Abstract
Viruses are implicated in the autoimmune destruction of the pancreatic islet β-cells that results in insulin deficiency and type 1 diabetes (T1D)1–4. Certain enteroviruses can infect β-cells in vitro5, have been detected in pancreatic islets of T1D patients6 and shown an association with T1D in meta-analyses4. However, establishing consistency of findings across studies has proved difficult. Obstacles to convincingly link RNA viruses to islet autoimmunity may be attributed to rapid viral mutation rates, cyclical periodicity of viruses7 and the selection of variants with altered pathogenicity and ability to spread in populations. β-cells strongly express cell surface Coxsackie and adenovirus receptor (CXADR) genes, which can facilitate enterovirus infection8. Studies of human pancreata and cultured islets have shown significant variation in enteroviral virulence to β-cells between serotypes and within the same serotype9–10. In this large-scale study of known eukaryotic DNA and RNA viruses in stools from children, we evaluated fecally shed viruses in relation to islet autoimmunity and T1D. This study demonstrated prolonged Enterovirus B (EV-B) rather than independent, short duration EV-B infections may be involved in the development of islet autoimmunity, but not T1D, in some young children. Furthermore, we found fewer early life Human mastadenovirus C infections and CXADR rs6517774 independently correlated with islet autoimmunity.
Keywords: type 1 diabetes, islet autoimmunity, T1D, autoantibody, autoantibodies, virus, virome, enterovirus, adenovirus, coxsackievirus
The Environmental Determinants of Diabetes in the Young (TEDDY) is the largest prospective observational cohort study of newborns with increased genetic risk for T1D followed closely in several countries with diverse exposures, including viruses. Two nested-matched case-control studies within TEDDY were designed, with islet autoimmunity and T1D as the respective outcomes. Longitudinal stool samples were examined for virome content prior to outcomes. We hypothesized that increased prevalence of enterovirus associated with risk of either islet autoimmunity and/or T1D in young children. We further explored the known human fecal virome for other viral associations with islet autoimmunity and T1D.
Metagenomic sequencing was performed on fecal specimens from 383 islet autoimmunity and 112 T1D case-control matched pair children from six TEDDY study sites distributed among U.S., Germany, Sweden and Finland (Supplementary Table 1). Samples were collected approximately monthly, from age 3 months until case event-time, totaling 8,654 stools for the islet autoimmunity and 3,380 stools for the T1D nested-matched case-control studies. To detect known RNA and DNA viruses, total nucleic acid was extracted from stools, reverse transcribed and subjected to next generation sequencing. Additionally, samples of each stool were cultivated on virus-susceptible cells to amplify enteroviruses before nucleic acids were extracted and subjected to the same analysis using VirMAP11 in parallel. The proportion of stools with viruses were calculated from merged primary and cultured virome output containing presence-absence data. Overall, the virome content among the included stools consisted of 621 taxa representing 96 genera of known eukaryotic viruses and 57 genera of bacteriophage, as defined by the International Committee on Taxonomy of Viruses (2017 release). The relative proportion of viruses in the virome included bacteriophage (72%), mammalian viruses (20%) and those associated with the food stream, which were mostly plant viruses (8%), Figure 1a. Of note, 55.8% of the samples were positive for any mammalian virus. Some abundant mammalian viruses listed (circovirus, gyrovirus) originate in food sources; however, it is unclear if these can replicate in humans. The most abundant human virus serotypes found in the stool of the children are shown in Supplementary Table 2.
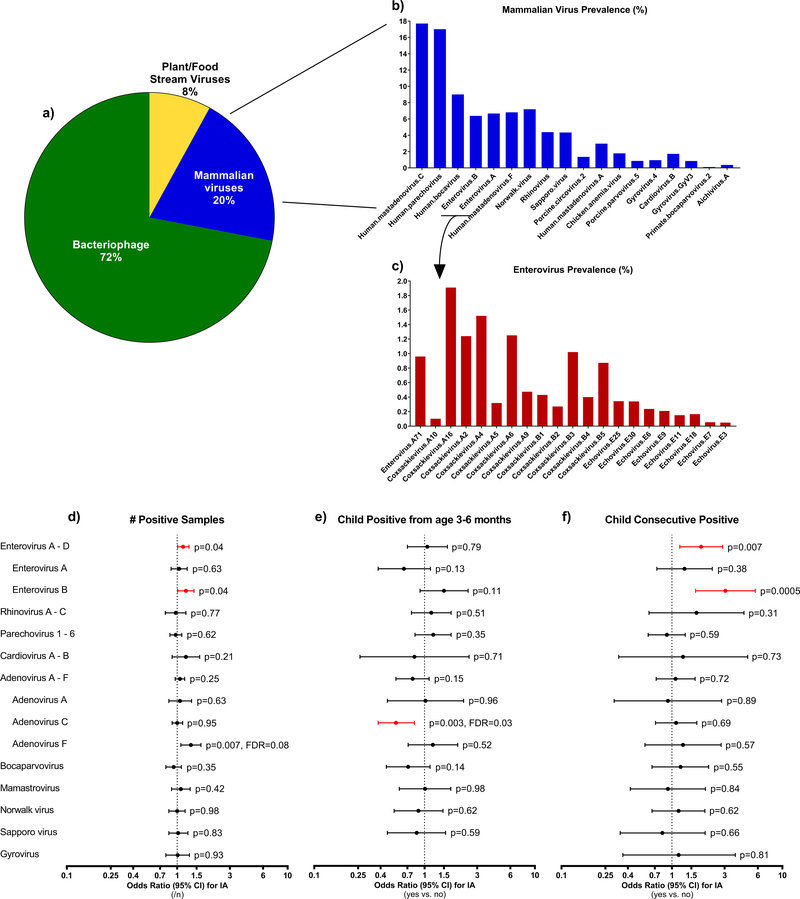
Figure 1 (a-f). Stool virome composition up to 36 months of age and common human viruses related to islet autoimmunity.
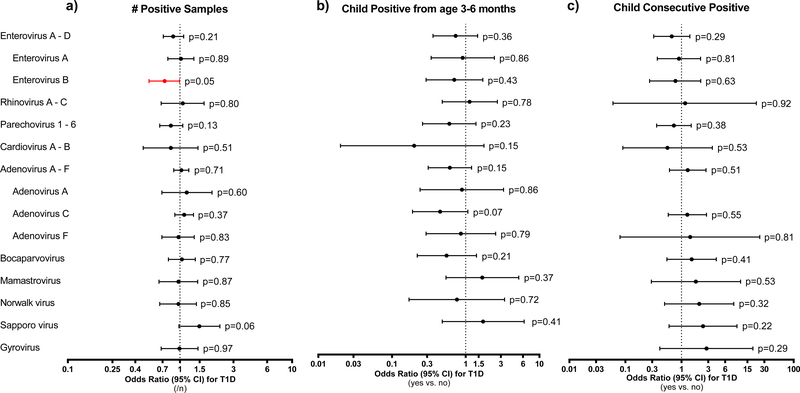
The pie chart (a) indicates the relative proportion of major classes of viruses found in the stool virome (n=5,725 samples, % positive overall, combined primary and cultured stools) in the first three years of life. Bar graphs indicate the estimated prevalence in the TEDDY cohort (n=6,890 children observed at risk for islet autoimmunity and type 1 diabetes up to 36 months of age) of the 20 most abundant mammalian viruses (b) and enterovirus serotypes found among the two most abundant enterovirus species (Enterovirus A, Enterovirus B) within the same stool dataset (c). The cohort prevalence were estimated by weighting the proportion of stool samples that were positive for the virus to account for how the children were selected as controls (n=495) into the two nested-matched case-control studies. The three forest plots (d-f) show how common human viruses relate to the odds of developing islet autoimmunity. The results were shown as odds ratios (OR, circle) and 95% confidence intervals (CI, bars) and were calculated using conditional logistic regression models with adjustment for HLA-DR-DQ genotype. All p-values were two-sided. OR>1 indicated a positive correlation between virus pattern and development of islet autoimmunity, OR<1 indicated an inverse correlation. Plot (d) examined if an increase in the number of samples positive for virus (n=4,327 matched pair samples) correlated with islet autoimmunity. Plot (e) examined if children positive for the virus between age 3 and 6 months (n=370 matched pair children) were related to islet autoimmunity. Plot (f) examined if children positive for the common virus in at least two consecutive samples (n=383 matched pair children) were related to islet autoimmunity. Black circles and CI bars represent non-significant associations. Red circles and CI bars represent statistical significant association for the a prior enterovirus at p<0.05 or for other viruses that showed a false discovery rate (FDR) at <0.05.
