Abstract
Histiocytoses are clonal hematopoietic disorders frequently driven by mutations in BRAF and MEK1/2 kinases. Currently, however, the developmental origins of histiocytoses in patients are not well understood, and clinically meaningful therapeutic targets outside of BRAF and MEK are undefined. Here we uncover activating mutations in CSF-1R, as well as rearrangements in RET and ALK which confer dramatic responses to selective inhibition of RET (selpercatinib) and crizotinib, respectively, in histiocytosis patients.
Genomic analyses of histiocytic neoplasms (including Langerhans Cell Histiocytosis (LCH) and Erdheim-Chester Disease (ECD)) have revolutionized our understanding of these disorders as clonal hematopoietic malignancies driven by MAPK signaling.1–3 These findings have also led to FDA approval of vemurafenib for BRAFV600E-mutant ECD and identification of clinical efficacy of MEK1/2 inhibition in BRAFV600E wild-type (WT) patients.4,5 Despite these advances, the cell-of-origin of the histiocytoses is not definitively known. In addition, histiocytoses represent a spectrum of diseases, and genetic alterations across histiocytosis subtypes have not been comprehensively evaluated. Finally, although the histiocytoses most commonly occur as sporadic, non-hereditary disorders, familial clustering has been well documented and occurs most often in monozygotic twins.6,7 This phenomenon has been understood to suggest a hereditary component of the disease, but germline genetic causes of histiocytoses are not known. Here we performed comprehensive genomic analyses of 270 patients with all subsets of histiocytoses, including monozygotic twins (Supplementary Tables 1–6). In so doing, we uncovered a novel series of activating receptor tyrosine kinase (RTK) alterations, including the first example in any disease of recurrent, activating mutations in CSF-1R (macrophage-CSF receptor (M-CSF-R)), the RTK required for monocyte/macrophage development8, as well as the clinical importance of targeting rearrangements in RET and ALK.
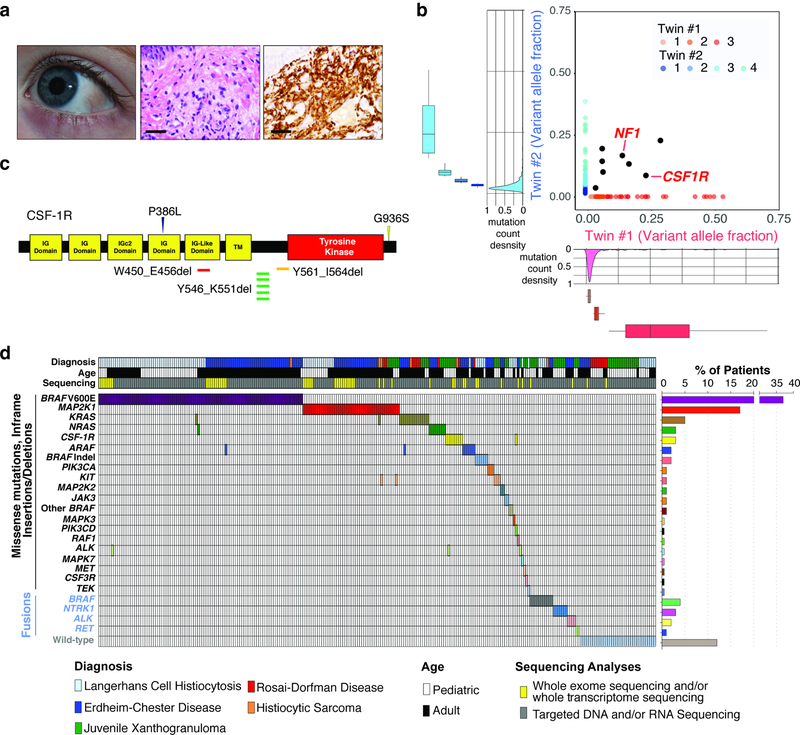
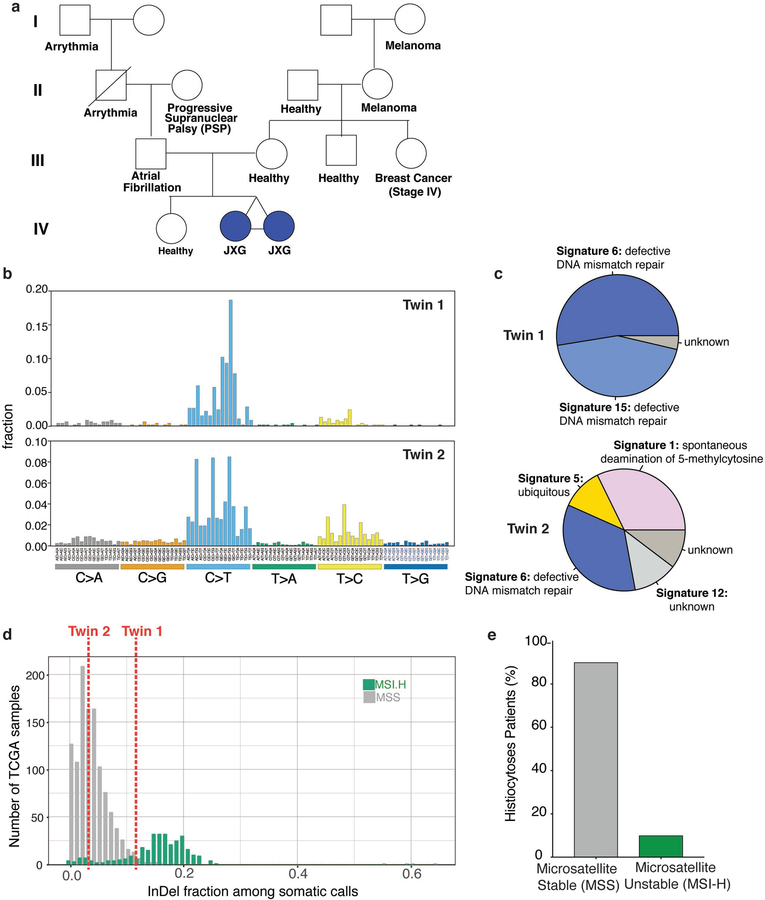
We performed whole exome sequencing (WES) of skin lesions, blood, and fingernails from monozygotic, monochorionic, diamniotic one-year-old twins with synchronous diagnoses of systemic juvenile xanthogranuloma (JXG) and the fingernails of each parent (Fig. 1a and Extended Data Fig. 1a). At one year of age, both twins developed skin lesions on their forehead, which over the course of four months grew in number and diameter extending to the scalp and upper chest. Skin biopsies of the scalp from each child were consistent with a diagnosis of JXG and ophthalmic examination of both children identified that twin 1 also had multiple, bilateral subconjuctival JXG lesions. Although no known germline pathogenic variants were found, we surprisingly found eight identical somatic mutations in skin lesions, but not blood or fingernails, across both twins. These included identical in-frame deletions in CSF-1R (CSF-1RY546_K551del) and nonsense mutations in NF1 (NF1E19X), each of which were in the same major clone in each twin (Fig. 1b). Beyond this set of shared mutations, however, each child harbored 195 and 816 distinctive protein-coding mutations unique to the tumor of Twin 1 (JXG-10) and Twin 2 (JXG-11), respectively (Fig. 1b; Supplementary Table 4). Both lesions shared a DNA mismatch repair mutational signature in addition to microsatellite instability, the latter of which was otherwise rare in sporadic histiocytoses (Extended Data Fig. 1b–e). There are two potential explanations for this scenario of a common clonal somatic origin of histiocytosis across identical twins. Disease causing somatic mutations may arise during early development within CSF-1R-expressing extra-embryonic, yolk-sac Erythro-Myeloid Progenitors that give rise to macrophages, as recently shown in mice.9 Alternatively, the malignant clone could have initiated in bone-marrow derived myeloid cells within one fetus and spread to the co-twin via vascular anastomoses. However, the same CSF-1R mutation was present in the lesions from both twins but was absent from blood or fingernails, favoring a single shared CSF-1R-mutant yolk sac precursor of both tumors.
