for 1 h. The extract was purified with Ni-NTA column (Roche) and was further purified with anti-FLAG-M2 resin (Sigma) (Xue et al., 2019). RPA was purified in E. coli strain BL21DE3 using construct p11d-tscRPA-30MxeHis6. E. coli was grown at 37°C and induced overnight at 16°C with 0.5 mM IPTG when OD600 reached 0.4–0.6. Cells were harvested by centrifugation and lysed by sonication. The extract was purified through a Ni-NTA column (Roche) and was further purified through a chitin column (NEB). To express the full-length yeast Exo1 in E. coli, the coding sequence of the initial 348 amino acids was optimized (sequence available upon request) and fused to the rest of wild-type ExoI sequence plus the coding sequence of six histidines at the 3′ end of the gene. Exo1 was expressed in Rosetta (DE3)pLys (Novagen) cells using pET21a-Exo1. Cultures were grown to 0.6–0.8 OD600 at 37°C before an overnight induction at 16°C with 0.5 mM IPTG. Cells were then harvested by centrifugation and stored at −80°C for future use. All experiments were conducted with a custom-built prism-type total internal reflection fluorescence (TIRF) microscope (Nikon) equipped with a 488 nm laser (Coherent Sapphire, 200 mW) and a 561 nm laser (Coherent Sapphire, 200 mW)(De Tullio et al., 2018; Ma et al., 2017). Lipid bilayers were prepared with 91.5% DOPC (Avanti Polar Lipids), 0.5% biotinylated–PE (Avanti Polar Lipids), and 8% mPEG 2000–DOPE (Avanti Polar Lipids), and deposited onto to the surface of a flowcell sample chamber containing nanofabricated barriers to lipid diffusion prepared by electron beam lithography (De Tullio et al., 2018; Ma et al., 2017). In brief, quartz slides (Finkenbeiner) were cleaned in piranha solution (a 3:1 mixture of sulfuric acid (97%) and 30% hydrogen peroxide) and coated with a layer of 3% polymethylmethacrylate (PMMA 25 kDa) (Polymer Source), a layer of 1.5% PMMA (495 kDa) (MICROCHEM), and a final layer of AquaSave (Mitsubishi Rayon Co., Ltd.). Then, patterns were written using an FEI scanning transmission electron microscope with equipped for e-beam lithography. The slides were developed by removing the PMMA from the areas exposed to the e-beam. Then a thin layer of chrome was deposited onto the slides by vapor depositing using a Angstrom Evovac Deposition System. Finally, the slides were cleaned with acetone and patterned slides are read to use.
All single molecule assays were performed at 30°C in resection buffer containing 20 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 1 mM DTT and 0.2 mg/mL BSA (Xue et al., 2019). Single-tethered dsDNA curtains were prepared using a λ-dsDNA (NEB) substrate bearing a 30-nt 3° end overhang (Xue et al., 2019). Sgs1 DNA binding activity (in the absence of Rad52) was measured by injecting 150 μL GFP-Sgs1 (0.2 nM,) in reaction buffer into a sample chamber containing dsDNA at a flow rate of 0.05 mL/min. The unbound GFP-Sgs1 was flushed away by washing with 2–5 mL reaction buffer at 0.5 mL/min, and the binding distribution of the remaining GFP-Sgs1 on the DNA was then measured at a flow rate of 0.15 mL/min. To test whether Rad52 could dislodge pre-bound GFP-Sgs1 from DNA ends, 500 μL Alexa546-Rad52 (0.1 nM) was injected into a sample chamber with end-bound GFP-Sgs1 at 0.15 ml/min in reaction buffer. Unbound Alexa546-Rad52 was flushed away by washing with 2–5 mL reaction buffer at 0.5 mL/min. Then, the binding distributions of GFP-Sgs1 and Alexa546-Rad52 were measured at a flow rate of 0.15 mL/min. To test whether pre-bound Rad52 could block end-binding by Sgs1, 500 μL Alexa546-Rad52 (0.05 nM) in reaction buffer was first injected into the sample chamber at 0.15 ml/min. Unbound Alexa546-Rad52 was flushed away by washing with an additional 2–5 mL reaction buffer at 0.5 mL/min. Then, 150 μL GFP-Sgs1 (0.2 nM) in reaction buffer was introduced into the sample chamber at a flow rate of 0.05 mL/min. Unbound GFP-Sgs1 was flushed away by washing with 2–5 mL reaction buffer at 0.5 mL/min. Then, the binding distribution of GFP-Sgs1 on λ-DNA with Alexa546-Rad52 was measured at a flow rate of 0.15 mL/min. For all dsDNA curtain experiments, images were acquired at a frequency of 1 frame per 10 s with 0.1 s integration time using two EMCCD cameras for the GFP and Alexa546 signals, and the illumination lasers were shuttered between each acquired image to minimize photo-bleaching (De Tullio et al., 2018). The resulting raw TIFF image files were imported as image stacks into NIH Image and any image drift was corrected using the StackReg function in ImageJ (De Tullio et al., 2018). The position of GFP-Sgs1 and Alexa546-Rad52 were determined based on the individual DNA-bound fluorescent protein complexes.