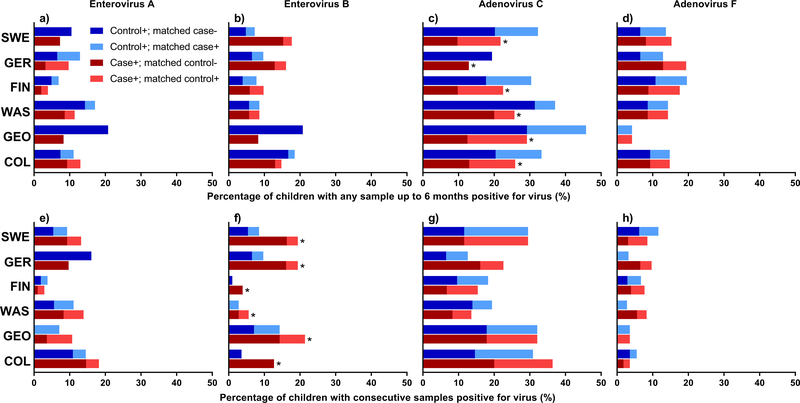
The most prevalent viruses at the species level (>2% of positive stools) were Human adenovirus, Parechovirus, Bocavirus and Enterovirus A (EV-A) and Enterovirus B, Figure 1b, 1c, Supplementary Table 3. Enterovirus (EV-A or EV-B) was detected in 12.8% of the 8,654 stools and in at least one stool for 55.4% of the 766 islet autoimmunity nested-matched case-control children. An EV-B in at least one stool was observed in 36.6% (140/383) cases and 37.1% (142/383) controls. The number of stools positive for EV-B positive (each additional positive sample, OR=1.20, 95%CI=1.01–1.42, p=0.04, Figure 1d) was associated with developing islet autoimmunity. EV-A and EV-B were lower in Finland (Figure 2, Panels a and b, p<0.001) compared to other sites. Children with consecutive EV-B positive stools were significantly more likely to be an islet autoimmunity case (OR=3.05, 95%CI=1.64–5.69, p=0.0005, Figure 1f). This association was similar across all matching strata, including across sites (ORs ≥ 1.7, Figure 2, Panel f). Adenovirus (39.6%), Parechovirus (25.1%), and Enterovirus (19.9%) were detected more frequently in stools from very young children age 3–6 months (Supplementary Table 4).
Figure 2 (Panels a-h). The percentage of children positive for a specific virus between ages 3 and 6 months and percentage of children with consecutive positive samples before islet autoimmunity development by clinical site.
Panels a-d show the percentage positive for virus between ages 3 and 6 months (Panel a – Enterovirus A (EV-A), Panel b – Enterovirus B (EV-B), Panel c – Human mastadenovirus C (HAdV-C), and Panel d – Human mastadenovirus F (HAdV-F)). Panels e-h show the percentage consecutive positive before islet autoimmunity (Panel e – EV-A, Panel f – EV-B, Panel g – HAdV-C, and Panel h – HAdV-F). The red and blue bars represent cases and matched controls (by clinical site, gender and family history of type 1 diabetes), respectively. The darker color show the percentage of children positive for virus pattern with matching child negative. The light blue and red bars represent percentage of children positive for virus pattern with a concordant result in the matching child. Specifically, dark blue denotes control is positive for the virus pattern and matching case is negative. Light blue denotes both control and matching case are positive for virus pattern. Dark red denotes case is positive for virus pattern and matching pair control is negative. Light red denotes both case and matching control are positive for virus pattern. Asterisk (*) denotes statistically significant difference overall of discordant cases positive for virus (dark red) compared to discordant controls positive for virus (dark blue) across clinical sites (matched pair children in US-Colorado, n=55; US-Georgia/Florida, n=28; US-Washington, n=36; Finland, n=104; Germany, n=31; and, Sweden, n=129). Significance was assessed using conditional logistic regression adjusted for HLA-DR-DQ genotype. All p-values are two-sided.
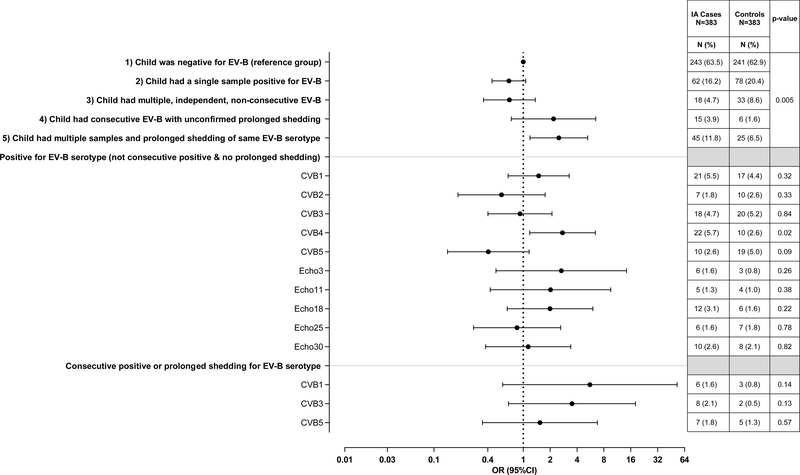
Longitudinal analysis of stools across children before islet autoimmunity revealed variable patterns of EV-B infection and shedding that associated with islet autoimmunity (Figure 3, p=0.005). Sequence data from virus capsid regions enabled identification of the exact serotype of EV-B in 81.2% of positive samples. A single EV-B infection (i.e., child with only one positive stool) was observed in 16.2% (n=62/383) of cases and 20.4% (78/383) of controls, Figure 3. We next examined children with multiple EV-B infections (i.e., >1 positive sample) and asked if these are multiple positive stools for the same strain of one serotype, which would indicate a prolonged shedding period lasting more than 30 days. The definition of “same virus strain” within a serotype was set as a RNA sequence homology >98% (i.e., evolution rate of acquired mutations during chronic EV-B infection12). The same EV-B serotype strain in more than one positive sample was observed in 11.8% (45/383) of cases and 6.5% (25/383) of controls. A majority of cases (77.8%, n=35/45) and controls (64%, n=16/25) with prolonged shedding were consecutively positive for a specific serotype. The median (interquartile, IQR) months of shedding the same virus was 6.0(1.5–13.1) for cases and 4.1(1.8–15.9) for controls. We also identified 3.9% (15/383) of case and 1.6% (6/383) of control children who were consecutive positive for EV-B in 2+ stools where virus read homology was just below 98% (homology 95–97%). This was usually due to lack of sufficient overlapping sequence reads in common regions. These children were likely shedding the same virus, but unconfirmed by our definition and were included as prolonged shedding in further analysis. The remaining children with multiple, independent, non-consecutive EV-B infections (n=51) were similar in islet autoimmunity association as children having a single infection. Children with prolonged shedding of same EV-B (OR=2.50, 95%CI=1.19–5.26) or children with consecutive positive with different serotype (OR=2.18, 95%CI=0.74–6.46) had a similar higher odds of being an islet autoimmunity case compared to children negative for an EV-B, while children having a single (OR=0.69, 95%CI=0.45–1.06) or multiple non-consecutive (OR=0.70, 95%CI=0.30–1.36) did not.
Figure 3. Children single positive, multiple positive, consecutively positive and/or prolonged shedding for Enterovirus B (EV-B) on risk of islet autoimmunity.
The pattern groups were the following: 1) child was negative for EV-B (reference group), 2) child had a single sample positive for EV-B, 3) child had multiple, independent, non-consecutive samples positive for EV-B, 4) child had consecutive EV-B, but with unconfirmed prolonged shedding, and 5) child had multiple and prolonged shedding of the same EV-B serotype. Consecutive positive allowed for no more than one missed monthly stool sample. Odds ratios (OR, circle) and 95% confidence intervals (CI, bars) were calculated using conditional logistic regression models with adjustment for HLA-DR-DQ genotype. All p-values were two-sided.
The children with prolonged shedding of Coxsackie B virus (CVB) were more likely to develop islet autoimmunity (OR=2.49, 95%CI=1.12–5.54, p=0.03). However, when individual serotypes were studied, the children positive for CVB4 with no evidence of prolonged shedding were also more likely to develop islet autoimmunity (OR=2.75, 95%CI=1.18–6.40, p=0.02, Figure 3).
There was no association between age and the timing of the first appearing EV-B infection nor was there an association with time of infection prior to developing islet autoimmunity (Extended Data Figure 1, Panels a-d). There was no overall difference between enterovirus infections and insulin or glutamic acid decarboxylase autoantibodies as the first appearing autoantibody.
