Abstract
BCL-XL is a well-validated cancer target. However, the on-target and dose-limiting thrombocytopenia limits the use of BCL-XL inhibitors such as ABT263 as safe and effective anticancer agents. To reduce the toxicity of ABT263, we converted it into DT2216, a BCL-XL proteolysis targeting chimera (PROTAC), that targets BCL-XL to the Von Hippel-Lindau (VHL) E3 ligase for degradation. We found that DT2216 was more potent against various BCL-XL-dependent leukemia and cancer cells but significantly less toxic to platelets than ABT263 in vitro because VHL is poorly expressed in platelets. In vivo, DT2216 effectively inhibits the growth of several xenograft tumors as a single agent or in combination with other chemotherapeutic agents, without causing significant thrombocytopenia. These findings demonstrate the potential to use PROTAC technology to reduce on-target drug toxicities and rescue the therapeutic potential of previously undruggable targets. Furthermore, DT2216 may be developed as a safe first-in-class anticancer agent targeting BCL-XL.
The evasion of apoptosis is a key hallmark of cancer1, which is in part attributable to the overexpression of anti-apoptotic proteins in the BCL-2 family, including BCL-2, BCL-XL, and MCL-12, 3. Inhibition of these BCL-2 family proteins with small molecules has been extensively investigated as a therapeutic strategy for cancers4–9, resulting in the discovery of ABT263 (navitoclax, a BCL-2 and BCL-XL dual inhibitor), ABT199 (venetoclax, a BCL-2 selective inhibitor) and several BCL-XL and MCL-1 monoselective inhibitors as promising anticancer drug candidates10–16. Currently, ABT199 is the only FDA-approved antitumor agent targeting the BCL-2 family proteins17, 18, whereas ABT263 is not because inhibition of BCL-XL induces on-target and dose-limiting thrombocytopenia 19–21.
Although ABT199 is useful for the treatment of certain hematological malignancies such as chronic lymphocytic leukemia and acute myeloid leukemia, it has limited utility for the treatment of solid tumors22–24, because most solid tumor cells are not dependent on BCL-2 for survival23, 24. In contrast, BCL-XL is predominantly overexpressed in many solid tumor cells and also in a subset of leukemia cells23, 24, and its expression is highly correlated with resistance to cancer therapy, independent of p53 mutational status25, 26. To date, BCL-XL stands as one of the most important validated cancer targets without a safe and effective therapeutic. In this context, development of a platelet-sparing BCL-XL targeting agent has the potential to transform the treatment of BCL-XL-dependent malignancies.
Proteolysis targeting chimeras (PROTACs) are bivalent small-molecules containing a ligand that recognizes the target protein linked to an E3 ligase ligand. Such molecules can recruit the target protein to the E3 ligase, promote proximity-induced ubiquitination of the target protein, and lead to its degradation through the ubiquitin proteasome system (UPS). PROTACs act catalytically to induce protein degradation in a sub-stoichiometric manner. Their effect is not limited by equilibrium occupancy, and therefore requires less total drug exposure27–31. They generally have longer-acting activity but reduced toxicity than traditional occupancy-driven protein inhibitors and are increasingly used to develop more effective antitumor agents32–38. More importantly, because PROTACs rely on E3 ligases to induce protein degradation, it is possible for them to achieve cell/tissue selectivity even when their target proteins are ubiquitously expressed. They can achieve such selectivity by targeting the proteins to an E3 ligase that is differentially expressed in tumor cells compared with normal tissues. Therefore, we hypothesized that PROTAC technology can be exploited to reduce the thrombocytopenia induced by BCL-XL inhibition, by converting a BCL-XL inhibitor into a BCL-XL PROTAC that targets BCL-XL to an E3 ligase that is minimally expressed in platelets. Here we report first proof-of-concept evidence using PROTAC technology to generate a cell selective BCL-XL PROTAC, termed DT2216, which has improved antitumor activity but reduced platelet toxicity compared with ABT263. This is achieved by targeting BCL-XL to the Von Hippel-Lindau (VHL) E3 ligase, which is minimally expressed in platelets. In addition, DT2216 does not degrade BCL-2, yet can synergistically kill a variety of cancer cells that are not solely dependent on BCL-XL for survival when combined with a BCL-2 inhibitor, an MCL-1 inhibitor or a chemotherapeutic agent. These findings support the potential of DT2216 to be developed as a safe first-in-class BCL-XL-targeting antitumor agent. More broadly, our results define a new general strategy to convert antitumor agents with on target tissue-specific and dose-limiting toxicities to tumor-selective, less toxic PROTACs by engaging a tumor- or tissue/cell-specific E3 ligase.
Results
DT2216 exhibits improved antitumor potency but reduced platelet toxicity by targeting BCL-XL to the VHL E3 ligase for degradation
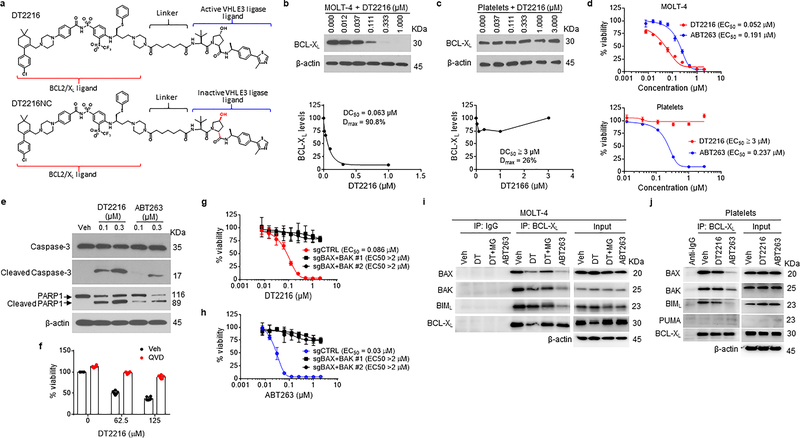
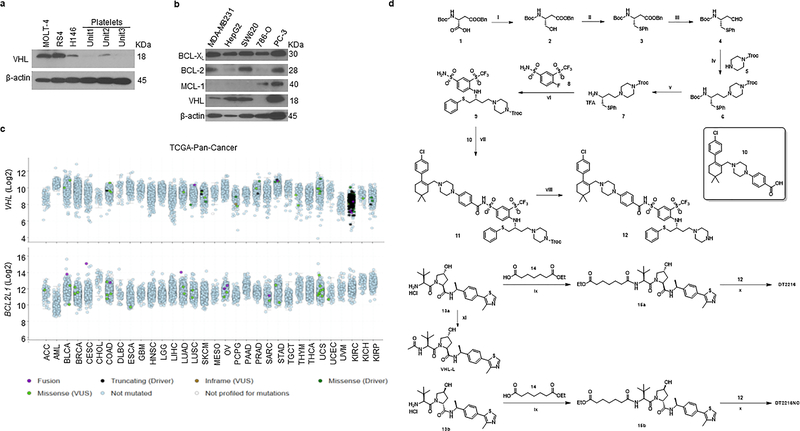
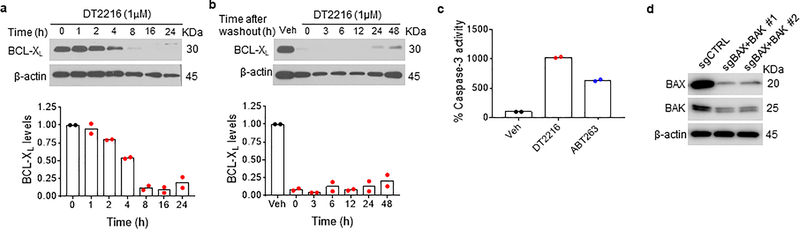
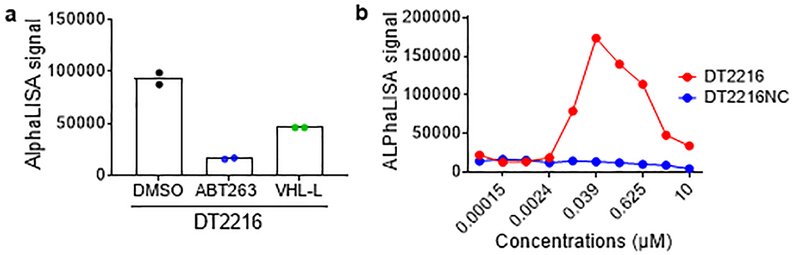
By analyzing human platelet RNA sequencing data, we identified VHL as an E3 ligase minimally expressed in human platelets39, 40. We confirmed this finding by immunoblotting, which showed that VHL expression was barely detectable in platelets. However, high levels of expression of VHL and BCL-XL were detected in multiple human tumor cells with the exception of VHL-null 786-O renal cell carcinoma (RCC) cell line (Extended Data Fig. 1a,b). This was consistent with high levels of VHL and BCL2L1 mRNA expression in a variety of human malignancies as we analyzed using The Cancer Genome Atlas (TCGA) database (Extended Data Fig. 1c)41, 42. Based on this finding, we rationally designed and synthesized a series of BCL-XL PROTACs that target BCL-XL to VHL for ubiquitination and degradation by linking the BCL-2/BCL-XL binding moiety (BCL-2/XL-L) derived from ABT263 to a VHL ligand (VHL-L) (Fig. 1a and Extended Data Fig. 1d). In addition, a BCL-XL PROTAC negative control (DT2216NC) compound that cannot bind to VHL was synthesized as a control. Among these BCL-XL PROTACs, DT2216 was selected as a lead because of its high potency in inducing BCL-XL degradation in MOLT-4 T-cell acute lymphoblastic leukemia (T-ALL) cells with the half-maximal degradation concentration (DC50) of 63 nM and maximum degradation (Dmax) of 90.8% (Fig. 1b). Notably, we observed no significant reduction in BCL-XL levels in platelets after incubation with up to 3 μM of DT2216 (Fig. 1c). The induction of BCL-XL degradation by DT2216 in MOLT-4 cells was rapid and long-lasting (Extended Data Fig. 2a,b). Because both MOLT-4 cells and platelets are solely dependent on BCL-XL for survival19, 24, 43, we next evaluated the effects of DT2216 on the viability of MOLT-4 cells and platelets in comparison with ABT263. As previously reported, ABT263 was highly toxic to both MOLT-4 cells and platelets (Fig. 1d)24, 43. In contrast, DT2216 (EC50 = 0.052 μM) was about 4-fold more cytotoxic to MOLT-4 cells than ABT263 (EC50 = 0.191 μM), and had minimal effect on the viability of platelets even at 3 μM (Fig. 1d). Both DT2216 and ABT263 killed MOLT-4 cells by caspase 3-mediated induction of apoptosis in a BAK- and BAX-dependent manner (Fig. 1e–h and Extended Data Fig. 2c,d). However, ABT263 functions as a BCL-XL inhibitor that inhibits the interaction of BCL-XL with BAK, BAX and BIM indiscriminately in both MOLT-4 cells and platelets, whereas DT2216 acts as a BCL-XL PROTAC that degrades BCL-XL selectively in MOLT-4 cells but not in platelets (Fig. 1i,j). These findings confirm that DT2216 is a BCL-XL PROTAC that has improved antitumor potency and reduced toxicity to platelets compared with ABT263.
Figure 1. DT2216, a BCL-XL PROTAC, selectively induces BCL-XL degradation and apoptosis in BCL-XL-dependent MOLT-4 T-ALL cells but not in platelets.
a, Chemical structures of DT2216 and its negative-control DT2216NC showing a BCL-2/-XL ligand linked to a VHL ligand via an optimized linker. DT2216NC has the inactive VHL ligand that does not bind to VHL. b, c, DT2216 selectively degrades BCL-XL in MOLT-4 cells but not in platelets after treatment with increasing concentrations of DT2216 as indicated for 16 h. A representative immunoblot is presented on the top panel. Densitometric analyses of BCL-XL expression are presented on the bottom panel as mean (n = 2 and 3 independent experiments for MOLT-4 and platelets, respectively). DC50, the drug concentration causing 50% protein degradation; Dmax, the maximum level of degradation. d, Viability of MOLT-4 cells and human platelets were determined after they were incubated with increasing concentrations of DT2216 and ABT263 for 72 h. The data are presented as mean ± SD from six and three replicate cell cultures in a representative experiment for MOLT-4 and platelets, respectively. Similar results were also observed in two additional independent experiments. For platelet viability assay, each experiment used platelets from one individual donor. EC50 values are the average of three independent experiments. e, A representative of two independent immunoblot analyses of cleaved and full-length caspase-3 and PARP1 in MOLT-4 cells 24 h after they were treated with vehicle (Veh), DT2216, or ABT263. f, Cell viability of MOLT-4 cells was determined after the cells were pretreated with the pan-caspase inhibitor Q-VD-OPh (QVD, 10 μM) and then treated with DT2216 for 72 h at indicated concentrations. Data are presented as mean ± SD from six replicate cell cultures in a representative experiment. Each symbol represents data from an individual replicate. Similar results were also observed in one additional independent experiment. g, h, Viability of non-targeting sg-RNA-transfected (sgCTRL) and Bax/Bak double knockout (KO; represented as sgBAX+BAK) H146 cells was determined after they were incubated with increasing concentrations of DT2216 or ABT263 for 72 h. Data are presented as mean ± SD from three replicate cell cultures in a representative experiment. Similar results were also observed in one additional independent experiment. i, j, MOLT-4 cells and human platelets were treated with either DT2216 (DT, 1 μM) or ABT263 (1μM) for 6 h. MOLT-4 cells were pretreated with MG132 (MG, 1 μM) for 1 h in order to block BCL-XL degradation. Immunoblots after immunoprecipitation with BCL-XL and in whole cell lysates (Input) are shown from a single experiment. β-actin was used as an equal loading control in all immunoblot analyses shown in Fig. 1b, c, e, i and j. The uncropped immunoblot images related to this figure are provided in separate source data file.
DT2216 induces proteasomal degradation of BCL-XL via the VHL E3 ligase
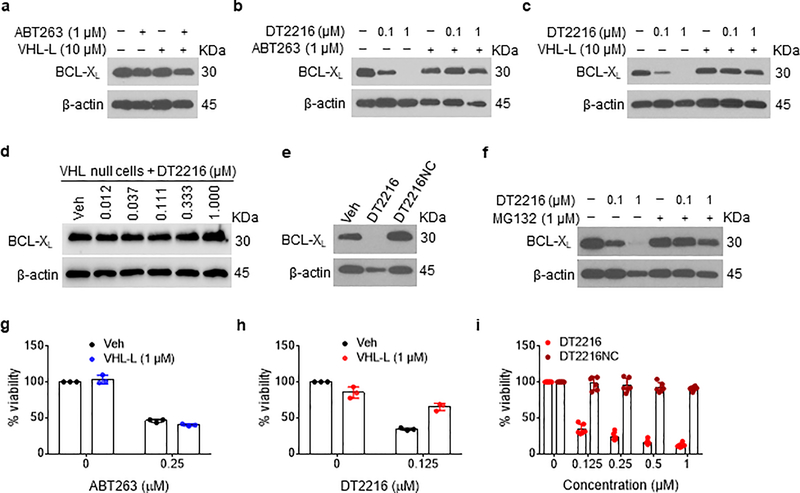
To further confirm that DT2216 degrades BCL-XL via VHL and proteasome, we first examined the effects of ABT263 and VHL-L alone and in combination on BCL-XL levels in MOLT-4 cells and found that none of these treatments affected the levels of BCL-XL (Fig. 2a). In addition, we found that pre-incubation of MOLT-4 cells with an excess amount of ABT263 or VHL-L inhibited the formation of the BCL-XL-DT2216-VHL ternary complex (Extended Data Fig. 3a) and DT2216-induced BCL-XL degradation (Fig. 2b,c). However, DT2216 had no effect on the levels of BCL-XL in VHL-null 786-O renal cell carcinoma (RCC) cells (Fig. 2d). Furthermore, DT2216NC which lacks VHL binding failed to form the BCL-XL-DT2216-VHL ternary complex (Extended Data Fig. 3b) and degrade BCL-XL (Fig. 2e) and inhibition of proteasome activity with MG132 abolished the degradation of BCL-XL induced by DT2216 (Fig. 2f). Moreover, the effect of DT2216 on MOLT-4 cell viability was also dependent on its PROTAC activity because VHL-L alone was not cytotoxic to the cells nor did it have any additive or synergistic effect on MOLT-4 cell viability when combined with ABT263 (Fig. 2g), but it did reduce the cytotoxicity of DT2216 (Fig. 2h). Finally, DT2216NC showed no cytotoxicity against MOLT-4 cells (Fig. 2i). Collectively, these data confirm that DT2216 acts as a PROTAC that depends on the VHL E3 ligase and proteasomes to degrade BCL-XL and induce apoptosis in MOLT-4 cells.
