Abstract
Opioid use kills tens of thousands of Americans each year, devastates families and entire communities, and cripples the healthcare system. Exposure to opioids causes long-term changes to brain regions involved in reward processing and motivation, leading vulnerable individuals to engage in pathological drug-seeking and drug-taking that can remain a lifelong struggle. The persistence of these neuroadaptations is mediated in part by epigenetic remodeling of gene expression programs in discrete brain regions. Although the majority of work examining how epigenetic modifications contribute to addiction has focused on psychostimulants like cocaine, research into opioid-induced changes to the epigenetic landscape is beginning to emerge. This review summarizes our knowledge of opioid-induced epigenetic modifications and their consequential changes to gene expression. Current evidence points towards opioids promoting higher levels of permissive histone acetylation and lower levels of repressive histone methylation, as well as alterations to DNA methylation patterns and non-coding RNA expression, throughout the brain’s reward circuitry. Additionally, studies manipulating epigenetic enzymes in specific brain regions are beginning to build causal links between these epigenetic modifications and changes in addiction-related behavior. Moving forward, studies must leverage advanced next-generation sequencing approaches and chromatin purification techniques combined with bioinformatics analyses to identify novel gene networks regulated by particular epigenetic modifications. Improved translational relevance will also require increased focus on volitional drug-intake models and standardization of exposure paradigms. Such work will significantly advance our understanding of how opioids cause persistent changes to brain function, and provide a platform on which to develop interventions for treating opioid addiction.
Keywords: Opioids, Addiction, Epigenetics, Transcription, Histone Acetylation, Histone Methylation
INTRODUCTION
Opioids claim the lives of nearly 50,000 Americans each year, and leave tens of thousands more struggling with addiction (1). Nevertheless, initiation of opioid use has been increasing at an alarming rate over the last decade (2–4), and users often shift from ingestion of prescription painkillers to intravenous injection of heroin - a significantly more addictive form of drug intake (5). Opioids induce physical dependence, which promotes short-term desire to re-use, in addition to motivational disturbances underlying key aspects of a longer-lasting addiction syndrome. With repeated exposure, positive subjective effects of opioids become integrated with internal states and external cues associated with drug-taking, creating triggers that can instigate drug-seeking even long after terminating use (6–8).
Long-term vulnerability to addiction is mediated by persistent alterations to the function of reward-processing networks in the brain, namely the mesocorticolimbic dopamine system (9). This system is comprised of dopamine neurons projecting from the ventral tegmental area (VTA) that terminate in the nucleus accumbens (NAc) as well as cortical regions including the amygdala, prefrontal cortex (PFC), and hippocampus (HPC). Opioids appear to produce their rewarding and reinforcing effects by activating mu-opioid receptors (MORs) within the VTA, causing disinhibition of dopamine neurons firing and, consequently, elevated dopamine neurotransmission within the NAc (10–16). Opioids also directly activate MORs on neurons of the NAc and other forebrain structures, and elicit distinct neuronal responses throughout the brain via signaling at kappa-ORs and delta-ORs (17). Additionally, these three types of ORs are expressed on non-neuronal cells, the function of which can be altered by opioid exposure (18–22). These diverse actions of opioids can alter various intracellular signaling cascades that drive long-lasting adaptations to cellular function which may promote behavioral and psychological abnormalities underlying addiction.
One persistent adaptation, commensurate with the long-lasting and experience-dependent nature of addiction, involves stable epigenetic modifications to the DNA of affected neurons (23). These modifications modulate transcription, without directly altering the nucleotide sequence, via conformational changes to chromatin structure and accessibility. Consequences of these modifications include altered basal levels of gene transcription, priming/desensitization of particular genes for activation or repression in response to drug or other stimuli, or regulation of splice variant expression (24; 25). Resultant transcriptional changes influence effector genes involved in diverse cellular functions, ultimately causing enduring changes to signaling cascades, cellular structure, and synaptic activity. Epigenetic modifications are critical for the formation and recall of long-term memory (26; 27), and their influence on gene expression programs likely play a critical role in the context of addiction in linking the rewarding experience of drugs with external and internal cues that generate craving and relapse (28).
Although our understanding of how epigenetic modifications promote addiction is growing for psychostimulants such as cocaine (29; 30), opioids have been largely left behind. All drugs of abuse produce some similar changes in reward system function and behavior, but this effect is mediated by divergent mechanisms of action distributed across different brain regions and cell types (31). Thus, we cannot assume transcriptional or epigenetic changes will generalize from one drug class to another. Advancing our knowledge of how opioids produce persistent epigenetic changes will be crucial to gain a better understanding of how opioid addiction develops, how it is maintained, and how to treat it. Here, we outline current knowledge of opioid-induced alterations to the epigenetic landscape of the brain’s reward circuitry, and point to functional consequences. Primary focus is given to studies of human addicts and rodent studies employing long-term experimenter-administered or self-administered opioid exposure paradigms which, although inherently limited in their direct clinical relevance, effectively model several aspects of drug addiction. We conclude by outlining what next steps should be taken to advance our understanding in hopes of fighting the opioid epidemic.
I. EPIGENETIC MODIFICATIONS PRODUCED BY LONG-TERM OPIOID EXPOSURE
Histone modifications
Gene expression depends on the ability of transcriptional machinery to access DNA, which is tightly packed into chromatin. To condense genetic material, DNA strands are wound around protein octamer spools known as histones. Histones are formed from combinations of four proteins: H2A, H2B, H3, and H4 (32; 33). These proteins’ N-terminal “tails” undergo extensive covalent modifications that either loosen or tighten the histone’s grip on DNA. Such modifications are the most studied in the context of addiction, and this is particularly true for opioids (Table 1).
Table 1. Epigenetic marks induced by opioid exposure.
Current clinical and preclinical evidence for changes to histone acetylation, histone methylation, DNA methylation, and expression of non-coding RNAs following repeated opioid exposure are summarized. Abbreviations: BLA, basolateral amygdala; CeA, central nucleus of the amygdala; CPP, conditioned reinforcement; dStri, dorsal striatum; FC, frontal cortex; HPC, hippocampus; IVSA, intravenous self-administration; LC, locus coeruleus; NAc, nucleus accumbens; PFC, prefrontal cortex; VLO, ventolateral orbitofrontal cortex; VTA, ventral tegmental area.
| Epigenetic Mark | Change | Brain Region | Model Examined | Gene targets identified? | Reference |
|---|---|---|---|---|---|
| Pan-H3ac | ↑ | Striatum | Human heroin addicts Rat IVSA | (36) | |
| Pan-H3phosphoac | ↑ | NAc (not PFC) | Mouse, repeated heroin in CPP | (34) | |
| H3K9ac | ↑ | LC, VLO | Rat, repeated morphine | Bdnf | (37; 38) |
| H3K14ac | ↑ | BLA | Mouse, repeated morphine in CPP, normalized during extinction | Bdnf, FosB, Creb | (39) |
| H3K18ac | ↑ | NAc | Rat, drug-primed reinstatement of heroin-seeking | (40) | |
| H3K23ac | ↑ | NAc/dStri | Human heroin addicts (not correlated with use history) | (36) | |
| H3K27ac | ↑ | NAc/dStri | Human heroin addicts Rat IVSA | Glutamate signaling, GRIA1 | (36) |
| H4K5ac | ↑ | NAc | Rat, drug-primed reinstatement of heroin-seeking | (40) | |
| H4K8ac | ↑ | NAc | Rat, drug-primed reinstatement of heroin-seeking | (40) | |
| H3K9me1 | No Δ | NAc | Mouse, repeated morphine | (41) | |
| H3K9me2 | ↓ | dStri | Mouse, repeated morphine | FosB, Bdnf, glutamate signaling genes | (41) |
| ↓ | CeA | Mouse, repeated morphine in CPP | Bdnf, glutamate signaling genes | (42) | |
| H3K9me3 | No Δ | NAc | Mouse, repeated morphine | (41) | |
| ↓ | VTA, LC | Mouse, 7d withdrawal from escalating morphine | Bdnf | (37) | |
| H3K27me3 | No Δ | NAc | Mouse, repeated morphine | (41) | |
| DNA Methylation | No Δ | Whole-brain | Mouse, repeated heroin or morphine | (48; 49) | |
| No Δ | VTA, NAc, PFC | Rat, heroin IVSA Mouse, repeated morphine in CPP |
(50; 51) | ||
| ↓ or No Δ | In vitro | Cell Lines | (117; 118) | ||
| ↑ exons ↑ gene body ↓ promoters |
OFC neurons (nuclei isolated by FACS) | Human heroin addicts | Differential effects in gene ontology analysis | (45) | |
| ↑ LINE-1, OPRM1 CpG islands | Thalamus, somato-sensory cortex, blood | Human heroin addicts | (44; 46; 47) | ||
| Region-specific changes | 10 brain regions | Rat, acute and chronic morphine | Bdnf, Nr3c1, Il6, Il1b | (55) | |
| Let-7 | ↑ | Whole-brain | Mouse, morphine pellets | (56) | |
| miR-339–3p | ↑ | HPC | Mouse, repeated morphine or fentanyl | Oprm1 mRNA | (57) |
| miR-133b | No Δ | VTA, NAc | Mouse, morphine pellets Mouse, morphine IVSA |
(58; 119) | |
| miR-218 | ↓ | NAc (not PFC, HPC) | Mouse, repeated (not acute) heroin | Gabrb3, Mecp2, Nrxn1, Gng3, Ube3a | (59) |
| H19, miR-675 | ↓ | vStri | Mouse, morphine IVSA | (58) | |
| Mirg, miR-154 | ↓ | vStri | Mouse, morphine IVSA | Fxyd4, Grm3, Odf21, Slc4a4; Oprm1 | (58) |
Histone Acetylation
Most studies of drug-induced epigenetic modifications have focused on acetylation of lysine residues on histone tails. Acetylation reduces electrostatic tension between histones and wound genes, creating an “open” chromatin state that facilitates gene transcription (Figure 1). In the context of opioids, this has primarily been studied on histone H3 tails. Preclinical work has found that repeated experimenter-administered or self-administered opioids increase global H3 acetylation within the mesolimbic dopamine system (34; 35), and this effect is consistent with findings in postmortem tissue from human heroin users (36). Strikingly, global H3 hyperacetylation in the striatum of heroin users appears to correlate with years of heroin use (36), suggesting that levels of heroin exposure may scale with stabilization of this chromatin modification.
