ABSTRACT
The endocytic protein EHD1 plays an important role in ciliogenesis by facilitating fusion of the ciliary vesicle and removal of CP110 (also known as CCP110) from the mother centriole, as well as removal of Cep215 (also known as CDK5RAP2) from centrioles to permit disengagement and duplication. However, the mechanism of its centrosomal recruitment remains unknown. Here, we address the role of the EHD1 interaction partner MICAL-L1 in ciliogenesis. MICAL-L1 knockdown impairs ciliogenesis in a similar manner to EHD1 knockdown, and MICAL-L1 localizes to cilia and centrosomes in both ciliated and non-ciliated cells. Consistent with EHD1 function, MICAL-L1-depletion prevents CP110 removal from the mother centriole. Moreover, upon MICAL-L1-depletion, EHD1 fails to localize to basal bodies. Since MICAL-L1 localizes to the centrosome even in non-ciliated cells, we hypothesized that it might be anchored to the centrosome via an interaction with centrosomal proteins. By performing mass spectrometry, we identified several tubulins as potential MICAL-L1 interaction partners, and found a direct interaction between MICAL-L1 and both α-tubulin–β-tubulin heterodimers and γ-tubulin. Our data support the notion that a pool of centriolar γ-tubulin and/or α-tubulin–β-tubulin heterodimers anchor MICAL-L1 to the centriole, where it might recruit EHD1 to promote ciliogenesis.
KEY WORDS: EHD1, MICAL-L1, Centriole, Cilia, Ciliogenesis, Tubulin
Highlighted Article: A pool of centriolar γ-tubulin and/or α/β-tubulins anchor MICAL-L1 to the centriole, where it might recruit EHD1 to promote ciliogenesis.
INTRODUCTION
Cilia are unique organelles that regulate a multitude of signaling pathways (Breslow and Holland, 2019), and defects in ciliogenesis lead to a wide range of ciliopathies that include Bardet–Biedl syndrome (Novas et al., 2015), Joubert syndrome (Brancati et al., 2010), polycystic kidney disease (Ghata and Cowley, 2017), retinitis pigmentosa (Megaw and Hurd, 2018) and others (Fabbri et al., 2019). Ciliogenesis occurs by two different processes, likely dependent on cell and tissue type: (1) the extracellular pathway, and (2) the intracellular pathway (Sorokin, 1962, 1968). In the extracellular pathway, before axonemal growth is initiated, the mother centriole (m-centriole) first docks with the plasma membrane and the axonemal microtubules extend and deform the membrane (Wang and Dynlacht, 2018). During intracellular ciliogenesis, vesicles dock at the distal appendages of the m-centriole and fuse to form the ciliary vesicle (CV) (Garcia et al., 2018). The CV recruits transition zone and intraflagellar transport proteins, and is deformed and extended by axonemal growth, ultimately leading to fusion with the plasma membrane (Lu et al., 2015).
Membrane trafficking plays a key role in the intracellular ciliogenesis pathway that characterizes fibroblasts and retinal pigment epithelium (RPE-1) cells (Sorokin, 1962, 1968). One of the first steps of ciliogenesis is the docking of myosin Va-containing preciliary vesicles derived from the endocytic pathways on the distal appendages of the m-centriole. Through an unknown mechanism, the Eps15 homology domain protein, EHD1, is recruited and interacts with SNAP29 to catalyze the fusion of the preciliary vesicles to form the ciliary vesicle (Lu et al., 2015). Shortly afterward, Arl13B (Caspary et al., 2007; Hori et al., 2008) and Rab11 are observed at the ciliary membrane, leading to the recruitment of the Rab11 effector, RABIN8 (also known as RAB3IP), which acts as a guanine nucleotide exchange factor (GEF) for Rab8 (Knodler et al., 2010), and ultimately ciliary regulatory proteins such as CP110 (also known as CCP110) are removed to promote generation of the ciliary sheath and its fusion with the plasma membrane upon axoneme growth (Lu et al., 2015).