Figure 1. Genomic analysis of 270 patients with sporadic and familial histiocytoses.
(a) Subconjunctival and skin lesions from one-year-old, monozygotic twins with JXG (middle: hematoxylin and eosin stain; right: CD68 immunohistochemistry; bars: 50μM). (b) Whole exome sequencing of histiocytosis lesions compared with fingernails from the twins in (a) reveal concordant somatic mutations (black) in both twins including shared CSF-1RY546_K551del and NF1E19X mutations. In addition, each child’s tumor harbors a set of unique genetic alterations (in blue and red, respectively; n=195 and n=816 mutations detected in twin 1 and 2 respectively). The density distribution of the variant allele frequency (VAF) of mutations is depicted outside of the axes with number of clones estimated in the inset (median VAF is shown within box, box edges represent 25th and 75th percentile values, and errors bars depict minimum and maximum values). (c) Location of somatic mutations in CSF-1R in histiocytoses. (d) Oncoprint of mutated kinases and their frequencies across the 270 patient cohort.
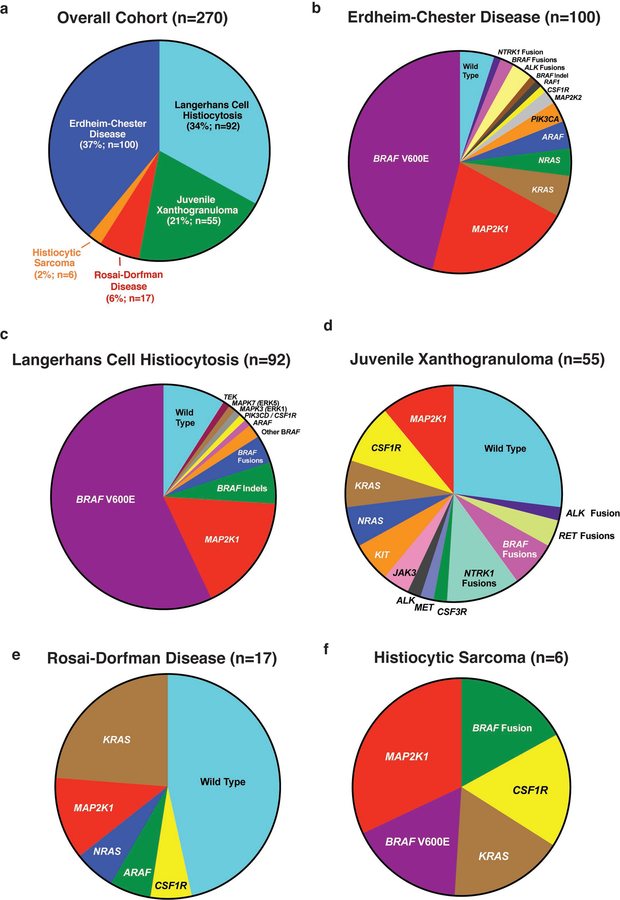
While an association between NF1 mutations and JXG is known based on their co-occurrence in neurofibromatosis10, mutations in CSF-1R have not been previously described in histiocytosis. We therefore sought to determine if mutations in CSF-1R exist in sporadic histiocytosis and sequenced 100 ECD (37%), 92 LCH (34%), 55 JXG (21%), 17 RDD (6%), and 6 histiocytic sarcoma (HS; 2%) lesions using WES, targeted DNA sequencing, and/or targeted RNA-sequencing for fusions (Extended Data Fig. 2a–f). This identified recurrent BRAFV600E, MAP2K1 (encoding MEK1), N/KRAS, and ARAF mutations as well as BRAF, NTRK1, and ALK fusions (Fig. 1). Additionally, CSF-1R mutations were found in nine patients (Fig. 1c–d).
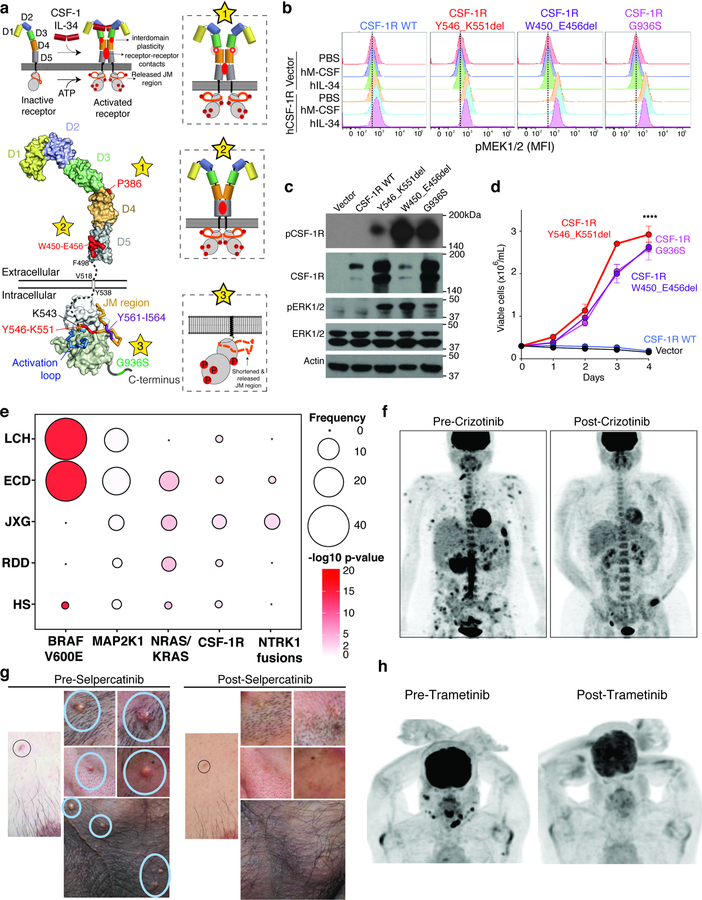
Over the last decade, structural and mechanistic studies of human CSF-1R have delineated each step in its activation.11–15 The mutations in CSF-1R discovered here are categorized into those that might enhance CSF-1R dimerization (Fig. 2a, star 1–2), and those that might promote its kinase activity (Fig. 2a, star 3). Both CSF-1RP386L and CSF-1RW450-E456del belong to the first class of mutations and are located in the extracellular region of CSF-1R. In contrast, CSF-1RY546-K551del and CSF-1RY561-I564del affect intracellular regions of CSF-1R critical to enforcing the inactive state of the kinase in the absence of ligand.15 Intracellular mutations in CSF-1R leading to receptor activation have never been described before. Ectopic expression of WT and mutant forms of CSF-1R in cells lacking endogenous CSF-1R confirmed that each CSF-1R mutant localized to the cell surface (Extended Data Fig. 3a). Moreover, expression of CSF-1R mutants, but not CSF-1RWT, conferred cytokine-independent growth to cells normally dependent on cytokines (Ba/F3 cells; Fig. 2b–d). Consistent with this, each mutation increased CSF-1R, MEK1/2, and ERK1/2 activation in cytokine-deprived conditions, as well as in response to the CSF-1R ligands IL-34 or M-CSF (Fig. 2c–d, Extended Data Fig. 3b, and Supplementary Fig. 1). Additionally, CSF-1R activating mutations sensitized cells to the CSF-1R-specific small-molecule inhibitors pexidartinib and BLZ945 (Extended Data Fig. 3c).