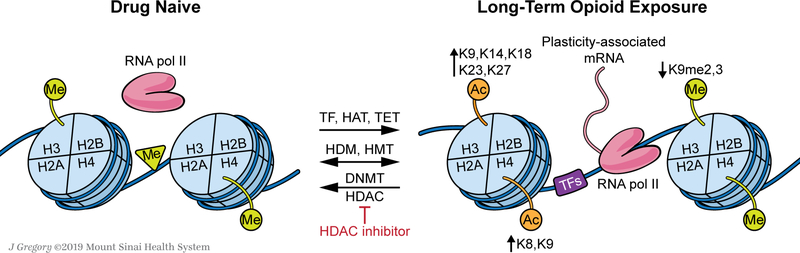
Figure 1. Repeated opioid exposure alters the epigenetic landscape of brain cells, causing persistent changes to transcriptional activity and cellular physiology.
Opioids induce a host of changes to the function of epigenetic editors and transcription factors (TFs) which remodel chromatin structure and DNA accessibility within various regions of the brain’s reward circuitry. Acetylation of histones H3 and H4 at several N-terminal tail lysine (K) residues is enhanced by opioid exposure (listed), promoting a more open, accessible chromatin state. Additionally, the repressive influence of histone methylation appears to be relieved by opioid exposure (listed). Opioids also induce complex changes to DNA methylation in a gene locus-specific manner. These epigenetic modifications are mediated by diverse interactions between TFs and epigenomic “editors” including histone acetyltransferases (HATs), histone methyltransferases (HMTs), DNA methyltransferases (DNMTs), histone deacetylases (HDACs), histone demethylases (HDMs), and DNA oxidases or demethylases (e.g., ten eleven translocation proteins, TETs). Several classes of non-coding RNAs are also involved (not shown in the figure). Studies manipulating the function of these enzymes, particularly HDACs or G9a (an H3K9me2 HMT), suggest that opioid-induced histone acetylation or reduced repressive histone methylation drives heightened behavioral responses to opioids, indicative of a “poised” state within the reward circuitry. Such changes are likely mediated by greater levels of transcriptional activity at genes critical for neuroplasticity and synaptic physiology which promote behavioral abnormalities underlying addiction.
Further dissection of the specific amino acid tail residues acetylated has demonstrated hyperacetylation at H3K9 (37; 38), H3K14 (39), H3K18 (40), and H3K27 (36) following repeated morphine or heroin exposure in either experimenter-administration or self-administration models (see Table 1). Of these marks, H3K27ac has received the most extensive characterization for its role in opioid addiction, predominantly from a single study combining clinical and preclinical approaches. Egervari and colleagues (36) demonstrate elevated H3K27ac levels in striatum of human heroin users as well as heroin self-administering rats, and observe a striking positive correlation between H3K27ac and years of use. Further, chromatin accessibility mapping with ATAC-seq confirmed that this mark induces an open chromatin state. Since H3K27ac is typically enriched at enhancer regions of DNA (41; 42), these results suggest that higher levels of this mark may facilitate persistent amplification of gene expression systems underlying opioid addiction.”
These authors also found H3K23ac to be upregulated in striatum of heroin users, but since this did not correlate with use patterns it was not further examined. One study examined histone H4 and, consistent with studies of H3, showed hyperacetylation at H4K5 and H4K8 within the NAc of rats displaying heroin-seeking behavior (40). Together, these studies provide converging evidence that opioids promote a more open, accessible chromatin state via histone acetylation, ultimately permitting higher levels of transcriptional activity critical for induction of plasticity-related gene expression.
Histone Methylation
Compared to acetylation, far less is known about how opioids alter histone methylation. To date, the few studies examining this epigenetic modification have only identified changes to the methylation state of a specific histone tail residue, H3K9, following opioid treatment (Table 1). Sun and colleagues (41) show that repeated morphine treatment reduces NAc H3K9 di-methylation (H3K9me2), but not mono- or tri-methylation. A similar reduction in H3K9me2 has been observed within the central nucleus of the amygdala following repeated opioid treatment (42). This reduction of H3K9me2 appears to promote transcriptional activity, and depends on the chronic nature of drug exposure (41). Using ChIP-sequencing to assess genome-wide deposition of H3K9me2 within the NAc, Sun and colleagues identified several genic loci showing differential enrichment of H3K9me2 following opioid exposure. Of particular interest was a reduction in enrichment of H3K9me2 throughout the FosB gene – a critical transcription factor that promotes drug addiction (see below; 24; 43). This suggests that chronic morphine may release repression of FosB by reduced H3K9me2 deposition at the gene. Further, Ingenuity Pathway Analysis identified changes in methylation at genes related to glutamatergic signaling, consistent with a role for H3K9me2 in regulating plasticity-associated transcriptional programs. Interestingly, opioid-induced transcriptional regulation via H3K9 methylation might not be specific to the NAc, as one study has also found reductions in H3K9 tri-methylation within the VTA and locus coeruleus after one week of withdrawal following an escalating dosing regimen (37).
DNA methylation
Methylation of DNA, primarily 5’-methylation of cytosines at cytosine-guanine dinucleotides (5mC), generally silences genes by physically blocking RNA polymerase II and thus gene transcription. Alternative forms of DNA methylation, such as 5-hydroxymethylation of cytosine (5hmC), which is enriched in brain, associate more frequently with transcriptional activation. Studies of DNA methylation in the context of opioid addiction have mostly been limited to (5mC) and to blood from clinical populations, post-mortem human brain tissue, and a small number of rodent opioid exposure paradigms (Table 1).
Genome-wide DNA methylation appears elevated by long-term heroin use based on higher levels of methylation at LINE-1 retrotransposon sites in blood leukocytes from heroin addicts compared to controls (44). Selective analysis of neurons within the frontal cortex of heroin users has identified differential methylation patterns between intragenic regions (45). Higher levels of methylation also occur at CpG-rich islands within the MOR gene, OPRM1, in both blood and brain tissue from heroin addicts (44; 46; 47). Intriguingly, this effect is also observed in patients receiving long-term opioid painkiller treatment compared to un-medicated patients, indicative of a pharmacological influence of chronic opioid exposure (44).