Despite the significant role for EHD1 in the generation of the ciliary vesicle and subsequent removal of CP110 from the m-centriole (Lu et al., 2015), the mechanism of recruitment of EHD1 to the centrosome remains unknown. Herein, we identify MICAL-L1, a central protein that is required for the biogenesis of tubular recycling endosomes and endocytic recycling (Giridharan et al., 2012, 2013; Rahajeng et al., 2012; Sharma et al., 2010, 2009), as a novel regulator of primary cilia, and provide evidence that it functions by recruiting EHD1 to the cilium, similar to its role in recruitment of EHD1 to tubular recycling endosomes (Sharma et al., 2009). Moreover, we demonstrate that MICAL-L1 localizes to centrioles even in non-ciliated cells, through a direct interaction with α-tubulin–β-tubulin heterodimers and/or γ-tubulin. Overall, our findings highlight MICAL-L1 as a novel regulator of ciliogenesis and resident of the centrosome whose role includes the recruitment of EHD1 to the nascent cilium.
RESULTS AND DISCUSSION
MICAL-L1 regulates ciliogenesis and localizes to the primary cilium
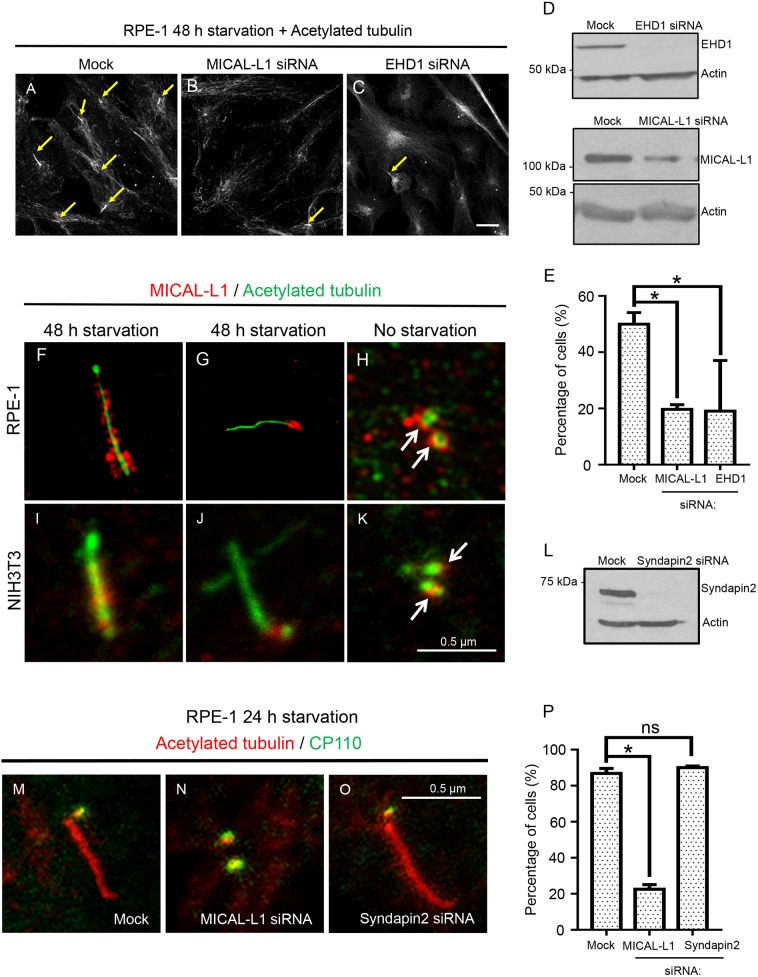
To determine the potential role of MICAL-L1 in ciliogenesis, we mock-treated RPE-1 cells, or treated them with siRNA specific for either MICAL-L1 or EHD1, followed by 48 h serum starvation, prior to detection of cilia with antibodies against acetylated tubulin (Fig. 1A–E). As expected, ∼50% of mock-treated RPE-1 cells were ciliated (Fig. 1A,E). However, knockdown of either MICAL-L1 or EHD1 (Fig. 1D) led to only ∼20% of RPE-1 cells being detected with cilia (Fig. 1B,C, and quantified in E). Moreover, whereas ∼60% of mock-treated serum-starved NIH3T3 cells generated cilia, upon MICAL-L1 knockdown, the percentage of ciliated cells was reduced to below 25% (Fig. S1A). Knockdown of MICAL-L1 also led to loss of anti-MICAL-L1 antibody staining at the basal body, demonstrating specificity of the antibody (Fig. S1B). In addition, ‘rescue’ of MICAL-L1-knockdown cells by transfection of siRNA-resistant GFP–MICAL-L1 led to dramatically increased percentages of ciliated cells (Fig. S2A,B). These data are consistent with a previous study demonstrating the involvement of EHD1 in ciliogenesis (Lu et al., 2015), but also show for the first time that the EHD1-binding partner and recycling endosome constituent MICAL-L1 is also required for ciliogenesis.