Figure 2. Activating mutations in CSF-1R and benefit of ALK and RET inhibition in histiocytosis.
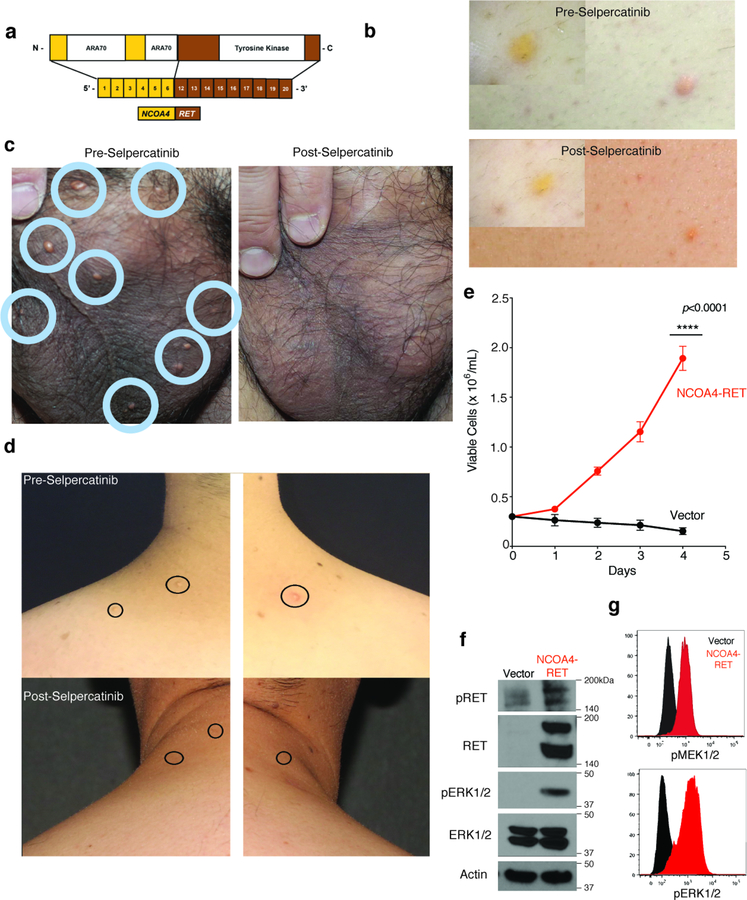
(a) Structural mapping of CSF-1R mutations and proposed impact on CSF-1R activation. Top left: Binding of CSF-1 or IL-34 to autoinhibited CSF-1R leads to receptor dimerization and activation hallmarked by conformational switching in the extracellular domains and receptor-receptor contacts to elicit phosphorylation of intracellular domains. Expulsion of the juxtamembrane (JM) region is a prerequisite to activate CSF-1R. Lower left: Structural mapping of mutations to crystal structures of the extracellular and intracellular segments of human CSF-1R (stars 1–3). Proposed mechanistic consquences of these mutations are elaborated via three insets with the corresponding star number. (b) Representative histograms of the median fluorescent intensity (MFI) of pMEK1/2 in BaF/3 cells expressing empty vector (shown in top three rows of each plot) or WT or mutant CSF-1R (each shown in the bottom three rows of each plot). Cells were grown in absence of cytokines and then analyzed following 60 minutes of stimulation with PBS, human M-CSF (100ng/mL), or hIL-34 (100ng/mL). Experiment performed with three independent biological experiments with similar results. (c) Representative western blot of phospho-CSF-1R (Tyr723), phospho-ERK1/2, and loading controls of cells from (b) in cytokine starvation conditions. Experiment performed with three independent biological exeperiments with similar results. (d) Cell numbers from (c) following IL-3 withdrawal (mean value from n=3 biological, independent experiments ± standard deviation). Calculation of p-values performed using the two-way ANOVA; ****p<0.0001. (e) Frequency and statistical enrichment of kinase mutations (x-axis) across histologic subtypes of histiocytic neoplasms (y-axis; LCH (n=67); ECD (n=80); JXG (n=24); RDD (n=8); HS (n=5)). Two-sided p-values, Fisher’s exact test. (f) Maximum Intensity Projection (MIP) images from 18F-Fluorodeoxyglucose (FDG) positron emission tomography (PET) scans pre- and 64-weeks post-crizotinib in a KIF5B-ALK ECD patient. (g) Photographs of NCOA4-RET xanthogranuloma skin lesions pre- and 12-weeks post selpercatinib. (h) MIP images from FDG PET scan pre- and 12-weeks post-Trametinib in a BICD2-BRAF LCH patient.