For ssDNA curtain assays, the ssDNA substrate was generated by rolling circle replication with a biotinylated primer, a circular M13 ssDNA template, and phi29 DNA polymerase (De Tullio et al., 2018; Ma et al., 2017). In brief, the biotinylated primer and M13 ssDNA was annealed through the PCR machine (95°C for 5 mins and ramp cool to 25°C within 90 minutes). Biotinylated ssDNA substrate was generated by adding phi29 into the annealing product for 30 min at 30°C. Sgs1 measurements were conducted at 30°C in reaction buffer supplemented with RPA (unlabeled) and 2.5 mM ATP. After the flow cell was attached the microfluidic device the ssDNA was aligned at a flow rate of 0.5 mL/min in reaction buffer (30 mM Tris–Cl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1.5 mM CaCl2, 1 mM DTT, 0.2 mg/ml BSA) lacking RPA. After 1 min the flow rate was adjusted to 1 mL/min and the ssDNA extended with 500 μl of 7 M Urea followed by a buffer change to reaction buffer containing 0.1 nM RPA for 5 min. After 5 min the buffer was switched to buffer reaction buffer containing ATP and after 3 min 1 nM Alexa546-Rad52 was injected into the flow cell. Rad52 was incubated for 5 min and then the flow cell was washed at 1 ml/min for 3 min. Samples containing either 10 nM GFP–Sgs1 + 0.1 nM RPA were injected into the flow cell at a rate of 1.0 mL per minute, flow then was stopped and the activity of Sgs1 was monitored for 20–25 minutes. All data were collected at one frame per 10 s with 100 ms integration time and the laser was shuttered between each acquired image to minimize photo–bleaching (Crickard et al., 2019; De Tullio et al., 2018). Raw TIFF images were imported as image stacks into ImageJ, and kymographs were generated from the image stacks by defining a 1–pixel wide region of interest (ROI) along the long–axis of the individual ssDNA molecules.
Exo1 purification
To express the full-length yeast Exo1 in E. coli, the coding sequence of the initial 348 amino acids was optimized (sequence available upon request) and fused to the rest of wild-type ExoI sequence plus the coding sequence of six histidines at the 3′ end of the gene. Exo1 was expressed in RosettaTM(DE3)pLys cells (Novagen) using pET21a-Exo1. Cultures were grown to 0.6–0.8 OD600 at 37°C before an overnight induction at 16°C with 0.5 mM IPTG. Cells were then harvested by centrifugation and stored at −80°C for future use. Exo1 purification was performed at 4°C. Cells were first resuspended in lysis buffer (40 mM KH2PO4 [pH 7.4], 150 mM KCl, 0.5 mM EDTA, 10% glycerol, 0.01% NP-40, 1 mM β-ME (2-mercaptoethanol), 1 mM PMSF (phenyl-methylsulfonyl fluoride), 5 mg/mL leupeptin, 5 mg/mL chymostatin, 5 mg/mL pepstatin, 2.5 mg/mL aprotinin) and lysed by sonication. The lysate was collected and centrifuged for 20 min at 20,000 g at 4°C. The supernatant was loaded onto a 5 mL SP column. The column was washed by 50 mL washing buffer (40 mM KH2PO4 [pH 7.4] 150 mM KCl, 0.5 mM EDTA, 10% glycerol, 0.1% NP-40 and 1 mM β-ME). Exo1 was eluted from the column by washing with 50 mL elution buffer (40 mM KH2PO4 [pH 7.4], 500 mM KCl, 0.5 mM EDTA, 10% glycerol, 0.01% NP-40, 0.2 mM β-ME). The eluant was further incubated with Ni-NTA resin for 1.5 hours. The column was then washed with 30 mL washing buffer (40 mM KH2PO4 [pH 7.4], 500 mM KCl, 0.5 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM β-ME) plus 15 mM imidazole and then Exo1 was eluted by a stepped gradient of 50, 100, 200 mM imidazole. The purified Exo1 was stored frozen at −80°C in small aliquots.