Further discovery of other associated viruses found the number of samples positive with Human mastadenovirus F (HAdV-F) up to seroconversion showed evidence of correlation with developing islet autoimmunity (OR=1.33, 95%CI=1.08–1.54, p=0.007, false discovery rate=0.08, Figure 1d). All but eleven of the samples positive for HAdV-F had serotype HAdV-F41. HAdV-F40 was additionally found in seven islet autoimmunity case samples and four controls. One child was positive for both serotypes. The direction of the association was not consistent across all study sites (Figure 2, Panels d and h).
We next examined stools before the earliest seroconversion at age 6 months on islet autoimmunity based on previous TEDDY publications linking this time period to increased risk of islet autoimmunity and T1D. No association was observed between enterovirus and islet autoimmunity in this timeframe (Figure 1e, Supplementary Table 4). However, Human mastadenovirus C (HAdV-C) was detected in fewer islet autoimmunity cases as compared to controls (OR=0.55, 95%CI=0.38–0.81, p=0.003, false discovery rate=0.03), Figure 1e, Supplementary Table 4. The direction of this association was consistent across all study sites (Figure 2, Panel c).
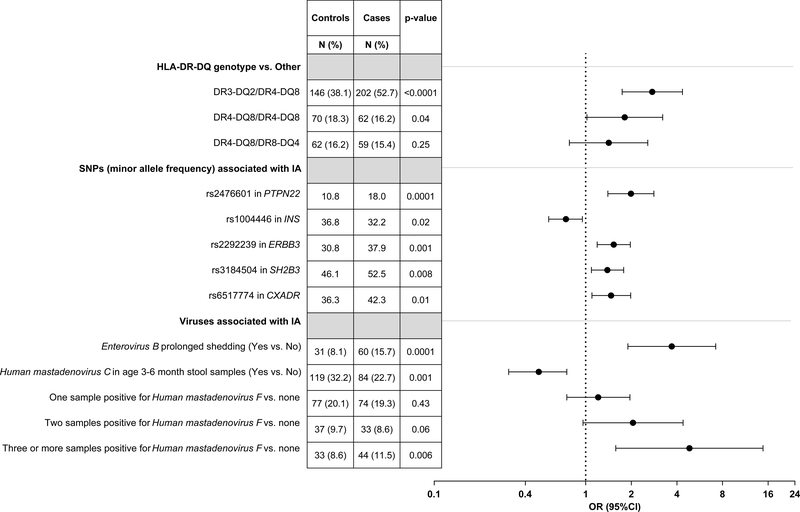
The propensity for finding EV-B in stools was strongly associated with age of the child and month the sample was collected (Supplementary Table 5). Nevertheless, prolonged shedding for EV-B (OR=3.70, 95%CI=1.90–7.22, p=0.0001, Figure 4, Supplementary Table 6) remained strongly associated with islet autoimmunity when controlling for the child’s EV-B propensity and their genetic islet autoimmunity risk. After controlling for EV-B, HAdV-F, HLA and specified single nucleotide polymorphisms (SNP), children with positive HAdV-C in early samples had a lower risk for islet autoimmunity (OR=0.49, 95%CI=0.31–0.75, p=0.001), Figure 4, Supplementary Table 6. The number of positive samples with HAdV-F showed a weak increased risk for islet autoimmunity (OR=4.84, 95%CI=1.58–14.80, p=0.04).
Figure 4. Multivariable conditional logistic regression of Enterovirus B (EV-B), Human mastadenovirus C (HAdV-C) and F (HAdV-F) on islet autoimmunity case status (n=378 matched pair children).
The model shown in figure includes HLA-DR-DQ genotype, a single nucleotide polymorphism (SNP) in PTPN22, INS, ERBB3 and SH2B3 (all previously reported associated with islet autoimmunity in the TEDDY cohort40 and confirmed associated with islet autoimmunity in this nested-matched case-control study), a SNP in the Coxsackie and adenovirus receptor gene (CXADR), prolonged shedding or consecutive positive samples for EV-B, HAdV-C detected in stool samples up to age 6 months and number of HAdV-F detected in stools during follow-up, on islet autoimmunity case status. In addition, the model included the first two principal components describing ancestry40 and a propensity score for a stool to be EV-B positive. Odds ratios (black circle), 95% confidence intervals bars, and test of significant difference between cases and matched controls are estimated from multivariable conditional logistic regression model. All p-values are two-sided.
We examined the association of EV-B and islet autoimmunity with a panel of SNPs reported to regulate antiviral responses, but these were not associated with islet autoimmunity, although rs2304256 in TYK2 was associated with positive EV-B in stools (OR=1.36, 95%CI=1.04–1.78, p=0.02, Supplementary Table 5). Based on the strong EV-B and HAdV-C findings with islet autoimmunity, five SNPs typed in the CXADR region (Chr21q21.1) were examined. Their chromosomal position order was rs6517774, rs1967939, rs1539798, rs2824400 and rs282440413. SNPs rs1539798 (minor allele frequency (MAF) <1%) and rs2824400 (r>0.99 with rs2824404) were excluded. Any child having the minor rs6517774-G allele correlated strongly with a lower propensity to be EV-B positive in a stool (OR=0.66, 95%CI=0.50–0.87, p=0.003, Supplementary Table 5). In contrast, each additional minor rs6517774-G allele increased the odds for islet autoimmunity (OR=1.47, 95%CI=1.10–1.98, p=0.01, Figure 4, Supplementary Table 6), particularly among first degree relatives (OR=3.76, 95%CI=1.62–8.73, interaction, p=0.003). SNP rs2824404 was associated with islet autoimmunity only in Finland (per additional minor C allele, HLA-adjusted, OR=1.68, 95%CI=1.09–2.57, p=0.02; Finland*SNP interaction, p=0.03). No significant interactions between EV-B and SNPs, either in CXADR, other gene regions, or HLA were observed.
The T1D nested-matched case-control study had a higher frequency of first degree relatives with T1D (35.7%) compared to the islet autoimmunity nested-matched case-control (21.9%), Supplementary Table 1. In all, 77.7% (87/112) T1D cases and 5.4% (6/112) controls developed islet autoimmunity. The number of positive stools for EV-B was lower among T1D cases compared to controls (OR=0.73, 95%CI=0.53–0.99, p=0.05, Extended Data Figure 2a, Supplementary Table 7). This was due to a lower frequency of multiple EV-B infections in cases (Supplementary Table 8). Eight T1D cases who developed islet autoimmunity, one T1D control who seroconverted for islet autoimmunity before their matching case event-time and one T1D control who did not have islet autoimmunity had prolonged shedding for the same EV-B serotype. All eight T1D cases and one control who developed islet autoimmunity started their prolonged shedding before seroconversion. Additionally, HAdV-C, similar to islet autoimmunity cases, was less likely to be detected in early stools (age ≤6 months) compared to the matched control for T1D cases (OR=0.45, 95%CI=0.19–1.07, p=0.07, Extended Data Figure 2b, Supplementary Table 4). No association with HAdV-F or other viruses was observed.
Lastly, further examination of the CXADR region and T1D showed a similar association as it did with islet autoimmunity in the islet autoimmunity nested-matched case-control (HLA and islet autoimmunity-adjusted OR=1.83, 95%CI=0.80–4.20, p=0.15). There were 27 T1D case-control pairs who had the same islet autoimmunity status during follow-up (23 islet autoimmunity negative and 4 positive) and 85 who had a different islet autoimmunity status (83 T1D cases and 2 T1D controls had islet autoimmunity, but matching child was negative).
This is the first report of a large-scale study using next generation sequencing to analyze the whole known virome in children with increased genetic risk for T1D, including analysis of genetically-linked SNPs associated with islet autoimmunity. Here, we report that children who exhibited prolonged shedding of the same EV-B serotype showed a strong association with islet autoimmunity, but not T1D. Whereas, children with independent, short duration EV-B infections did not associate with islet autoimmunity or T1D, indicating that the type and duration of an infection may be critical. Additionally, our results showed that having a HAdV-C infection in very early life was associated with a decreased risk of islet autoimmunity and T1D. Independently, children who carried the minor SNP allele rs6517774 in the CXADR gene region were more likely to develop islet autoimmunity.
Previous work examining the stool virome in both small retrospective case-control and larger prospective-based nested case-control studies have been inconclusive2,4,14–16. The Finnish DIPP study has repeatedly reported an association between enterovirus infections and subsequent initiation of islet autoimmunity. This association was recently observed by detecting enterovirus RNA in stools16 and enterovirus RNA and antibodies in follow-up sera3,17,18. Likewise, the Norwegian MIDIA (environmental trigger(s) of type 1 diabetes) study reported enterovirus RNA in the blood at the time of islet autoantibody appearance19. Other smaller prospective studies14,16 have not shown an association.