In addition to activating mutations in CSF-1R, we also uncovered mutations in CSF-3R, KIT, ALK, MET, JAK3, and RAF1, as well as RET fusions, none of which have previously been described in histiocytoses (Fig. 1d and Extended Data Fig. 4). There was highly significant mutual exclusivity in kinase alterations across patients (p=4.39×10−54; Fig. 1d) in addition to a significant association between specific kinase alterations and clinicopathologic histiocytoses subtypes. For example, the BRAFV600E mutation was significantly enriched in LCH and ECD (56% and 46% of patients, respectively; -log10p=16) but less common in JXG, RDD, or HS (0%, 0%, and 17% of patients, respectively; -log10p= 0, 0, and 2, respectively) (Fig. 2e and Extended Data Fig. 4–9). In contrast, mutations in CSF-1R, as well as NTRK1 fusions were significantly enriched in JXG (10% and 12% of patients; -log10 p=3 and 3, respectively).
Currently, the efficacy of MEK inhibition or inhibitors beyond RAF/MEK1/2 in kinase fusion-expressing histiocytoses is not defined. To this end, in the course of this study, patients bearing BICD2-BRAF, KIF5B-ALK, and NCOA4-RET rearranged histiocytoses were treated with the MEK1/2 inhibitor trametinib, the ALK inhibitor crizotinib, and the RET inhibitor selpercatinib, respectively. For example, a 45-year-old woman with extensive osseous, cutaneous, soft tissue, and leptomeningeal involvement of KIF5B-ALK ECD had progressive disease despite treatment with interferon-alpha. Treatment with crizotinib resulted in a marked therapeutic and radiologic response that has been sustained for 25 months (Fig. 2f). Another patient with disseminated, cutaneous xanthogranuloma-family non-LCH had an NCOA4-RET rearrangement (Extended Data Fig. 10a–d). This rearrangement potently transformed hematopoietic cells (Extended Data Fig. 10e–g and Supplementary Fig. 2). Treatment with selpercatinib resulted in dramatic resolution of disfiguring lesions maintained now for 6 months of treatment (Fig. 2g and Extended Data Fig. 10a–d). Finally, a patient with refractory, multisystem LCH harboring a BICD2-BRAF fusion had a dramatic response to the MEK inhibitor trametinib and has remained on treatment for 6 months (Fig. 2h).
The discovery of recurrent, activating mutations in RTKs highlights the potential for targeted inhibition of RTKs such as CSF-1R in histiocytoses, as well as the use of inhibitors of ALK and RET fusions in subsets of patients bearing these alterations. In addition, the presence of a CSF-1R mutation in lesions from two monozygotic twins but absent from their blood or fingernails, suggests that histiocytoses may arise in some cases from a mutation in extra-embryonic macrophage progenitors.9
METHODS
Patients
The study was conducted according to the Declaration of Helsinki, and human tissues were obtained with patient-informed consent under approval by the Institutional Review Boards of Memorial Sloan Kettering Cancer Center, St. Jude Children’s Research Hospital, the National Human Genome Research Institute, the Children’s Hospital of Pittsburgh of the University of Pittsburgh Medical Center, the Hospital Sant Joan de Déu, and Pitié-Salpêtrière Hospital. The patient treated with selpercatinib was treated on MSK IRB protocol 17–256 while the patient with a BICD2-BRAF fusion treated with trametinib, and the patient with an ALK fusion treated with crizotinib were treated off-label and not on protocols or single-patient compassionate-use protocols.
Excised lesions were either flash-frozen for DNA/RNA extraction and/or fixed in 4% neutral-buffered formalin, embedded in paraffin, and processed by routine histologic methods. For patients undergoing WES and targeted exon sequencing, DNA extracted from peripheral blood mononuclear cells (PB MNCs) or fingernail clippings was utilized as a paired normal germline control. In total, specimens from 270 patients with diverse histiocytoses subtypes were analyzed.
Genomic Analysis
Genomic analyses were performed on DNA extracted from histiocyte tissue biopsies using a variety of assays—most commonly, targeted exon sequencing using MSK-IMPACT or HemePACT.16,17 Prior to DNA extraction, FFPE samples from all cases were reviewed to confirm that the tissue was of sufficient size to generate a minimum of 50 ng of 20% histiocyte nucleic acid. DNA was isolated from 40-μm-thick sections of FFPE tissue. Targeted RNA sequencing was also used for the purposes of detecting gene fusions using the MSK-Fusion targeted RNA-seq assay.18 WES was also performed, based on DNA adequacy from fresh-frozen tissue biopsies or targeted sequencing libraries, using DNA from PB MNCs or fingernails as a germline control as previously described.3
Analysis of WES data, which includes mapping, coverage and quality assessment, single-nucleotide variant (SNV)/indel detection, tier annotation for sequence mutations, and prediction of deleterious effects of missense mutations, has been described previously19,20. Approximately 250 ng of DNA from each sample was sheared to an average of 150 bp in a Covaris instrument for 360 seconds (duty cycle, 10%; intensity, 5; cycles/burst, 200). Bar-coded libraries were prepared using the Kapa Low-Throughput Library Preparation Kit Standard (Kapa Biosystems), amplified using the KAPA HiFi Library Amplification Kit (Kapa Biosystems; 8 cycles), and quantified using Qubit Fluorimetric Quantitation (Invitrogen) and Agilent Bioanalyzer. An equimolar pool of the 4 bar-coded libraries (300 ng each) was used as input to capture the exome using one reaction tube of the Nimblegen SeqCap EZ Human Exome Library v3.0 (Roche; cat no. 06465684001), according to the manufacturer’s protocol. The pooled capture library was quantified by Qubit (Invitrogen) and Bioanalyzer (Agilent) and sequenced on an Illumina HiSeq 2500 using a paired end, 100 nucleotide in length run mode, to achieve an average of 100x coverage.
Histiocytic neoplasms are routinely characterized by low tumour cellularity, which often results in low variant-allele fractions for established driver mutations such as BRAFV600E.3 As such, for samples in which no MAPK-pathway mutations were called using established pipelines developed and optimized for use in more-cellular solid tumours, sequences were manually curated, and mutations with lower read support were salvaged.
Mutations identified by whole exome sequencing were validated using a custom-designed, TruSeq Custom Amplicon probe. Design Studio (Illumina) was used to design amplicons covering the regions of interest. The regions were amplified using 250 ng of template genomic DNA, using the manufacturer’s instructions, with 25 cycles of amplification, and were run on an Illumina MiSeq 2 × 250 cartridge.
Mutational signatures were generated using the R package deconstructSig.21 This package infers the weighted contributions of a large set of estimated signatures found across tumor types22 to the patterns of mutations observed in a single tumor sample.
MSI status classification was performed using a random forest classifier trained on 999 exome-sequenced tumor samples from four different tumor types from the TCGA (colon, rectum, stomach, and uterine corpus endometrial adenocarcinoma), and validated on a 30% hold-out set (427 tumor samples comprising MSI high and microsatellite stable samples from colon, rectum, stomach, and endometrium) for the test set. Positive predictive value and negative predictive value in the test set was 1 and 0.988 respectively (all as described previously23,24).