Fig. 1.
MICAL-L1 regulates ciliogenesis. (A–E) RPE-1 cells were mock-treated (with Lipofectamine in the absence of the siRNA oligonucleotide), treated with MICAL-L1 siRNA or with EHD1 siRNA for 72 h. During the last 48 h, the cells were serum-starved to induce ciliogenesis. Cilia were detected by immunostaining with an anti-acetylated tubulin antibody (marked by yellow arrows in A–C). Compared to mock-treated cells (A), fewer cilia were generated upon MICAL-L1 depletion (B), and upon EHD1 depletion (C). MICAL-L1 and EHD1 knockdown efficacies (>90%) were verified by immunoblotting (D). The percentage of ciliated cells following serum-starvation was quantified and presented as a bar graph (E). Scale bar: 10 μm. (F–K) RPE-1 (F–H) or NIH3T3 (I–K) cells were fixed or treated with medium containing 0.2% serum for 48 h. Ciliated (F,G,I,J) or non-ciliated cells (H,K) were immunostained with antibodies to MICAL-L1 (red) and acetylated tubulin (green). MICAL-L1 localized to the cilia (F,I) and the basal body (G,J) upon serum starvation, and the centrosome (H,K) even in the absence of ciliogenesis. Micrographs in F–H were obtained by sr-SIM, whereas micrographs in I–K were from confocal microscopy. Scale bar: 0.5 μm. (L–P) RPE-1 cells were mock-treated (M), or subjected to MICAL-L1 depletion (N) or syndapin 2 siRNA-mediated depletion (O), followed by serum starvation for 24 h, and immunostained with acetylated tubulin (red) and CP110 (green) antibodies. CP110 removal from the m-centriole was impaired upon MICAL-L1 depletion (N; green dots co-localized with red acetylated tubulin; quantified in P). Upon syndapin 2 depletion, CP110 was released from the m-centriole (O, quantified in P). The efficacy of syndapin 2 knockdown was determined by immunoblotting (L). Graph shows the percentage of cells displaying CP110 removal from the m-centriole (P). Scale bar: 0.5 μm. *P<0.001; ns, not significant (P>0.05), for comparison between mock- and siRNA-treated cells; n=3 experiments (>100 cells quantified for each experiment). Error bars denote standard deviation.
Given our finding that MICAL-L1 regulates ciliogenesis, we asked whether a pool of MICAL-L1 localizes to cilia and/or the ciliary pocket membrane (hereafter collectively referred to as cilia). To this aim, we used serum-starved RPE-1 and NIH3T3 cells immunostained with antibodies to both acetylated tubulin (to detect cilia) and endogenous MICAL-L1 (Fig. 1F,G, and I,J, respectively). In ciliated cells from both cell lines, we detected MICAL-L1 along the length of the cilium (Fig. 1F,I), or localized at the base of the cilium, potentially at the ciliary pocket membrane (Fig. 1G,J). Although we have yet to understand why MICAL-L1 is observed exclusively at the base of some cilia, it is possible that this represents an early stage in ciliogenesis and additional experimentation will be required to determine whether this is the case. Three-dimensional super-resolution structured illumination microscopy (sr-SIM) suggested that MICAL-L1 might localize to the distal side of the m-centriole, based on its localization with respect to the centrin 1 distal centriole marker (Fig. S2C and D); however, in some instances the proximal centriolar marker C-Nap 1 (also known as CEP250) could also be observed juxtaposed with MICAL-L1 (data not shown), highlighting the need for single-molecule super-resolution or electron microcopy to ultimately resolve this issue. Importantly, MICAL-L1 was also observed at the acetylated tubulin-marked centrosomes of non-ciliated cells (Fig. 1H,K, see arrows), suggesting it plays an earlier role than EHD1 in ciliogenesis. However, consistent with findings that EHD1 is not required for the transport and recruitment of distal appendage vesicles to distal appendages (Insinna et al., 2019), MICAL-L1-depletion had little effect on myosin Va-positive vesicles and their arrival and docking at the centriolar distal appendages (Fig. S3).