Figure 2. DT2216 degrades BCL-XL in a VHL- and proteasome-dependent manner.
a, ABT263 and/or VHL-L cannot induce BCL-XL degradation in MOLT4 cells. An immunoblot analysis of BCL-XL in MOLT-4 cells is shown after the cells were treated with ABT263 or VHL-L or the combination of both for 16 h. b, Pretreatment with ABT263 blocks the BCL-XL degradation by DT2216. An immunoblot analysis of BCL-XL in MOLT-4 cells after they were either left untreated or pretreated with ABT263 for 1 h and then treated with or without DT2216 as indicated for 16 h before being assayed. c, Pretreatment with VHL-L blocks the BCL-XL degradation by DT2216. An immunoblot analysis of BCL-XL in MOLT-4 cells is shown. The cells were either left untreated or pretreated with VHL-L for 1 h and then treated with or without DT2216 as indicated for 16 h before being assayed. d, An immunoblot analysis of BCL-XL in VHL-null 786-O cells treated with vehicle (Veh) or increasing concentrations of DT2216 for 16 h. e, An immunoblot analysis of BCL-XL in MOLT-4 cells is shown. The cells were treated with 1 μM of DT2216 and its negative-control DT2216NC for 16 h before being assayed. f, Proteasome inhibition blocks the BCL-XL degradation by DT2216. A representative of two immunoblot analyses of BCL-XL in MOLT-4 cells after they were either left untreated or pretreated with the proteasome inhibitor MG132 for 1 h, and then treated with or without DT2216 for 16 h. g, Cell viability of MOLT-4 cells treated with or without ABT263 in the presence or absence of VHL-L for 72 h. h, Cell viability of MOLT-4 cells after they were either left untreated or pretreated with VHL-L for 1 h, and then treated with DT2216 for 72 h. The data presented in g and h are mean ± SD from three replicate cell cultures in one representative experiment. Each symbol represents data from an individual replicate. Similar results were obtained in an additional independent experiment. i, Cell viability of MOLT-4 cells after treatment with increasing concentrations of DT2216 or its negative-control DT2216NC for 72 h. The data represent mean ± SD from six replicate cell cultures in one representative experiment. Each symbol represents data from an individual replicate. Similar results were obtained in one additional independent experiment. β-actin was used as an equal loading control in immunoblot analyses shown in Fig. 2a–f. The uncropped immunoblot images related to this figure are provided in separate source data file.
DT2216 is a specific BCL-XL PROTAC
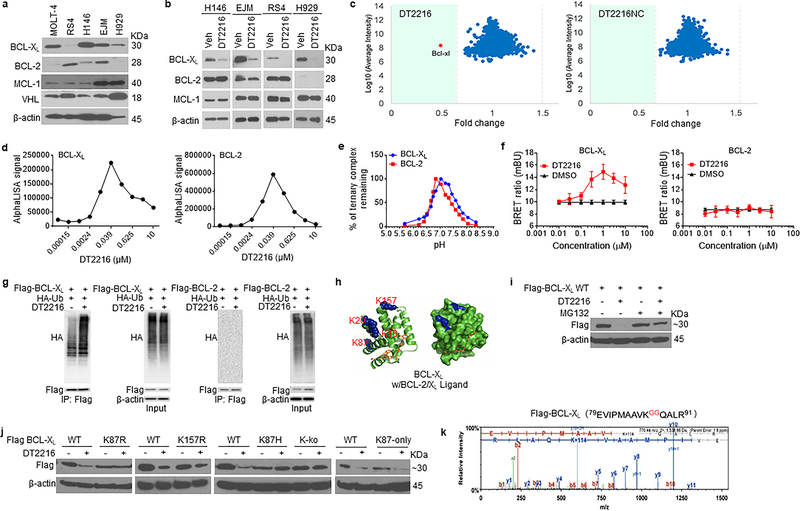
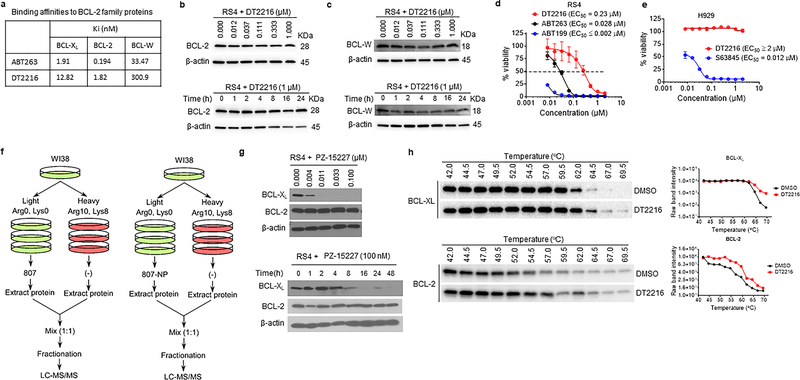
ABT263 has a high binding affinity for BCL-XL and BCL-2 but binds more weakly to other members in the BCL-2 family such as BCL-W or MCL-1 (Extended Data Fig. 4a)13, 24. The binding affinities of DT2216 for BCL-XL, BCL-2, and BCL-W were all reduced about 7- to 9-fold compared to that of ABT-263 but remained high for BCL-XL and BCL-2. To investigate the ability of DT2216 to degrade not only BCL-XL but also BCL-2 and other anti-apoptotic BCL-2 family members, we selected tumor cell lines that express high levels of BCL-XL, BCL-2, BCL-W and/or MCL-1 (Fig. 3a and Extended Data Fig. 4b,c). The analyses revealed that DT2216 selectively degraded BCL-XL but did not change the levels of BCL-2 in all cells examined even though it has a higher binding affinity for BCL-2 than BCL-XL. In addition, DT2216 had no effect on the levels of BCL-W and MCL-1 in these cells (Fig. 3b and Extended Data Fig. 4b,c). These findings are in agreement with the observations that DT2216 exhibited an 8-fold reduction in its potency against RS4 B-cell ALL cells, which primarily depend on BCL-2 for survival, compared to ABT26324 (Extended Data Fig. 4d and Supplementary Table 1). DT2216 was also not cytotoxic to MCL-1-dependent H929 myeloma cells16 (Extended Data Fig. 4e and Supplementary Table 1). To further validate the specificity of DT2216, we used the stable isotope labeling with amino acids in cell culture (SILAC) and liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics to analyze the changes in proteins in cells after DT2216 and DT2216NC treatment (Extended Data Fig. 4f). The SILAC/LC-MS/MS results show that DT2216, but not DT2216NC, reduced the levels of BCL-XL, but both agents did not affect expression of any other proteins (Fig. 3c), demonstrating that DT2216 is a specific BCL-XL PROTAC. The lack of BCL-2 degradation by BCL-XL PROTACs is not unique to DT2216 because the other BCL-XL PROTAC we synthesized that is similar to DT2216 but target a different E3 ligase (such as cereblon), degraded BCL-XL but not BCL-2 (Extended Data Fig. 4g).
Figure 3. DT2216 is a BCL-XL-specific PROTAC and induces BCL-XL degradation through K87 ubiquitination.
a, A representative of two immunoblot analyses of BCL-XL, BCL-2, MCL-1 and VHL in distinct tumor cell lines. b, Representative immunoblot analysis of BCL-XL, BCL-2 and MCL-1 in H146 SCLC after they were treated with 0.1 μM DT2216 for 48 h and in RS4 B-ALL cells and EJM and H929 multiple myeloma cells after they were treated with 1 μM DT2216 for 16 h. Similar results were obtained in one additional independent experiment. c, Proteomic analysis showing specificity of DT2216 on BCL-XL degradation in comparison with its negative-control DT2216NC in WI-38 cells. d, Ternary complex formation of BCL-XL or BCL-2 with DT2216 and VHL determined by AlphaLISA assay. Data are expressed as mean of a single experiment (n = 2 technical replicates). Similar results were obtained in two more independent assays performed with BCL-XL. e, pH stability of ternary complex formed by DT2216 and VHL complex and BCL-XL or BCL-2 as measured by AlphaLISA assay. Data are expressed as mean (n = 2 technical replicates). Similar results were obtained in one additional independent experiment. f, NanoBRET ternary complex formation of BCL-XL and BCL-2. Ternary complex formation was determined in 293T cells after they transiently expressed HiBit-BCL-XL, LgBit, and HaloTag-VHL or HiBit-BCL-2, LgBit, and HaloTag-VHL and then treated with a serial dilution of DT2216. Data are expressed as mean ± SEM of three independent experiments. g, Representative immunoblot of HA, Flag, β-actin following Flag immunoprecipitation of protein extracts from 293T cells cotransfected as indicated with Flag-BCL-XL and HA-Ub or Flag-BCL-2 and HA-Ub plasmids, then the cells were treated with or without DT2216 (1 μM) and MG132 (10 μM) as indicated for 4 h. Data are representative of three independent experiments. h, Crystal structure of BCL-XL with ABT263 (BCL-2/XL ligand). The lysines are colored in blue. i, A representative immunoblot analysis of BCL-XL showing that DT2216 induces wild-type (WT) BCL-XL degradation in a proteasome-dependent manner. Flag-BCL-XL-WT plasmid was transfected into 293T cells for 40 h and then the cells were treated with or without DT2216 (1 μM) and MG132 (10 μM) as indicated for 6 h. Similar results were obtained in two more independent experiments. j, Representative immunoblot analysis of BCL-XL showing that DT2216 induces BCL-XL degradation dependent on K87 ubiquitination. For the analysis, Flag-BCL-XL-WT, K87R and K157R (lysine to arginine), K87H (lysine to histidine), K-ko (all the lysines in BCL-XL were mutated to arginines) and K87-only (all the lysines in BCL-XL were mutated to arginines except K87) mutant plasmids were transfected into 293T cells for 40 h, and then the cells were treated with or without 1 μM DT2216 for 6 h. Similar results were obtained in two more independent experiments. k, K87 is the only ubiquitination site triggered by DT2216. 293T cells were co-transfected with Flag-BCL-XL and HA-Ub vectors. Extracts were immunoprecipitated with anti-Flag Affinity Resin, followed by trypsin and AspN digestion and tandem mass spectrometry, as described in Material and methods. A fragmentation spectrum of ubiquitinated EVIPMAAVkQALR peptide (ubiquitinated K87 residue) of BCL-XL. Parent ion corresponding to this peptide has been subjected to higher-energy collisional dissociation in mass spectrometer. The detected b- and y-fragment ion series have been labeled. The results were obtained from a single experiment. β-actin was used as equal loading control in immunoblotting experiments shown in Fig. 3a, b, h, i and j. The uncropped immunoblot images related to this figure are provided in separate source data file.
The selectivity of a PROTAC is largely determined by its ability to form a stable and cooperative ternary complex with its targeted protein(s) and E3 ligase44, 45. To elucidate the mechanism of selectivity of DT2216, we first measured the formation of the BCL-2- or BCL-XL-DT2216-VHL ternary complexes in vitro using the AlphaLISA assay44. These analyses revealed that both BCL-2 and BCL-XL can form stable ternary complexes with DT2216 and VHL in cell-free conditions (Fig. 3d,e). However, only BCL-XL, but not BCL-2, seems to be able to form stable ternary complexes with DT2216 and VHL in live cells as determined by nanoBRET assay46 even though both proteins could bind to DT2216 in cellular thermal shift assay (Fig. 3f and Extended Data Fig. 4h). These results are consistent with the ability of DT2216 to selectively induce BCL-XL, but not BCL-2, ubiquitination and subsequent degradation by proteasomes (Fig. 3g) and are in agreement with previous reports demonstrating that the specificity of a PROTAC can be determined in part by its ability to form ternary complexes with its target proteins44, 45. The differential formation of ternary complexes in vitro and in cell may be attributable to numerous factors including the inherent differences between these two assays. For example, the full length BCL-XL and BCL-2 were expressed in cells for the nanoBRET assay, whereas recombinant transmembrane domain-deleted proteins were used for the AlphaLISA assay. In addition, some other proteins may interfere with the interaction between BCL-2 and DT2216/VHL in live cells. However, the exact reason for the limited capacity of DT2216 to form ternary complexes with BCL-2 in cells in not known at this point and would be an important topic for future research.
Inspection of BCL-XL/ABT263 X-ray crystal structures (PDB entry 4QNQ) (Fig. 3h)14 revealed K16, K20, K87, and K157 in BCL-XL as solvent exposed residues and potential ubiquitination sites. The conformations of other two lysine residues (K205 and K233) are not resolved probably because they are buried in the transmembrane region of mitochondria, which precludes them as the potential ubiquitination sites47. Analyses of lysine to arginine BCL-XL mutants revealed that DT2216 can degrade Flag-tagged wild-type BCL-XL and the K157R mutant but not the BCL-XL mutants with all lysines mutated to arginines (K-ko) or K87R or K87H single mutant (Fig. 3i,j). These results support that K87 is required for the degradation of BCL-XL by DT2216. Consistently, only K87-ubiquinated BCL-XL could be detected in DT2216-treated cells by proteomics. Moreover, BCL-XL which only retains K87 (K87-only, i.e. all the lysines are mutated to arginines except K87) could be degraded by DT2216 (Fig. 3j,k). These findings suggest that DT2216 degrades BCL-XL in a K87 ubiquitination dependent manner, which is in agreement with a recent report indicating that the formation of ternary complex is necessary but not sufficient for a PROTAC to induce its target ubiquitination and degradation48.
DT2216 is a more potent antitumor agent than ABT263 with reduced platelet toxicity in vivo
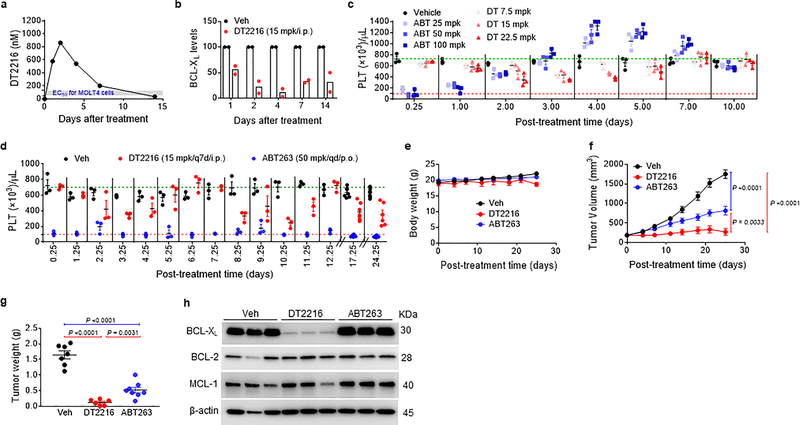
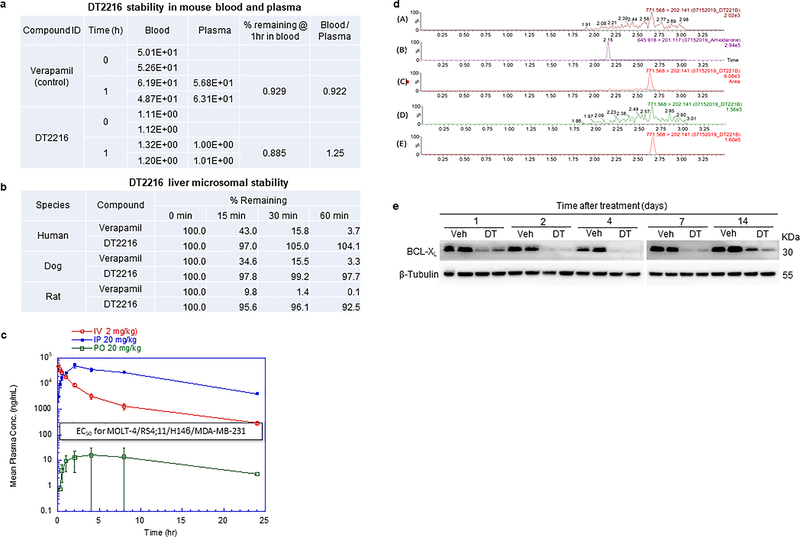
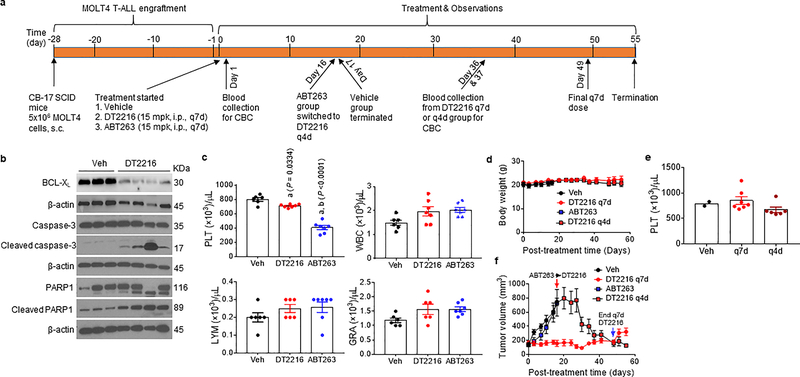
DT2216 is metabolically stable and has a favorable pharmacokinetic property for in vivo study via either i.p. or i.v. injection, but is not bioavailable by p.o. administration (Extended Data Fig. 5a–c and Supplementary Tables 2,3). A single dose of i.p. injection of DT2216 at 15 mg/kg (mpk) produced an intratumoral concentration of DT2216 in MOLT-4 T-ALL xenografts significantly greater than the cellular EC50 values of DT2216 for the cells for more than a week, which led to a sustained reduction in BCL-XL expression (Fig. 4a,b and Extended Data Fig. 5d,e and Supplementary Table 4). Next, we examined the effects of different doses of DT2216 on platelet levels in mice in comparison with the therapeutically equivalent doses of ABT263 considering that DT2216 is about 4-fold more potent than ABT263 against MOLT-4 cells in vitro (Fig. 1d)13. ABT263 doses ranging from 25 to 100 mpk caused severe thrombocytopenia in mice after 6 hours of administration (Fig. 4c), which raises the risk of spontaneous hemorrhage as reported previously49. Platelet counts gradually recovered 3 days after ABT263 administration and then exceeded normal values thereafter, before coming back to normal levels 10 days after the treatment. This rebound can potentially increase the risk of thrombosis50. In contrast, platelet counts after treatment with DT2216 were mildly reduced and were not followed by reactive thrombocytosis (Fig. 4c). These results demonstrate that, at the therapeutically equivalent doses, DT2216 is less toxic to platelets than ABT263 in mice.