Plasmids
To investigate the functional roles and the activation of oncogenic signalling pathways, we cloned the human CSF-1RY546_K551del, CSF-1RW450_E456del, CSF-1RY561_I564del, CSF-1RG936S, and CSF-1RP386L mutations, as well as the NCOA4-RET fusion and expressed them in Ba/F3 cells. Murine stem-cell virus (MSCV)-based expression vectors with GFP and the full-length CSF-1R wild-type were used as controls. Mutational constructs were cloned into the MSCV-IRES-GFP backbone (MIGII), and checked by digestion and sequencing.
Western Blotting
Anti-phospho-p44/42 MAPK (ERK1 and ERK2) (Thr202/Tyr204) (no. 9101), anti-p44/42 MAPK (ERK1 and ERK2) (137F5) (no. 4695), anti-MEK1 and MEK2 (47E6) (no. 9126), anti-CSF-1R (M-CSF Receptor) (D309X) (no. 67455), anti-phospho-CSF-1R (Tyr723) (49C10) (no. 3155), anti-RET (D3D8R)(no.14698), and anti-phospho-RET (Tyr905)(no. 3221), as well as the secondary antibodies anti-rabbit IgG-HRP (no. 7076) and anti-mouse IgG-HRP (no. 7074) were purchased from Cell Signaling Technology. Anti-β-actin (A5441) was purchased from Sigma-Aldrich. Cell lysates were prepared in RIPA buffer supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Equal amounts of protein, as measured by the Bradford protein assay, were resolved in 4–12% Bis-Tris NuPage gradient gels (Life Technologies), and transferred electrophoretically on a polyvinylidene difluoride 0.45-m membrane. Membranes were blocked for 1 h at room temperature in 5% bovine serum albumin (BSA) in TBST before being incubated overnight at 4 °C with the primary antibodies. All primary antibodies were diluted 1:1,000 in 5% BSA in TBST, except anti-β-actin, which was diluted 1:5,000 in 5% BSA in TBST. After three washes of 10 min in TBST, secondary antibodies were diluted 1:2,000 in 5% BSA in TBST and incubated for 1 h at room temperature. After another three washes in TBST, detection of the signal was achieved by incubating the membrane on an ECL solution from Millipore and exposure on autoradiography films from Denville Scientific.
Phospho-Flow Cytometry
Fluorescence-activated cell-sorted DAPI−eGFP+ Ba/F3 cells that stably expressed the MIGIIempty vector, MIGII-CSF-1R wild type, MIGII-CSF-1R (Y546_K551del), MIGII-CSF-1R (W450_E456del), CSF-1R (Y561_I564del), CSF-1R (G936S), and MIGII-NCOA4-RET constructs were all grown in RPMI + 10% FBS + penicillin and streptomycin medium, without mouse IL-3 for at least 24 hours prior to the phospho-flow cytometry experiments. Each construct was stimulated with human-M-CSF (hM-CSF) (100 ng/mL), human-IL34 (hIL34) (100 ng/mL), or PBS for 60 minutes. Afterwards, the cells were fixed in 2.1% paraformaldehyde (PFA) and then permeated with 90% methanol and water as per routine. Then, the cells were stained with the following primary antibodies at 1:50 for 30 minutes at room temperature. Afterwards, the cells are stained with SCBT secondary, goat anti-rabbit F’Ab2 APC at 1:1000 for 30 minutes at room temperature. The data is then collected from a Fortessa II flow cytometer and analyzed using FlowJo version 9.9 software.
Drug Studies
Pexidartinib and BLZ945 were purchased from Selleckchem. LOXO-292 (Selpercatinib) was purchased from Active Biohem. Drug studies were conducted in vitro using fluorescence-activated cell-sorted DAPI−eGFP+ Ba/F3 cells that stably expressed the MIGII-empty vector, MIGII-CSF-1R (Y546_K551del), MIGII-CSF-1R (W450_E456del), CSF-1R (Y561_I564del), and CSF-1R (G936S) and MIGII-NCOA4-RET constructs using the CellTiter-Glo Luminescent Cell Viability Assay from Promega Corporation, according to the manufacturer’s instructions. The MIGII CSF-1R (Y546_K551del), MIGII-CSF-1R (W450_E456del), CSF-1R (Y561_I564del), and CSF-1R (G936S) and MIGII-NCOA4-RET, fluorescence-activated cell-sorted Ba/F3 cells were maintained in RPMI + 10% FBS + penicillin and streptomycin medium, without mouse IL-3. MIGII-EV was maintained in RPMI + 10% FBS + penicillin and streptomycin with recombinant mouse IL-3 (1 ng/ml).
Structural analysis
Crystal structures of the extracellular domain of human CSF-1R (Protein Data Bank entry 4WRM)13 and of the intracellular kinase domains (Protein Data Bank entry 2OGV)15 were visualized and analyzed using PyMOL v. 2.0.3 (Schrödinger).
Statistics
Statistical significance was analysed with GraphPad Prism Pro 7 using one-way and two-way ANOVA as indicated in the figure legends. Significance was considered at p < 0.05. For representative images of Western blots and flow cytometry plots, experiments were performed three independent times using distinct biological isolates. Cell culture experiments were repeated in independent biological triplicates to ensure reproducibility of the observations. Equal variance was assumed for cell-counting experiments. Fig. 1b and Fig. 2e were generated using R (version 3.5.1) and the R-packages ggplot2 (version 3.1.0) and dplyr_ (version 0.8.0.1)
Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary linked to this article.
Data Availability
Whole-exome sequencing data are deposited in DBGAP project accession number phs001864.v1.p1.
Extended Data
Extended Data Fig. 1. Pedigree and genomic analyses of monozygotic twins with CSF-1R mutant Juvenile Xanthogranuloma (JXG).
(a) Pedigree of twins with JXG. Twin 1 and 2 are monochorionic, diamniotic identical twins born to an otherwise healthy 35-year-old mother. At one year of age, both twins developed small skin lesions on their forehead, which over the course of four months grew in number and diameter extending to the scalp and upper chest. Scalp biopsies were consistent with a diagnosis of JXG. Ophthalmic examination of both twins also identified multiple, bilateral subconjunctival lesions compatible with JXG in twin 1 only. Both twins were monitored in our institution until 2.5 years-of-age with stable, persistent JXG with no visual compromise or neurologic signs or symptoms, and therefore no indication for therapy. Complete blood counts and comprehensive metabolic profiles of both twins have been within normal limits since birth. (b) Mutational profile of the JXG lesion from each twin displaying the fraction of mutations found in each trinucleotide context (performed using deconstructSigs26). (c) Piechart showing the relative contribution of each mutational signature in the JXG lesion from (b) in each twin (based on the 30 mutational signatures detected by Sanger/COSMIC30). Signatures 6 and 15 are associated with defective DNA mismatch repair. (d) Histogram illustrating the fraction of indels among all mutations identified in the JXG lesions of twins 1 and 2 (red dashed lines) along with the distribution of indel fractions for TCGA samples (colorectal, endometrial, ovarian, stomach) with known microsatellite instability (MSI) status assayed from mononucleotide markers (as described previously29; MSI-H: high microsatellite instability; MSS: microsatellite stable). (e) Bar graph depicting proportion of histiocytosis samples that are MSS (90%; n=28/31) versus MSI-H (10%; n=3/31) based on WES.