We found that prolonged shedding after the acute phase of EV-B infection preceded subsequent initiation of islet autoimmunity. This finding was consistent across geographic areas. Children with multiple, independent EV-B infections without prolonged shedding were less likely to be diagnosed with T1D. This result is surprising and may be due to two possibilities acting together or separately; firstly, EV-B is associated with T1D only through its association with islet autoimmunity, and, secondly, the CXADR rs6517774 SNP is associated with T1D independent of how it relates with islet autoimmunity risk. Approximately a quarter of the cases in the T1D nested-matched case-control rapidly progressed to disease without capture of prior islet autoimmunity development20. Among this group, the CXADR G-minor allele appeared to both associate with an increased T1D risk and a lower detection of EV-B. The reason why CXADR is associated with lower EV-B will need further investigation.
EV-B are enteric viruses that replicate in the respiratory and gut mucosa and mucosal immune system. The viral subgroup found significantly linked to islet autoimmunity was the CVB, consisting of five of six detected serotypes (CVB1–5). TEDDY showed an association with CVB4 and islet autoimmunity; whereas, earlier findings linking CVB1 to T1D were not confirmed. These previous studies used serology and detected more CVB1 infections in cases compared to the present study3,21. The cyclic periodicity of enteroviruses may cause variation in detection in certain time periods and geographic location.
Taken together with the published literature, our results support CVB as a candidate virus associated with islet autoimmunity3,22. Here, consecutive positive or prolonged shedding of CVB was associated with islet autoimmunity. The underlying mechanisms remain unknown. Speculatively, this phenomenon could be related to the persistence of more virulent virus strains with higher replication capacity23 or altered efficacy in controlling innate immune responses. A wide variation has been observed in the ability of different CVB1 strains to stimulate innate immune system responses in dendritic cells24. Another mechanism could be that EV-B are prone to 5’ terminal genomic deletions, leading to prolonged non-lytic infections25. Alternatively, prolonged shedding could be caused by weaker immune protection in case children. Prolonged enterovirus shedding, previously recognized26–28, might be a biomarker for defective innate immune defenses against certain viruses or immune dysregulation that leads to autoimmunity. Only a small proportion of islet autoimmunity cases with EV-B resulted in prolonged shedding, approximately 16% detected in this study. Overall, the role of enterovirus in T1D is likely prior to islet autoantibody seroconversion.
These results suggested that EV-B, particularly CVB, could have properties that make them potentially diabetogenic. CVB have a tropism to the pancreas5,29. They infect pancreatic islets during systemic infection in infants, while the exocrine pancreas is less affected. Studies of pancreas tissue of T1D patients have shown enterovirus protein almost exclusively in the β-cells29. There has been detection of low grade enteroviral infection in islets of T1D patients6,30,31. Thus, functionally ‘defective’ enterovirus might focally attack and persist within the β-cells with low levels of viral replication6. Viral tropism depends largely on the expression of viral receptors by the cell. Interestingly, among enteroviruses, only CVB use the CXADR as the main receptor to infect cells, and this tight-junction type molecule is strongly expressed by β-cells, particularly in the membrane of insulin granules32. CXADR is also used by adenoviruses. There is enhanced expression of the CXADR gene in the pancreas of both islet autoimmunity and T1D subjects32. This receptor is important for virus internalization into cells33 and might induce inflammation and tissue damage34. As supportive analysis, we found that children carrying the minor rs6517774-G allele within the CXADR region had an increased risk for islet autoimmunity, but lower number of EV-B infections. Children who have both minor alleles for rs6517774 and rs2824404 (primarily, Finland) were strongly associated with islet autoimmunity in absence of EV-B, suggesting greater penetrance of these SNPs in low threshold exposure to EV-B. Caution is advised in over-interpretation of these SNP findings, as they need to be independently examined.
We report a new and potentially interesting finding that adenovirus was associated with islet autoimmunity. Both coxsackievirus and adenovirus use the CXADR to infect cells. And, to our knowledge, no human study has shown a link between adenovirus and risk of islet autoimmunity or T1D. Nevertheless, we found a protective association with HAdV-C. The mechanism of action for this association is unknown. However, early HAdV-C infections before the age of 6 months were associated with a low risk of islet autoimmunity. No other detected virus was associated with low risk of islet autoimmunity, which suggests that some HAdV-C factors may be involved. Since HAdV-C and CVB use the same receptor, shown to associate with increased islet autoimmunity risk, we speculate that competition in receptor binding might be a mechanism for protection by HAdV-C and CVB risk. Conversely, HAdV-C infections were frequent and they are known to persist long periods, for example, in tonsillar tissues36, where they may cause long-lasting activation in the innate immune system of the respiratory track. This could protect against other viruses that replicate in the same tissues, including islet autoimmunity-associated enteroviruses. In fact, this viral interference has been suggested between adenoviruses and rhinoviruses36 and interferon-lambda signaling in the gut activated by astrovirus can protect against norovirus infection in the mouse37. However, in the present study we did not find significant interactions between HAdV-C and EV-B; thus, further studies are needed to determine whether viral interference could explain our adenovirus finding.
Although, our sample size was large, there were limitations in power, especially evaluating virus serotypes. This may be attributed to the constant geographic movement and evolution of enteroviral serotypes which shift between variants with altered pathogenicity21,38,39. Notably, the association with EV-B prolonged shedding on islet autoimmunity risk was observed among stools with higher compliance in children up to age 36 months. We were unable to adequately measure virus abundance due to the timing of infection onset being unknown relative to when the stool was collected and the abundance measures being inappropriate for the sequencing methodology. We deliberately cultivated on cells to increase the enterovirus hits by amplifying the virus signal, but all viruses were considered. Both primary and cultured virome data were combined before analysis, but also showed the same association between EV-B and islet autoimmunity when analyzed separately. Stool collection began at age 3–4 months and compliance declined from age two years onward. Therefore, the number of children with consecutive positive/shedding may be underestimated.
Overall, the observed virome composition aligned with the existing knowledge about viral epidemiology in children. The most frequently detected human viruses (Human mastadenovirus, Parechovirus, Bocavirus, Mamastrovirus, Norovirus and Enterovirus) were common in previous studies using similar technologies. It will be important to identify different immunological response profiles to viral infection. While, some EV-B serotypes were variably associated with islet autoimmunity, future studies will require more targeted and sensitive approaches to detect prolonged shedding, such as Ampliseq. Both epidemiological and bench investigations are needed to identify the serotypes that are most prone to trigger islet autoimmunity or protect against it and to discern the host and viral mechanisms that lead to prolonged infections and lack of clearing virus in the gut. Although coxsackievirus vaccines are already in development, more experimental work is needed to elucidate the mechanisms of how long-term or persistent virus infections may promote immunological dysregulation leading to islet autoimmunity.
Methods
Six clinical research centers - three in the U.S. (Colorado, Georgia/Florida, Washington), and three in Europe (Finland, Germany, and Sweden) participated in a population-based HLA screening of newborns between 2004 and 2010. Children with high risk HLA genotypes as described41 were enrolled (n=8,676) and prospectively followed from three months of age to 15 years with study visits that include a blood draw every three months until four years and every three or six months thereafter depending on islet autoimmunity positivity. HLA-DR-DQ genotypes were confirmed by reverse blot hybridization at the central HLA Reference Laboratory at Roche Molecular Systems, Oakland, CA41. Single nucleotide polymorphisms (SNP) were genotyped by the Center for Public Health Genomics at the University of Virginia using the Illumina ImmunoChip, which is a custom array for genotyping of SNPs selected from regions of the human genome firmly associated with autoimmune diseases42. The final selection of SNPs, including 186,000 SNPs in 186 regions for 12 autoimmune diseases, was decided by the ImmunoChip Consortium. Stool samples were collected monthly from ages 3 to 48 months and then quarterly until the age of 10 years as previously described43,44.
Persistent confirmed autoimmunity was defined by the presence of a confirmed islet autoimmunity (glutamic acid decarboxylase (GADA), insulinoma-associated 2 (IA-2A) or insulin (IAA)) at each of the two TEDDY laboratories on two or more consecutive visits45. T1D diagnosis was defined according to American Diabetes Association criteria 46. The detailed study design and methods have been previously published47–50. The protocol was approved by each local Institutional Review Board and all participants provided written informed consent prior to study participation.