Since EHD1 is responsible for removal of the capping protein CP110 from the m-centriole to facilitate ciliary growth (Lu et al., 2015), we asked whether MICAL-L1 mediates CP110 removal in RPE-1 cells. Indeed, whereas ∼90% of mock-treated cells displayed CP110 loss from the m-centriole (Fig. 1M, quantified in P), upon MICAL-L1 depletion, only ∼20% of cells displayed CP110 loss from the m-centriole (Fig. 1N, quantified in P). Syndapin 2 (also known as PACSIN2) depletion (Fig. 1L), another EHD1 interaction partner that plays a role in the generation of tubular recycling endosomes (TRE) along with MICAL-L1 (Giridharan et al., 2013), had no effect on CP110 retention on the m-centriole (Fig. 1O, quantified in P). This is consistent with the recent findings of Insinna et al., who demonstrated that whereas syndapin 1 (also known as PACSIN1) controls ciliogenesis in RPE-1 cells, in this cell line syndapin 2 is superfluous for ciliogenesis (Insinna et al., 2019).
MICAL-L1 recruits EHD1 to the primary cilium
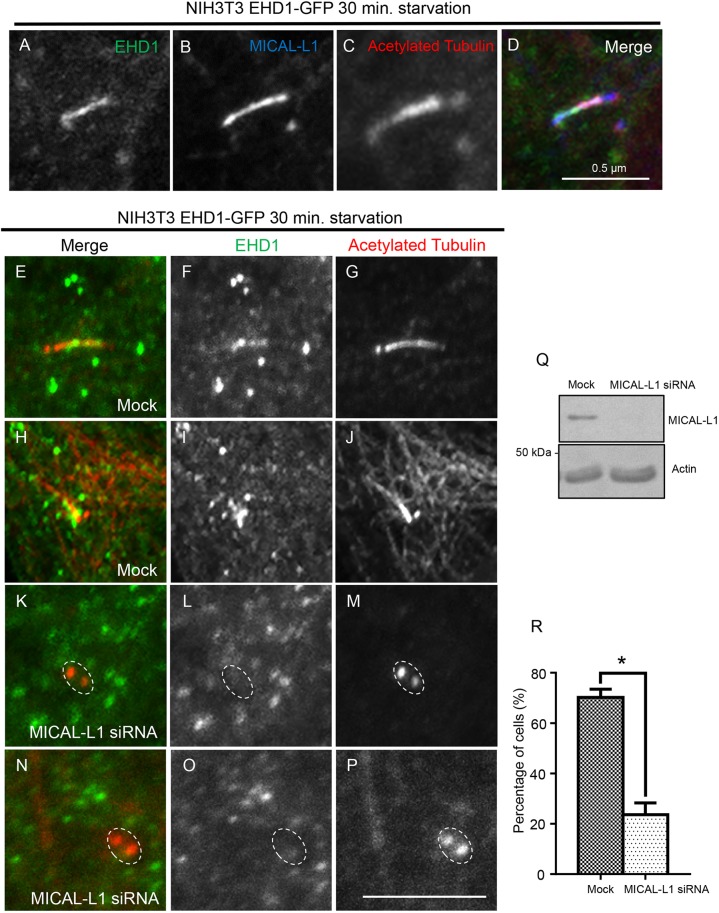
A central unanswered question is how EHD1, which is required both for SNAP29 recruitment and CV fusion, as well as CP110 removal from the m-centriole (Lu et al., 2015), is recruited to the basal body and cilium/ciliary pocket. Given its role in recruitment of EHD1 to TRE (Giridharan et al., 2013; Sharma et al., 2010, 2009), and the localization of MICAL-L1 to the centrosome in non-ciliated cells (Fig. 1H,K), we hypothesized that MICAL-L1 is involved in EHD1 recruitment to the cilium. Accordingly, we serum-starved CRISPR/CAS9 gene-edited NIH3T3 cells expressing endogenous levels of EHD1 as a GFP fusion protein (Xie et al., 2018; Yeow et al., 2017), and immunostained them for EHD1, MICAL-L1 and acetylated tubulin. Under these serum-starved conditions, both EHD1 and MICAL-L1 could be detected along acetylated tubulin-stained cilia (Fig. 2A–D). To assess whether MICAL-L1 recruits EHD1 to cilia, we again used our NIH3T3 cells with EHD1–GFP. Upon serum starvation, in mock-treated cells, EHD1–GFP localized to acetylated tubulin-decorated cilia in ∼70% of the cells (Fig. 2E–J, quantified in R). However, upon MICAL-L1 depletion (Fig. 2Q), in the majority of cells, EHD1 was absent from the acetylated tubulin-stained centrioles (only ∼20% had EHD1 localization to the centrioles) (Fig. 2K–P, quantified in R). These data suggest that MICAL-L1 likely precedes EHD1 in the regulation of ciliogenesis by recruiting EHD1 to centrioles.