Compared to work with clinical populations, preclinical studies have had difficulty recapitulating the effects of opioids on DNA methylation, particularly within the reward system. Chronic morphine or heroin treatment with stable or escalating doses did not alter whole-brain DNA methylation (48; 49). Further, neither morphine treatment nor heroin self-administration altered DNA methylation in the mesocorticolimbic dopamine system of rodents (50; 51), which contrasts with evidence that cocaine alters global DNA methylation in the PFC and NAc (51–53). However, one study identified several changes to global or promoter-specific 5mC and 5hmC levels across multiple brain regions following chronic morphine exposure in rats (54). Whether these changes have functional consequences at the level of gene expression or behavior remains to be determined. Unraveling the complexities of DNA methylation in opioid addiction will require more effective and consistent preclinical studies, as well as those focusing on locus- specific changes in DNA methylation. Once identified, genes affected by DNA methylation can be cross-referenced against loci affected by histone modifications to generate a comprehensive understanding of how these opioid-induced epigenetic changes interact to control gene expression.
Non-coding RNA
In addition to histone modifications and DNA methylation, gene expression is regulated by noncoding RNAs at the level of transcription and translation including microRNAs (miRs) and long non-coding RNAs. Although much more work has been done for psychostimulants, especially cocaine (55), some studies have identified changes in miRNA activity following opioid exposure as summarized in Table 1. Region-specific increases have been observed for miR-339–3p and the Let-7 family (56; 57), while decreases have been observed for miR-154, miR-675, and miR-218 following chronic opioid exposure (58; 59). Although no studies have directly examined changes in long non-coding RNAs following opioid exposure, preliminary evidence suggests that these are also regulated in heroin addicts (60). Thus, non-coding RNAs are emerging as important regulators of transcriptional activity in opioid addiction, but more studies are required to understand the functional consequences of these epigenetic changes.
II. ROLE OF EPIGENETIC AND TRANSCRIPTIONAL REGULATORS IN RESPONSE TO OPIOIDS
Most of the evidence for a relationship between opioid use and epigenetic alterations discussed above is correlational. However, studies directly manipulating enzymes responsible for epigenetic modifications are beginning to build causal relationships between specific forms of epigenetic regulation and opioid-induced behavioral abnormalities. Numerous enzymes contribute to establishing the epigenetic landscape at a given locus, and can serve one of three functions: “writers” which place marks, “readers” which recognize and facilitate transcriptional modulation at marks, and “erasers” which remove marks (Figure 1).
The most common manipulation of epigenetic editors in the context of behavioral responses to opioids has been the use of non-specific pharmacological inhibitors of histone deacetylases (HDACs), which “erase” acetyl groups from histone tails (Table 2). Systemic treatment with different HDAC inhibitors can potentiate morphine-induced locomotor sensitization, and promote the formation of a morphine-conditioned place preference (CPP; 61; 62). Additionally, HDAC inhibitors can blunt reinstatement of a CPP but enhance drug-primed reinstatement of heroin-seeking behavior (40), indicative of a dynamic, stimulus-dependent response. Similar potentiation is observed for morphine-induced locomotor sensitization when the inhibitor is infused into the ventrolateral orbitofrontal cortex (38), and for heroin- or morphine-induced CPP when HDAC inhibitors are infused in the amygdala or NAc (34; 39). Only two studies have suggested opposite changes in behavior following HDAC treatment, but these authors used unconventional approaches to examine behavioral responses to opioids, such as sensitization to a single morphine injection (63), or intracerebroventricular delivery of conditioning doses of morphine (64). Further, intra-striatal infusion of a bromodomain inhibitor, which blocks the ability of “readers” to identify acetylated lysine residues, blunts heroin self-administration and heroin-seeking behavior (36). Taken together, these studies suggest that opioid-induced histone acetylation puts the reward system in a “poised” state, potentiating behavioral responses to opioids. These actions are similar to those observed for cocaine (23).
Table 2. Opioid-induced behavioral outcomes following manipulation of epigenetic editors.
Summary of studies examining behavioral responses to opioids following pharmacological or genetic manipulations of epigenetic enzyme function. Abbreviations: BLA, basolateral amygdala; CeA, central nucleus of the amygdala; CPP, conditioned reinforcement; dStri, dorsal striatum; HDAC, histone deacetylase; HMT, histone methyltransferase; HPC, hippocampus; ICV, intracereboventricular; IVSA, intravenous self-administration; NaBut, sodium butyrate; NAc, nucleus accumbens; PFC, prefrontal cortex; TSA, trichostatin A; VLO, ventolateral orbitofrontal cortex; VPA, valproic acid; VTA, ventral tegmental area.
| Chromatin State | Manipulation | Location of Treatment | Result | Reference |
|---|---|---|---|---|
| Open | Pan-HDAC inhibitor (NaBut) | Systemic | ↑ Morphine-induced locomotor sensitization, CPP | (61) |
| Pan-HDAC inhibitor (NaBut) | Systemic | ↑ Morphine-induced CPP ↓ Reinstatement of CPP |
(62) | |
| Pan-HDAC Inhibitor (VPA + NaBut) | Systemic | ↓ Morphine-induced locomotor sensitization (acute, single-dose) | (63) | |
| Pan-HDAC inhibitor (NaBut) | Both systemic and ICV | No Δ, Heroin IVSA ↑ Drug-primed reinstatement of heroin-seeking |
(40) | |
| Pan-HDAC inhibitor (VPA) | ICV | ↓ Morphine-induced CPP (morphine delivered ICV) | (64) | |
| Pan-HDAC inhibitor (TSA) | NAc (but not PFC) | ↑ Heroin-induced CPP | (34) | |
| Pan-HDAC inhibitor (TSA) | BLA | ↑ Morphine-induced CPP, ↓ CPP extinction | (39) | |
| Pan-HDAC inhibitor (TSA) | VLO | ↑ morphine-induced locomotor sensitization | (38) | |
| G9a (HMT) knockdown | NAc | ↑ Morphine-induced CPP, locomotor sensitization | (41) | |
| G9a inhibitor | CeA | ↑ Morphine-induced CPP | (42) | |
| SIRT1 (HDAC) KO | NAc (but not dstri) | ↓ Morphine-induced CPP | (66) | |
| Closed | G9a overexpression | NAc | ↓ Morphine-induced CPP, locomotor sensitization | (41) |
| SIRT1 overexpression | NAc (but not dstri) | ↑ morphine-induced CPP | (66) | |
To our knowledge, only two epigenetic editors have been selectively manipulated in the context of behavioral responses to opioids - exceptionally few when compared to the extensive literature for cocaine and other psychostimulants (30; 65). Ferguson and colleagues (66) examined the sirtuin family of HDACs, and found that repeated morphine treatment selectively upregulates SIRT1 in the NAc, without altering other sirtuins. Overexpression of SIRT1 within the NAc potentiated morphine-induced CPP, while knockdown of SIRT1 has the opposite effect. These results contrast with global HDAC inhibition studies described above and suggest that sirtuins may have a different role from the canonical HDACs. These actions of morphine also contrast with those of cocaine, which induces both SIRT1 and SIRT2 in this brain region (66). The histone methyltransferase G9a, known to be critical for aspects of cocaine reward (67), has also been manipulated in models of opioid addiction. Sun and colleagues showed that G9a is likely responsible for decreased H3K9me2 levels in the NAc following chronic opioid exposure, consistent with other reports in the CeA (42). Indeed, overexpression of G9a within the NAc increased H3K9me2 and blunted both morphine-induced CPP and locomotor sensitization, while knockdown or pharmacological inhibition within the amygdala has the opposite effect (41; 42). These studies parallel results with cocaine and suggest that G9a typically exerts a repressive influence on behavioral responses to drugs of abuse, and repeated exposure to opioids (or cocaine) relinquishes this control. Additionally, some emerging work points to an important role for chromatin remodelers such as BRG1 (SMARCA4) in mediating reinforcing effects of opioids (68). Interestingly, these latter effects may be mediated by altered function of non-neuronal cells, particularly oligodendrocytes, which exhibit significant gene regulation in response to opioids (69) and appear compromised in human heroin users (70–72).
Transcription Factors
Both epigenetic changes and regulation of editing enzymes rely on drug-induced activation of intracellular signaling pathways, which couple synaptic activity with transcriptional regulation through downstream activation of transcription factors – proteins that bind directly to DNA in a sequence-specific manner. Thus, the development and expression of epigenetic changes depend on iterative interactions between intracellular signaling cascades and the marks themselves (73).