Nested-matched case-control population
A nested-matched case-control was conducted through risk-set sampling using metadata and islet autoimmunity sample results as of 31 May 2012, as previously detailed47. In a separate nested-matched case-control, each child diagnosed with T1D had a control selected based on their event time from birth (Supplementary Table 1). The details of the nested-matched case-control studies and sampling structure have been published47. A 1:1 matched nested-matched case-control was used for the viral metagenomics samples. There were 418 children with incident persistent confirmed islet autoimmunity (cases) that were matched to a selected control of the same sex, clinical site and family history with T1D (general population or first degree relative) at the time of islet autoimmunity development; and, 114 children who were newly diagnosed with T1D with similar matched combination to a control at the time of diagnosis. In all, 95 of the 418 islet autoimmunity cases were included in the T1D nested-matched case-control, 323 islet autoimmunity cases did not develop T1D and 19 T1D cases who did not develop persistent confirmed islet autoimmunity prior to diagnosis47. Due to lack of matching stool sampling availability, there was a 9% reduction in the number of pairs for the 1:1 viral metagenomic study for the islet autoimmunity nested-matched case-control, which left 383 pairs (n = 4,327 age matched stool samples in each group) and a 2% reduction in the number of pairs for the 1:1 T1D nested-matched case-control resulting in 112 pairs (n=1,690 age matched stool samples in each group). All incidence cases were at least 6 months of age or older at time of risk set. There were 93 of the 383 children who developed islet autoimmunity included in the T1D nested-matched case-control (87 cases seroconverted to islet autoimmunity and were diagnosed with T1D and 6 matched controls who seroconverted for islet autoimmunity, but not diagnosed with T1D as of 31 May 2012). The islet autoimmunity and T1D nested-matched case-control were analyzed separately with paired controls specific to each nested-matched case-control. As a result, the viral metagenomic analysis for the two nested-matched case-controls will have 80% or greater power at a significance level of 5% to detect an odds ratio (OR) >2.28 with the 383 pairs and an OR >4.58 with the 112 pairs if the exposure proportion is 10%. The selection of samples sent to the Alkek Center for Metagenomics and Microbiome Research at the Baylor College of Medicine, Houston, TX, were determined by the TEDDY Data Coordinating Center (USF Health Informatics Institute, Tampa, FL) without the laboratory knowing the case-control status. Otherwise, there was no randomization, and the investigators were not blinded during outcome assessment.
Overall, only 6.8% of the controls (n=26) in the islet autoimmunity nested-matched case-control went on to develop persistent islet autoimmunity at a median of 2.4 years after time of the risk set in which they were selected as a control and 5.4% (n=6) of the controls in the T1D nested-matched case-control progressed to T1D as of 31 December 2018.
Stool Samples and Metadata
Stool samples were collected monthly from 3 to 48 months of life, then every three months thereafter until the age of 10, into three plastic stool containers provided by the clinical center. Children who were antibody negative after 4 years of age were encouraged to submit 4 times a year even though after 4 years their visit schedule changed to biannual. Parents sent the stool containers at either ambient or +4°C temperature with guaranteed delivery within 24 hours in the appropriate shipping box to the NIDDK Repository if residing in the U.S. or their affiliated clinical center if residing in Europe. The European clinical centers stored the stool samples and sent monthly bulk shipments of frozen stool to the NIDDK Repository. Detailed study design and methods have been previously published43,47–49.
Metadata were collected using validated questionnaires that have been either published or extensively scrutinized by experts. TEDDY provides many tools, such as ‘The TEDDY book’, to the parents to assist in real-time collection of all events in their child’s life to ensure bias and error are minimized. At each visit the study personnel go over the TEDDY book with the primary caretaker and extract pertinent information using standardized study forms. Data are extracted by trained staff members during scheduled visits every 3 months starting at 3 months of age and entered directly via standard forms (web forms or teleforms), which are transmitted electronically. Front-end constraints are employed in the web application to prevent the entry of invalid data and the TEDDY Error Reporting and Verification System consists of a set of programs that conduct automated quality control on the data, report and resolve errors, an integrated database for storing error data, and a set of programs that generate reports for monitoring data cleaning efforts. The details of the system have been published50. Additionally, systematic differences in sample collection between sites may exist, although procedures set up to assess and preserve viral material in the stool samples were uniform50.
All parents or guardians provided written informed consent before participation in genetic screening and enrollment. The study was performed in compliance with all relevant ethical regulations.
Metagenomic whole genome shotgun (WGS) sequencing and analysis
The nested-matched case-control populations for both the islet autoimmunity and T1D designs included 8,654 and 3,380 matched total stool samples that passed quality filtering and were included in subsequent blinded testing as previously published43,44. Filtrates of stools (~0.15–0.2 mg dispersed in 100–200 μl saline and passed through 1.2 μM filter) were directly extracted for total nucleic acids and also incubated on mixtures of four cell lines (Hela, Vero, RD cells expressing Coxsackievirus-adenovirus receptor, HEK-293; 25% each, plated at 40% overall confluency) in DMEM containing 2% calf serum for six days to amplify viruses present at very low levels. Cell lines were chosen for breadth of virus replicative efficiency of type A, B, C enteroviruses, as well as, other common viruses. Infected cultures were not passaged. Cells and supernatants were collected for total nucleic acid extraction and analysis.
Total nucleic acids were extracted and sequenced independently in parallel from both primary stool and cultured stool samples (cells and supernates) using the MagMax Viral RNA Isolation Kit (Cat # AM1939, Thermo Fisher Scientific, Waltham, MA) without employing DNAse to prevent DNA extraction. Extracted viral RNA was reverse transcribed using SuperScript II RT (Cat # 18064014, Thermo Fisher, Waltham, MA) and random hexamers. After short molecule and random hexamer removal with ChargeSwitch (Cat # CS12000, Thermo Fisher, Waltham, MA), molecules were amplified and tagged with a 12 base-pair barcode tag containing a V8A2 semi-random primer (BC12-V8A2 construct using AccuPrimeTM Taq polymerase and cleaned with ChargeSwitch kit). Tags were attached via a barcoded, semi-random primer construct51 resulting in dual barcoded (same barcode on both sides) amplified fragments. The indexes used were 12 bp golay codes. Uniquely barcoded amplicons derived from each sample were pooled at 30 per sequencing lane and a single WGS library was generated on the pool without shearing. Separate negative controls were introduced during extraction, amplification, and library preparation steps. Positive controls (a mix of four lab strain viruses, poliovirus, simian rotavirus, adenovirus, mouse herpesvirus) were included blindly in certain wells of all sample plates. We performed a single WGS library prep per sequencing lane of pooled, pre-barcoded samples, minimizing carry-over as each lane only had a single index. Additionally, because the samples all carried secondary internal barcodes, they were not subject to carry-over or cross-bleed sometimes observed from run to run with library indexes on the Illumina platform. The size of the library was verified via bioanalyzer to ensure appropriate range for the platform (~200–1000 bp) then the library was loaded in an Illumina HiSeq2000 (Illumina, Carlsbad, CA) and sequenced using the 2×100bp chemistry at the Human Genome Sequencing Center at Baylor College of Medicine. Stool samples were received blinded, without metadata, from the NIDDK repository in defined “runs” that were generally sequenced on the same sequencing flowcell in different lanes. Reads were demultiplexed into a sample bin using the barcode prefixing read-1 and read-2, allowing zero mismatches. Demultiplexed reads were further processed by trimming off barcodes, semi-random primer sequences, and Illumina adapters. This process utilized a custom demultiplexer and the BBDuk algorithm included in BBMap52.
Virome Analysis
The resulting trimmed primary and cultured virome datasets were analyzed using a pipeline created at the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine, employing a clustering algorithm (VirMAP) that reconstructs putative viral genomes using a mapping assembly strategy that leverages both nucleotide and translated nucleotide alignment information11. VirMAP uses a custom formatted version of gbvrl (GenBank virus) and gbphg (GenBank Phage), however, to filter out false positives all other Genbank Organismal divisions were used as a master database. Viral taxonomies were assigned using a scoring system that incorporates nucleotide and translated nucleotide alignment results in a bits-per-base fashion and optimizes for the highest resolution taxonomic rank, generating a taxonomy ID aggregate bit score output. VirMAP testing and evaluation with in-house and public metagenomic viral datasets indicated the 300 aggregate bit-score threshold provides superior accuracy and sensitivity11. VirMAP has undergone extensive validation utilizing mock viral communities and publically available reference data sets that also report PCR analysis11.