Figure 4. DT2216 is more potent against MOLT-4 T-ALL xenografts and less toxic to platelets than ABT263 in mice.
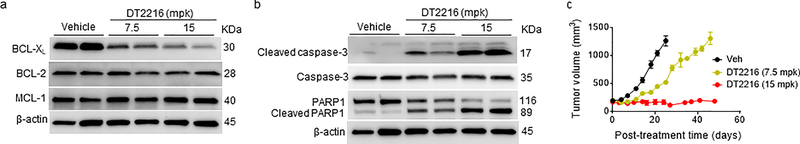
a, Concentration of DT2216 in MOLT-4 tumors after a single DT2216 administration (15 mpk/i.p.). Data represents average of two mice in a group at each time point. b, Densitometric analysis of BCL-XL protein levels in tumors (mean, n = 2 mice in a group at each time point) at different durations after a single Vehicle (Veh) or DT2216 administration (15 mpk/i.p.). Each symbol represents data (% of Veh) from an individual animal. The representative immunoblots are shown in Extended Data Fig. 5e. c, Numeration of platelets (PLT) 0.25, 1, 2, 3, 4, 5, 7 and 10 days after a single i.p. injection with DT2216 or p.o. dosing with ABT263 at indicated doses. Data are represented as mean ± SEM (n = 3 mice in each group). Each symbol represents data from an individual animal. d, Numeration of PLT after the mice were continuously treated with DT2216 or ABT263 as indicated. Data are represented as mean ± SEM (n = 3 mice in each group till day 12.25; n = 7 mice in Veh, 7 mice in DT2216, and 8 mice in ABT263 at day 17.25; n = 7 mice in Veh, 6 mice in DT2216, and 8 mice in ABT263 at day 24.25). Each symbol represents data from an individual animal. e, f, Body weight and tumor volume changes in mice after the start of treatment with vehicle (Veh), DT2216 (15 mpk/q7d/i.p.) or ABT263 (50 mpk/qd/p.o.). Data presented are mean ± SEM (n = 7 mice in Veh, 7 mice in DT2216 and 8 mice in ABT263 at the start of treatment). Statistical significance was determined by two-sided unpaired Student’s t-test. g, MOLT-4 tumor bearing mice were sacrificed 25 days after treatment initiation (four days and one day after last dose of DT2216 and ABT263, respectively). Tumor weights at the end of study are shown. Data are presented as mean ± SEM (n = 7 mice in Veh, 6 mice in DT2216 and 8 mice in ABT263). Each symbol represents data from an individual animal and the middle horizontal line represents mean. Statistical significance was determined by two-sided unpaired Student’s t-test. h, Immunoblot analysis of BCL-XL, BCL-2 and MCL-1 in MOLT-4 tumors (n = 3 mice in each group). Mpk, mg/kg; q7d, once a week treatment; qd, daily treatment. The uncropped immunoblot images related to this figure are provided in separate source data file.
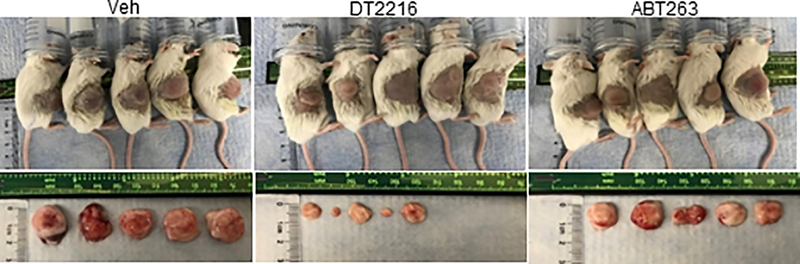
Dose-escalation studies showed that the antitumor effect of DT2216 after weekly administration at 15 mpk DT2216 was more effective in suppressing the growth of MOLT-4 T-ALL xenografts in mice than 7.5 mpk DT2216 (Extended Data Fig. 6a–c). We next compared the effects of repeated treatments with DT2216 (15 mpk/q7d/i.p.) and ABT263 (50 mpk/qd/p.o.) on blood platelet counts and MOLT-4 T-ALL xenograft growth (Fig. 4d–g and Extended Data Fig. 7). In these analyses, daily ABT263 treatment induced severe and persistent thrombocytopenia with only moderate effects on MOLT-4 T-ALL xenograft growth as shown previously13. In contrast, once weekly dosing of DT2216 almost completely inhibited MOLT-4 T-ALL xenograft growth with only moderate and transient reduction in platelet counts and no significant change in body weight. MOLT-4 T-ALL xenografts progressing under ABT263 treatment at a lower dose level (15 mpk/q7d/i.p.) that did not cause severe thrombocytopenia, effectively responded to treatment with DT2216 (15 mpk/q4d/i.p.) without significant changes in body weight and blood platelet counts (Extended Data Fig. 8a–f). The potent antitumor activity of DT2216 was correlated with its ability to induce BCL-XL degradation (Fig. 4h). These findings support that DT2216 is a safer and more potent antitumor agent than ABT263. It also demonstrated that DT2216 has the potential to be used as a single agent to treat tumors solely dependent on BCL-XL.
Increased antitumor activity of DT2216 in combination with other BCL-2 family protein inhibitors
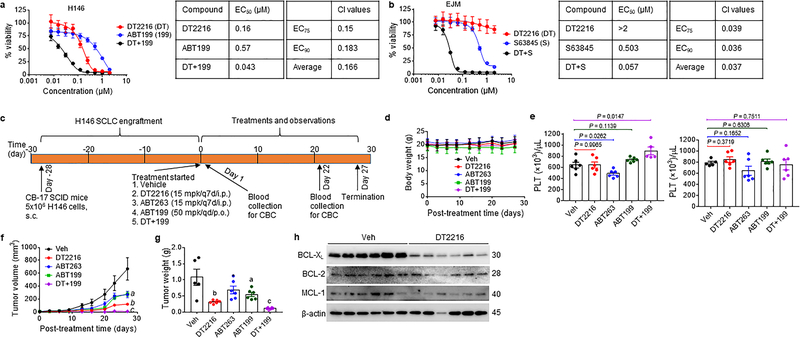
Hematological and solid tumors frequently engage more than one member of the BCL-2 family proteins for survival24, 51, 52. For example, NCI-H146 (H146) human small cell lung cancer (SCLC) cells are dependent on both BCL-XL and BCL-2 for survival, whereas EJM myeloma cells depend on MCL-1 and BCL-XL for survival16 (Fig. 5a,b and Supplementary Table 1). Inhibition of BCL-2 and MCL-1 with their respective inhibitors ABT199 and S63845 had moderate effects on the viability of H146 and EJM cells, respectively. However, their effect was significantly augmented by the addition of DT2216, resulting in synergistically killing of both cell lines (Fig. 5a,b).
Figure 5. Synergy of DT2216 with other BCL-2 family protein inhibitors.
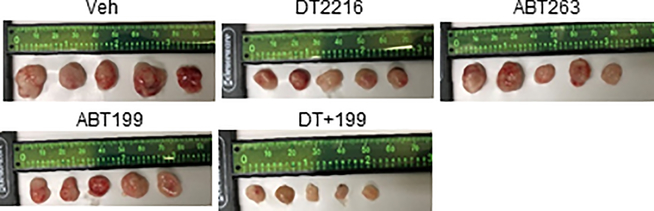
a, Percentage of viable H146 SCLC cells after 72 h treatment with increasing concentrations of DT2216 (DT) and ABT199 (199, a BCL-2 specific inhibitor) alone or the combination of these two at equimolar concentrations (1:1) as indicated is presented on the left panel. EC50 values for each of these treatments, the combination index (CI) at EC75 and EC90 values, and the average CI are presented in the table on the right panel. Data are presented as mean ± SD from six replicate cell cultures in one representative experiment. Similar results were obtained in one additional independent experiment. b, Percentage of viable EJM multiple myeloma cells after 72 h treatment with increasing concentrations of DT2216 (DT) and S63845 (S, a MCL-1 specific inhibitor) alone or the combination of these two at equimolar concentrations (1:1) is presented on the left panel. EC50 values for each of these treatments, CI at EC75 and EC90 values, and the average CI are presented in the table on the right panel. Data are presented as mean ± SD from six replicate cell cultures in one representative experiment. Similar results were obtained in one additional independent experiment. c, Illustration of the experimental design of H146 SCLC xenograft model. d, Body weight changes in H146 SCLC tumor bearing mice after the start of treatment with vehicle (Veh), DT2216, ABT263 or ABT199 alone or the combination of DT2216 (DT) and ABT199 (199) as shown in c. Data are presented as mean ± SEM (n = 6 mice per group at the start of treatment). e, Blood platelets (PLT) were numerated one day after first treatment with all the agents (left panel), and one day after last treatment with DT2216 or ABT263 and 22nd dose of ABT199 (right panel) as shown in c. Data are presented as mean ± SEM (n = 5 mice each for DT+199 in left panel and Veh in right panel, n = 6 mice each in other groups). Each symbol represents data from an individual animal. Statistical significance was determined by two-sided unpaired Student’s t-test. f, Changes in tumor volume over time after the start of treatment as shown in c. Data are presented as mean ± SEM (n = 6 mice per group at the start of treatment). a (P = 0.0381 ABT263 vs. Veh, P = 0.0400 ABT199 vs. Veh); b ( P = 0.0069 vs. Veh, P = 0.0152 vs. ABT263, P = 0.0044 vs. ABT199); and c (P = 0.0022 vs. Veh, P = 0.0001 vs. DT2216, P = 0.0003 vs. ABT263, P <0.0001 vs. ABT199) determined by two-sided unpaired Student’s t-test at post-treatment day-27. g, The average wet weight of excised tumors from each group. Data are presented as mean ± SEM (n = 5 mice in Veh, and 6 mice each in other groups). Each symbol represents data from an individual animal. a (P = 0.0524 vs. Veh); b (P = 0.0086 vs. Veh, P = 0.0116 vs. ABT263, P = 0.0072 vs. ABT199); and c (P = 0.002 vs. Veh, P <0.0001 vs. DT2216, P = 0.0006 vs. ABT263, P <0.0001 vs. ABT199) determined by two-sided unpaired Student’s t-test. h, A representative of two immunoblot analyses of BCL-XL, BCL-2 and MCL-1 in the H146 SCLC tumors excised at the end of experiment (n = 6 mice in each group). β-actin was used as a loading control. The uncropped immunoblot images related to this figure are provided in separate source data file.
Next, we examined whether DT2216 can be combined with ABT199 to more effectively eradicate BCL-XL- and BCL-2-co-dependent tumors such as H146 SCLC xenografts compared to ABT263 (Fig. 5c). Treatment of H146 SCLC xenografts with DT2216, ABT199, ABT263 and DT2216 plus ABT199 induced no significant changes in body weight (Fig. 5d). Administration of DT2216 at 15 mpk/week by i.p. injection alone or in combination with ABT199 at 50 mpk/day by p.o. administration did not cause any significant reduction in platelets, whereas i.p. administration of 15 mpk/week of ABT263 did (Fig. 5e). Treatment with DT2216, ABT199, or ABT263 alone significantly inhibited the tumor growth, resulting in 81.6%, 57.9%, and 59.5% mean tumor growth inhibition (TGI) at the end of the experiment (Fig. 5f,g and Extended Data Fig. 9), respectively. Moreover, the combination of DT2216 and ABT199 induced markedly increased antitumor effects resulting in almost complete suppression of tumor growth (mean 98.2% TGI). The TGI induced by DT2216 was also associated with a significant reduction in BCL-XL expression in the tumors harvested at the end of DT2216 treatment (Fig. 5h). These findings suggest that DT2216 can be combined with ABT199 to more effectively treat BCL-XL- and BCL-2-co-dependent tumors than either agent alone or ABT263 without causing significant platelet toxicity. Of note, the combination of DT2216 and S63845, a selective MCL-1 inhibitor, was lethal to mice because the liver cells are also dependent on both BCL-XL and MCL-1 for survival53, which limits the use of this combination as a systemic therapy to treat cancer.
Synergy between DT2216 and conventional chemotherapy
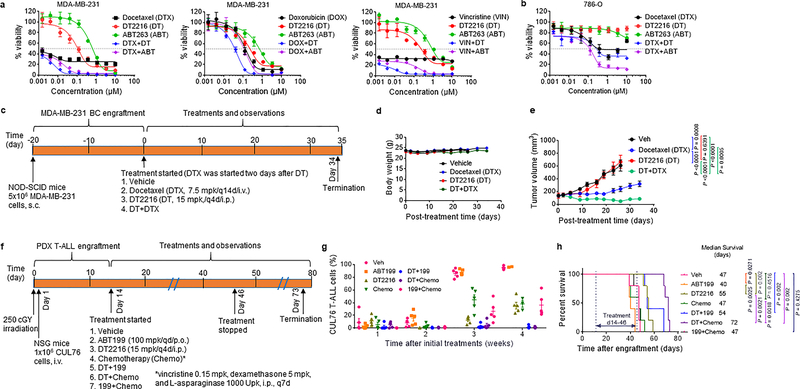
High levels of BCL-XL expression are associated with chemotherapy resistance across multiple tumor types25, 26, and inhibition of BCL-XL with ABT263 can improve the therapeutic efficacy of various chemotherapeutic agents24, 54, 55. However, the on-target thrombocytopenia resulting from BCL-XL inhibition prevents the use of combination therapy of ABT263 with other cytotoxic chemotherapeutic agents in clinic24, 54, 55. To evaluate the potential of DT2216 to overcome chemoresistance, we evaluated the ability of this agent to enhance the effects of chemotherapy. In these experiments, DT2216 sensitized drug-resistant triple negative MDA-MB-231 breast cancer (BC) cells to docetaxel, doxorubicin, and vincristine in vitro (Fig. 6a). A similar chemosensitizing effect was also observed in PC-3 prostate, HepG2 liver, and SW620 colon cancer cells (Supplementary Table 5), but not in 786-O RCC cells (Fig. 6b), which lack VHL expression (Fig. 2d and Extended Data Fig. 1b).
Figure 6. Synergy of DT2216 with chemotherapy.
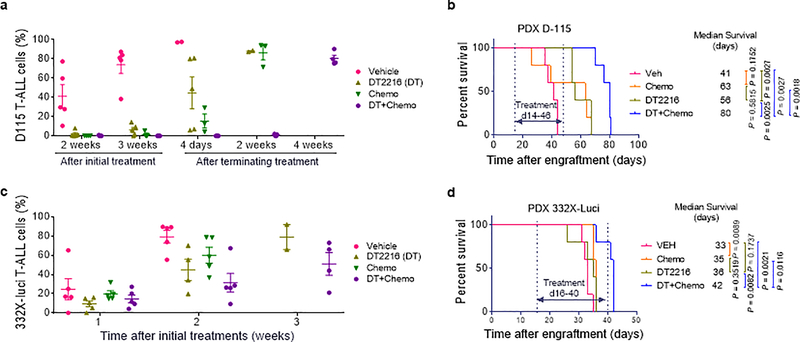
a, Percentage of viable MDA-MB-231 triple negative breast cancer cells after 72 h treatment with increasing concentrations of DT2216, ABT263, docetaxel (DTX), doxorubicin (DOX), or vincristine (VIN) alone or the combination of DT2216 (DT) or ABT263 (ABT) with one of these chemotherapeutic agents. Data are presented as mean ± SD from three replicate cell cultures in one representative experiment. Similar results were obtained in two additional independent experiments for DTX. b, Percentage of viable VHL-null 786-O renal cell carcinoma cells after 72 h treatment with docetaxel (DTX), DT2216, or ABT263 alone, or the combination of DT2216 (DT) or ABT263 (ABT) with DTX as indicated. Data are presented as mean ± SD from three replicate cell cultures of a single experiment. For the combination treatment, cells were treated with equimolar ratio of two drugs (i.e. 1:1) except the combination of DTX with DT2216 or ABT263 in MDA-MB-231 cells where the ratios of DTX and DT2216 or ABT263 were 1:10. c, Illustration of the experimental design of MDA-MB-231 breast cancer xenograft model. d, e, Body weight and tumor volume changes in MDA-MB-231 tumor bearing mice after the start of treatment with vehicle (Veh), DTX, DT2216, or the combination of these two as shown in c. Data are presented as mean ± SEM (n = 10 mice in each group at the start of treatment). Statistical significance was determined by two-sided unpaired Student’s t-test at post-treatment day-26. f, Illustration of the experimental design of CUL76 T-ALL patient-derived xenograft (PDX) model. g, Percentage of CUL76 T-ALL cells in mouse blood collected at various times after the initiation of the treatments as shown in f. Data are presented as mean ± SEM (n = 5 mice in each group at the start of treatment). Each symbol represents data from an individual animal, and the middle horizontal line represents mean. h, Kaplan-Meier survival curve showing the survival of mice after CUL76 T-ALL engraftment. Medium survival time within each treatment group is presented along with statistical analysis results (n = 5 mice in each group at the start of treatment). Statistical significance was determined by Log-rank (Mantel-Cox) test.