One critical regulator of transcription is cyclic AMP response element-binding protein (CREB). CREB is activated downstream of multiple signaling pathways and integrates the transcriptional response to a broad range of cellular stimuli (74). Once activated, CREB promotes transcriptional activation by binding to cyclic AMP response elements (CREs) throughout the genome, many of which activate gene expression programs relevant to addiction (75–78). In the context of opioids, CREB has primarily been studied for its role in the aversive state produced by withdrawal (79), but some work suggests a dynamic regulation of CREB at different stages of drug-taking in a region-specific manner. For example, chronic morphine treatment enhances CREB signaling and activity in the NAc (80; 81). Overexpression of CREB specifically within the NAc reduces morphine reward (81), which is similar to effects observed with cocaine (82). Thus, persistent CREB signaling within the NAc may be a mechanism of tolerance to the rewarding effects of drugs of abuse. However, whole-brain knockdown or systemic pharmacological inhibition of CREB reduces morphine CPP and heroin-seeking, respectively (83; 84). These effects are opposite to findings with cocaine, and suggest that brain regions other than NAc have a significant impact on CREB-mediated transcriptional responses to opioids.
Another important transcription factor mediating responses to drugs of abuse is activator protein- 1 (AP-1). AP-1 is comprised of heterodimers of Fos family (c-Fos, FosB, Fral, and Fra2) and Jun family (c-Jun, JunB, JunD) members, which are expressed rapidly and transiently following acute drug exposure. AP-1 mediated gene expression promotes plasticity within reward regions via transcriptional activation or repression of genes involved in the behavioral and pharmacological response to cocaine (75). While typically short-lived, AP-1 activity can be extended by drug-induced expression of ΔFosB – a truncated splice variant of FosB. ΔFosB is exceptionally stable, enabling it to persist long after induction and accumulate with repeated drug exposure (85). While most work to date has focused on cocaine-induced potentiation of ΔFosB expression, particularly within the NAc and dorsal striatum (86), parallel changes are observed with opioids. Treatment with a sensitizing regimen of morphine increases ΔFosB in the NAc and ventral pallidum, and persists for over 3d into withdrawal (87). Global knockout of ΔFosB blunts morphine reward (88), while this is potentiated by upregulating ΔFosB within the NAc or striatum (89). Thus, ΔFosB may be an important mediator of prolonged transcriptional activity that sets the stage for long-term epigenetic modifications induced by opioids that promote addiction. Induction of ΔFosB by drugs of abuse shows an interesting pattern of cell-type specificity: all drugs of abuse induce ΔFosB solely within the D1 subtype of NAc medium spiny neuron, whereas opioids alone induce it in both D1 and D2 subtype neurons (90). This holds for both investigator-administered as well as self-administered drug, and points to some unique effects of opioids on gene expression in this brain region.
III. TRANSCRIPTIONAL CONSEQUENCES OF LONG-TERM OPIOID EXPOSURE
Exposure to drugs of abuse or to drug-associated stimuli elicits multiple waves of transcription. The rapid and transient induction of multiple immediate early genes (IEGs) sets the stage for persistent changes to the expression of effectors genes critical for long-term plasticity. These waves of gene expression regulate, and are regulated by, epigenetic modifications.
Immediate early genes
IEG induction couples rapid synaptic activation and intracellular signaling with long-term changes in neurons. Many IEGs encode transcription factors and contribute to epigenetic depositions onto chromatin (91; 92). Reciprocally, epigenetic modifications alter the pattern of IEG expression upon drug exposure. Like other drugs of abuse, opioids cause rapid induction of many IEGs after acute treatment (93). However, regulation patterns of IEG induction following chronic opioid exposure are still unclear. Some studies suggest increases in Arc expression in the PFC and striatum following repeated experimenter-administered morphine (94; 95). Self-administration studies have identified increases in expression of Fos, Arc, Egr1, and Egr2 in the PFC that persist for at least 24 hr (96), while Egr1 and Egr2 are reported to be decreased within the NAc, possibly mediated by changes in methylation state of these genes (50). Drug-seeking behavior is also associated with changes in IEG expression. For example, Egr1 is differentially regulated between the NAc and PFC following cue-induced heroin-seeking (97). This same paradigm also elicits distinct IEG profiles specifically within PFC neurons that are tagged as “activated” by cue and context re-exposure compared to surrounding non-activated neurons, with increases in FosB, Arc, Egr1, and Egr2 (98). The pattern of IEG expression produced specifically in neurons responsive to drug-associated stimuli suggests that there may be a specific molecular signature defining a population of neurons that drive craving and contribute to relapse.
Effector genes
Early studies into opioid-induced gene expression have typically taken a candidate gene approach, noting significant alterations to genes encoding opioid receptors, transcriptional regulators, and proteins that control structural and functional aspects of the cell, as reviewed extensively elsewhere (17; 79). However, candidate gene studies have limited potential to harvest a comprehensive understanding of the transcriptional landscape associated with opioid addiction. To assess this, studies employ whole-genome sampling techniques such as microarray profiling and next-generation RNA sequencing (RNA-seq). Although these studies are emerging for opioids, the results are difficult to synthesize. Comparisons between studies are challenging due to variability of exposure paradigms (e.g., drug treatment schedules, doses, self-administration designs), and the surprising lack of overlap in differentially regulated genes (see Table 3). Although this work suggests that several distinct pathways contribute to the development of opioid addiction, the lack of consistency must be addressed through standardization of experimental design and greater use of comprehensive RNA-seq approaches combined with advanced bioinformatics pipelines. These strategies are being extensively employed to identify transcriptomic and epigenomic changes to the reward system in cocaine addiction (53; 67; 99–104). Considering evidence from humans that cocaine and heroin elicit drastically distinct transcriptional profiles (105; 106), contrasting cocaine and opioid datasets will be critical to pinpoint unique or common patterns that promote addiction to these two classes of abused drugs.
Table 3. Summary of studies examining transcriptome-wide changes in gene expression following long-term opioid exposure.
Microarrays and RNA-seq approaches are beginning to be employed to identify opioid-induced changes to gene regulation within the brain’s reward circuitry. However, few studies have performed network analyses using advanced bioinformatics approaches, and commonalities between experimental design are lacking. Abbreviations: CPu, caudate/putamen; dStri, dorsal striatum; FC, frontal cortex; GO, gene ontology; HPC, hippocampus; IVSA, intravenous self-administration; NAc, nucleus accumbens; OFC, orbitofrontal cortex; PFC, prefrontal cortex; qPCR, quantitative polymerase chain reaction; vMB, ventral midbrain; vStri, ventral striatum; VTA, ventral tegmental area.