Extended Data Fig. 2. Pie charts of histologic subsets of histiocytoses analyzed with kinase mutations identified in each subset.
(a) Numbers and percentage of patients with each histological subtype of histiocytosis sequenced in this study. Frequency of kinase mutations in the subset of patients with (b) Erdheim-Chester Disease, (c) Langerhans Cell Histiocytosis, (d) Juvenile Xanthogranuloma, (e) Rosai-Dorfman Disease, and (f) Histiocytic Sarcoma.
Extended Data Fig. 3. CSF-1R mutants are expressed on the cell surface and result in phosphorylation of CSF-1R.
(a) Representative anti-CSF-1R and GFP flow cytometry analysis of Ba/F3 cell expressions of human CSF-1R constructs from an MSCV-IRES-GFP construct. Experiments were performed with n=3 independent biological replicates. (b) Representative histograms of the median fluorescent intensity (MFI) of phospho-CSF-1R Tyrosine 723 intracellular flow cytometry in cells from (a) in cytokine depletion conditions following 60 minutes of stimulation with PBS, human M-CSF (hM-CSF; 100ng/mL), or hIL-34 (100ng/mL). Cells expressing empty vector (“vector”) are shown in the top three rows of each plot) and those expressing wild-type (WT) or mutant CSF-1R constructs are shown in the bottom three rows of each plot. Experiments were performed with n=3 independent biological experiments. (c) Bar graphs of IC50 values of Ba/F3 cells expressing an empty vector control or CSF-1R mutations in response to Pexidartinib (left) or BLZ945 (right). Mean values of n=3 biologically independent experiments ± standard deviation is shown. Calculation of p-values performed using ordinary one-way ANOVA; ****p<0.0001.
Extended Data Fig. 4.
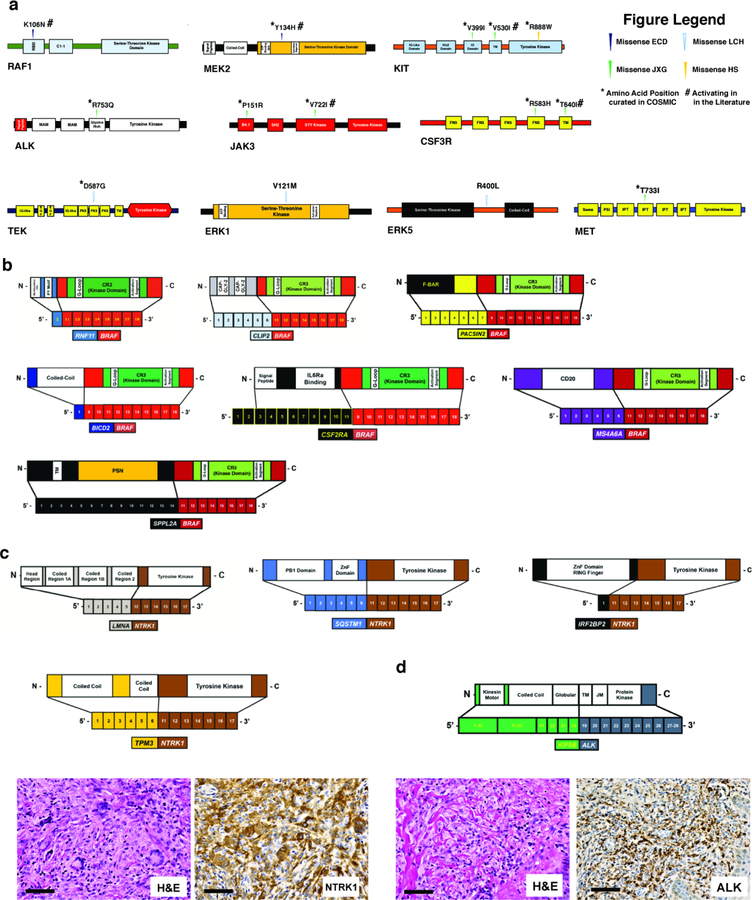
Novel kinase mutations and fusions identified in patients with histiocytic neoplasms in this study. (a) Protein diagrams of novel, somatic point mutations uncovered in kinases in histiocytoses in this study, as well as whether they have been previously described as somatic in cancer (noted by an asterisk, “*”) and/or functionally characterized (noted by pound sign, “#”). Of the 14 mutations illustrated, the RAF1 K106N5, MEK2 Y134H5, JAK3 V722I31, KIT V530I32, and CSF3R T64033 mutations have been shown to be activating previously. Also, four co-existed with other kinase alterations in the following combinations: CSF3R R583H mutation co-occurred with CSF3R T640I in a JXG patient, KIT R888W mutation co-occurred with MEK1 F53L in a histiocytic sarcoma patient, KIT V530I mutations co-occurred with a MEK1 V93I mutation in a JXG patient. (b) BRAF fusions, (c) NTRK1 fusions, (d) and ALK fusions identified. Hematoxylin and eosin (H&E) and anti-NTRK1 and anti-ALK immunohistochemical staining shown in TPM3-NTRK1-rearranged juvenile xanthogranuloma and KIF5B-ALK-rearranged Erdheim-Chester Disease tumor biopsies, respectively.