To determine whether DT2216 can sensitize MDA-MB-231 BC cells to docetaxel in vivo, we generated MDA-MB-231 BC xenografts in mice and then treated the mice with vehicle, docetaxel and/or DT2216 as shown in Fig. 6c. We found that DT2216 alone had minimal effect on the tumor growth, while docetaxel was able to substantially inhibit the growth of the tumor. However, the combination of docetaxel and DT2216 was more effective in suppressing the growth of the tumor than docetaxel alone without causing significant changes in body weight (Fig. 6d,e).
Patient-derived xenograft (PDX) tumor models can better recapitulate tumor biology of human diseases than conventional tumor xenograft models employing cancer cell lines. They are also more predictive of clinical outcomes of experimental therapeutic agents than the latter56. Therefore, we used CUL76 T-ALL PDX to validate whether DT2216 can be combined with ABT199 or chemotherapy to more effectively inhibit the growth of tumor cells from relapsed and refractory T-ALL patients. CUL76 T-ALL PDX was highly resistant to conventional chemotherapy and expressed high levels of BCL-XL, BCL-2 and MCL-1 (Fig. 6f and Supplementary Fig. 1a,c). This PDX grew rapidly after being transplanted into NOD-SCID IL2Rgnull (NSG) mice (Fig. 6g and Supplementary Fig. 1b). Mice with CUL76 T-ALL PDX had a median survival time of 47, 40 and 47 days after the initiation of treatments with vehicle, ABT199 and a standard T-ALL chemotherapy regimen consisting of vincristine, dexamethasone and L-asparaginase (VDL), respectively (Fig. 6h). Treatment with DT2216 alone or DT2216 plus ABT199 moderately prolonged the survival of the mice, whereas the combination of DT2216 with VDL chemotherapy substantially increased the survival of the mice (Fig. 6h). The extension of survival correlated with reduced circulating tumor burden of CUL76 T-ALL cells serially measured by flow cytometry in the blood (Fig. 6g and Supplementary Fig. 1c). Similar results were also observed in two additional T-ALL PDX models (Extended Data Fig. 10a–d). These findings confirm that DT2216 can sensitize drug resistant tumor cells to chemotherapy in vivo.
Discussion
Here we report a novel strategy to reduce the on-target and dose-limiting normal tissue toxicity of an antitumor agent using PROTAC technology. Specifically, we show that we can convert a non-selective BCL-2/BCL-XL inhibitor that is highly toxic to platelets into a BCL-XL-specific PROTAC with significantly reduced platelet toxicity. This was achieved by linking the BCL-2/BCL-XL binding moiety derived from ABT263 to a VHL-L with an empirically optimized linker. The resulting BCL-XL-specific PROTAC lead compound, DT2216, can selectively induce BCL-XL degradation in various tumor cells, but not in platelets, because it targets BCL-XL to the VHL E3 ligase that is minimally expressed in platelets.
These findings may have important clinical implications. First, BCL-XL is a well-validated cancer target. However, the on-target and dose-limiting thrombocytopenia induced by BCL-XL inhibition has limited the clinical use of BCL-XL inhibitors2, 4, 20, 21. By converting a BCL-2/BCL-XL dual inhibitor into a BCL-XL-specific PROTAC, we developed a safer and more effective BCL-XL-targeting antitumor agent. Our lead BCL-XL degrader, DT2216, significantly reduces the on-target and dose-limiting platelet toxicity resulting from conventional BCL-XL inhibition. In addition, BCL-XL-specific PROTACs should have another advantage compared to BCL-2/BCL-XL dual inhibitors or BCL-XL mono-inhibitors, because PROTACs act catalytically to induce protein degradation in a sub-stoichiometric manner, and their effect is not limited by equilibrium occupancy27–31. Therefore, it is not surprising that DT2216 is more potent than ABT263 in inducing tumor cell apoptosis in vitro and inhibiting tumor growth in vivo. These findings suggest that DT2216 has greater clinical potential than ABT263 or other BCL-XL inhibitors. However, it is unlikely that DT2216 will be totally devoid of platelet toxicity, because it retains a relatively reasonable binding affinity to BCL-XL, and thus can still function as a moderate BCL-XL inhibitor that could potentially induce mild thrombocytopenia if concentrations are high enough. Therefore, identifying effective and tolerable doses and schedule of DT2216 administration will be an important task in future clinical oncology trials.
Interestingly, while ABT263 inhibits both BCL-XL and BCL-2, DT2216 is BCL-XL-specific and does not induce BCL-2 degradation. In fact, proteomic analysis demonstrated that DT2216 is highly specific for BCL-XL degradation. Similarly, the pan-bromodomain and extra-terminal (BET) inhibitor JQ1, which binds and inhibits the BET proteins BRD2, BRD3 and BRD4, has been converted into a selective BRD4 PROTAC57. Others have shown that a promiscuous ligand for multi-kinases could induce only a subset of its targets for degradation when converted into PROTACs38, 58. As seen with other selective PROTACs reported previously, the lack of BCL-2 degradation by DT2216 is at least partially attributed to its inability to form a stable target protein-PROTAC-E3 ligase ternary complex in cells44, 45. We anticipate that because DT2216 is a specific BCL-XL PROTAC, it will have limited effect on cancers that depend on both BCL-XL and BCL-2 for survival24. However, we showed that this hurdler can be overcome by combining DT2216 with other selective inhibitors of BCL-2 family proteins (such as ABT199) or with standard chemotherapy.
Our results present the proof-of-principle for the use of tissue specific E3 ligases to direct tissue- or disease-specific degradation of a target protein using PROTAC technology. We demonstrate here that this strategy could rescue BCL-XL as an anti-cancer target from its on-target and dose-limiting toxicity. This approach could also be applied to convert other toxic antitumor agents into tumor-selective PROTACs by engaging E3 ligases more abundantly expressed in tumor cells than in normal tissues.
Online Methods
Chemical Synthesis
The chemical structures and synthetic schemes for DT2216 and DT2216NC are presented in Extended Data Fig. 1d. Detailed synthetic procedures are provided below.
General Methods
Tetrahydrofuran (THF), dichloromethane (DCM), toluene, and acetonitrile were obtained via a solvent purification system by filtering through two columns packed with activated alumina and 4 Å molecular sieve, respectively. All other chemicals obtained from commercial sources were used without further purification. Flash chromatography was performed using silica gel (230–400 mesh) as the stationary phase. Reaction progress was monitored by thin layer chromatography (silica-coated glass plates) and visualized by UV light, and/or by LC-MS. NMR spectra were recorded in CDCl3 at 400 MHz for 1H NMR. Chemical shifts δ are given in ppm using tetramethylsilane as an internal standard. Multiplicities of NMR signals are designated as singlet (s), broad singlet (br s), doublet (d), doublet of doublets (dd), triplet (t), quartet (q), and multiplet (m). All final compounds for biological testing were of ≥98.0% purity as analyzed by LC–MS, performed on an Advion AVANT LC system with the expression CMS using a Thermo Accucore™ Vanquish™ C18+ UHPLC Column (1.5 μm, 50 × 2.1 mm) at 40 °C. Gradient elution was used for UHPLC with a mobile phase of acetonitrile and water containing 0.1% formic acid.
Preparation of (R)-3-((tert-butoxycarbonyl)amino)-4-hydroxybutyric acid benzyl ester (2)
N-Methylmorpholine (4.41 mL, 40.1 mmol) and isobutyl chloroformate (4.43 mL, 34.2 mmol) were added into a stirred solution of N-Boc-D-aspartic acid 4-benzyl ester (1) (10.0 g, 30.9 mmol) in THF (250 mL) at −25 °C. The resulting mixture was stirred at −25 °C for 30 min and allowed to warm to −15 °C. A solution of NaBH4 (2.94 g, 77.7 mmol) in water (100 mL) was then added to the mixture in one portion resulting in evolution of gas. The mixture was stirred for 30 min at 15 °C and quenched with 1N HCl (aq.). The solution was extracted with ethyl acetate three times and the combined organic layers were washed with brine, dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. The crude product was used directly in the next step. 1H NMR (400 MHz, CDCl3) δ 7.48–7.29 (m, 5H), 5.17 (br s, 1H), 5.12 (s, 2H), 4.05–3.93 (m, 1H), 3.69 (t, J = 5.2 Hz, 2H), 2.67 (d, J = 6.1 Hz, 2H), 2.38 (br s, 1H), 1.42 (s, 9H). LC-MS (ESI): m/z 310.3 [M+H]+.
Preparation of benzyl (R)-3-((tert-butoxycarbonyl)amino)-4-(phenylthio)butanoate (3)
A mixture of compound 2 (30.9 mmol), diphenyl disulfide (8.8 g, 40.2 mmol), and Bu3P (9.9 mL, 40.2 mmol) in toluene (150 mL) was heated at 80 °C under N2 overnight. The mixture was cooled to room temperature and concentrated under reduced pressure. The crude product was purified by silica gel flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound (7.1 g, yield 57% in two steps). 1H NMR (400 MHz, CDCl3) δ 7.44–7.09 (m, 10H), 5.15 (br s, 1H), 5.08 (s, 2H), 4.24–3.97 (m, 1H), 3.23 (dd, J = 13.7, 5.5 Hz, 1H), 3.08 (dd, J = 13.6, 7.3 Hz, 1H), 2.80 (dd, J = 16.2, 5.1 Hz, 1H), 2.67 (dd, J = 16.4, 5.7 Hz, 1H), 1.40 (s, 9H). LC-MS (ESI): m/z 402.2 [M+H]+.
Preparation of tert-butyl N-[(2R)-4-oxo-1-(phenylsulfanyl)butan-2-yl]carbamate (4)
DIBAL-H (1.2 M in toluene, 34.0 mL, 40.8 mmol) was added dropwise to a solution of compound 3 (7.1 g, 17.7 mmol) in toluene (80 mL) at −78 °C and stirred for 3 h. The reaction mixture was then quenched with NH4Cl (aq.) and diluted with ethyl acetate. The resulting mixture was filtered and the filtrate was poured into water, extracted with ethyl acetate. The combined organic phases were washed with brine, dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. The crude product was purified by silica gel flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound (4.16 g, yield 80%). LC-MS (ESI): m/z 296.2 [M+H]+.
Preparation of 2,2,2-trichloroethyl (R)-4-(3-((tert-butoxycarbonyl)amino)-4-(phenylthio)butyl)piperazine-1-carboxylate (6)
To a mixture of compound 4 (592 mg, 2.00 mmol), compound 5 (753 mg, 2.88 mmol) and triethylamine (TEA) (1.12 mL, 8.05 mmol) in DCM (15 mL) was added NaBH(OAc)3 (638 mg, 3.00 mmol). The resulting solution was stirred at room temperature overnight before being poured into water and extracted with DCM. The combined organic phases were washed with brine, dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. The crude product was purified by silica gel flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound (733 mg, yield 68%). 1H NMR (400 MHz, CDCl3) δ 7.43–7.36 (m, 2H), 7.32–7.27 (m, 2H), 7.19 (t, J = 7.3 Hz, 1H), 5.44 (br s, 1H), 4.76 (s, 2H), 3.99–3.84 (m, 1H), 3.72–3.49 (m, 4H), 3.23 (dd, J = 13.3, 4.6 Hz, 1H), 3.10–2.95 (m, 1H), 2.61–2.31 (m, 6H), 1.96–1.61 (m, 2H), 1.43 (s, 9H). LC-MS (ESI): m/z 540.1 [M+H]+.
Preparation of 2,2,2-trichloroethyl (R)-4-(3-amino-4-(phenylthio)butyl)piperazine-1-carboxylate trifluoroacetic acid (TFA) salt (7)
To a mixture of compound 6 (733 mg, 1.36 mmol) in DCM (5 mL) was added TFA (2.0 mL, 26.1 mmol). The mixture was stirred at room temperature for 1 h and solvents were removed under reduced pressure. The solid residue was washed with diethyl ether to afford the title compound 7 (752 mg, yield 100%) as a white solid. 1H NMR (400 MHz, CDCl3): δ 7.41–7.33 (m, 2H), 7.31–7.26 (m, 2H), 7.23–7.15 (m, 1H), 4.74 (s, 2H), 3.73–3.41 (m, 4H), 3.20–2.66 (m, 5H), 2.58–2.28 (m, 6H), 1.84–1.57 (m, 2H). LC-MS (ESI): m/z 440.1 [M+H]+.
Preparation of 2,2,2-trichloroethyl (R)-4-(4-(phenylthio)-3-((4-sulfamoyl-2-((trifluoromethyl)sulfonyl)phenyl)amino)butyl)piperazine-1-carboxylate (9)
A mixture of compound 7 (752 mg, 1.36 mmol), 8 (417 mg, 1.36 mmol), and TEA (945 μL, 6.80 mmol) in acetonitrile (20 mL) was stirred under reflux for 4 h. Solvents were evaporated under reduced pressure and the crude product was purified by silica gel flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound (780 mg, yield 79%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 2.2 Hz, 1H), 7.84 (d, J = 9.1 Hz, 1H), 7.42–7.37 (m, 2H), 7.36–7.27 (m, 3H), 7.05 (d, J = 8.6 Hz, 1H), 6.65 (br s, 1H), 5.13 (br s, 2H), 4.76 (s, 2H), 4.02–3.88 (m, 1H), 3.75–3.40 (m, 4H), 3.16–2.97 (m, 2H), 2.82–2.26 (m, 6H), 2.19–2.05 (m, 1H), 1.85–1.77 (m, 1H). LC-MS (ESI): m/z 727.0 [M+H]+.
Preparation of 2,2,2-trichloroethyl (R)-4-(3-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazine-1-carboxylate (11)
A mixture of compound 9 (780 mg, 1.07 mmol), 10 (470 mg, 1.07 mmol), EDCI (411 mg, 2.14 mmol) and DMAP (262 mg, 2.14 mmol) in DCM (40 mL) was stirred at room temperature overnight. Solvent was evaporated under reduced pressure and the crude product was purified by silica gel flash column chromatography using DCM and methanol (MeOH) as eluents to afford the title compound (859 mg, yield 70%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 8.37 (d, J = 2.0 Hz, 1H), 8.13 (d, J = 9.2 Hz, 1H), 7.62 (d, J = 8.9 Hz, 2H), 7.41–7.27 (m, 6H), 7.12 (d, J = 8.7 Hz, 1H), 6.98 (d, J = 8.3 Hz, 2H), 6.79 (d, J = 9.0 Hz, 2H), 6.58 (d, J = 9.4 Hz, 1H), 4.75 (s, 2H), 3.95–3.81 (m, 1H), 3.63–3.38 (m, 4H), 3.33–3.23 (m, 4H), 3.11 (dd, J = 13.8, 4.9 Hz, 1H), 3.00 (dd, J = 13.9, 7.5 Hz, 1H), 2.81 (s, 2H), 2.46–2.01 (m, 15H), 1.75–1.65 (m, 1H), 1.46 (t, J = 6.3 Hz, 2H), 0.98 (s, 6H) ppm. LC-MS (ESI): m/z 1147.1 [M+H]+.
Preparation of (R)-4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-((1-(phenylthio)-4-(piperazin-1-yl)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (12)
Zinc powder (960 mg, 14.8 mmol) was added to a mixture of compound 11 (316 mg, 0.28 mmol) and acetic acid (600 μL, 10.5 mmol) in THF (20 mL). The reaction mixture was stirred at room temperature for 5 h. The solid was removed by filtration and the filtrate was poured into water and extracted with ethyl acetate. The combined organic phases were washed with brine, dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. The crude product was purified by silica gel flash column chromatography using DCM, MeOH, and TEA as eluents to afford the title compound (210 mg, yield 78%). 1H NMR (400 MHz, CDCl3) δ 8.21 (s, 1H), 7.93 (d, J = 9.2 Hz, 1H), 7.85 (d, J = 8.6 Hz, 2H), 7.33–7.24 (m, 2H), 7.22–7.08 (m, 5H), 6.92 (d, J = 8.3 Hz, 2H), 6.77 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.7 Hz, 2H), 6.46 (d, J = 9.3 Hz, 1H), 3.83–3.67 (m, 1H), 3.17–3.08 (m, 4H), 3.02–2.92 (m, 5H), 2.89–2.78 (m, 1H), 2.72 (s, 2H), 2.64–2.13 (m, 12H), 2.04–1.91 (m, 3H), 1.62–1.49 (m, 1H), 1.39 (t, J = 6.3 Hz, 2H), 0.91 (s, 6H) ppm. LC-MS (ESI): m/z 973.2 [M+H]+.