Comparative analysis of differences in determination of virus positivity were tested at four levels of VirMAP aggregate bit-scores of 100, 400, 700, 1000, which returned 1487, 1280, 1078, 971 enterovirus-positive stools, respectively. From this analysis, an aggregate bit-score of 400 was chosen as most conservative for accuracy which eliminated false positives, reduced risk of low level cross sample contamination, and increased the accuracy of enterovirus serotype determinations, yet maintained sensitivity for output for statistical analysis. The distribution of enterovirus reads in enterovirus positive stools at the 400 aggregate bit-score level ranged from 12 to 27,396,504 reads with a median of 13,117 and mean of 15,233 reads. VirMAP analysis returned Enterovirus A (EV-A) or B (EV-B) present in 12.8% of stools, which compares well with PCR analysis of stools in a stool sample pilot study conducted on a subset of control TEDDY children who were 14.6% positive for the same viruses53. This published TEDDY pilot study of PCR on stools contained an overlap in stool population (n=111) with those samples in this current study using next generation sequencing (NGS). A comparison of results was performed at the TEDDY Data Coordinating Center with the PCR and NGS laboratories blinded to the overlapped samples. By design, the PCR study included samples from 3 to 18 months of age. The median (interquartile range, IQR) age of the child when stool samples were taken was 10.5 (7.9–14.0) months. NGS identified EV-B in 8 of 111 stools and PCR in 2 of 111 of the samples. The two samples detected by PCR were also detected by NGS (both primary and cultured results) and both had the same serotypes. An additional 6 EV-B were detected by NGS, 3 were from the primary only and 3 from the cultured only results. In the total stool population of the respective studies (PCR study and current NGS study), aggregate level comparison showed the EV-B serotype prevalence across all stool samples was similar. This comparison of detection methods, NGS versus PCR, was only done for enteroviruses, as they were the main a priori hypothesis. In the current NGS analysis, a comparison of primary and cultured EV-B results showed no difference in association with islet autoimmunity. The combined (i.e., primary or cultured) EV-B consecutive positive results in association with islet autoimmunity, as shown in Figure 1f, revealed HLA-adjusted OR=3.05, 95%CI=1.64–5.69, p=0.0005; the primary EV-B association was OR=2.95, 95%CI=1.49–5.83, p=0.002; and, the cultured enterovirus association was OR=2.34, 95%CI=1.08–5.07, p=0.03.
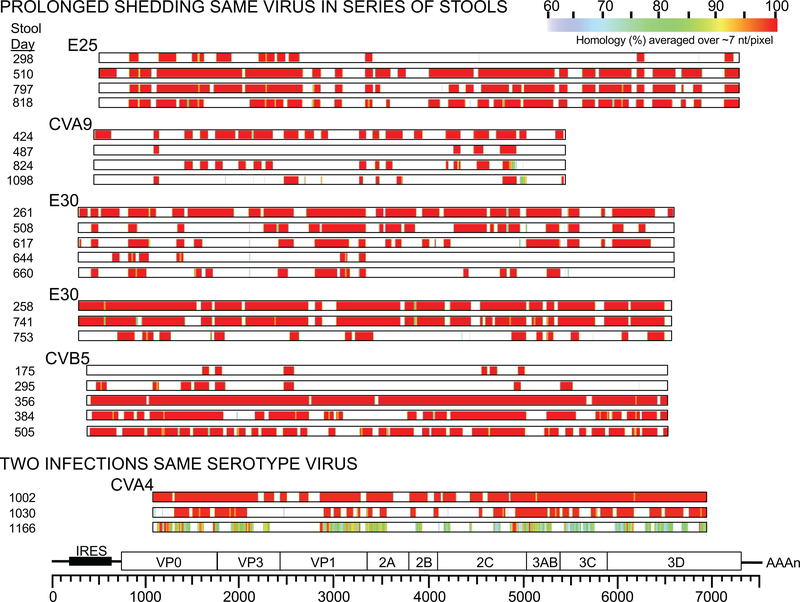
For determination if successive stools contained enterovirus from the same infection or a new infection with a different virus of the same serotype, the threshold of homology among virus contigs was taken at 98% identity or greater. Heatmaps of contig alignments of successive stools from six children are shown (Extended Data Figure 3) indicating >98% homology exists across the virus genome regions mapped in most cases (522 of 554 serotyped enteroviruses in successive stools), individual mutations appearing in successive months are indicated by narrow yellow lines. It was determined that of 189 occurrences of the same serotype enterovirus occurring in successive stools, only 14 were below 98% homology, 6 of those were possibly successive infections with different viruses of the same serotype (<95% homology). In the islet autoimmunity nested-matched case-control, length of prolonged shedding period (duration) was not significantly associated with case status in the children with consecutive positive EV-B (Extended Data Figure 4).
For analysis, the viral positive reads above the VirMAP 400 aggregate bit-score level were converted to yes or no scores (1 vs 0). Virus read counts could not be linked to intensity of infection since the origin date of infections of virus in stools are unknown, thus we could only reliably determine if the virus was present in the particular stool or not. The reduction in the information content and dimensionality of the data was carried out in order to quantify the association between the presence of viral taxa and the development of islet autoimmunity. The methods employed, starting from sample collection and ending with sequencing, preclude a quantitative analysis of the viral taxa found mainly due to the use of the un-targeted approaches in the data generation workflow. Briefly, the extraction process aims to produce clean nucleic acids indiscriminately from any source present. Although the PCR amplification process uses a semi-random primer, it does not provide enough specificity to distinguish viral from non-viral genomes. Moreover, the sequencing step indiscriminately sequences the DNA molecules present in the sequencing library. Finally, the data analysis included a series of filtering steps that aim to discard non-viral sequences. The fraction of viral reads differed across samples and its size didn’t correlate with the amount of data generated. As a result, the number of sequencing reads assigned to specific viral taxa were not representative of true viral load.
Statistical Methods
The islet autoimmunity nested-matched case-control included 383 case-control paired subjects with 4,327 paired monthly stool samples available prior to islet autoimmunity. Median (IQR) months of age of islet autoimmunity, number of stool samples available per subject, time between consecutive stool samples and the duration of shedding for those who were prolonged were 21.8(13.3,33.1), 9.0(5,16), 1.1(0.7,3.4), and 5.1(1.4–15.1) months, respectively. The T1D nested-matched case-control included 112 paired case-controls across 1,690 paired monthly stool samples with an average 13.0(8,22) stool samples available per subject. A distribution of the matching factors among the cases are shown in Supplementary Table 1.
The nested-matched case-control represents a biased sample of the TEDDY cohort due to the sampling of a disproportional number of controls correlated with a case. To estimate the cohort prevalence of viruses in monthly stool samples up to 36 months of age, the strata proportions were first estimated within the 24 matching strata (six sites*gender*family history) using stool samples of controls selected into the islet autoimmunity and T1D nested-matched case-control studies (n=5,725 samples). Prevalence of viruses within strata were estimated by inverse probability weighting. Each control selected within each risk set (time of case) was taken as a sample of stools and the sample prevalence was weighted by the inverse probability of being selected into the risk set. The target population was all stool samples from all children (n=6,890/8,676) available to be selected as a control at the earliest risk set (age of first case) within each strata, Median (IQR) age at earliest risk across strata was 9.0(6.7–9.9) months. The remaining children in TEDDY dropped out of the study before the first case was observed. Strata weights were calculated as the proportion of children in the target population that were included in the matching strata. The strata weights were then applied to the strata proportions to estimate the cohort prevalence. The estimated prevalence of mammalian viruses in stools of children up to 36 months of age are displayed in Figure 1b.
Discordance between cases and matched controls, both in the number of stool samples positive for each common virus (positive >2% of samples) and in the presence of consecutive samples positive for the virus (allowing for one missed monthly visit), were evaluated using conditional logistic regression models adjusting for HLA genotypes. The magnitude of the associations were assessed by odds ratios with 95% confidence intervals. It was an a priori hypothesis to examine the role of enteroviruses given evidence in prior literature. For the remaining common viruses (n=12) and with consideration of multiple comparisons, a false discovery rate of less than 10% was the criteria for further evaluation.
Significant viral associations were next examined adjusting for genetic factors by including associated SNP risk factors in the conditional logistic model, SNPs in MDA5 (IFIH1), PTPN2, TYK2, BACH2, as well as, SNPs previously associated with either islet autoimmunity or T1D40,54 were considered. The influence of potential non-genetics confounders at a sampling and subject level were controlled for by, including in the model, a propensity score for whether a child will have a stool sample positive for the virus. The propensity score was created to adjust for risk factors associated with the probability of EV-B and to better estimate the average effect of EV-B on risk of islet autoimmunity without making strong assumptions on how islet autoimmunity is related to the risk factors associated with EV-B. The virus propensity score was calculated from a logistic random effects model that regressed stool sample virus positivity on age of child, year and month of stool sample collection, matching factors, case status and other associated factors (p<0.05). The score for each stool sample was estimated using the fixed effects in the final model, assuming all children were controls, and then calculating the marginal prediction. The scores were then averaged across samples of the child to create subject level propensity scores.