DNA curtain assays for Exo1
Exo1-mediated single molecule experiments were performed at 30°C in Exo1 reaction buffer (40 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.1 nM YOYO1, and 0.2 mg/mL BSA). For experiments without Rad52, DNA end resection was initiated by injection 150 μL of 4 nM Exo1 at a flowrate of 0.15 ml/min. Image acquisition was initiated immediately before the protein injections. To test the effect of Rad52 on Exo1-mediated end resection, 500 μL Alexa546-Rad52 (1 nM) was injected into a sample chamber at0.15 ml/min in Exo1 reaction buffer. Unbound Alexa 546-Rad52 was then flushed away by washing with 2–5 mL Exo1 reaction buffer at 0.5 mL/min. DNA end resection was initiated by injection 150 μL of 4 nM Exo1 at 0.15 ml/min. Image acquisition was initiated immediately before the injection of Exo1.
ATP hydrolysis assays
ATP hydrolysis assays were performed in reaction buffer (30 mM Tris–Cl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1.5 mM CaCl2, 1 mM DTT, 0.2 mg/ml BSA) in the presence of M13 ssDNA (2.5 μM nucleotides total concentration; NEB) with 0.8 μM RPA 1 mM ATP and trace amounts of γ32P-ATP (3000 Ci/mmol). Unlabeled Rad52 was titrated as described in figure legends. Reactions were performed at 30°C. Aliquots were removed at specified time points and quenched by mixing with an equal volume of 25 mM EDTA and 1% SDS. The quenched reactions were spotted on TLC plates (Millipore) and resolved in 0.5 M LiCl plus 1 M Formic acid. Dried TLC plates were exposed to phosphor–imaging screen, and scanned with a Typhoon platform (GE Healthcare).
QUANTIFICATION AND STATISTICAL ANALYSIS
ChIP analysis
For statistical analysis, Prism 8 GraphPad software was used. Significance was determined using the Mann-Whitney test. Statistical significance was shown in figures (*p < 0.05, **p < 0.01, ***p < 0.001).
Measurement of DSB ends resection and SSA
Experiments were repeated at least three times for each mutant and the average resection rate was plotted with error bars showing standard deviation.
DATA AND CODE AVAILABILITY
Single molecule data acquisition was preformed using commercially available software (NIS-Element software). Data analysis was preformed using FIJI and Prism6. All raw data has been deposited to Mendeley Data and is available at https://doi.org/10.17632/g566xfgd96.1.
Supplementary Material
Highlights.
Rad52 attenuates resection of DNA double-strand break (DSB) ends
Rad52 prevents hyper-resection by Rqh1 (S. pombe) or Sgs1 (S. cerevisiae)
Rad52 interferes with Sgs1 and Dna2-mediated resection in vitro
Rad52 inhibits Sgs1 loading at DSB ends and its translocation along DNA
ACKNOWLEDGMENTS
We thank S. Sanders, A. Aguilera, J. Haber, and T. Fischer for the gifts of strains and plasmids. This work was funded by grants from the National Institute of Health (GM080600 and GM125650 to G.I., ES007061 to P.S., R35GM124765 to H.N., R35GM118026 and R01CA217973 to E.C.G., and P01CA92584 to P.S. and E.C.G.) and the Cancer Prevention and Research Institute of Texas (REI award RR180029 to P.S.). J.B.C. is the Mark Foundation for Cancer Research Fellow for the Damon-Runyon Cancer Research Foundation (award DRG 2310–17).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2019.08.017.