Preparation of Ethyl 7-(((S)-1-((2S,4R)-4-hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-7-oxoheptanoate (15a)
Compounds 13a and 13b were prepared according to reported procedures in the literature35. A mixture of compound 13a (100 mg, 0.207 mmol), acid 14 (44 mg, 0.234 mmol), HATU (82 mg, 0.216 mmol), and TEA (160 μL, 1.15 mmol) in DCM was stirred at room temperature for 1 h. The reaction mixture was poured into water and extracted with DCM. The combined organic layers were washed with aq. NH4Cl solution and saline, dried over Na2SO4, and concentrated under vacuum. The crude product was purified by silica gel column chromatography to afford the title compound (105 mg, yield 83%). 1H NMR (400 MHz, CDCl3) δ 8.68 (s, 1H), 7.50–7.32 (m, 5H), 6.26 (d, J = 8.7 Hz, 1H), 5.14–5.03 (m, 1H), 4.72 (t, J = 7.9 Hz, 1H), 4.59–4.46 (m, 2H), 4.14–4.08 (m, 3H), 3.61 (dd, J = 11.3, 3.7 Hz, 1H), 2.57–2.45 (m, 4H), 2.32–2.18 (m, 4H), 2.11–2.05 (m, 1H), 1.66–1.58 (m, 4H), 1.48 (d, J = 6.9 Hz, 3H), 1.36–1.22 (m, 5H), 1.04 (s, 9H) ppm. LC-MS (ESI): m/z 615.4 [M+H]+.
Preparation of ethyl 7-(((S)-1-((2R,4S)-4-hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-7-oxoheptanoate (15b)
Compound 15b was prepared using the procedure described for the synthesis of compound 15a by using compound 13b instead of compound 13a. Yield 80%. 1H NMR (400 MHz, CDCl3) δ 8.65 (s, 1H), 7.35 (d, J = 9.2 Hz, 5H), 6.13–6.03 (m, 1H), 5.11–5.00 (m, 1H), 4.68 (dd, J = 8.5, 3.8 Hz, 1H), 4.55–4.44 (m, 1H), 4.33 (d, J = 7.1 Hz, 1H), 4.13–4.04 (m, 3H), 3.59 (dd, J = 10.4, 5.3 Hz, 1H), 2.51 (s, 3H), 2.47–2.38 (m, 1H), 2.27–2.15 (m, 4H), 2.11–2.01 (m, 1H), 1.62–1.53 (m, 4H), 1.40 (d, J = 7.0 Hz, 3H), 1.32–1.19 (m, 5H), 1.06 (s, 9H) ppm. LC-MS (ESI): m/z 615.5 [M+H]+.
Preparation of (2S,4R)-1-((S)-2-(7-(4-((R)-3-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-7-oxoheptanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (DT2216)
Compound 15a (105 mg, 0.171 mmol) was dissolved in MeOH (5 mL) and treated with LiOH monohydrate (43 mg, 1.02 mmol) in water (0.5 mL) for 2 h. The pH of the reaction mixture was slowly adjusted to 5–6 with 1.0 N HCl. The mixture was concentrated directly under reduced pressure to afford the corresponding acid, which was dissolved in DCM (5 mL) and mixed with compound 12 (158 mg, 0.162 mmol), HATU (68 mg, 0.179 mmol), and TEA (113 μL, 0.810 mmol). The resulting solution was stirred at room temperature for 1 h before being poured into water and extracted with DCM. The combined organic layers were washed with aq. NH4Cl solution and saline, dried over Na2SO4, and concentrated under reduced pressure. The crude product was purified by silica gel flash column chromatography to afford DT2216 (115 mg, yield 46%). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.34 (d, J = 2.3 Hz, 1H), 8.08 (d, J = 9.1 Hz, 1H), 7.69 (d, J = 8.6 Hz, 2H), 7.46–7.27 (m, 12H), 7.14–6.93 (m, 3H), 6.76 (d, J = 8.6 Hz, 2H), 6.61 (d, J = 9.4 Hz, 1H), 6.34 (d, J = 8.7 Hz, 1H), 5.13–5.00 (m, 1H), 4.79–4.69 (m, 1H), 4.60 (d, J = 8.7 Hz, 1H), 4.49 (s, 1H), 4.10 (d, J = 11.4 Hz, 1H), 3.96–3.83 (m, 1H), 3.72–2.81 (m, 13H), 2.55–2.00 (m, 24H), 1.77–1.30 (m, 12H), 1.03 (s, 9H), 0.97 (s, 6H) ppm. LC-MS (ESI): m/z 1542.0 [M+H]+.
Preparation of (2R,4S)-1-((S)-2-(7-(4-((R)-3-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-7-oxoheptanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (DT2216NC)
DT2216NC was prepared using the procedure described for the synthesis of DT2216 by using compound 15b instead of compound 15a. Yield 40%. 1H NMR (400 MHz, CDCl3) δ 8.65 (s, 1H), 8.36 (d, J = 1.9 Hz, 1H), 8.11–8.04 (m, 1H), 7.69 (d, J = 8.8 Hz, 2H), 7.44–7.26 (m, 12H), 7.08 (d, J = 8.5 Hz, 1H), 6.98 (d, J = 8.3 Hz, 2H), 6.76 (d, J = 9.0 Hz, 2H), 6.62 (d, J = 9.4 Hz, 1H), 6.17 (d, J = 6.9 Hz, 1H), 5.13–5.02 (m, 1H), 4.68 (dd, J = 8.5, 3.9 Hz, 1H), 4.54–4.44 (m, 1H), 4.34 (d, J = 6.9 Hz, 1H), 4.13–4.02 (m, 1H), 3.97–3.84 (m, 1H), 3.72–3.56 (m, 2H), 3.43–3.22 (m, 7H), 3.09–2.79 (m, 4H), 2.51–2.00 (m, 24H), 1.73–1.24 (m, 12H), 1.07 (d, J = 4.8 Hz, 9H), 0.98 (s, 6H) ppm. LC-MS (ESI): m/z 1541.8 [M+H]+.
Preparation of (2S,4R)-1-((S)-2-acetamido-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (VHL-L)
To a solution of compound 13a (52 mg, 0.108 mmol) and TEA (42 μL, 0.302 mmol) in DCM (5 mL) was added acetic anhydride (10.4 μL, 0.110 mmol). The resulting solution was stirred at room temperature for 1 h. The reaction was quenched with water (5 mL) and extracted with DCM. The organic layer was washed with aq. NH4Cl solution and saline, dried over Na2SO4, and concentrated under reduced pressure. The crude product was purified by silica gel flash column chromatography to afford VHL-L (41 mg, yield 78%). 1H NMR (400 MHz, CDCl3) δ 8.69 (s, 1H), 7.47 – 7.34 (m, 5H), 6.34 (d, J = 8.8 Hz, 1H), 5.17 – 5.03 (m, 1H), 4.70 (t, J = 7.9 Hz, 1H), 4.61 – 4.45 (m, 2H), 4.06 (d, J = 11.3 Hz, 1H), 3.70 – 3.52 (m, 1H), 2.57 – 2.42 (m, 4H), 2.10 – 2.03 (m, 1H), 1.98 (s, 3H), 1.48 (d, J = 6.8 Hz, 3H), 1.05 (s, 9H) ppm. LC-MS (ESI): m/z 487.2 [M+H]+.
Chemical compounds and antibodies
The information for commercially available chemical compounds, anticancer drugs and antibodies are provided in Supplementary Tables 6 & 7.
Cell lines and cell culture
Human T-ALL MOLT-4 (Cat. No. CRL-1582), B-ALL RS4;11 (RS4, Cat. No. CRL-1873), SCLC NCI-H146 (H146, Cat. No. HTB-173), breast cancer MDA-MB-231 (Cat. No. HTB-26), prostate cancer PC-3 (Cat. No. CRL-1435), hepatocellular carcinoma HepG2 (Cat. No. HB-8065), colorectal carcinoma SW620 (Cat. No. CCL-227), renal cell carcinoma 786-O (Cat. No. CRL-1932), lung fibroblasts WI-38 (Cat. No. CCL-75) and epithelial kidney HEK 293T (293T, Cat. No. ACS-4500) cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Human multiple myeloma cell lines [EJM and NCI-H929 (H929)] were kind gift from Dr. Erming Tian at the Winthrop P. Rockefeller Cancer Institute at University of Arkansas for Medical Sciences. MOLT-4, RS4 and H146 cell lines were cultured in RPMI 1640 medium (Cat No. 22400–089, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Cat. No. S11150H, Atlanta Biologicals, Flowery Branch, GA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Pen-Strep, Cat. No. 15140122, Thermo Fisher Scientific). EJM cells were cultured in RPMI media with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin supplemented with 1% MarrowMAX Bone Marrow Medium (Cat. No. 12260014, Thermo Fisher Scientific). H929 cells were cultured in RPMI media with 10% FBS and 100 U/mL penicillin and 100 μg/mL streptomycin supplemented with 0.05 mM 2-mercaptoethanol. All other cell lines were cultured in complete Dulbecco’s modified Eagle medium (DMEM, Cat. No. 12430054, Thermo Fisher Scientific) with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. All the cell lines were maintained in a humidified incubator at 37 °C and 5% CO2.
Immunoblotting
Cells were treated with DT2216 and other compounds at indicated concentrations and durations. Untreated and treated cells were harvested and washed once with ice-cold phosphate buffered saline, pH 7.2 (PBS, Cat. No. 20012027; Thermo Fisher Scientific). Adherent cells were harvested using 0.25% Trypsin-EDTA solution (Cat. No. 25200056, Thermo Fisher Scientific) and washed with ice-cold PBS. The cell pellets were lysed in Radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, 5 mM EDTA, 1 mM EGTA; Cat. No. BP-115DG, Boston Bio Products, Ashland, MA, USA) supplemented with 1% protease inhibitor cocktail (Cat. No. P8340, Sigma-Aldrich, St. Louis, MO, USA) and 1% phosphatase inhibitor cocktail (Cat. No. P0044, Sigma-Aldrich). Protein lysates were incubated on ice for 15 min and then kept at −80 °C overnight to allow for complete lysis. The samples were thawed on ice and then centrifuged at maximum speed/14000 rpm for 15 min in a refrigerated centrifuge. The protein concentration in the supernatants was determined using the Pierce BCA protein Assay kit (Cat. No. 23225, Thermo Fisher Scientific). The protein concentration was normalized and the samples were reduced in 4X Laemmli’s SDS-sample buffer (Cat. No. BP-110R, Boston Bio Products) and denatured at 95°C in a heated-block. An equal amount of protein samples (20–40 μg/lane) were resolved using precast 4–20% Tris-glycine gels (Mini-PROTEAN TGX™, Cat. No. 456–1094, Bio-Rad, Hercules, CA, USA), and resolved proteins were transferred onto 0.2 μm pore size PVDF blotting membranes (Cat. No. LC2002, Thermo Fisher Scientific) using mini Trans-blot electrophoretic transfer cell (Bio-Rad). The membranes were blocked with non-fat dry milk (5% w/v) in 1X Tris-buffered saline-Tween-20 (TBST, Cat. No. J77500, Affymetrix, Santa Clara, CA, USA) for 1 h at room temperature, and subsequently probed with primary antibodies at a predetermined optimal concentration in non-fat dry milk (5% w/v in TBST) overnight at 4°C. The membranes were washed three times (5–10 min each) in TBST and then incubated with horse radish peroxidase (HRP)-conjugated secondary antibodies for 1–1.5 h at room temperature. Following sufficient washing with TBST, the membranes were exposed with chemiluminescent HRP substrate (Cat. No. WBKLS0500, MilliporeSigma, Billerica, MA, USA), and the signal was detected using autoradiography (SRX-101, Konica, Shinjuku, Tokyo, Japan) or the ChemiDoc MP Imaging System (Bio-Rad). The immunoblots were quantified by densitometry using ImageJ software (NIH) and the data were expressed as relative band intensities normalized to equal loading control.
Viability assays in cancer cells
Cancer cells in complete cell culture medium were seeded in 96-well plates (100 μL/well) at the optimized densities (50,000–100,000 suspension cells, 3,000–5,000 adherent cells). Suspension cells were treated 30 min after seeding, whereas adherent cells were allowed to adhere overnight and then treated. Compound treatments were prepared in complete cell culture media and 100 μL of 2X treatment-containing media were added to each well. Complete cell culture media without treatment was added in control wells, and medium alone wells were included and served as background control. The outer wells of 96-well plate were not used for treatment and were filled with 200 μL of PBS to reduce evaporation of media from inner wells. Each compound/combination was tested at nine different concentrations with three to six replicates, unless otherwise specified. For combination treatments with DT2216 and ABT199 or DT2216 and S63845, cells were treated at equimolar concentrations in two-fold serial dilutions. For combination treatment with DT2216 and chemotherapeutics, cells were treated at equimolar concentrations and three-fold serial dilutions, unless otherwise specified. The cell viability was measured after 72 h by Tetrazolium-based MTS assay. MTS reagent (2 mg/mL stock; Cat. No. G1111, Promega, Madison, WI, USA) was freshly supplemented with Phenazine methosulfate (PMS, 0.92 mg/mL stock, Cat. No. P9625, Sigma-Aldrich) in 20:1 ratio, and 20 μL of this mixture was added to each control and treatment well. The cells were incubated for four hours at 37°C and 5% CO2, and then the absorbance was recorded at 490 nm using Biotek’s Synergy Neo2 multi-mode plate reader (Biotek, Winooski, VT, USA). The average absorbance value of background control wells was subtracted from absorbance value of each vehicle-control and treatment wells and percent cell viability [(At/A0) × 100)] was determined in each treatment well, where At is the absorbance value of treatment well and A0 is the average absorbance value of control wells after background subtraction. The data were expressed as average % cell viability and fitted in non-linear regression curves using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA).
Platelet viability assays
Human platelet rich plasma (PRP) was purchased from Zenbio (Cat. No. SER-PRP-SDS, Research Triangle Park, NC, USA). PRP was used for experiments immediately after delivery. PRP was transferred into 50 mL polypropylene tubes each containing 5 mL acid citrate buffer (Cat. No. sc-214744, Santa Cruz Biotechnology, Dallas, TX, USA). To prevent clotting, prostaglandin E1 (PGE1, Cat. No. sc-201223A, Santa Cruz Biotechnology) and apyrase (Cat. No. A6237, Sigma-Aldrich) were added to final concentrations of 1 μM and 0.2 units/mL, respectively. After gently mixing the solution, platelets were pelleted by centrifugation at 1200 g for 10 min. Pelleted platelets were gently washed without disrupting them in 2 mL HEPES-buffered Tyrode’s solution (10 mM HEPES, 135 mM NaCl, 2.8 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 12 mM NaHCO3, 0.4 mM NaH2PO4, 0.25% BSA and 5.5 mM glucose, pH 7.4; Cat. No. PY-921WB, Boston BioProducts) containing 1 μM PGE1 and 0.2 units/mL apyrase. After washing, pellets were slowly resuspended in 10 mL HEPES-buffered Tyrode’s solution containing 1 μM PGE1 and 0.2 units/mL apyrase. The number of platelets was counted using a HEMAVET 950FS hematology analyzer (Drew Scientific, Inc., Miami Lakes, FL, USA). For viability assays, platelet number was adjusted to 2 × 108/mL in HEPES-buffered Tyrode’s solution containing 1 μM PGE1, 0.2 units/mL apyrase and 10% FBS. Each treatment was given in 2 mL platelet suspension in 15 mL polypropylene tubes for 72 h. The tubes were placed on a rotating platform at room temperature for the duration of treatment. After 72 h treatment, 200 μL of untreated or treated platelets were plated in each well of 96-well plates and the viability was measured by MTS assay as described in the previous method.
Determination of EC50 and combination index
Dose-response curves (Variable slope-four parameters) were generated for each test compound and their half maximal effective concentration (EC50) values were determined using GraphPad Prism 7. The combination indices (CIs) were determined using Compusyn version 1.0 software (http://www.combosyn.com). CI<0.3 was considered as strong synergistic effect induced by two compounds when they were treated in combination. The average of CI at EC75 (Concentration with 75% loss of cell viability) and EC90 (concertation with 90% loss of cell viability) are presented in Fig. 5 and Supplementary Table 5 as a more accurate measure of synergy by different combinations.
Caspase-3 activity assay
The caspase-3 activity in untreated and treated cells was measured using EnzChek caspase-3 assay kit #2, Z-DEVD-R110 substrate (Cat. No. E13184, Thermo Fisher Scientific) following manufactures’ instructions. Briefly, MOLT-4 cells were seeded in 60 mm dishes (2.5 × 106 cells in 5 mL complete cell culture medium/dish), and then treated with indicated concentrations of DT2216 or ABT263 for 24 h. Cells were lysed in 1X cell lysis buffer by subjecting them to a freeze-thaw cycle in ice-ethanol bath or by 30 min incubation on ice. After centrifugation at 5000 rpm/5 min, 50 μL of supernatant from each sample was transferred into duplicate microplate wells and 50 μL of 2X substrate solution was added to each well. 50 μL of lysis buffer + 50 μL of 2X substrate solution was added to background wells. After incubation for 30 min at room temperature, fluorescence was measured (excitation/emission ~ 496/520 nm) using a Biotek’s Synergy Neo2 multi-mode plate reader. The data were expressed as percent caspase-3 activity [(Ft/F0) × 100)], where Ft is the fluorescence value of treatment well and F0 is the average fluorescence value of control wells after background subtraction.