| Papers with gene lists | Paradigm | Regions | Genes/pathways differentially regulated |
|---|---|---|---|
| (105) | Human heroin addicts, microarray | NAc | 1050 transcripts altered, 10x more than cocaine observed in previous companion paper, with only 25 overlapping differentially regulated transcripts, and only 10 oppositely regulated No change in myelin-related genes (unlike cocaine; potentially region specific) ↓ Presynaptic machinery genes (neurotransmitter release: vesicle storage, release, recycling; not observed for cocaine), synaptic function genes ↑ TRKB (opposite to cocaine), ↑ FAS, ↓ prodynorphin (opposite to cocaine) |
| (45) | Human heroin addicts, microarray | Isolated nuclei from OFC neurons | GO terms associated with hypermethylation: axons, synaptic compartments, synaptic membrane, transmission of nerve impulse, axonogenesis, cell-cell-signaling Networks associated with hypomethylation: gene expression and regulation, regulation of neuron differentiation Differentially methylated genes: SLC17A7, OPRL1, TET3, ARC |
| (120) | Oxycodone IVSA, RNA-seq | vStri, dStri | Inflammation/immune pathways: vStri: 126 ↑, 15 ↓; dStri: 54 ↑, 1 ↓ Linked to glial responses to morphine |
| (121) | Oxycodone IVSA, RNA-seq | vStri, dStri | Opioid signaling, stress pathways, neurotransmission, serotonin signaling, kinases and TFs vStri qPCR confirmation: Pomc ↑, Htr1b ↑, Fkbp3 ↑, Htr7 ↓, Grin3a ↓, dStri qPCR confirmation: Gabr2b ↓, Gabra1 ↓ |
| (122) | Oxycodone IVSA, RNA-seq | NAc, CPu | NAc: 6 ↑, 8 ↓, CPu: 3 ↑, 2 ↓. Focus on structural markers: integrins, axon guidance factors. |
| (58) | Morphine IVSA, long-access, yoked design | vStri, vMB | 21000 differentially regulated genes, large GO lists across exposure paradigms Gene regulation due to morphine exposure: cell differentiation, cell-cell signalling, immune response, oxidative stress signaling Gene regulation specific to morphine-reinforced behavior: neuroplasticity, axonal guidance, miRNA pathways |
| (123) | Repeated morphine, microarray | vStri, PFC | Large list of altered genes, ingenuity pathway analysis to identify GO networks affected; Identified chromatin remodeling genes pFc, plasticity-related genes in NAc Ingenuity pathway analyses: neuroadaptive processes (long-term potentiation, axonal guidance, ephrin, and neuregulin pathways) |
| (124) | Repeated heroin, microarray | NAc | Comparison with methamphetamine treatment, 21 genes differentially regulated by heroin Focus on circadian genes (Gm129, Dbp, Per1, Per2) |
| (125) | Heroin IVSA rat, yoked design subtractive hybridization | NAc core and shell subregions | Active vs. passive drug intake causes major transcriptional differences in the NAc shell, with minimal differences in NAc core NAc shell pathways affected: transcription, translation, and cell metabolism 25 ↓, including TFs (Hnrp, Tbp-1), signaling (Chn1, Limk1, 14–3–3), |
| (61) | Repeated morphine, with challenge, microarray | dStri | Morphine: Arc, Nfkbia, Ttr, Kcnj13 Potentiated by co-administration of HDAC inhibitor: rhythm genes (Per1, Rev-erba, Cry1), addiction genes (Fos, Nr4a1, Zbtb16, FosB) |
| (126) | Acute morphine, heroin microarray | dStri | Extensive gene lists, hierarchical clustering Identify different patterns of induction based on gene identities and timing of tissue extraction following injection |
| (95) | Escalating morphine treatment, microarray | FC | Heat shock pathways (Hsp70, Hsp27, Hsp40, Hsp105, Cryab, BiP), circadian rhythm, synaptic activity pathways, Arc, nucleoporin p2 |
| (127) | Heroin IVSA, yoked design, microarray | PFC | Intracellular signaling genes; physiology related genes; GO pathways included developmental processes |
| (128) | Oxycodone IVSA, microarray | HPC | Microarray for synaptic plasticity (84 genes examined) Compared adolescent vs. adult, many age-dependent changes ↑ Cadherin2 (Cdh2), ↑ CREM, |
| (129) | Repeated morphine, subtractive hybridization | HPC | 6 ↑: vesicular transport, heat shock, steroid synthesis, oxidoreductase activity 5 ↓: cellular processes (cytoskeletal organization, vesicular transport, cell adhesion, iron transport, growth receptor binding, transaminase activity) |
| (130) | Repeated morphine, cocaine, RNA-seq | VTA | 152 ↑, 35 ↓ for morphine 5 ↑ 28 ↓ for cocaine Focused on Sgk1 |
Glutamate signaling and synaptic remodeling
Despite inconsistencies within the transcriptional literature, glutamate signaling and associated synaptic remodeling pathways have emerged as critical targets for opioid-induced epigenetic and transcriptional changes. Abnormalities in glutamate signaling support behavioral disturbances underlying addiction (107–109), and such changes may be present in heroin users (110; 111). Correspondingly, several studies have identified opioid-induced epigenetic modifications to glutamatergic transcriptional networks in human heroin addicts and preclinical models. These include enhanced chromatin accessibility surrounding glutamatergic genes (36), DNA methylation at key genes involved in glutamate plasticity (45), and changes to glutamatergic gene expression, particularly the GluA1 receptor (36; 41; 112). Thus, epigenetic modifications to glutamatergic signaling may be a critical mechanism that supports opioid addiction.
Conclusions and Future Directions
Our understanding of how opioids induce persistent neuroplastic changes within the brain’s reward circuitry is growing. Several epigenetic changes have been identified and linked to changes in gene expression programs that interact with the physiology of neurons, including higher levels of permissive histone acetylation and lower levels of repressive histone methylation. Manipulations of epigenetic editors suggests that these modifications potentiate behavioral responses to opioids. Complex changes in DNA methylation state have been identified in humans, and additional preclinical work is required to determine consequences of these changes. Emerging evidence for a role of non-coding RNAs in opioid addiction will be important to identify post-transcriptional regulation of gene expression induced by opioids. An array of transcriptional changes are induced by opioid exposure, but systematic patterns are currently unclear. Overall, current evidence suggests opioid-induced epigenetic modifications switch the reward system into a hyperresponsive state promoting future drug-seeking and drugtaking. However, it is difficult at this stage to build a conclusive narrative as to how opioids reprogram the epigenetic and transcriptional landscape of the brain.
To elaborate the epigenetic mechanisms underlying opioid addiction, several experimental aims and technological advancements are necessary. Much more consistency in opioid exposure paradigms is required, and studies should employ human-relevant dosing regimens, or shift to volitional models of opioid exposure, to promote translational relevance. To determine the stability of opioid-induced epigenetic modifications, changes to these marks should be characterized following both short- and long-term withdrawal periods. Most studies have also focused on exposure to a single opioid compound. Considering differences in their potency, signaling properties, and use patterns, systematic comparisons of multiple opioids and their epigenetic consequences should be performed. Additionally, front-line treatment of opioid addiction with MOR agonists or partial agonists, such as methadone or buprenorphine, are known to produce epigenetic modifications (113), and future work should explore how such changes interact with those induced by abused opioids. Preclinical studies have also examined male rodents almost exclusively, and greater efforts are needed to understand how sex differences influencing opioid-induced epigenetic modifications (114).
Furthermore, the opioid field must embrace cutting-edge epigenomic and transcriptomic techniques to generate the richest datasets possible that tell conclusive stories as to how opioids fundamentally alter the brain’s reward system. Opioid models should leverage next-generation sequencing technologies more extensively. Examples include RNA-seq to identify novel gene targets regulated by opioids, ATAC-seq to search the genome for regions of opened or closed chromatin following long-term opioid exposure, and ChIP-seq to link epigenetic changes with affected gene loci. Further, functional consequences of epigenetic modifications should be identified by manipulating these targets in a cell-type specific manner using recently developed locus-specific epigenetic editing tools (115). Work in the cocaine field is beginning to examine transcriptional regulation in a cell-type-specific manner (116), and parallel studies should be performed with opioids for both neuronal and non-neuronal cells. These novel approaches should be combined with bioinformatics analyses to identify differentially regulated gene networks and novel targets for opioid addiction. Concerted effort on these fronts will form a strong foundation on which to generate more effective treatment strategies and preventative measures in the fight against opioid addiction.
Acknowledgements
This work was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada to CJB, a doctoral fellowship from Boehringer Ingelheim Fonds to AG, and National Institutes of Health grants (P01DA047233 and R37DA007359) to EJN. The authors thank Jill Gregory for assistance with figure preparation.
Footnotes
Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Center for Disease Control, Prevention (2017): Centers for Disease Control and Prevention: Provisional Counts of Drug Overdose Death (As of August 6, 2017). Retrieved from https://wonder.cdc.gov/.