Fig. 2.
MICAL-L1 recruits EHD1 to the primary cilium. (A–D) CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1–GFP were immunostained for MICAL-L1 (blue) and acetylated tubulin (red). (E–R) CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1–GFP were mock-treated (with Lipofectamine in the absence of the siRNA oligonucleotide; E–J) or treated with MICAL-L1-siRNA (K–P), serum starved for 30 min, and immunostained for acetylated tubulin (red) or with anti-GFP antibodies (green). EHD1 was detected on acetylated tubulin-labeled cilia in mock-treated cells (E–J), but in the absence of MICAL-L1, EHD1 was not detected on basal bodies positive for acetylated tubulin (K–P; dashed ovals). MICAL-L1 siRNA efficacy (>90%) was confirmed by immunoblotting (Q). The bar graph indicates the percentage of cells showing recruitment of EHD1 onto cilia or the m-centriole (R). *P<0.001; n=3 independent experiments (>30 cells per experiment were quantified). Error bars denote standard deviation. Scale bars: 0.5 μm.
MICAL-L1 interacts directly with tubulins at the centrosome
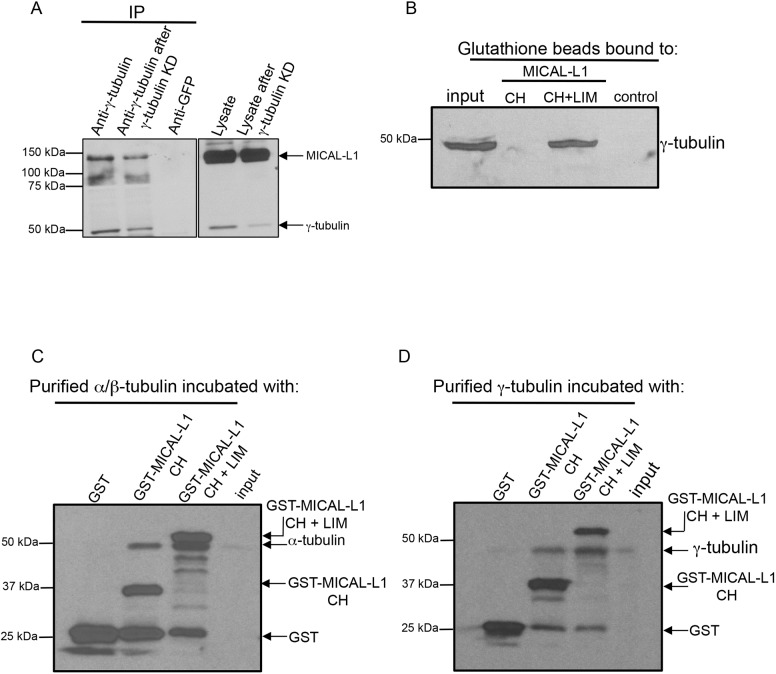
The residence of MICAL-L1 at the centrosome even in non-ciliated cells suggests a potential interaction with a centrosomal protein(s). Immunoprecipitations of full-length MICAL-L1 and GST pulldown experiments with the MICAL-L1 calponin homology (CH) and ‘Lin11, Isl-1 and Mec-3’ (LIM) domains coupled with mass spectrometry elicited a variety of tubulin species, hinting at a potential microtubule interaction. To test for interactions with tubulins, we performed immunoprecipitations with γ-tubulin from cell lysates and immunoblotted with antibodies to MICAL-L1 and γ-tubulin (Fig. 3A). MICAL-L1 bands (and γ-tubulin bands) were detected by precipitating with γ-tubulin antibodies, but not with a control immunoglobulin (Fig. 3A, anti-GFP). Moreover, the level of MICAL-L1 immunoprecipitated by γ-tubulin was decreased when lysates from cells treated with γ-tubulin siRNA were used (Fig. 3A, γ-tubulin, ‘anti-γ-tubulin after γ-tubulin KD’). In addition, purified GST-tagged MICAL-L1 CH and LIM domains alone precipitated γ-tubulin (Fig. 3B). To assess whether the MICAL-L1–tubulin interactions were direct, we next incubated the MICAL-L1 CH and LIM domains, the MICAL-L1 CH domain and GST-only controls with either purified α-tubulin--β-tubulin heterodimers (Fig. 3C) or γ-tubulin (Fig. 3D). As demonstrated, in these in vitro assays, both the α-tubulin–β-tubulin heterodimers and γ-tubulins interacted robustly with the MICAL-L1 CH and LIM domains, and to a lesser extent, the CH domains alone, but displayed no interactions with the control GST. These findings suggest that MICAL-L1 is initially recruited to centrosomes in non-ciliated cells through a direct interaction with γ-tubulin and/or α-tubulin–β-tubulin heterodimers.