Bak and Bax double knockout by CRISPR/Cas9 genomic editing
To deplete Bak and Bax, the sgRNAs targeting human Bak and Bax were designed and cloned into lentiCRISPR v2 vector (a gift from Feng Zhang; Addgene plasmid # 52961). Packaging 293T cells were transfected with Bak, Bax sgRNAs or negative control (non-targeting sgRNA-sgCTRL)59 and helper vectors (pMD2.G and psPAX2; Addgene plasmid #s 12259 and 12260) using Lipofectamine 2000 reagent (Cat# 11668019, Life Technologies). Medium containing lentiviral particles and 8 μg/mL polybrene (Sigma-Aldrich) was used to infect H146 cells. Infected cells were selected in medium containing 1 μg/mL puromycin. The target guide sequences are as follows: Bak-sg1: forward (5’-CACCGTCATCGGGGACGACATCAAC-3’) and reverse (5’-AAACGTTGATGTCGTCCCCGATGAC-3’); Bak-sg2: forward (5’-CACCGCTGCAACCTAGCAGGTGAGC-3’) and reverse (5’- AAACGCTCACCTGCTAGGTTGCAGC-3’); Bax-sg1: forward (5’-CACCGGGATCGAGCAGGGCGAATGG-3’) and reverse (5’-AAACCCATTCGCCCTGCTCGATCCC-3’); Bax-sg2: forward (5’-CACCGAGCGAGTGTCTCAAGCGCAT-3’) and reverse (5’- AAACATGCGCTTGAGACACTCGCTC-3’).
Immunoprecipitation
Human cancer cells or platelets were lysed in Pierce IP lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol; Cat. No. 87787; Thermo Fisher Scientific) supplemented with 1% (v/v) protease and phosphatase inhibitor cocktail (Cat. No. PPC1010; Sigma-Aldrich). Lysates were precleared by incubating with 1 μg of mouse IgG (Cat. No. sc-2025; Santa Cruz Biotechnology) and 20 μL of protein A/G-PLUS agarose beads (25% v/v; Cat. No. sc-2003; Santa Cruz Biotechnology) for 30 min at 4°C. 1000 μg of cell lysates were incubated with 2 μg of anti-BCL-XL (Cat. No. sc-56021; Santa Cruz Biotechnology) or anti-IgG for 1 h followed by addition of 25 μL of protein A/G-PLUS agarose beads and incubated overnight at 4°C. Immunoprecipitates were collected by centrifugation and washed 2–3 times with IP lysis buffer. Samples were mixed with Laemmli’s SDS-buffer, denatured and analyzed by SDS-PAGE and immunoblotting. An anti-rabbit HRP-conjugated Fc fragment specific secondary antibody directed against primary antibodies was used to detect immune complexes in immunoblotting.
AlphaScreen for the determination of DT2216-BCL-XL, BCL-2 and BCL-W binding affinity
To evaluate the binding affinities of DT2216 and ABT263 towards BCL-XL, BCL-2 and BCL-W the AlphaScreen competitive binding assays were performed. The assay was performed at room temperature and reagents were diluted in a buffer containing 250 mM HEPES pH 7.5, 1 M NaCl, 1% BSA, and 0.05% Tween-20. Purified recombinant His-tagged BCL-XL (0.1 nM, Cat. No. SRP0187, Sigma-Aldrich), BCL-2 (0.2 nM, Cat. No. SRP0186, Sigma-Aldrich) or BCL-W (0.2 nM, Cat. No. B1059, Sigma-Aldrich) were incubated with increasing concentrations of DT2216 or ABT263 and 15 nM biotin-tagged BAD (Biotin-LWAAQRYGRELRRMSDEFEGSFKGL N-terminal, AnaSpec, Fremont, CA) or 30 nM BIM peptides (Biotin-MRPEIWIAQELRRIGDEFNA N-terminal, AnaSpec) to a final volume of 40 μL in 96-well PCR plate. BAD peptide was used to assess the binding affinity of compounds towards BCL-XL whereas BIM peptide was used for BCL-2 and BCL-W. After 24 h incubation, 5 μL 6X His-acceptor beads (Cat. No. AL128M, Perkin Elmer, Houston, TX, USA) were added to each well at 20 μg/mL final concentration and incubated for 1 h. Thereafter, 5 μL streptavidin-donor beads were added (Cat. No. 6760002, Perkin Elmer) to each well at 20 μg/mL final concentration and incubated for 30 min. At the end of the incubation period, 17 μL of each sample was transferred in adjacent wells of 384-well proxy plate (Cat. No. 6008280, Perkin Elmer). The plate was scanned using Alpha program on Biotek’s Synergy Neo2 multi-mode plate reader. The inhibition constant (Ki) was calculated using non-linear regression, one site, competitive binding, Fit Ki function on GraphPad Prism 7 software based on experimentally determined Kd for each protein/peptide pair.
Proteomics analysis
Sample preparation and LC-MS/MS analysis
To label cells with stable isotopic amino acids (SILAC), WI-38 cells were propagated in DMEM SILAC media deficient in both L-lysine and L-arginine (Cat. No. 88364, Thermo Fisher Scientific) and supplemented with light lysine (12C614N2-K) and arginine (12C614 N4-R) for light state (Cat. No. 89987 and # 89989; Thermo Fisher Scientific), and 13C615N2-K and 13C615N4-R for heavy state labeling (Cat. No. 88209 and 89990, Thermo Fisher Scientific). Cells were cultured for at least six doubling times for complete incorporation. The light-labeled WI-38 cells were untreated (DMSO) and the heavy-labeled WI-38 cells were treated with 1 μM DT2216 or DT2216NC for 6 h, respectively. Reverse labeling was used in the second biological replicate. The untreated and DT2216 or DT2216NC-treated cells were harvested by centrifugation at 500 g for 5 min. Pellets were washed twice by resuspending in 1 mL of ice cold PBS. The cell pellets were resuspended in 20 mL freshly prepared lysis buffer (2% SDS, 100 mM Tris/HCl pH 7.6) containing MS-SAFE Protease and Phosphatase Inhibitor (Cat. No. MSSAFE-5VL, Sigma-Aldrich) for sonication. The lysate was centrifuged at 15,000 g for 10 min at 18 °C. The supernatant was stored at −80 °C for proteomic analysis. Protein concentration was measured by BCA assay (Cat. No. 23227, Thermo Fisher Scientific) and SILAC pairs were mixed in equimolar amounts. Purified proteins were reduced, alkylated, and digested using filter-aided sample preparation60. Tryptic peptides were separated into 36 fractions on a 100 × 1.0 mm Acquity BEH C18 column (Cat. No. 186002350, Waters, Milford, MA, USA) using an UltiMate 3000 UHPLC system (Thermo Fisher Scientific) with a 40 min gradient from 99:1 to 60:40 buffer A (0.1% formic acid, 0.5% acetonitrile):B (0.1% formic acid, 99.9% acetonitrile) ratio under basic pH conditions, and then consolidated into 12 super-fractions. Each super-fraction was then further separated by reverse phase XSelect CSH C18 2.5 μm resin (Cat. No. 186006109, Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Peptides were eluted using a 60 min gradient from 97:3 to 60:40 buffer A:B ratio. Eluted peptides were ionized by electrospray (2.15 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) in top-speed data-dependent mode. MS data were acquired using the FTMS analyzer in profile mode at a resolution of 240,000 over a range of 375 to 1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0.
Raw data processing
Proteins were identified and SILAC ratios determined using MaxQuant with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da. The derived peak list was searched with the built-in Andromeda search engine against the reference human proteome downloaded from Uniprot (http://www.uniprot.org/) on 03–13-2018. The search parameters for both algorithms included carbamidomethylation of cysteine residues as a fixed modification and N-terminal acetylation, oxidation at methionine, and SILAC labeling 13C615N2-K and 13C615N4-R as variable modifications. Trypsin was specified as the protease and a maximum of two missed cleavages were allowed. The data were screened against a target decoy database and the false discovery rate (FDR) was set to 1% at the peptide level and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm61. Statistical analysis was performed using Perseus software62. A threshold with SILAC ratio greater than 1.5-fold and q-value of 0.05 for the Benjamini-Hochberg false discovery rate were used to identify the proteins with significant changes. The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository63 with the dataset identifier PXD010878.
Cellular thermal shift assay (CETSA)
CETSA assay was adapted from Smith et al48. Briefly, 2.5 × 107 MOLT-4 or RS4 cells were treated with 1 μM DT2216 or DMSO for 6 h, then harvested, washed with PBS, and resuspended in PBS containing protease and phosphatase inhibitors. For MOLT-4 cells, 10 μM MG132 was added in to prevent the degradation of BCL-XL. The resuspended cells were freeze–thawed four times with liquid nitrogen. After each freeze–thaw cycle, lysate was vortexed briefly to ensure homogenous thawing. The soluble fraction was separated from cell debris by centrifugation at 20,000 ×g for 30 min at 4 °C and then heated at gradient temperature from 42 °C to 69.5 °C for 3 min and cooled down to 25 °C for another 3 min. The treated samples were centrifugated at 20,000 ×g for 30 min at 4 °C to remove the denatured proteins and then analyzed by immunoblotting.
AlphaLISA assay for ternary complex formation
We used AlphaLISA assay to monitor ternary complex formation between target protein, PROTAC and E3 ligase, as described previously44, 58. To validate the formation of the ternary complex of BCL-XL/BCL-2, DT2216 and the VHL E3 ligase, a fixed concentration of His-tagged recombinant proteins (100 nM BCL-XL and10 nM BCL-2) and recombinant active GST-tagged VHL/Elongin B/C complex (50 nM for BCL-XL, 5 nM for BCL-2; Cat. No. 029641, US Biological, Salem, MA, USA) was incubated with varying concentrations of test compounds in 4-fold serial dilutions to a final volume of 40 μL in 96-well PCR plate. After 30 min incubation at room temperature, 5 μL Alpha Glutathione-donor beads (Cat. No. 6765300, Perkin Elmer) were added to each well at 20 μg/mL final concentration and incubated for 15 min. Thereafter, 5 μL 6X His-acceptor beads were added to each well at 20 μg/mL final concentration and incubated for an additional 45 min at room temperature. Thereafter, 17 μL of each sample was transferred in adjacent wells of 384-well proxy plate and the plate was scanned using Alpha program on Biotek’s Synergy Neo2 multi-mode plate reader. The data were expressed as average AlphaLISA signal and plotted against different concentrations of compounds.
nanoBRET ternary complex formation assay
Plasmids were purified on miniprep columns according to the manufacturer’s protocol (Cat. No. 27106 Qiagen, Germantown, MD, USA). Plasmids HaloTag-VHL (Cat. No. CS1679C155) and CMV LgBit (Cat. No. CS1956B03) were purchased from Promega. HiBit-BCL-XL (pDL2288) and HiBit-BCL-2 (pDL2306) were constructed based on pBiT3.1 N-HiBiT (Cat. No. N2361, Promega), pLX304-BCL-XL (DNASU plasmid Cat. No. HsCD00437924, Tempe, AZ, USA) and Flag-BCL-2 (a gift from Clark Distelhorst, Addgene plasmid #18003, Watertown, MA, USA) plasmids through Gibson assembly method (Gibson Assembly Master Mix, Cat. No. E2611S, NEB, Ipswich, MA, USA) using the primers as follow. pBiT3.1-N-Bcl-XL-FP1: TGGCTCGAGCGGTGGGAATTCTGGTATGTCTCAGAGCAACCGGGAGCTGGTG; pBiT3.1-N-Bcl- XL-RP1: TCTTCCGCTAGCTCCACCGGATCCTCCTCATTTCCGACTGAAGAGTGAGCC; pBiT3.1-N-Bcl- XL-Vector-FP1: GGCTCACTCTTCAGTCGGAAATGAGGAGGATCCGGTGGAGCTAGCGGAAGA; pBiT3.1-N-Bcl- XL-Vector-RP1: CACCAGCTCCCGGTTGCTCTGAGACATACCAGAATTCCCACCGCTCGAGCCA; pBiT3.1-N-Bcl-2-FP1: TGGCTCGAGCGGTGGGAATTCTGGTATGGCGCACGCTGGGAGAACAGGGTAC; pBiT3.1-N-Bcl-2-RP1: TCTTCCGCTAGCTCCACCGGATCCTCCTCACTTGTGGCCCAGATAGGCACCCAG; pBiT3.1-N-Bcl-2-Vector-FP1: CTGGGTGCCTATCTGGGCCACAAGTGAGGAGGATCCGGTGGAGCTAGCGGAAGA; pBiT3.1-N-Bcl-2-Vector-RP1: GTACCCTGTTCTCCCAGCGTGCGCCATACCAGAATTCCCACCGCTCGAGCCA. DNA sequences in these plasmids were authenticated by automatic sequencing. 293T cells (8 ×105) were transfected with Lipofectamine 2000 reagent (Cat # 11668019, Thermo Fisher Scientific) and 1 μg HaloTag-VHL, 10 ng HiBit-BCL-XL and 10 ng LgBit or 1 μg HaloTag-VHL, 10 ng HiBit-BCL-2 and 10 ng LgBit. After 24 h, 2 × 104 transfected cells were seeded into white 96-well tissue culture plates in FluoroBrite™ DMEM (Cat. No. A18967–02, Thermo Fisher Scientific) containing 4% FBS with or without HaloTag NanoBRET 618 Ligand (Cat. No. PRN1662, Promega) and incubated overnight at 37 °C, 5% CO2. The following day, different doses of DT2216 were added into the medium and plates were incubated at 37 °C, 5% CO2, for 6 h. After treatment, NanoBRET™ Nano-Glo® Substrate (Cat. No. N1662, Promega) were added into the medium and the contents were mixed by shaking the plate for 30 s before measuring donor and acceptor signals on Biotek plate reader. Dual filtered luminescence was collected with a 450/50 nm bandpass filter (donor, NanoBiT-BCL-XL protein or NanoBiT-BCL-2 protein) and a 610 nm long pass filter (acceptor, HaloTag NanoBRET ligand) using an integration time of 0.5 s. mBRET ratios were calculated following the NanoBRET™ Nano-Glo® Detection System (Cat. No. N1662, Promega).
Co-immunoprecipitation assay for the determination of BCL-XL and BCL-2 ubiquitination
Plasmid pSG5-Flag-BCL-XL (pDL2009) was constructed based on pSG5-Flag vector (a gift from Diane Hayward in Johns Hopkins University, which originally generated by Stratagene) and pLX304-BCL-XL (DNASU plasmid Cat. No. HsCD00437924, Tempe, AZ, USA) through Gibson assembly method and the following two primers sets: primer set-1, forward (5’-GACTACAAGGACGACGATGACAAGGGATCTATGTCTCAGAGCAACCGGGAGCTGGTG-3’) and reverse (5’-GTTCTGCTTTAATAAGATCTGGATCTTCATTTCCGACTGAAGAGTGAGCCCAGCAGAAC-3’); primer set-2, forward (5’-GTTCTGCTGGGCTCACTCTTCAGTCGGAAATGAAGATCCAGATCTTATTAAAGCAGAAC-3’) and reverse (5’-CACCAGCTCCCGGTTGCTCTGAGACATAGATCCCTTGTCATCGTCGTCCTTGTAGTC-3’). DNA sequence of the plasmid was authenticated by automatic sequencing. 293T cells were co-transfected with Flag-BCL-XL and HA-Ub (a gift from Ted Dawson, Addgene, Plasmid #17608, Watertown, MA, USA) or Flag-BCL-2 (a gift from Clark Distelhorst, Addgene plasmid #18003, Watertown, MA, USA) and HA-Ub for 40 h and then treated with DMSO or 1 μM DT2216 for 4 h, and 10 μM MG132 was added to prevent protein degradation. Proteins were extracted by using IP lysis buffer (Cat. No. 87788, Thermal Fisher Scientific) and then subjected to immunoprecipitation using Anti-FLAG M2 Magnetic Beads (Cat. No. M8823, Sigma-Aldrich) according to the manufacturer’s protocol. Anti-FLAG M2 Magnetic Beads was washed with 1X TBS three times and then added to protein samples, and the mixture was incubated at 4 °C with rotation overnight. The magnetic beads were collected and then washed three times with 1X TBS. Immunoprecipitated samples were eluted with 2X SDS sample buffer and boiled 5 min at 95 °C.