- 2.Health F (2016): The impact of the Opioid Crisis on the Healthcare System: A Study of Privately Billed Services. New York. [Google Scholar]
- 3.Center for Behavioral Health Statistics and Quality Substance Abuse and Mental Health Services Administration (2018): Key substance use and mental health indicators in the United States: Results from the 2017 National Survey on Drug Use and Health. Rockville, MD: Retrieved from https://www.samhsa.gov/data. [Google Scholar]
- 4.Cicero TJ, Inciardi JA, Muñoz A (2005): Trends in abuse of Oxycontin and other opioid analgesics in the United States: 2002–2004. JPain. 6: 662–672. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee G, Edelman EJ, Barry DT, Becker WC, Cerdá M, Crystal S, et al. (2016): Non-medical use of prescription opioids is associated with heroin initiation among US veterans: a prospective cohort study. Addiction. 111: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koob GF, Volkow ND (2010): Neurocircuitry of addiction. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 35: 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koob GF (2015): The dark side of emotion: the addiction perspective. Eur J Pharmacol. 753: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson TE, Berridge KC (1993): The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res Brain Res Rev. 18: 247–291. [DOI] [PubMed] [Google Scholar]
- 9.Hyman SE, Malenka RC, Nestler EJ (2006): Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 29: 565–598. [DOI] [PubMed] [Google Scholar]
- 10.Johnson SW, North RA (1992): Opioids excite dopamine neurons by hyperpolarization of local interneurons. J Neurosci. 12: 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrowski NL, Hatfield CB, Caggiula AR (1982): The effects of low doses of morphine on the activity of dopamine-containing cells and on behavior. Life sciences. 31: 2347–2350. [DOI] [PubMed] [Google Scholar]
- 12.Di Chiara G, Imperato A (1988): Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proceedings of the National Academy of Sciences of the United States of America. 85: 5274–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spanagel R, Herz A, Shippenberg TS (1990): The effects of opioid peptides on dopamine release in the nucleus accumbens: an in vivo microdialysis study. Journal of Neurochemistry. 55: 1734–1740. [DOI] [PubMed] [Google Scholar]
- 14.Spagnolo PA, Kimes A, Schwandt ML, Shokri-Kojori E, Thada S, Phillips KA, et al. (2019): Striatal Dopamine Release in response to Morphine: a [11C]-raclopride positron emission tomography study in Healthy Men. Biological Psychiatry. doi: 10.1016/j.biopsych.2019.03.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zito KA, Vickers G, Roberts DC (1985): Disruption of cocaine and heroin self-administration following kainic acid lesions of the nucleus accumbens. Pharmacology Biochemistry and Behavior. 23: 1029–1036. [DOI] [PubMed] [Google Scholar]
- 16.Shippenberg TS, Elmer GI (1998): The neurobiology of opiate reinforcement. Crit Rev Neurobiol. 12: 267–303. [DOI] [PubMed] [Google Scholar]
- 17.Przewlocki R (2004): Opioid abuse and brain gene expression. Eur J Pharmacol. 500: 331–349. [DOI] [PubMed] [Google Scholar]
- 18.Oguri K, Lee NM, Loh HH (1976): Apparent protein kinase activity in oligodendroglial chromatin after chronic morphine treatment. Biochem Pharmacol. 25: 2371–2376. [DOI] [PubMed] [Google Scholar]
- 19.Stiene-Martin A, Zhou R, Hauser KF (1998): Regional, developmental, and cell cycle-dependent differences in mu, delta, and kappa-opioid receptor expression among cultured mouse astrocytes. Glia. 22: 249–259. [PMC free article] [PubMed] [Google Scholar]
- 20.Hauser KF, Gurwell JA, Bhat NR (1993): Endogenous opioid systems and the growth of oligodendrocyte progenitors: paradoxical increases in oligodendrogenesis as an indirect mechanism of opioid action. Glia. 9: 157–162. [DOI] [PubMed] [Google Scholar]
- 21.Knapp PE, Maderspach K, Hauser KF (1998): Endogenous opioid system in developing normal and jimpy oligodendrocytes: mu and kappa opioid receptors mediate differential mitogenic and growth responses. Glia. 22: 189–201. [DOI] [PubMed] [Google Scholar]
- 22.Auvity S, Saba W, Goutal S, Leroy C, Buvat I, Cayla J, et al. (2017): Acute Morphine Exposure Increases the Brain Distribution of [18F]DPA-714, a PET Biomarker of Glial Activation in Nonhuman Primates. Int J Neuropsychopharmacol. 20: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nestler EJ (2014): Epigenetic mechanisms of drug addiction. Neuropharmacology. 76 Pt B: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robison AJ, Nestler EJ (2011): Transcriptional and epigenetic mechanisms of addiction. Nature reviews Neuroscience. 12: 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaenisch R, Bird A (2003): Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 33 Suppl: 245–254. [DOI] [PubMed] [Google Scholar]
- 26.Guan Z, Giustetto M, Lomvardas S, Kim J-H, Miniaci MC, Schwartz JH, et al. (2002): Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 111: 483–493. [DOI] [PubMed] [Google Scholar]
- 27.Dulac C (2010): Brain function and chromatin plasticity. Nature. 465: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everitt BJ (2014): Neural and psychological mechanisms underlying compulsive drug seeking habits and drug memories--indications for novel treatments of addiction. Eur J Neurosci. 40: 2163–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker DM, Cates HM, Heller EA, Nestler EJ (2015): Regulation of chromatin states by drugs of abuse. Curr Opin Neurobiol. 30: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson EM, Penrod RD, Barry SM, Hughes BW, Taniguchi M, Cowan CW (2018): It is a complex issue: emerging connections between epigenetic regulators in drug addiction. Eur J Neurosci. 133: 978. [DOI] [PubMed] [Google Scholar]
- 31.Badiani A, Belin D, Epstein D, Calu D, Shaham Y (2011): Opiate versus psychostimulant addiction: the differences do matter. Nature reviews Neuroscience. 12: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg RD (1974): Chromatin structure: a repeating unit of histones and DNA. Science (New York, NY). 184: 868–871. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997): Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389: 251–260. [DOI] [PubMed] [Google Scholar]
- 34.Sheng J, Lv ZG, Wang L, Zhou Y, Hui B (2011): Histone H3 phosphoacetylation is critical for heroin-induced place preference. Neuroreport. 22: 575–580. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Yan P, Hui T, Zhang J (2014): Epigenetic upregulation of PSD-95 contributes to the rewarding behavior by morphine conditioning. Eur J Pharmacol. 732: 123–129. [DOI] [PubMed] [Google Scholar]
- 36.Egervari G, Landry J, Callens J, Fullard JF, Roussos P, Keller E, Hurd YL (2017): Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target. Biological Psychiatry. 81: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashayekhi FJ, Rasti M, Rahvar M, Mokarram P, Namavar MR, Owji AA (2012): Expression levels of the BDNF gene and histone modifications around its promoters in the ventral tegmental area and locus ceruleus of rats during forced abstinence from morphine. Neurochem Res. 37: 1517–1523. [DOI] [PubMed] [Google Scholar]
- 38.Wei L, Zhu Y-M, Zhang Y-X, Liang F, Barry DM, Gao H-Y, et al. (2016): Microinjection of histone deacetylase inhibitor into the ventrolateral orbital cortex potentiates morphine induced behavioral sensitization. Brain research. 1646: 418–425. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Lai J, Cui H, Zhu Y, Zhao B, Wang W, Wei S (2015): Inhibition of histone deacetylase in the basolateral amygdala facilitates morphine context-associated memory formation in rats. J Mol Neurosci, 2nd ed. 55: 269–278. [DOI] [PubMed] [Google Scholar]
- 40.Chen W-S, Xu W-J, Zhu H-Q, Gao L, Lai M-J, Zhang F-Q, et al. (2016): Effects of histone deacetylase inhibitor sodium butyrate on heroin seeking behavior in the nucleus accumbens in rats. Brain research. 1652: 151–157. [DOI] [PubMed] [Google Scholar]
- 41.Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, et al. (2012): Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 32: 17454–17464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Tao W, Hou Y-Y, Wang W, Kenny PJ, Pan ZZ (2014): MeCP2 repression of G9a in regulation of pain and morphine reward. J Neurosci. 34: 9076–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nestler EJ, Barrot M, Self DW (2001): DeltaFosB: a sustained molecular switch for addiction. Proceedings of the National Academy of Sciences of the United States of America. 98: 11042–11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doehring A, Oertel BG, Sittl R, Lotsch J (2013): Chronic opioid use is associated with increased DNA methylation correlating with increased clinical pain. Pain. 154: 15–23. [DOI] [PubMed] [Google Scholar]