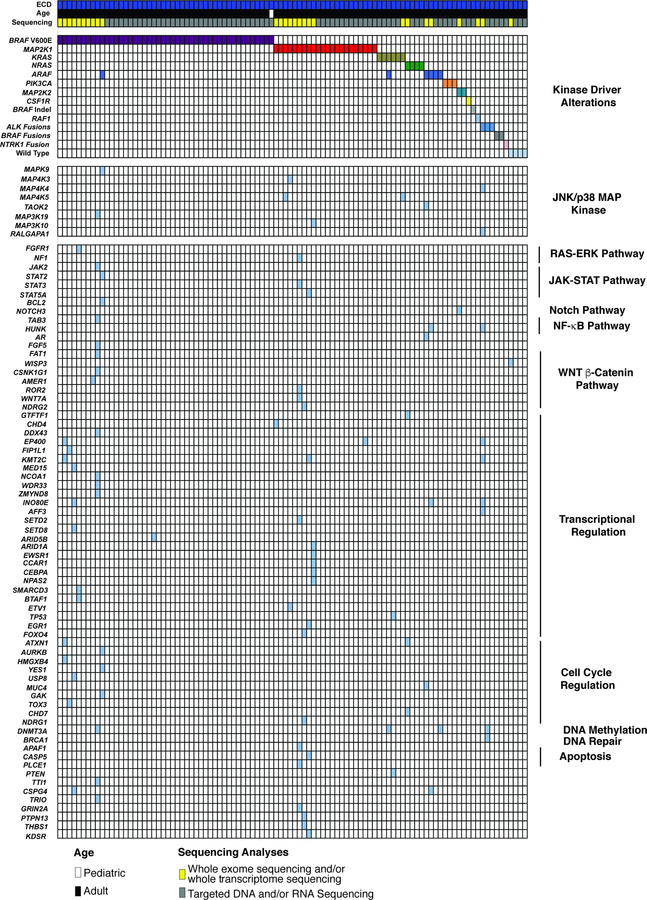
Extended Data Fig. 5. Oncoprint of mutations identified in the Erdheim-Chester Disease cohort (n=100 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (ECD), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
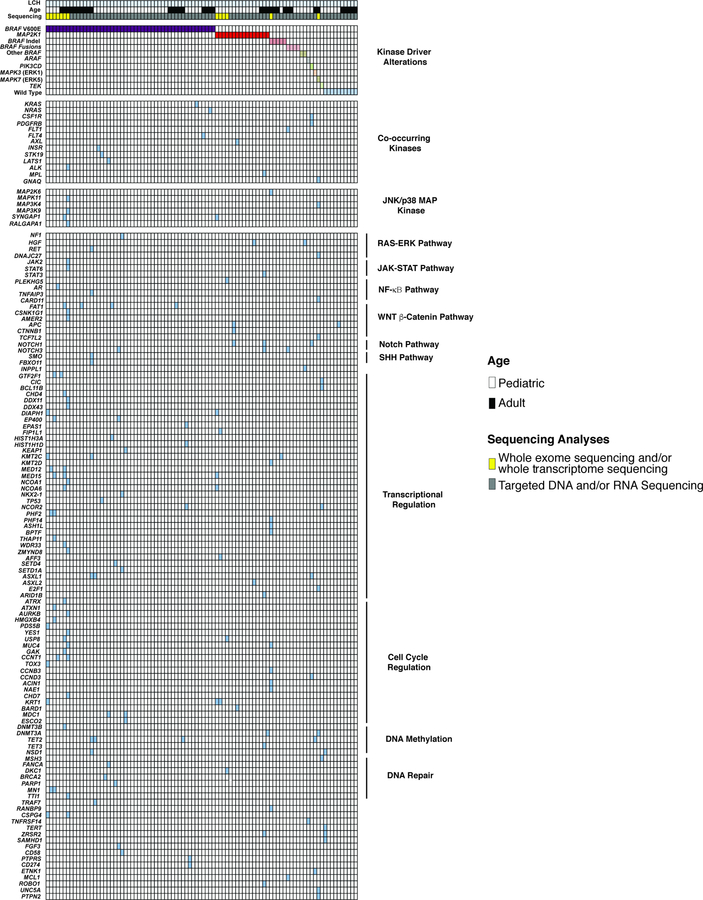
Extended Data Fig. 6. Oncoprint of mutations identified in the Langerhans Cell Histiocytosis cohort (n=92 patients).
Results of whole exome and targeted DNA and RNA sequencing of LCH neoplasms. Each patient is represented in one column. Diagnosis (LCH), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
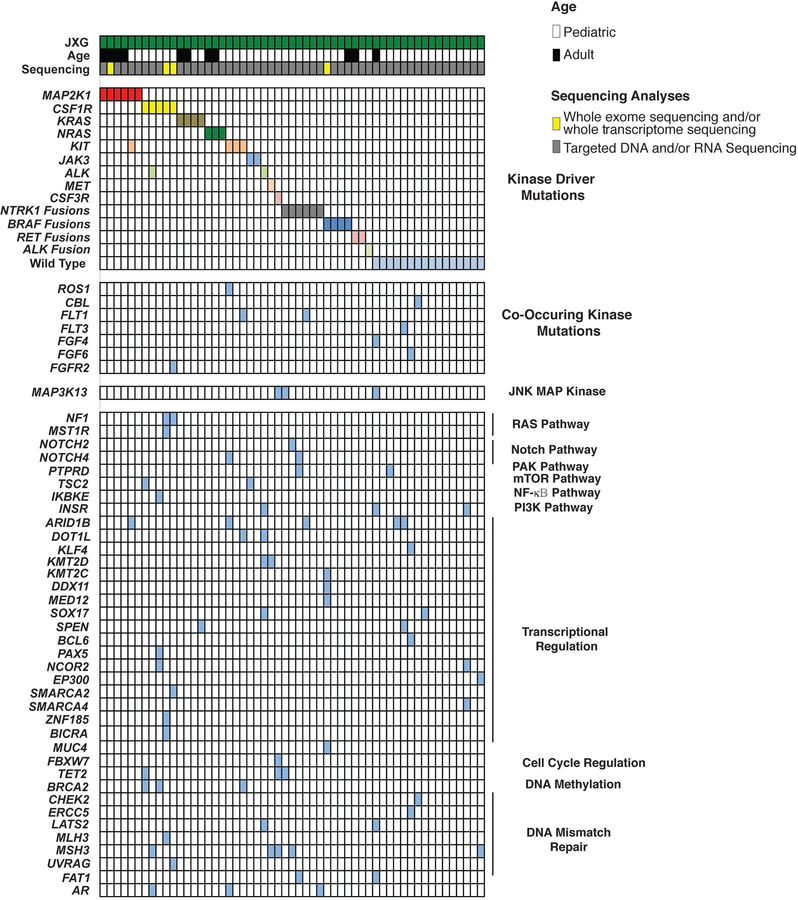
Extended Data Fig. 7. Oncoprint of mutations identified in the Juvenile Xanthogranuloma cohort (n=55 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (JXG), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 8. Oncoprint of mutations identified in the Rosai Dorfman Disease cohort (n=17 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (RDD), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases or involve a diverse array of co-occurring pathways (shown on the right).