Fig. 3.
MICAL-L1 interacts directly with tubulins. (A) Mock-treated (lysate) or γ-tubulin knockdown (lysate after γ-tubulin knockdown) NIH3T3 cells were lysed and subjected to immunoblotting (right panel). Immunoprecipitations (IP) were undertaken using anti-γ-tubulin, or rabbit anti-GFP (control) antibodies. Immunoprecipitated proteins were immunoblotted with anti-MICAL-L1 and anti-γ-tubulin antibodies (left panel). (B) 500 ng of purified GST alone, GST–MICAL-L1-CH domain or GST-MICAL-L1-CH+LIM domains were bound to GST beads, and incubated with NIH3T3 cell lysates. Precipitated proteins were eluted from the beads and immunoblotted using anti-γ-tubulin antibody. (C,D) 500 ng of purified GST, GST–MICAL-L1-CH domain or GST–MICAL-L1-CH+LIM domains (bait) were bound to GST beads, and treated with 1 U of micrococcal nuclease. 0.1 µg purified α-tubulin–β-tubulin heterodimers (C) or γ-tubulin (D) were treated with 1 U of micrococcal nuclease. Immobilized GST–bait proteins were incubated with α-tubulin–β-tubulin heterodimers or γ-tubulin, and bound protein was immunoblotted.
MICAL-L1 displays a centriolar localization
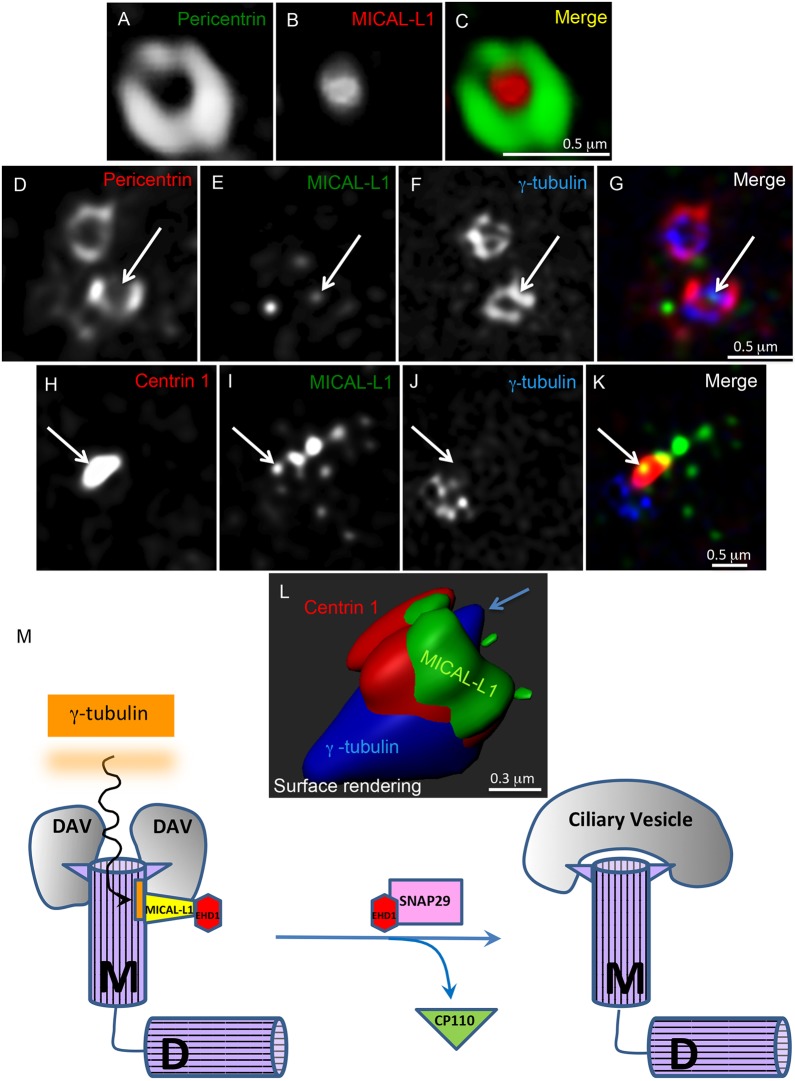
Given that α/β-tubulins comprise the centriolar wall (Breslow and Holland, 2019), and that γ-tubulins localize to the centrioles in addition to their well-characterized localization to the pericentriolar matrix (PCM) (Fuller et al., 1995; Sonnen et al., 2012), we asked whether MICAL-L1 displays a centriolar or PCM localization pattern. Using sr-SIM, we imaged cells for the PCM marker, pericentrin, together with MICAL-L1 (Fig. 4A–C). Surprisingly, MICAL-L1 (red) displayed little overlap with the typical peripheral pericentrin ring (green), and instead appeared closer to the center of the ring. We next immunostained the cells in three channels, including a γ-tubulin stain, in addition to that for pericentrin and MICAL-L1 (Fig. 4D–G). As expected, γ-tubulin displayed partial overlap in the peripheral PCM with pericentrin, but consistent with the findings of Fuller et al. (1995) and Sonnen et al. (2012), γ-tubulin also partially localized to a more central region of the centrosome, likely corresponding with the centrioles. Indeed, the MICAL-L1 signal displayed partial overlap with the small pool of centriolar γ-tubulin, but little or no overlap with pericentrin or γ-tubulin in the PCM (Fig. 4D–G, arrows mark the overlap). Moreover, when