REFERENCES
- Bai Y, Davis AP, and Symington LS (1999). A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics 153, 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JR, Cuella-Martin R, Barazas M, Liu R, Oliveira C, Oliver AW, Bilham K, Holt AB, Blackford AN, Heierhorst J, et al. (2018). The ASCIZDYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat. Commun 9, 5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JC, and Kowalczykowski SC (2016). Mechanics and single-molecule interrogation of DNA recombination. Annu. Rev. Biochem 85, 193–226. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, and West SC (1998). Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch LH, Yang Y, Sterling JF, Roberts SA, Chao FG, Xu H, Zhang L, Walsh J, Resnick MA, Mieczkowski PA, and Gordenin DA (2011). Damage-induced localized hypermutability. Cell Cycle 10, 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P (2015). DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem 290, 22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, and Kowalczykowski SC (2010). The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem 285, 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, and Kowalczykowski SC (2010). DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, and Gordenin DA (2015). Clusters of multiple mutations: incidence and molecular mechanisms. Annu. Rev. Genet 49, 243–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, and Gordenin DA (2012). Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 8, e1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, and Ira G (2012). The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Yu Y, Wang J, Zhu S, Zhou J, Papusha A, Cui D, Pan X, Kwon Y, et al. (2016). Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 44, 2742–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. (2012). The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Xue C, Wang W, Kwon Y, Sung P, and Greene EC (2019). The RecQ helicase Sgs1 drives ATP-dependent disruption of Rad51 filaments. Nucleic Acids Res. 47, 4694–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2018). Shelterin-mediated telomere protection. Annu. Rev. Genet 52, 223–247. [DOI] [PubMed] [Google Scholar]
- De Tullio L, Kaniecki K, and Greene EC (2018). Single-stranded DNA curtains for studying the Srs2 helicase using total internal reflection fluorescence microscopy. Methods Enzymol. 600, 407–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari M, Dibitetto D, De Gregorio G, Eapen VV, Rawal CC, Lazzaro F, Tsabar M, Marini F, Haber JE, and Pellicioli A (2015). Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 11, e1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Piedrafita L, Aldea M, and Herrero E (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837–848. [DOI] [PubMed] [Google Scholar]
- Gibb B, Ye LF, Kwon Y, Niu H, Sung P, and Greene EC (2014). Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol 21, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA (2018). Assembling a protective shield. Nat. Cell Biol 20, 862–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Longerich S, and Rosenberg SM (1994). Recombination in adaptive mutation. Science 264, 258–260. [DOI] [PubMed] [Google Scholar]
- He YJ, Meghani K, Caron MC, Yang C, Ronato DA, Bian J, Sharma A, Moore J, Niraj J, Detappe A, et al. (2018). DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 563, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her J, Ray C, Altshuler J, Zheng H, and Bunting SF (2018). 53BP1 mediates ATR-Chk1 signaling and protects replication forks under conditions of replication stress. Mol. Cell. Biol 38, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Akagawa R, Yamasaki T, Hirota K, Sasa K, Natsume T, Kobayashi J, Sakuma T, Yamamoto T, Komatsu K, et al. (2015). Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells 20, 1059–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, and Kowalczykowski SC (2010). Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL, Baĉinskaja G, Che J, Cheblal A, Elango R, Epshtein A, Fitzgerald DM, Gómez-González B, Khan SR, Kumar S, et al. (2019). Guidelines for DNA recombination and repair studies: Cellular assays of DNA repair pathways. Microb. Cell 6, 1–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, and Russell P (2011). Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, and Muzi-Falconi M (2008). Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27, 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, and Haber JE (1998). Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409. [DOI] [PubMed] [Google Scholar]
- Lee M, Lee CH, Demin AA, Munashingha PR, Amangyeld T, Kwon B, Formosa T, and Seo YS (2014). Rad52/Rad59-dependent recombination as a means to rectify faulty Okazaki fragment processing. J. Biol. Chem 289, 15064–15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M, Pinto C, and Cejka P (2017). The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 31, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, and Yan H (2008). Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 36, 6091–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, and Russell P (2007). Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, and Heyer WD (2010). Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol 17, 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CJ, Steinfeld JB, and Greene EC (2017). Single-stranded DNA curtains for studying homologous recombination. Methods Enzymol. 