Specifically, the regression adjustment and calculation of the propensity score from the case-control data were performed by fitting a model for P(E=1| C, D) and predicting exposure for each stool sample as if they were a control. D indicated whether a child was an islet autoimmunity case (D=1) or a control (D=0). E was the exposure of interest and C was a set of covariates sufficient to adjust for confounding55. The covariates considered in addition to the matching variables and case status were age of child, year of sample, month of sample, lifestyle factors (maternal smoking, alcohol consumption during pregnancy, working mother), demographic variables (parental age and education, mothers first child, living status, household crowding, ethnic minority) and factors related to the child (birth weight and early weight gain, start of daycare, illness and conditions early in life and SNPs including those reported to regulate antiviral responses). Also, probiotic use in early life that TEDDY has shown associated with islet autoimmunity56, exclusive breast feeding and gestational age at birth were included. The propensity score for a stool sample to be positive for EV-B was calculated from a logistic random effects model that regressed EV-B on covariates. The final model adjusting for case status is shown in Supplementary Table 5. Samples taken in the fall, children aged 18 to 24 months, sites other than Washington and Finland, child not having a minor allele for SNP-rs6517774 in CXADR, but having a minor allele of SNP-rs23042556 in TYK2 showed higher propensity to have a positive EV-B in stool sample. Assuming all children were controls, the marginal prediction was calculated from this model and used as an estimation of the propensity score for each stool sample. The average was taken across stool samples of the child to use as an overall propensity score. Supplementary Table 9 shows how the EV-B group distributed across the overall propensity score ranges. As expected, there were more children with consecutive positive EV-B at higher propensity score ranges. This propensity was included in the conditional logistic model to adjust for non-genetic T1D risk factors that may confound the association.
Viruses showing an association with islet autoimmunity (or T1D, if appropriate) were further characterized by testing for interactions with HLA and SNPs. All p-values were two-sided, and less than 0.05 was considered significant unless otherwise stated. SAS 9.4 (SAS Institute, Cary, NC, USA) was used for the statistical analysis and GraphPad Prism 8.0 (La Jolla, CA, USA) was used for the figures.
Life Sciences Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data Availability
TEDDY virome sequencing data that support the findings of this study have been deposited in the NCBI database of Genotypes and Phenotypes (dbGaP) with the primary accession code phs001442.v1.p1, in accordance with the dbGaP controlled-access authorization process. Clinical metadata and virome results data analyzed for the current study will be made available in the NIDDK Central Repository at https://www.niddkrepository.org/studies/teddy.
Code Availability
VirMAP was used to generate the virome data and has been deposited in GitHub, https://github.com/cmmr/virmap. All of the software code and dependencies are listed on the GitHub site. SAS 9.4 (SAS Institute, Cary, NC, USA) was used for the statistical analysis and GraphPad Prism 8.0 (San Diego, CA, USA) was used for the figures.
Extended Data
Extended Data Fig. 1. Percentage of stool samples at age of first appearance of Enterovirus B.
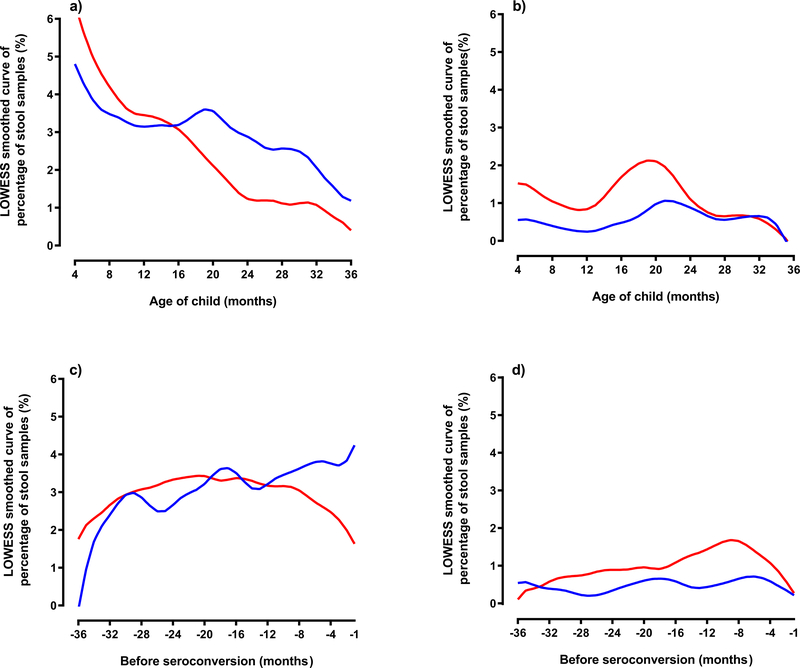
Extended Data Figure 1 (Panels a-d). Percentage of stool samples at age of first appearance of Enterovirus B. Panel a shows sample positivity and Panel b sample consecutive positivity. Panels c and d show months prior to autoantibody seroconversion of Enterovirus B for sample positivity (c) and sample consecutive positivity (d) by autoantibody case status (n=383 matched pair children). Blue line represents control samples and red line represents case samples. The timing of the first appearance of an Enterovirus B infection from enrollment (3 months of age) or months prior to islet autoimmunity showed no obvious trend by age of child.
Extended Data Fig. 2. Common human viruses related to type 1 diabetes (T1D).
Extended Data Figure 2 (Panels a-c). Common human viruses related to type 1 diabetes. The three forest plots (a-c) show how common human viruses relate to the odds of children being diagnosed with T1D. The results were shown as odds ratios (OR, circle) and 95% confidence intervals (CI, bars) and were calculated using conditional logistic regression models with adjustment for HLA-DR-DQ genotype. OR>1 indicated a positive correlation between virus pattern and diagnosis with T1D, OR<1 indicated an inverse correlation. Plot (a) examined if an increase in the number of samples positive for virus was correlated with T1D (n=112 matched pair children). Plot (b) examined if children positive for the virus between 3 and 6 months of age were related to T1D (n=103 matched pair children). Plot (c) examined if children positive for the common virus in at least two consecutive samples (yes versus no) were related to T1D (n=112 matched pair children). Black circles and CI bars represent non-significant associations. Red circles and CI bars represent significant association with T1D. The number of positive stool samples for Enterovirus B was lower among T1D cases compared to matched controls. Human mastadenovirus C, similar to islet autoimmunity cases, was less likely to be detected in early stool samples (3–6 months of age) compared to the matched control for T1D cases. All p-values were two-sided.
Extended Data Fig. 3. Heatmaps of contig alignments of successive stools (n=6 children).
Extended Data Figure 3. Heatmaps of contig alignments of successive stools (n=6 children). Heatmaps showing percent homology of alignments of enterovirus contigs isolated from successive stools from the same child. Stool collection date (successive days in the study) are shown, the serotype for the enterovirus aligned, all are aligned to an enterovirus genome map with scale of nucleotides at the bottom. Heatmap color is assigned on ~7 nt/pixel, heatmap color scale of percent homology is shown at the top.
Extended Data Fig. 4. Children consecutive positive for Enterovirus B with prolonged shedding of same serotype.
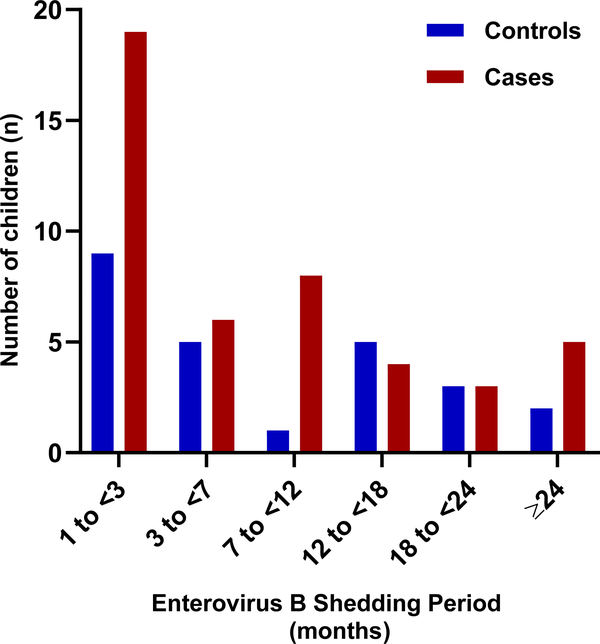
Extended Data Figure 4. Children consecutive positive for Enterovirus B with prolonged shedding of same serotype. Categorical months of shedding by number of children for islet autoimmunity cases (n=45) and controls (n=25). Red bars denote cases and blue bars denote controls. Length of prolonged shedding period (duration) was not associated with case status in the children with consecutive positive Enterovirus B. Conditional logistic regression was used to evaluate significance; test was two-sided.