Mutational analysis
Flag-BCL-XL-K87R, Flag-BCL-XL-K87H, Flag-BCL-XL-K157R, Flag-BCL-XL-K-ko, and Flag-BCL-XL-K87-only were constructed based on pSG5-Flag-BCL-XL (pDL2009) mentioned above by using QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) and the following primer sets: BCL-XL-K87R forward (5’-GTGATCCCCATGGCAGCAGTAAGGCAAGCGCTGAGGGAGGCAGGC-3’) and reverse (5’-GCCTGCCTCCCTCAGCGCTTGCCTTACTGCTGCCATGGGGATCAC-3’); BCL-XL-K87H forward (5’-CCCATGGCAGCAGTACATCAAGCGCTGAGGGAG-3’) and reverse (5’-CTCCCTCAGCGCTTGATGTACTGCTGCCATGGG-3’); BCL-XL-K157R forward (5’-TGTGCGTGGAAAGCGTAGACAGGGAGATGCAGG-3’) and reverse (5’-CCTGCATCTCCCTGTCTACGCTTTCCACGCACA-3’). BCL-XL-K-ko was constructed by mutating each of the six K (K16, K20, K87, K157, K205, and K233) to R by the primers mentioned above and the following primer sets: BCL-XL-K16R & K20R forward (5’-GTGGTTGACTTTCTCTCCTACAGGCTTTCCCAGAGAGGATACAGCTGGAGTCAGT-3’) and reverse (5’-ACTGACTCCAGCTGTATCCTCTCTGGGAAAGCCTGTAGGAGAGAAAGTCAACCAC-3’); BCL-XL-K205R forward (5’-CAATGCAGCAGCCGAGAGCCGAAGGGGCCAGGAACGCTTCAACCGC-3’) and reverse (5’-GCGGTTGAAGCGTTCCTGGCCCCTTCGGCTCTCGGCTGCTGCATTG-3’); BCL-XL-K233R forward (5’-CTGGGCTCACTCTTCAGTCGGAGATGAAGATCCAGATCTTATTAA-3’) and reverse (5’-TTAATAAGATCTGGATCTTCATcTCCGACTGAAGAGTGAGCCCAG-3’). BCL-XL-K87-only was constructed by mutating each of the five K (K16, K20, K157, K205, and K233) besides K87 to R by the primers mentioned above. DNA sequences in all these plasmids were authenticated by automatic sequencing.
Identification of ubiquitination site in BCL-XL
Enrichment of FLAG-tagged protein for mass spectrometry
293T cells were co-transfected with Flag-BCL-XL (pDL2009) and HA-Ub vectors, or Flag-BCL-2 and HA-Ub vectors. Proteins were extracted by using IP lysis buffer and then subjected to immunoprecipitation using Pierce™ Anti-DYKDDDDK Affinity Resin (Cat. No. A36801, Thermal Fisher Scientific) according to the manufacturer’s protocol. Anti-DYKDDDDK Affinity Resin was washed with 1X TBS three times and then added to protein samples, and the mixture was incubated at 4 °C with rotation for 4 h. The immunoprecipitated samples were centrifuged at 1000×g for 1 min at 4 °C, and then washed three times with 1X TBS. Immunoprecipitated samples were eluted with 0.1 M glycine HCl (pH 2.8).
Sample preparation and LC-MS/MS
Purified proteins were reduced, alkylated, and digested using filter-aided sample preparation60. Resulting peptides were then separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Peptides were eluted using a 90 min gradient from 97:3 to 60:40 buffer A:B ratio A (buffer A: 0.1% formic acid and 0.5% acetonitrile in water, buffer B: 0.1% formic acid in acetonitrile). Eluted peptides were ionized by electrospray (2.15 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) in top-speed data-dependent mode. MS data were acquired using the FTMS analyzer in profile mode at a resolution of 240,000 over a range of 375 to 1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0.
Proteomics data analysis
Proteins were identified by database search against human Uniprot database (73911 proteins, March 16, 2019) using Mascot with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da. Methionine oxidation (+15.99492 Da), protein N-terminal acetylation (+42.03670) and lysine ubiquitination (+114.04293 Da) were variable modifications; cysteine was assigned a fixed carbamidomethyl modification (+57.021465 Da). Percolator was used to filter the peptide spectrum matches to a false discovery rate of 1%. Scaffold (Proteome Software, Portland, Oregon, USA) was used to verify MS/MS based peptide and protein identifications. Protein identifications were accepted if they could be established with less than 1.0% false discovery and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm61. The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository61 with the dataset identifier PXD015454.
Mice
Female CB17/Icr-Prkdcscid/IcrIcoCrl (CB-17 SCID), CB17.Cg-PrkdcscidLystbg-J/Crl (CB-17 SCID-beige) or NOD.CB17-Prkdcscid/NCrCrl (NOD-SCID) mice aged 5–6 weeks were purchased from Charles River Laboratories (Wilmington, MA, USA), and were housed in the AAALAC-accredited animal facilities at University of Arkansas for Medical Sciences (UAMS) or University of Florida (UF) under pathogen free conditions. Female NOD-SCID IL2Rgnull (NSG) mice aged 6–7 weeks were purchased from Jackson Laboratory (Bar Harbor, Main, USA) and were housed in the AAALAC-accredited animal facilities at MD Anderson Cancer Center (MDACC) under pathogen free conditions. Mice received food and water ad libitum and wereallowed to acclimatize for 1–2 weeks before being used for experiments at the age of 6–8 weeks. All animal work was approved and done in accordance with the approvals from the Institutional Animal Care and Use Committees of UAMS, UF, and MDACC with the exception of the pharmacokinetic (PK) studies that were done by BioDuro (San Diego, CA, USA), a global contract research organization, through a contract. All animal studies were complied with the ethical regulations and humane endpoint criteria according to the NIH Guidelines for the Care and Use of Laboratory Animals.
Analysis of DT2216 in Tumor Samples
A bioanalytical method was developed for the quantification of DT2216 in tumor homogenates using a Waters Xevo TQ-S Micro triple quadrupole mass spectrometer and Acquity Class I UPLC (Milford, MA, USA). Chromatographic separation was achieved using a mobile phase consisting of aqueous ammonium acetate buffer (10 mM, pH 3.5) (pump A) and acetonitrile (pump B) on a Waters Acquity BEH C18 column (1.7 μm, 2.1 × 50 mm). A gradient method started with pump B supplying 15% of acetonitrile up to 0.5 min. The composition of acetonitrile was linearly increased to 80% until 1.5 min and kept constant until 3.0 min. The composition of acetonitrile was immediately decreased to 15% at 3.0 min and kept constant until 3.5 min. The flow rate of the mobile phase was 0.35 mL/min and the injection volume was 2 μL. Analysis of DT2216 was performed using electro spray ionization (ESI) in the positive mode. Capillary voltage, cone gas, desolvation gas, desolvation temperature and source temperature were optimized for maximum analyte response and held at 3.0 kV, 50 L/h, 900 L/h, 450°C and 450°C, respectively. The mass spectral analysis of DT2216 and internal standard (amiodarone) was achieved by multiple reaction monitoring (MRM). Compound parameters are detailed in Supplementary Table 8. Data acquisition and quantitation were performed using MassLynx and TargetLynx XS software version 4.2. For the bioanalysis of DT2216, pieces of tumor samples were weighted and homogenized in water (1:4, %w/v) using a tissue homogenizer (Dremel, BioSpec Products, Inc., Bartlesville, OK, USA). Sample cleanup (20 μL) was performed by a protein precipitation method using acetonitrile containing the internal standard (40 ng/mL) as a quenching solution (80 μL). Calibration (5, 10, 50, 100, 200, 400 and 500 ng/mL) and quality control standards (5, 7.5, 250 and 450 ng/mL) were prepared in drug-free tumor homogenate and analyzed using the UPLC-MS/MS method. The method was linear for a calibration range of 5–500 ng/mL. The accuracy and precision calculated for the method were within the specified limits64 Test samples were analyzed along with freshly prepared calibration and quality control standards (Extended Data Fig. 5d). Tumor concentration-time data was subjected to non-compartmental analysis and pharmacokinetic parameters were calculated using Phoenix WinNonlin, version 6.1.
Pharmacodynamics of BCL-XL degradation by DT2216
MOLT-4 xenografts were established in female CB-17 SCID-beige mice as described in following methods. Tumor-bearing mice were treated with single injection of vehicle or DT2216 (15 mpk/i.p.) when the tumors were ~600 mm3. Two mice each from vehicle and DT2216-treated groups were euthanized at each time point as indicated in the figure legend of Fig. 4b and tumors were harvested. The proteins were extracted from tumors and used for BCL-XL degradation by immunoblot analysis. Some portion of these tumors were used for DT2216 analysis as described in the previous method.
Protein extraction from tumors for immunoblotting
Following resection, the tumors were snap frozen and stored at −80 °C until later use. Tumor tissue (~50 mg wet tissue) was lysed in 1 mL of RIPA lysis buffer supplemented with 1% protease inhibitor cocktail and 1% phosphatase inhibitor cocktail. The tissues were kept on ice and were homogenized with the help of a hand homogenizer. The samples were incubated on ice for 2–3 h and then kept at −80 °C overnight to allow for complete lysis. The subsequent sample preparation and immunoblotting were performed as described in the immunoblotting method.
In vivo platelet toxicity assays
Single dosing with DT2216 or ABT263
Female 5–6 weeks old CB-17 SCID-beige mice were treated with single i.p. doses of DT2216 (7.5, 15 and 25 mpk) or single p.o. doses of ABT263 (25, 50 and 100 mpk). Approximately 50 μL of blood was collected at different time points from each mouse as mentioned in Fig. 4c legend via submandibular plexus route in EDTA-tubes (RAM Scientific, Inc. Nashville, TN, USA) and platelets were enumerated using an automated hematology analyzer HEMAVET 950FS (Drew Scientific, Miami Lakes, FL, USA).
Daily dosing with ABT263 or once a week DT2216
Female CB-17 SCID-beige mice were treated with ABT263 (50 mpk/day/p.o.) or DT2216 (15 mpk/week/i.p.). Approximately 50 μL of blood was collected from each mouse 6 h (0.25 d) after each dose of ABT263 or collected from each mouse after 0.25 d, 1.25 d, 2.25 d, 3.25d, 4.25 d, 5.25 d, 6.25 d, 7.25 d, 8.25 d, 9.25 d, 10.25 d, 11.25 d, 12.25 d, 17.25 d and 24.25 d of once a week dosing with DT2216. Platelets were enumerated using HEMAVET 950FS.
Complete blood cell counts
Approximately 50 μL of blood was collected from each mouse in EDTA tubes via submandibular plexus or retro-orbital bleeding. The mice were anaesthetized with isoflurane inhalation for retro-orbital bleeding. The blood was immediately used for complete blood counts (CBCs) using an automated hematology analyzer HEMAVET 950FS. The data were expressed as number of different blood cells or platelets per μL of blood.
MOLT-4 T-ALL xenograft mouse model
To test the effect of DT2216 on tumor growth in MOLT-4 T-ALL xenografts, MOLT-4 T-ALL cells were harvested and suspended in regular RPMI medium and mixed with Matrigel (1:1) (Cat. No. 356231, Corning, Corning, NY, USA). The cells (5 × 106 cells) suspended in 100 μL of RPMI medium-Matrigel mixture were subcutaneously (s.c.) implanted in the right flank of CB-17 SCID mice. Tumor growth was monitored daily and tumors were measured twice a week using Vernier caliper or digital calipers. Tumor volume was determined using the formula; [(L × W2) × 0.5], where L is length/long dimension in millimeter (mm) and W is the width/short dimension in mm. The treatment started once the average tumor volume reached 150–200 mm3. The animals were randomly assigned into separate groups (n = 6–8) in a way that each group had nearly equal starting average tumor volume. Mice were weighed twice a week and the treatments were given according to average mouse weight within each group before initiation of treatment. DT2216 and ABT263 for i.p. administration were formulated in 50% PHOSAL 50 PG, 45% MIGLYOL® 810 N and 5% Polysorbate 80. DT2216 and ABT263 were administered via i.p. injection at 15 mpk/week in 100 μL vehicle (Extended Data Fig. 8). ABT263 for oral administration was formulated in 10% ethanol, 30% PEG 400 and 60% PHOSAL 50 PG (Fig. 4). Control mice received 100 μL vehicle via i.p. injection. The mice were euthanized when the maximum tumor size in a mouse reached the humane endpoint according to institutional policy concerning tumor endpoints in rodents. In addition, to prevent excessive pain or distress, the mice were euthanized if the tumors became ulcerated or the mice showed any signs of ill health. Mice were euthanized by CO2 suffocation followed by cervical dislocation and various tissues including tumors were harvested for further analyses.
H146 SCLC xenograft model
To test the effect of DT2216 alone or in combination with ABT199 on tumor growth in H146 SCLC xenografts, 5 × 106 H146 SCLC cells were suspended in regular RPMI medium, mixed with Matrigel, and s.c. implanted in the right flank of female CB-17 SCID mice as described in the previous method. Tumor growth was monitored, tumors were measured, and the tumor volume was determined as mentioned in the above method. The treatment started after four weeks of cell implantation as shown in Fig. 5c. The animals were randomly assigned into groups treated with vehicle, DT2216 15 mpk/week (i.p. injection), ABT263 15 mpk/week (i.p. injection), ABT199 50 mpk/day (oral administration), and DT2216 + ABT199. DT2216 and ABT263 were formulated as described above. ABT199 was formulated for oral dosing in 60% PHOSAL 50 PG, 30% polyethylene glycol (PEG) 400 and 10% ethanol. All the animals were euthanized in accordance with the institutional policy and various tissues were harvested. Tumors were weighed and used for BCL-XL, BCL-2 and MCL-1 expression by immunoblotting.
MDA-MB-231 BC xenograft model
To test the effect of DT2216 in combination with Docetaxel on tumor growth in MDA-MB-231 breast cancer xenografts, 5 × 106 MDA-MB-231 cells were suspended in regular RPMI medium, mixed with Matrigel, and s.c. implanted in the right flank of female NOD-SCID mice. The animals were randomly assigned to vehicle, DT2216 (15 mpk/q4d/i.p.), Docetaxel (7.5 mpk/q14d/i.v.) and DT2216 + Docetaxel groups when the average tumor volume reached ~ 130 mm3. Docetaxel was dissolved in 5% DMSO + 30% PEG 300 + 5% Tween 80 + 60% dH2O. The solution was filter sterilized to obtain clear solution for intravenous administration. DT2216 was administered two days before starting dosing with Docetaxel, as also mentioned in Fig. 6c.
T-ALL PDX models
CUL76 and 332X-Luci T-ALL PDX models were kind gifts from Dr. Adolfo Ferrando (Columbia University, New York, NY, USA) and Dr. Kim Yong-Mi (Children’s Hospital Los Angeles, Los Angeles, CA), respectively. D115 T-ALL PDX model was from Dr. Marina Konopleva’s Lab (UTMDACC, Houston, TX). The use of these models was in accord with the Declaration of Helsinki using an IRB-approved protocol from MD Anderson Cancer Center. To establish T-ALL PDX mouse models, 8 weeks old NSG mice were sublethally irradiated (0.25 Gy) 24 h prior to cell inoculation. PDX cells (1 × 106) were suspended in PBS and injected into mice through the tail vein. Tumor engraftment was determined by co-staining for human and murine anti-CD45 (BioLegend, San Diego, CA) in bone marrow aspiration samples (CUL76 and D115 T-ALL) or in peripheral blood (332X-Luci) at 10 days post injection. Mice were randomized into different groups. CUL76 PDX were treated with DT2216 (15 mpk/q4d/i.p.), ABT199 (100 mpk/day/p.o.), chemotherapy (vincristine 0.15 mpk + dexamethasone 5 mpk + L-asparaginase 1000 Units/kg, i.p., weekly), combination of DT2216 and ABT199 or chemotherapy, or ABT199 plus chemotherapy as shown in Fig. 6f. D115 and 332X-Luci PDXs were treated with DT2216, chemotherapy, or combination of DT2216 and chemotherapy as shown in Extended Data Fig. 10. Mice were monitored for disease progression by weekly assessing leukemic (hCD45+) cells in the peripheral blood or bone marrow aspirates and followed for survival.
Statistical analyses
All the graphs presented in this manuscript were made and statistical analyses were performed using GraphPad Prism 7 software. For analysis of means of three or more groups, analysis of variance (ANOVA) tests were performed. In the event that ANOVA justified post hoc comparisons between group means, the comparisons were conducted using Tukey’s multiple-comparisons test. Two-sided unpaired Student’s t-test was used for comparisons between the means of two groups. Kaplan-Meier test was used for the survival rate in Fig. 6h and Extended Data Fig. 10b and d, and the data were statistically analyzed using Log-rank (Mantel-Cox) test. P < 0.05 was considered to be statistically significant. The precise P values have been provided wherever possible and appropriate.
Life sciences reporting summary
Further information on experimental design and reagents is provided in the life sciences reporting summary of this article.
Data availability
The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository with the dataset identifiers PXD010878 and PXD015454, as mentioned in the methods also. The raw immunoblot images and statistical analyses files are supplied as source data. Other raw and analyzed data files are available from the corresponding author upon reasonable request. A data availability statement is also included in the reporting summary linked to this article.