582, 193–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malacaria E, Pugliese GM, Honda M, Marabitti V, Aiello FA, Spies M, Franchitto A, and Pichierri P (2019). Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun 10, 1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AS, Daley JM, Pham NT, Niu H, Xue X, Ira G, and Sung P (2017). A novel role of the Dna2 translocase function in DNA break resection. Genes Dev. 31, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, and Symington LS (2008). Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, and Symington LS (2010). Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, Bianchi A, Zimmermann M, Durocher D, and de Lange T (2018). 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Lisby M, and Rothstein R (2009). Rad52. Curr. Biol 19, R676–R677. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, and Kowalczykowski SC (1998). Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, and Paull TT (2010). Mre11-Rad50-Xrs2 and Sae2 promote 5′strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol 17, 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. (2010). Mechanism of the ATP-dependent DNA endresection machinery from Saccharomyces cerevisiae. Nature 467, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs F, Somyajit K, Altmeyer M, Rask MB, Lukas J, and Lukas C (2016). 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol 23, 714–721. [DOI] [PubMed] [Google Scholar]
- Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, and Fischer T (2016). Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–1013.e1007. [DOI] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, and Rosenberg SM (2005). A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19, 791–804. [DOI] [PubMed] [Google Scholar]
- Rickman K, and Smogorzewska A (2019). Advances in understanding DNA processing and protection at stalled replication forks. J. Cell Biol 218, 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, and Jackson SP (2007). Human CtIP promotes DNA end resection. Nature 450, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, and Jasin M (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, and Durocher D (2019). Shieldin—the protector of DNA ends. EMBO Rep. 20, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, and Sung P (2009). Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem 284, 33275–33284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, and Lee SE (2007). RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol 27, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, and Lee SE (2010). Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, and Ogawa T (1998). Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Sinha S, Li F, Villarreal D, Shim JH, Yoon S, Myung K, Shim EY, and Lee SE (2017). Microhomology-mediated end joining induces hypermuta-genesis at breakpoint junctions. PLoS Genet. 13, e1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M (2000). Where does fission yeast sit on the tree of life? Genome Biol. 1, REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, and McGill CB (1995). DNA synthesis errors associated with double-strand-break repair. Genetics 140, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, and Janscak P (2014). DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem 289, 27314–27326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunder S, Greeson-Lott NT, Runge KW, and Sanders SL (2012). A new method to efficiently induce a site-specific double-strand break in the fission yeast Schizosaccharomyces pombe. Yeast 29, 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P (1997). Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Symington LS (2016). Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol 51, 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkácĉ J, Xu G, Adhikary H, Young JTF, Gallo D, Escribano-Díaz C, Krietsch J, Orthwein A, Munro M, Sol W, et al. (2016). HELB is a feedback inhibitor of DNA end resection. Mol. Cell 61, 405–418. [DOI] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, and Burma S (2012). Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst.) 11, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Macris M, Sehorn MG, and Sung P (2006). Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 408, 445–463. [DOI] [PubMed] [Google Scholar]
- Villa M, Bonetti D, Carraro M, and Longhese MP (2018). Rad9/53BP1 protects stalled replication forks from degradation in Mec1/ATR-defective cells. EMBO Rep. 19, 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Warr N, Taylor DL, and Watts FZ (1999). The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27, 2655–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Wang W, Crickard JB, Moevus CJ, Kwon Y, Sung P, and Greene EC (2019). Regulatory control of Sgs1 and Dna2 during eukaryotic DNA end resection. Proc. Natl. Acad. Sci. USA 116, 6091–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, and Gordenin DA (2008). Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 4, e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gordenin DA, and Resnick MA (2010). A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst.) 9, 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, and Ira G (2008). Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single molecule data acquisition was preformed using commercially available software (NIS-Element software). Data analysis was preformed using FIJI and Prism6. All raw data has been deposited to Mendeley Data and is available at https://doi.org/10.17632/g566xfgd96.1.