Supplementary Material
Acknowledgements
The TEDDY Study is funded by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Centers for Disease Control and Prevention (CDC), and JDRF. This work supported in part by the NIH/NCATS Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR001082).
Members of the TEDDY Study Group are listed in the Supplementary File.
Footnotes
Competing Interests
H.H. is a shareholder and chairman of the Board of Vactech Ltd. and member of the Scientific Advisory Board of Provention Bio, Inc., which develop vaccines against picornaviruses and coxsackievirus B. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from the disclosed.
References
- 1.van der Werf N, Kroese FGM, Rozing J & Hillebrands JL Viral infections as potential triggers of type 1 diabetes. Diabetes-Metab Res 23, 169–183 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Stene LC, et al. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY). Diabetes 59, 3174–3180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laitinen OH, et al. Coxsackievirus B1 is associated with induction of beta-cell autoimmunity that portends type 1 diabetes. Diabetes 63, 446–455 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Yeung WC, Rawlinson WD & Craig ME Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ 342, d35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anagandula M, et al. Infection of human islets of Langerhans with two strains of Coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J Med Virol 86, 1402–1411 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Krogvold L, et al. Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes 64, 1682–1687 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Enterovirus and Human Parechovirus Surveillance — United States, 2009–2013. Morbidity and Mortality Weekly Report (MMWR) 64, 940–943 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ifie E, et al. Unexpected subcellular distribution of a specific isoform of the Coxsackie and adenovirus receptor, CAR-SIV, in human pancreatic beta cells. Diabetologia 61, 2344–2355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roivainen M, et al. Mechanisms of coxsackievirus-induced damage to human pancreatic beta-cells. J Clin Endocrinol Metab 85, 432–440 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Roivainen M, et al. Functional impairment and killing of human beta cells by enteroviruses: the capacity is shared by a wide range of serotypes, but the extent is a characteristic of individual virus strains. Diabetologia 45, 693–702 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Ajami NJ, Wong MC, Ross MC, Lloyd RE & Petrosino JF Maximal viral information recovery from sequence data using VirMAP. Nat Commun 9, 3205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laassri M, et al. Evolution of echovirus 11 in a chronically infected immunodeficient patient. PLoS Pathog 14, e1006943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genome Reference Consortium Human Build 38.
- 14.Tapia G, et al. Human enterovirus RNA in monthly fecal samples and islet autoimmunity in Norwegian children with high genetic risk for type 1 diabetes: the MIDIA study. Diabetes Care 34, 151–155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honkanen H, et al. Detection of enteroviruses in stools precedes islet autoimmunity by several months: possible evidence for slowly operating mechanisms in virus-induced autoimmunity. Diabetologia 60, 424–431 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Simonen-Tikka ML, et al. Human enterovirus infections in children at increased risk for type 1 diabetes: the Babydiet study. Diabetologia 54, 2995–3002 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Oikarinen S, et al. Enterovirus RNA in blood is linked to the development of type 1 diabetes. Diabetes 60, 276–279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salminen K, et al. Enterovirus infections are associated with the induction of beta-cell autoimmunity in a prospective birth cohort study. J. Med. Virol 69, 91–98 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Cinek O, et al. Enterovirus RNA in longitudinal blood samples and risk of islet autoimmunity in children with a high genetic risk of type 1 diabetes: the MIDIA study. Diabetologia 57, 2193–2200 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lee HS, et al. Next-generation sequencing for viruses in children with rapid-onset type 1 diabetes. Diabetologia 56, 1705–1711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sioofy-Khojine AB, et al. Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia 61, 1193–1202 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Oikarinen S, et al. Virus antibody survey in different European populations indicates risk association between coxsackievirus B1 and type 1 diabetes. Diabetes 63, 655–662 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Bessaud M, Joffret ML, Blondel B & Delpeyroux F Exchanges of genomic domains between poliovirus and other cocirculating species C enteroviruses reveal a high degree of plasticity. Sci Rep 6, 38831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamalainen S, et al. Coxsackievirus B1 reveals strain specific differences in plasmacytoid dendritic cell mediated immunogenicity. J Med Virol 86, 1412–1420 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Leveque N, et al. Functional Consequences of RNA 5’-Terminal Deletions on Coxsackievirus B3 RNA Replication and Ribonucleoprotein Complex Formation. J Virol 91(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung PW, Huang YC, Chang LY, Lin TY & Ning HC Duration of enterovirus shedding in stool. J Microbiol Immunol Infect 34, 167–170 (2001). [PubMed] [Google Scholar]
- 27.Alexander JP Jr., Gary HE Jr. & Pallansch MA Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis 175 Suppl 1, S176–182 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Melnick JL & Rennick V Infectivity titers of enterovirus as found in human stools. J Med Virol 5, 205–220 (1980). [DOI] [PubMed] [Google Scholar]
- 29.Ylipaasto P, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 47, 225–239 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Richardson SJ, Leete P, Bone AJ, Foulis AK & Morgan NG Expression of the enteroviral capsid protein VP1 in the islet cells of patients with type 1 diabetes is associated with induction of protein kinase R and downregulation of Mcl-1. Diabetologia 56, 185–193 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Busse N, et al. Detection and localization of viral infection in the pancreas of patients with type 1 diabetes using short fluorescently-labelled oligonucleotide probes. Oncotarget 8, 12620–12636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodik M, et al. Coxsackie-adenovirus receptor expression is enhanced in pancreas from patients with type 1 diabetes. BMJ Open Diabetes Res Care 4, e000219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafren DR, et al. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J Virol 69, 3873–3877 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito M, et al. Expression of coxsackievirus and adenovirus receptor in hearts of rats with experimental autoimmune myocarditis. Circ Res 86, 275–280 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Garnett CT, et al. Latent species C adenoviruses in human tonsil tissues. J Virol 83, 2417–2428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, et al. Broad spectrum respiratory pathogen analysis of throat swabs from military recruits reveals interference between rhinoviruses and adenoviruses. Microb Ecol 59, 623–634 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Ingle H, et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat Microbiol 4, 1120–1128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messacar K, Abzug MJ & Dominguez SR 2014 outbreak of enterovirus D68 in North America. J Med Virol 88, 739–745 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Greninger AL, et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012–14): a retrospective cohort study. Lancet Infect Dis 15, 671–682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krischer JP, et al. Genetic and Environmental Interactions Modify the Risk of Diabetes-Related Autoimmunity by 6 Years of Age: The TEDDY Study. Diabetes Care 40, 1194–1202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagopian WA, et al. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 12, 733–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkes M, Cortes A, van Heel DA & Brown MA Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 14, 661–673 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatanen T, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonifacio E, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J. Clin. Endocrinol. Metab 95, 3360–3367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 37, S81–S90 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Lee HS, et al. Biomarker discovery study design for type 1 diabetes in The Environmental Determinants of Diabetes in the Young (TEDDY) study. Diabetes Metab Res Rev 30, 424–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann. N. Y. Acad. Sci 1150, 1–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr. Diabetes 8, 286–298 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Vehik K, et al. Methods, quality control and specimen management in an international multicentre investigation of type 1 diabetes: TEDDY. Diabetes Metab Res. Rev 29, 557–567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clem AL, Sims J, Telang S, Eaton JW & Chesney J Virus detection and identification using random multiplex (RT)-PCR with 3 ‘-locked random primers. Virol J 4(2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bushnell B BBMap short read aligner. (University of California, Berkeley, California, 2016). [Google Scholar]
- 53.Sioofy-Khojine AB, et al. Molecular epidemiology of enteroviruses in young children at increased risk of type 1 diabetes. PLoS One 13, e0201959 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krischer JP, et al. The Influence of Type 1 Diabetes Genetic Susceptibility Regions, Age, Sex, and Family History on the Progression From Multiple Autoantibodies to Type 1 Diabetes: A TEDDY Study Report. Diabetes 66, 3122–3129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Handbook of Modern Statistical Methods (Taylor & Francis Group, Boca Raton, FL, 2018). [Google Scholar]
- 56.Uusitalo U, et al. Association of Early Exposure of Probiotics and Islet Autoimmunity in the TEDDY Study. JAMA Pediatr 170, 20–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TEDDY virome sequencing data that support the findings of this study have been deposited in the NCBI database of Genotypes and Phenotypes (dbGaP) with the primary accession code phs001442.v1.p1, in accordance with the dbGaP controlled-access authorization process. Clinical metadata and virome results data analyzed for the current study will be made available in the NIDDK Central Repository at https://www.niddkrepository.org/studies/teddy.