Extended Data
Extended Data Fig. 1. Expression of anti-apoptotic BCL-2 members and VHL in cancer, and the synthetic schemes of DT2216 and DT2216NC.
a, A representative of three immunoblot analyses of VHL in three different human tumor cell lines and platelets from three different individuals (indicated as units 1–3). b, Immunoblot analyses of the basal protein levels of BCL-XL, BCL-2, MCL-1 and VHL in different solid tumor cells. Data are a representative of two independent experiments. c, BCL2L1 and VHL mRNA expression (Log2 transformed) and mutational status were analyzed using TCGA PanCancer Atlas studies via cBioPortal. d, Synthetic schemes of DT2216, DT2216NC and VHL-L. Reagents and conditions: (i) (1) N-methylmorpholine, isobutyl chloroformate, THF, −25 °C then −15 °C; (2) NaBH4, H2O; (ii) Bu3P, diphenyl disulfide, toluene, 80 °C; (iii) DIBAL-H, toluene, −78 °C (iv) compound 5, NaBH(OAc)3, TEA, DCM; (v) TFA, DCM; (vi) TEA, acetonitrile, reflux; (vii) compound 10, EDCI, DMAP, DCM; (viii) Zn, HOAc, THF; (ix) HATU, TEA, DCM; (x) (1) LiOH monohydrate, MeOH, H2O; (2) compound 12, HATU, TEA, DCM; (xi) Ac2O, TEA, DCM.
Extended Data Fig. 2. The BCL-XL degradation by DT2216 is rapid and long lasting.
a, Immunoblot analysis of BCL-XL expression in MOLT-4 cells after they were treated with DT2216 for various durations as indicated. A representative immunoblot is presented on the top panel. Densitometric analysis of BCL-XL expression is presented on the bottom panel as mean of two independent experiments. Each symbol represents data (% of 0 h) from an individual experiment. b, Analysis of BCL-XL expression by immunoblot in MOLT-4 cells treated with DT2216 for 16 h followed by drug withdrawal and then cultured without DT2216 for 0 to 48 h as indicated. A representative immunoblot is on the top panel. Densitometric analysis of BCL-XL expression is presented on the bottom panel as the mean of two independent experiments. Each symbol represents data (% of Veh) from an individual experiment. c, Caspase-3 activity in MOLT-4 cells was measured 24 h after they were treated with 1 μM of DT2216 or ABT263. Data are presented as mean (n = 2 technical replicates) of a representative experiment. Each symbol represents data (% of Veh) from an individual replicate. Similar results were obtained in an additional independent experiment. d, Representative immunoblot to confirm CRISPR/Cas9-mediated double knockout of Bax and Bak in H146 cells. The experiment was repeated independently one more time with similar results.
Extended Data Fig. 3. ABT263 or VHL-L blocks the formation of ternary complex of BCL-XL, DT2216 and VHL.
a, Recombinant His-tagged BCL-XL protein (100 nM) was incubated with the VHL-Elongin B/C complex (50 nM) with 0.039 μM of DT2216. The ternary complex formation was abrogated in the presence of ABT263 (1 μM) or VHL-L (10 μM). Data are expressed as mean (n = 2 technical replicates). Each symbol represents data from an individual replicate. b, The negative-control of DT2216 (DT2216NC) cannot form ternary complex with BCL-XL and the VHL-complex. Recombinant His-tagged BCL-XL protein (100 nM) was incubated with the VHL-Elongin B/C complex (50 nM) with increasing concentrations of DT2216 or DT2216NC. Data are expressed as mean (n = 2 technical replicates).
Extended Data Fig. 4. DT2216 binds to both BCL-XL and BCL-2 but cannot degrade BCL-2.
a, The binding affinities of DT2216 and ABT263 towards BCL-XL, BCL-2 and BCL-W were measured by AlphaScreen and are represented in terms of inhibition constant (Ki). The data are average of two independent experiments each performed in duplicates. b, c, Immunoblot analyses of BCL-2 and BCL-W are shown after the RS4 cells were treated with indicated concentrations of DT2216 for 16 h (upper panels), or with 1 μM of DT2216 for indicated durations (lower panels). d, Cell viability of BCL-2-dependent RS4 cells after treatment with increasing concentrations of DT2216, ABT199 or ABT263 for 72 h. Data are presented as mean ± SD from six replicate cell cultures in one experiment. Similar results were obtained in two additional independent experiments for DT2216. The EC50 of DT2216 is the average of three independent experiments. e, Cell viability of MCL-1-dependent H929 cells after treatment with increasing concentrations of DT2216 or S63845 for 72 h. Data are presented as mean ± SD from six replicate cell cultures in one experiment. f, Schematic representation of proteomic assay shown in fig. 3d. g, Representative immunoblot analyses are shown after the cells were treated with indicated concentrations of PZ-15227 for 16 h (top panel) or with 0.1 μM of PZ-15227 for indicated time points (bottom panel). Similar results were obtained in one more independent experiment. h, CETSA assay for BCL-XL and BCL-2. MOLT-4 (for BCL-XL) or RS4 (for BCL-2) cells were treated with DMSO, or 1 μM of DT2216 for 6 h. Raw band intensities are mean of two independent experiments.
Extended Data Fig. 5. Drug metabolism and PK/PD profile of DT2216.
a, DT2216 stability in mouse blood and plasma. b, DT2216 liver microsomal stability. c, Plasma concentrations of DT2216 after a single administration of 2 mpk (i.v. injection), 20 mpk (i.p. injection) or 20 mpk (p.o. administration) are presented as Mean ± SD (n = 3 mice/group). These studies were done by BioDuro (San Diego, CA, USA), a global contract research organization, through a contract. d, MRM chromatograms of (A) DT2216 in drug-free brain homogenate, (B) Internal standard in spiked drug-free tumor homogenate (40 ng/mL), (C) DT2216 in spiked tumor homogenate (5 ng/mL; LLOQ), (D) DT2216 in tumor sample taken from vehicle dosed mouse at 24 h, (E) DT2216 in tumor sample taken at 24 h after 15 mpk/i.p. administration. e, Representative immunoblot analysis of BCL-XL in tumors at different durations after DT2216 (DT, 15 mpk/i.p.) administration (n = 2 mice in Veh and DT2216 groups at each time points). Similar results were obtained in two more immunoblot experiments. Mpk, mg/kg.
Extended Data Fig. 6. DT2216 induces BCL-XL degradation and apoptosis in MOLT-4 T-ALL xenografts and suppresses their growth in a dose-dependent manner.
a, b, T-ALL xenograft tumors were harvested two days after female CB-17 SCID mice received one i.p. injection of DT2216 at 7.5 mpk and 15 mpk. A single immunoblot analysis of BCL-XL, BCL-2, MCL-1, cleaved and full length caspase-3 and PARP1 in the tumors is shown. c, Changes in tumor volume over time after the start of treatment with vehicle (Veh), or DT2216 (7.5 or 15 mpk/q7d/i.p.). All the data presented are mean ± SEM (n = 8 mice in Veh, 8 mice in DT2216 7.5 mpk, and 7 mice in DT2216 15 mpk). The data from DT2216 15 mpk group are also presented in Extended Data Fig. 8f in which the tumor size in these mice were continuously monitored till day 55 post treatment.
Extended Data Fig. 7. Images of MOLT-4 tumor-bearing mice and excised tumors.
The images shown (quantification data are shown in Fig. 4f,g) were captured at the end of experiment when the mice were treated with Veh, DT2216 (15 mpk/q7d/i.p.) or ABT263 (50 mpk/qd/p.o.). The tumor-bearing mice (shown in the top panel) and harvested tumors (shown in the bottom panel) are not placed in an identical order.
Extended Data Fig. 8. DT2216 induces regression of MOLT-4 xenografts without causing thrombocytopenia.
a, Illustration of the experimental design of MOLT-4 T-ALL xenograft mouse model. b, Representative of two independent immunoblot analyses of BCL-XL, cleaved and full-length caspase-3 and PARP1 in MOLT-4 T-ALL xenografts harvested two days after tumor-bearing mice were treated with a single injection of vehicle (Veh) or DT2216 (15 mpk/i.p.). c, Blood platelets (PLT), white blood cells (WBC), lymphocytes (LYM) and granulocytes (GRA) were numerated one day after first treatment with vehicle (Veh), DT2216 (15 mpk/i.p.) or ABT263 (15 mpk/i.p.) as shown in a. a and b represents statistical significance vs. Veh and DT2216, respectively, determined by one-way ANOVA and Tukey’s multiple comparison test. d, Body weight changes in mice after the start of treatment with vehicle (Veh), DT2216 or ABT263 as shown in a. e, Numeration of PLT one day after the 6th dose of DT2216 (15 mpk/q7d/i.p. or 15 mpk/q4d/i.p.). Data are presented as mean ± SEM (n = 2 mice in Veh, 7 mice in DT2216 q7d, and 6 mice in DT2216 q4d). f, Changes in tumor volume over time after the start of treatment with vehicle (Veh), DT2216 or ABT263 as shown in a. Data presented in c, d and f are mean ± SEM (n = 6 mice for Veh group and 7 mice each for DT2216 and ABT263 groups). Each symbol in c and e represents data from an individual animal.
Extended Data Fig. 9. Images of excised H146 SCLC tumors.
The tumor images shown for the quantification data presented in Fig. 5g. Mice were treated as shown in Fig. 5c.
Extended Data Fig. 10. Anti-leukemic activity of DT2216 alone and in combination with chemotherapy in T-ALL PDX models.
a, 8 weeks old female NSG mice (n = 20 mice) were injected with 1 × 106 D115 cells 24 h after 0.25 Gy irradiation. Upon engraftment, mice were randomized to receive vehicle, DT2216 (15 mpk/i.p./q4d for 3 weeks), chemotherapy [Chemo (vincristine 0.15 mpk + dexamethasone 5 mpk + L-asparaginase 1000 U/kg, i.p., q7d for 3 weeks)], or the combination of DT2216 with chemotherapy. Disease burden was followed by engraftment in bone marrow by checking hCD45% in bone marrow aspiration samples through flow cytometry. Data are presented as mean ± SEM (n = 5 mice in each group). Each symbol represents data from an individual animal, and the middle horizontal line represents mean. b, Mice survival was followed and statistical significance was determined by Log-rank test (n = 5 mice in each group). c, 8 weeks old female NSG mice (n = 20 mice) were injected with 1 × 106 332X-luci cells 24 h after 0.25 Gy irradiation. Upon engraftment, mice were randomized to receive vehicle, DT2216, chemotherapy, or the combination of DT2216 with chemotherapy as mentioned in a. Disease burden was followed by checking hCD45% in peripheral blood samples (retro orbital) through flow cytometry. Data are presented as mean ± SEM (n = 5 mice in each group). Each symbol represents data from an individual animal, and the middle horizontal line represents mean. d, Mice survival was followed and statistical significance was determined by Log-rank test (n = 5 mice in each group).
Supplementary Material
Acknowledgments
This study was supported by US National Institutes of Health (NIH) grants R01 CA211963 (D.Z.), R01 CA219836 (D.Z.), R01 GM109645 (R.H.), R01 CA205224 (R.H.), R01 CA200673 (W.Z.), R01 CA203834 (W.Z.), R21 CA223371 (G.Z.), R35 CA210065 (A.F.), R01 CA172809 (Y.-M.K.), Department of Defense grant BC180227 (W.Z.), the Schwab Charitable fund (M.K.) and CPRIT grant RP160716 (P.J.H.). Mass spectrometric support was provided by Alan J. Tackett and Samuel G. Mackintosh in the University of Arkansas for Medical Sciences Proteomics Core Facility.
Footnotes
Competing interests: S.K., X.Z, Y.H., P.Z., G.Z., and D.Z. are inventors of two pending patent applications for use of BCL-XL PROTACs as senolytic and antitumor agents. R.H., G.Z., and D.Z. are co-founders of and have equity in Dialectic Therapeutics, which develops BCL-XL PROTACs to treat cancer.
References
- 1.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell. 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Letai A, & Sarosiek K Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 20, 175–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igney FH & Krammer PH Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2, 277–288 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi A, Fairbrother WJ, Leverson JD, & Souers AJ From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov. 16, 273–284 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Adams JM & Cory S The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 26, 1324–1337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed JC Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 111, 3322–3330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas S et al. Targeting the bcl-2 family for cancer therapy. Expert Opin Ther Targets. 17, 61–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opfermann JT Attacking cancer’s Achilles heel: antagonism of antiapoptotic BCL-2 family members. FEBS J. 283, 2661–2675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner TP, Lopez A, Reyna DE, Spitz AZ, & Gavathiotis E Progress in targeting the BCL-2 family of proteins. Curr Opin Chem Biol. 39, 133–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delbridge AR, Grabow S, Strasser A, & Vaux DL Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 16, 99–109 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Delbridge AR & Strasser A The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 22, 1071–1080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oltersdorf T et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 435, 677–681 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Tse C et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Research. 68, 3421–3428 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Souers AJ et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 19, 202–208 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Tao ZF et al. Discovery of a potent and selective BCL-XL inhibitor with in vivo activity. ACS Med Chem Lett. 5, 1088–1093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotschy A et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 538, 477–482 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Deeks ED Venetoclax: First Global Approval. Drugs. 76, 979–987 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Roberts AW et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 374: 311–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason KD et al. Programmed anuclear cell death delimits platelet life span. Cell. 128, 1173–1186 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Schoenwaelder SM et al. Bcl-xL–inhibitory BH3 mimetics can induce a transient thrombocytopathy that undermines the hemostatic function of platelets. Blood. 118: 1663–1674 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Kaefer A et al. Mechanism-based pharmacokinetic/pharmacodynamic meta-analysis of navitoclax (ABT-263) induced thrombocytopenia. Cancer Chemother Pharmacol. 74, 593–602 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Itchaki G & Brown JR The potential of venetoclax (ABT-199) in chronic lymphocytic leukemia. The Adv Hematol. 7, 270–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perini GF, Ribeiro GN, Neto JVP, Campos LT, & Hamerschlak N BCL-2 as therapeutic target for hematological malignancies. J Hematol Oncol. 11, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leverson JD et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci Transl Med. 7, 279ra40 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Amundson SA et al. An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res. 60, 6101–6110 (2000). [PubMed] [Google Scholar]
- 26.Vogler M Targeting BCL2-proteins for the treatment of solid tumours. Adv in Medicine. 2014, 943648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai AC, & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Runcie AC, Chan KH, Zengerle M, & Ciulli A Chemical genetics approaches for selective intervention in epigenetics. Curr Opin Chem Biol. 33, 186–194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshaies RJ Protein degradation: Prime time for PROTACs. Nat Chem Biol. 11, 634–635 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Churcher I Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J Med Chem. 61, 444–452 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Ohoka N, Shibata N, Hattori T, & Naito M Protein knockdown technology: application of ubiquitin ligase to cancer therapy. Curr Cancer Drug Targets. 16, 136–146 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondeson DP et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 11, 611–617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai AC et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed. 55, 807–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina K et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 113, 7124–7129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saenz DT et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang HT et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem Biol. 25, 88–99.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bray PF et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 14, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kissopoulou A, Jonasson J, Lindahl TL, & Osman A Next generation sequencing analysis of human platelet PolyA+ mRNAs and rRNA-depleted total RNA. PLoS One. 8, e81809 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogler M et al. BCL2/BCL-X(L) inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood. 117, 7145–7154 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowak RP et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol. 14, 706–714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riching KM et al. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem Biol. 13, 2758–2770 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Farmer T, O’Neill KL, Naslavsky N, Luo X, & Caplan S Retromer facilitates the localization of Bcl-xL to the mitochondrial outer membrane. Molecular biology of the cell, 30(10), 1138–1146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith BE et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun. 10, 131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morowski M et al. Only severe thrombocytopenia results in bleeding and defective thrombus formation in mice. Blood. 121, 4938–4947 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Rinder HM et al. Correlation of thrombosis with increased platelet turnover in thrombocytosis. Blood. 91, 1288–1294 (1998). [PubMed] [Google Scholar]
- 51.Koch R et al. Biomarker-driven strategy for MCL1 inhibition in T-cell lymphomas. Blood. 133, 566–575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger S et al. Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer. Elife. 5, pii: e20352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hikita H et al. Mcl-1 and Bcl-xL cooperatively maintain integrity of hepatocytes in developing and adult murine liver. Hepatology. 50, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J et al. The Bcl-2/Bcl-X(L)/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol Cancer Ther. 10, 2340–2349 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Ackler S et al. The Bcl-2 inhibitor ABT-263 enhances the response of multiple chemotherapeutic regimens in hematologic tumors in vivo. Cancer Chemother Pharmacol. 66: 869–880 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Pompili L, Porru M, Caruso C, Biroccio A, & Leonetti C Patient-derived xenografts: a relevant preclinical model for drug development. J Exp Clin Cancer Res. 35, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zengerle M, Chan KH, & Ciulli A Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol. 10, 1770–1777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bondeson DP et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem Biol. 25, 78–87.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References (for methods)
- 59.Lv D-W, Zhang K, & Li R Interferon regulatory factor 8 regulates aspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction. PLoS Pathog. 14, e1006868 ((2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiśniewski JR et al. Universal sample preparation method for proteome analysis. Nat Methods. 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Nesvizhskii AI, Keller A, Kolker E & Aebersold R A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods. 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Vizcaíno JA et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Food US and Administration Drug. “Bioanalytical Method Validation, Guidance for Industry.” US Department of Health and Human Services Food and Drug Administration, Rockville, MD: (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository with the dataset identifiers PXD010878 and PXD015454, as mentioned in the methods also. The raw immunoblot images and statistical analyses files are supplied as source data. Other raw and analyzed data files are available from the corresponding author upon reasonable request. A data availability statement is also included in the reporting summary linked to this article.