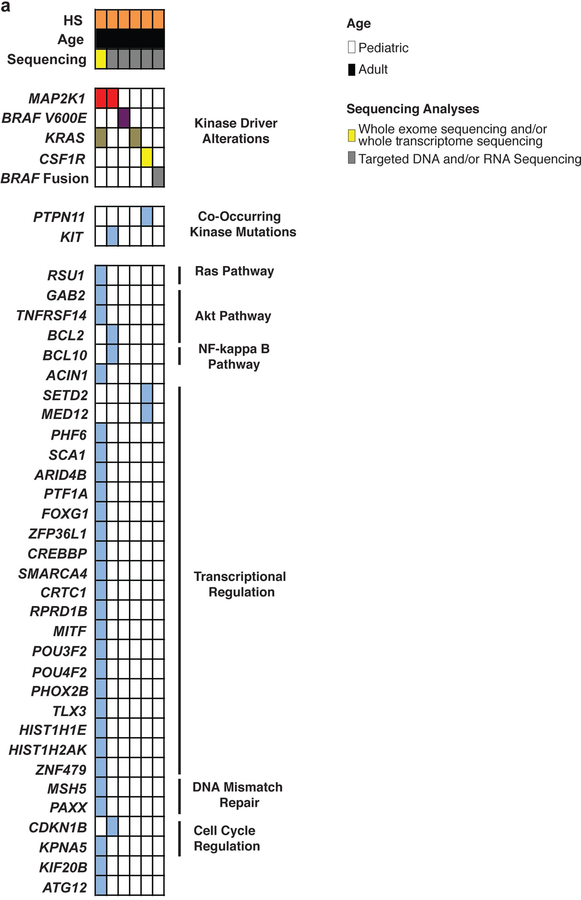
Extended Data Fig. 9. Mutations identified in histiocytic sarcoma (HS) (n=6 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (HS), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 10. Characteristics of RET fusion-driven histiocytosis and response to selpercatinib.
(a) Protein diagram of NCOA4-RET fusion identified in a cutaneous xanthogranuoma patient. Photographs of NCOA4-RET JXG skin lesions pre- and 12-weeks post selpercatinib on back (b), scrotum (c), and neck (d). (e) Number of Ba/F3 cells expressing an empty vector or the NCOA4-RET fusion following IL-3 withdrawal (mean of n=3 independent biological experiments ± standard deviation). Calculation of p-values performed using two-way ANOVA; ****p<0.0001. Representative western blotting (f) and phospho-protein flow cytometric analysis (g) of phospho-MEK1/2 and phosho-ERK1/2 in the cells from (e). Experiments performed in three independent biological experiments with similar results.
Supplementary Material
Acknowledgements
We thank the patients and their families for participating in this study as well as Lori Schmitt of UPMC Children’s Hospital of Pittsburgh for histological technical support. This work was supported by Genentech and grants from the Histiocytosis Association, the Erdheim-Chester Disease Global Alliance, the American Society of Hematology, the Leukemia & Lymphoma Society, the Pershing Square Sohn Foundation, the Functional Genomics Initiative of Memorial Sloan Kettering Cancer Center (MSK), the Society of MSK, the Translational and Integrative Medicine Award of MSK, the Geoffrey Beene Center of MSK, Frame Fund, Nonna’s Garden Foundation, the Flanders Institute for Biotechnology-Belgium (VIB), and National Institutions of Health (K08 CA218901, UL1TR001857, P30 CA008748, 1 R01 CA201247).
Footnotes
Competing financial interests
The authors declare the following competing interests: B.H.D.: grants from National Cancer Institute, American Society of Hematology, and Erdheim-Chester Disease Global Alliance Foundation. A.Y.: grants from the Leukemia & Lymphoma Society, Aplastic Anemia & MDS International Foundation, and the Lauri Strauss Leukemia Foundation. G.A.U.: personal fees from Sanofi and grants from Sanofi, Novartis, Genentech. M. Ladanyi: advisory board compensation from Merck, AstraZeneca, BristolMyers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare. D.B.S.: served as a consultant and received honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Vivideon Therapeutics and Illumina. Stock options from Loxo Oncology. M.F.B.: personal fees from Roche and research support from Illumina. D.M.H.: personal fees from Atara Biotherapeutics, Chugai Pharma, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, Debiopharm Group, and Genentech, and grants from National Cancer Institute, AstraZeneca, Puma Biotechnology, and Loxo Oncology. I.D.: served as a consultant for Apexigen, Bayer, and Celgene and research support from BMS and Novartis. S.N.S.: program grants from the Flanders Institute for Biotechnology, Belgium (VIB) and the special research fund of Ghent University, and grants for Research Foundation Flanders (FWO). M.E.L.: personal fees from LOXO, AstraZeneca, Roche/Genentech, Novartis. AD: Research fuding from Foundation Medicine and personal fees from Ignyta/Genentech/Roche, Loxo/Bayer/Lilly, Takeda/Ariad/Millenium, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Helsinn, Beigene, BergenBio, Hengrui Therapeutics, Exelixis, Tyra Biosciences, Verastem, and MORE Health. E.L.D.: grants from Erdheim-Chester Disease Global Alliance, The Histiocytosis Association, the Society of Memorial Sloan Kettering, the Frame Fund, the Joy Family West Foundation, and the American Society for Clinical Oncology. O.A.-W.: grants from National Cancer Institute, National Heart Lung and Blood Institute, Pershing Square Sohn Foundation, the Histiocyte Society, and H3B Biomedicine, and personal fees from H3B Biomedicine, Foundation Medicine Inc., Merck, and Jansen unrelated to this manuscript. The remaining authors have nothing to disclose.
References
- 1.Badalian-Very G, et al. Blood 116, 1919–1923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty R, et al. Blood 124, 3007–3015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamond EL, et al. Cancer discovery 6, 154–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyman DM, et al. The New England journal of medicine 373, 726–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond EL, et al. Nature 567, 521–524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz AM, et al. J Am Acad Dermatol 24, 32–37 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Chantorn R, Wisuthsarewong W, Aanpreung P, Sanpakit K & Manonukul J Pediatr Dermatol 25, 470–473 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Dai XM, et al. Blood 99, 111–120 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Mass E, et al. Nature 549, 389–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambiaghi S, Restano L & Caputo R Pediatr Dermatol 21, 97–101 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Elegheert J, et al. Structure 19, 1762–1772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felix J, et al. Structure 21, 528–539 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Felix J, et al. Structure 23, 1621–1631 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Elegheert J, et al. Nat Struct Mol Biol 19, 938–947 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Walter M, et al. J Mol Biol 367, 839–847 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Cheng DT, et al. The Journal of molecular diagnostics: JMD 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durham BH, et al. Blood (2017). [Google Scholar]
- 18.Benayed R, et al. Clinical cancer research: an official journal of the American Association for Cancer Research (2019). [Google Scholar]
- 19.Gruber TA, et al. Cancer Cell 22, 683–697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, et al. Nature 481, 157–163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal R, McGranahan N, Herrero J, Taylor BS & Swanton C Genome Biol 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, et al. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang MN, et al. Sci Rep 5, 13321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes-Ciriano I, Lee S, Park WY, Kim TM & Park PJ Nature communications 8, 15180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov LB, et al. bioRxiv (2018). [Google Scholar]
- 26.Walters DK, et al. Cancer Cell 10, 65–75 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Cammenga J, et al. Blood 106, 3958–3961 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Maxson JE, et al. Clinical cancer research: an official journal of the American Association for Cancer Research 22, 757–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-exome sequencing data are deposited in DBGAP project accession number phs001864.v1.p1.