- 45.Kozlenkov A, Jaffe AE, Timashpolsky A, Apontes P, Rudchenko S, Barbu M, et al. (2017): DNA Methylation Profiling of Human Prefrontal Cortex Neurons in Heroin Users Shows Significant Difference between Genomic Contexts of Hyper- and Hypomethylation and a Younger Epigenetic Age. Genes (Basel). 8: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen DA, Yuferov V, Hamon S, Jackson C, Ho A, Ott J, Kreek MJ (2009): Increased OPRM1 DNA methylation in lymphocytes of methadone-maintained former heroin addicts. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 34: 867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chorbov VM, Todorov AA, Lynskey MT, Cicero TJ (2011): Elevated levels of DNA methylation at the OPRM1 promoter in blood and sperm from male opioid addicts. J Opioid Manag. 7: 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fragou D, Zanos P, Kouidou S, Njau S, Kitchen I, Bailey A, Kovatsi L (2013): Effect of chronic heroin and cocaine administration on global DNA methylation in brain and liver. Toxicol Lett. 218: 260–265. [DOI] [PubMed] [Google Scholar]
- 49.Chao M-R, Fragou D, Zanos P, Hu C-W, Bailey A, Kouidou S, Kovatsi L (2014): Epigenetically modified nucleotides in chronic heroin and cocaine treated mice. Toxicol Lett. 229: 451–457. [DOI] [PubMed] [Google Scholar]
- 50.Imperio CG, McFalls AJ, Hadad N, Blanco-Berdugo L, Masser DR, Colechio EM, et al. (2018): Exposure to environmental enrichment attenuates addiction-like behavior and alters molecular effects of heroin self-administration in rats. Neuropharmacology. 139: 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian W, Zhao M, Li M, Song T, Zhang M, Quan L, et al. (2012): Reversal of cocaine- conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex. (Homberg J, editor) PloS one. 7: e33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright KN, Hollis F, Duclot F, Dossat AM, Strong CE, Francis TC, et al. (2015): Methyl supplementation attenuates cocaine-seeking behaviors and cocaine-induced c-Fos activation in a DNA methylation-dependent manner. J Neurosci. 35: 8948–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, et al. (2015): Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J Neurosci. 35: 8042–8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrow TM, Byun H-M, Li X, Smart C, Wang Y-X, Zhang Y, et al. (2017): The effect of morphine upon DNA methylation in ten regions of the rat brain. Epigenetics. 12: 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kenny PJ (2014): Epigenetics, microRNA, and addiction. Dialogues Clin Neurosci. 16: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Y, Yang C, Kirkmire CM, Wang ZJ (2010): Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci. 30: 10251–10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Q, Hwang CK, Zheng H, Wagley Y, Lin H-Y, Kim DK, et al. (2013): MicroRNA 339 down-regulates p-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J. 27: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tapocik JD, Luu TV, Mayo CL, Wang B-D, Doyle E, Lee AD, et al. (2013): Neuroplasticity, axonal guidance and micro-RNA genes are associated with morphine self-administration behavior. Addict Biol. 18: 480–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan B, Hu Z, Yao W, Le Q, Xu B, Liu X, Ma L (2017): MiR-218 targets MeCP2 and inhibits heroin seeking behavior. Sci Rep. 7: 40413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ (2011): Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. Journal of Neurochemistry. 116: 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchis-Segura C, Lopez-Atalaya JP, Barco A (2009): Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 34: 2642–2654. [DOI] [PubMed] [Google Scholar]
- 62.Wang R, Zhang Y, Qing H, Liu M, Yang P (2010): The extinction of morphine-induced conditioned place preference by histone deacetylase inhibition. Neuroscience letters. 483 : 137–142. [DOI] [PubMed] [Google Scholar]
- 63.Jing L, Luo J, Zhang M, Qin W-J, Li Y-L, Liu Q, et al. (2011): Effect of the histone deacetylase inhibitors on behavioural sensitization to a single morphine exposure in mice. Neuroscience letters. 494: 169–173. [DOI] [PubMed] [Google Scholar]
- 64.Mu P, Yu L-C (2007): Valproic acid sodium inhibits the morphine-induced conditioned place preference in the central nervous system of rats. Neuroscience letters. 426: 135–138. [DOI] [PubMed] [Google Scholar]
- 65.Maze I, Nestler EJ (2011): The epigenetic landscape of addiction. Annals of the New York Academy of Sciences. 1216: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, et al. (2013): Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J Neurosci. 33: 16088–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. (2010): Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science (New York, NY). 327: 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin JA, Caccamise A, Werner CT, Viswanathan R, Polanco JJ, Stewart AF, et al. (2018): A Novel Role for Oligodendrocyte Precursor Cells (OPCs) and Sox10 in Mediating Cellular and Behavioral Responses to Heroin. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 43: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avey D, Sankararaman S, Yim AKY, Barve R, Milbrandt J, Mitra RD (2018): Single-Cell RNA-Seq Uncovers a Robust Transcriptional Response to Morphine by Glia. Cell Rep. 24: 3619–3629.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu H, Li L, Hao Y, Cao D, Xu L, Rohrbaugh R, et al. (2008): Disrupted white matter integrity in heroin dependence: a controlled study utilizing diffusion tensor imaging. Am J Drug Alcohol Abuse. 34: 562–575. [DOI] [PubMed] [Google Scholar]
- 71.Bora E, Yucel M, Fornito A, Pantelis C, Harrison BJ, Cocchi L, et al. (2012): White matter microstructure in opiate addiction. Addict Biol. 17: 141–148. [DOI] [PubMed] [Google Scholar]
- 72.Li W, Zhu J, Li Q, Ye J, Chen J, Liu J, et al. (2016): Brain white matter integrity in heroin addicts during methadone maintenance treatment is related to relapse propensity. Brain Behav. 6: e00436–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narlikar GJ, Fan H-Y, Kingston RE (2002): Cooperation between complexes that regulate chromatin structure and transcription. Cell. 108: 475–487. [DOI] [PubMed] [Google Scholar]
- 74.Carlezon WA, Duman RS, Nestler EJ (2005): The many faces of CREB. Trends Neurosci. 28: 436–445. [DOI] [PubMed] [Google Scholar]
- 75.McClung CA, Nestler EJ (2003): Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 6: 1208–1215. [DOI] [PubMed] [Google Scholar]
- 76.Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW (2007): Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 10: 1029–1037. [DOI] [PubMed] [Google Scholar]
- 77.Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, Malenka RC (2006): CREB modulates excitability of nucleus accumbens neurons. Nat Neurosci. 9: 475–477. [DOI] [PubMed] [Google Scholar]
- 78.Huang YH, Lin Y, Brown TE, Han M-H, Saal DB, Neve RL, et al. (2008): CREB modulates the functional output of nucleus accumbens neurons: a critical role of N-methyl-D-aspartate glutamate receptor (NMDAR) receptors. J Biol Chem. 283: 2751–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mazei-Robison MS, Nestler EJ (2012): Opiate-induced molecular and cellular plasticity of ventral tegmental area and locus coeruleus catecholamine neurons. Cold Spring Harb PerspectMed. 2: a012070–a012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shaw-Lutchman TZ, Barrot M, Wallace T, Gilden L, Zachariou V, Impey S, et al. (2002): Regional and cellular mapping of cAMP response element-mediated transcription during naltrexone-precipitated morphine withdrawal. J Neurosci. 22: 3663–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barrot M, Olivier JDA, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ, et al. (2002): CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proceedings of the National Academy of Sciences of the United States of America. 99: 11435–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carlezon WA, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, et al. (1998): Regulation of cocaine reward by CREB. Science (New York, NY). 282: 2272–2275. [DOI] [PubMed] [Google Scholar]
- 83.Lai M, Zhu H, Sun A, Zhuang D, Fu D, Chen W, et al. (2014): The phosphodiesterase-4 inhibitor rolipram attenuates heroin-seeking behavior induced by cues or heroin priming in rats. Int J Neuropsychopharmacol. 17: 1397–1407. [DOI] [PubMed] [Google Scholar]
- 84.Walters CL, Blendy JA (2001): Different requirements for cAMP response element binding protein in positive and negative reinforcing properties of drugs of abuse. J Neurosci. 21: 9438–9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen J, Kelz MB, Hope BT, Nakabeppu Y, Nestler EJ (1997): Chronic Fos-related antigens: stable variants of deltaFosB induced in brain by chronic treatments. J Neurosci. 17: 4933–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perrotti LI, Weaver RR, Robison B, Renthal W, Maze I, Yazdani S, et al. (2008): Distinct patterns of DeltaFosB induction in brain by drugs of abuse. Synapse (New York, NY). 62: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDaid J, Dallimore JE, Mackie AR, Napier TC (2006): Changes in accumbal and pallidal pCREB and deltaFosB in morphine-sensitized rats: correlations with receptor-evoked electrophysiological measures in the ventral pallidum. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 31: 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Solecki W, Krowka T, Kubik J, Kaczmarek L, Przewlocki R (2008): Role of fosB in behaviours related to morphine reward and spatial memory. Behavioural Brain Research. 190: 212–217. [DOI] [PubMed] [Google Scholar]
- 89.Zachariou V, Bolanos CA, Selley DE, Theobald D, Cassidy MP, Kelz MB, et al. (2006): An essential role for DeltaFosB in the nucleus accumbens in morphine action. Nat Neurosci. 9: 205–211. [DOI] [PubMed] [Google Scholar]
- 90.Lobo MK, Zaman S, Damez-Werno DM, Koo JW, Bagot RC, DiNieri JA, et al. (2013): ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 33: 18381–18395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sheng M, Greenberg ME (1990): The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 4: 477–485. [DOI] [PubMed] [Google Scholar]
- 92.Weaver ICG, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, et al. (2007): The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 27: 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bisagno V, Cadet JL (2019): Expression of immediate early genes in brain reward circuitries: Differential regulation by psychostimulant and opioid drugs. Neurochem Int. 124: 10–18. [DOI] [PubMed] [Google Scholar]
- 94.Marie-Claire C, Courtin C, Roques BP, Noble F (2004): Cytoskeletal genes regulation by chronic morphine treatment in rat striatum. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 29: 2208–2215. [DOI] [PubMed] [Google Scholar]
- 95.Ammon S, Mayer P, Riechert U, Tischmeyer H, Hollt V (2003): Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Brain Res Mol Brain Res. 112: 113–125. [DOI] [PubMed] [Google Scholar]
- 96.Kuntz KL, Patel KM, Grigson PS, Freeman WM, Vrana KE (2008): Heroin self-administration: II. CNS gene expression following withdrawal and cue-induced drug-seeking behavior. Pharmacology Biochemistry and Behavior. 90: 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmidt ED, Voorn P, Binnekade R, Schoffelmeer ANM, De Vries TJ (2005): Differential involvement of the prelimbic cortex and striatum in conditioned heroin and sucrose seeking following long-term extinction. Eur J Neurosci. 22: 2347–2356. [DOI] [PubMed] [Google Scholar]
- 98.Fanous S, Guez-Barber DH, Goldart EM, Schrama R, Theberge FRM, Shaham Y, Hope BT (2013): Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. Journal of Neurochemistry. 124: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Damez-Werno DM, Sun H, Scobie KN, Shao N, Rabkin J, Dias C, et al. (2016): Histone arginine methylation in cocaine action in the nucleus accumbens. Proceedings of the National Academy of Sciences of the United States of America. 113: 9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. (2014): Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun H, Damez-Werno DM, Scobie KN, Shao N-Y, Dias C, Rabkin J, et al. (2017): Regulation of BAZ1A and nucleosome positioning in the nucleus accumbens in response to cocaine. Neuroscience. 353: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, et al. (2018): Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biological Psychiatry. 84: 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, et al. (2009): Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 62: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sadakierska-Chudy A, Frankowska M, Jastrz^bska J, Wydra K, Miszkiel J, Sanak M, Filip M (2017): Cocaine Administration and Its Withdrawal Enhance the Expression of Genes Encoding Histone-Modifying Enzymes and Histone Acetylation in the Rat Prefrontal Cortex. Neurotox Res. 32: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ (2006): Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 31: 2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ (2004): Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. Journal of Neurochemistry. 88: 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hearing M, Graziane N, Dong Y, Thomas MJ (2018): Opioid and Psychostimulant Plasticity: Targeting Overlap in Nucleus Accumbens Glutamate Signaling. Trends Pharmacol Sci. 39: 276–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolf ME (2016): Synaptic mechanisms underlying persistent cocaine craving. Nature reviews Neuroscience. 17: 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scofield MD, Heinsbroek JA, Gipson CD, Kupchik YM, Spencer S, Smith ACW, et al. (2016): The Nucleus Accumbens: Mechanisms of Addiction across Drug Classes Reflect the Importance of Glutamate Homeostasis. Pharmacological reviews. 68: 816–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC, et al. (2011): Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biological Psychiatry. 69: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jacobs MM, Okvist A, Horvath M, Keller E, Bannon MJ, Morgello S, Hurd YL (2013): Dopamine receptor D1 and postsynaptic density gene variants associate with opiate abuse and striatal expression levels. Mol Psychiatry. 18: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hou Y-Y, Cai Y-Q, Pan ZZ (2015): Persistent pain maintains morphine-seeking behavior after morphine withdrawal through reduced MeCP2 repression of GluA1 in rat central amygdala. JNeurosci. 35: 3689–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marie-Claire C, Jourdaine C, Lepine J-P, Bellivier F, Bloch V, Vorspan F (2017): Pharmacoepigenomics of opiates and methadone maintenance treatment: current data and perspectives. Pharmacogenomics. 18: 1359–1372. [DOI] [PubMed] [Google Scholar]
- 114.Mavrikaki M, Pravetoni M, Page S, Potter D, Chartoff E (2017): Oxycodone self-administration in male and female rats. Psychopharmacology (Berl). 234: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hamilton PJ, Lim CJ, Nestler EJ, Heller EA (2018): Neuroepigenetic Editing. Methods Mol Biol. 1767: 113–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. (2010): Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science (New York, NY). 330: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trivedi M, Shah J, Hodgson N, Byun H-M, Deth R (2014): Morphine induces redox-based changes in global DNA methylation and retrotransposon transcription by inhibition of excitatory amino acid transporter type 3-mediated cysteine uptake. Mol Pharmacol. 85: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Knothe C, Doehring A, Ultsch A, Lotsch J (2016): Methadone induces hypermethylation of human DNA. Epigenomics. 8: 167–179. [DOI] [PubMed] [Google Scholar]
- 119.Garcia-Pérez D, Lopez-Bellido R, Hidalgo JM, Rodriguez RE, Laorden ML, Nünez C, Milanés MV (2015): Morphine regulates Argonaute 2 and TH expression and activity but not miR-133b in midbrain dopaminergic neurons. Addict Biol. 20: 104–119. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Liang Y, Levran O, Randesi M, Yuferov V, Zhao C, Kreek MJ (2017): Alterations of expression of inflammation/immune-related genes in the dorsal and ventral striatum of adult C57BL/6J mice following chronic oxycodone self-administration: a RNA sequencing study. Psychopharmacology (Berl). 234: 2259–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y, Liang Y, Randesi M, Yuferov V, Zhao C, Kreek MJ (2018): Chronic Oxycodone Self-administration Altered Reward-related Genes in the Ventral and Dorsal Striatum of C57BL/6J Mice: An RNA-seq Analysis. Neuroscience. 393: 333–349. [DOI] [PubMed] [Google Scholar]
- 122.Yuferov V, Zhang Y, Liang Y, Zhao C, Randesi M, Kreek MJ (2018): Oxycodone Self-Administration Induces Alterations in Expression of Integrin, Semaphorin and Ephrin Genes in the Mouse Striatum. Front Psychiatry. 9: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tapocik JD, Letwin N, Mayo CL, Frank B, Luu T, Achinike O, et al. (2009): Identification of candidate genes and gene networks specifically associated with analgesic tolerance to morphine. J Neurosci. 29: 5295–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Piechota M, Korostynski M, Sikora M, Golda S, Dzbek J, Przewlocki R (2012): Common transcriptional effects in the mouse striatum following chronic treatment with heroin and methamphetamine. Genes Brain Behav. 11: 404–414. [DOI] [PubMed] [Google Scholar]
- 125.Jacobs EH, Spijker S, Verhoog CW, Kamprath K, De Vries TJ, Smit AB, Schoffelmeer ANM (2002): Active heroin administration induces specific genomic responses in the nucleus accumbens shell. FASEB J. 16: 1961–1963. [DOI] [PubMed] [Google Scholar]
- 126.Piechota M, Korostynski M, Solecki W, Gieryk A, Slezak M, Bilecki W, et al. (2010): The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 11: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuntz-Melcavage KL, Brucklacher RM, Grigson PS, Freeman WM, Vrana KE (2009): Gene expression changes following extinction testing in a heroin behavioral incubation model. BMC Neurosci. 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y, Brownstein AJ, Buonora M, Niikura K, Ho A, Correa da Rosa J, et al. (2015): Self administration of oxycodone alters synaptic plasticity gene expression in the hippocampus differentially in male adolescent and adult mice. Neuroscience. 285: 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marie-Claire C, Courtin C, Robert A, Gidrol X, Roques BP, Noble F (2007): Sensitization to the conditioned rewarding effects of morphine modulates gene expression in rat hippocampus. Neuropharmacology. 52: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heller EA, Kaska S, Fallon B, Ferguson D, Kennedy PJ, Neve RL, et al. (2015): Morphine and cocaine increase serum- and glucocorticoid-inducible kinase 1 activity in the ventral tegmental area. Journal of Neurochemistry. 132: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]