we immunostained cells for the centriolar marker centrin 1, together with MICAL-L1 and γ-tubulin, we found that MICAL-L1 partially overlapped with centrin 1 (Fig. 4H–K, see arrows), but not with pericentriolar γ-tubulin, and extended along an axis that likely represents the cilium (Fig. 4I,K). Surface rendering of these images consistently showed MICAL-L1 distal to γ-tubulin and the PCM, partially overlapping with centrin 1 and potentially centriolar γ-tubulin (Fig. 4L).
Fig. 4.
MICAL-L1 displays a centriolar localization. (A–C) NIH3T3 cells were serum starved for 30 min and immunostained for pericentrin (green) and MICAL-L1 (red). sr-SIM images were acquired. Scale bar: 0.5 μm (D–G) NIH3T3 cells were serum-starved for 30 min and immunostained for pericentrin (red), MICAL-L1 (green), and Alexa Fluor 488-conjugated anti-γ-tubulin antibody (blue). The cells were analyzed by sr-SIM imaging. Images were processed by 3D rendering using Imaris. MICAL-L1 partially colocalized with γ-tubulin in the central region of the centrosome (arrow), likely corresponding with the centrioles. Scale bar: 0.5 μm. (H–L) NIH3T3 cells were serum-starved for 30 min and immunostained for Centrin 1 (red), MICAL-L1 (green), and Alexa 488-conjugated anti-γ-tubulin antibody (blue) (H–K). Scale bar: 0.5 μm. Arrows show partial overlap between MICAL-L1 and centrin 1. 3D surface rendering suggests that MICAL-L1 displays partial overlap with centrin 1 and centriolar γ-tubulin, but not with pericentriolar γ-tubulin (L). Blue arrow marks centriolar γ-tubulin overlapping with MICAL-L1. Scale bar: 0.3 μm. (M) Simplified model for the involvement of MICAL-L1 in ciliogenesis. γ-tubulin and/or α/β-tubulin associated with the m-centriole recruit MICAL-L1 (primarily) to the distal m-centriole region. During the induction of ciliogenesis, MICAL-L1 recruits EHD1, which interacts with SNAP29 and facilitates CP110 removal from the m-centriole, leading to ciliary vesicle fusion and ciliogenesis. M, mother-centriole; D, daughter-centriole.
Given the interactions between MICAL-L1 and γ-tubulin, and the partial overlap observed at the centriole as seen with sr-SIM, we hypothesized that ciliogenesis may first require MICAL-L1 recruitment to centrioles by γ-tubulin, and in turn, the recruitment of EHD1 by MICAL-L1. To address this hypothesis of ‘serial recruitment’, we used our NIH3T3 CRISPR/Cas9 EHD1-knockout cells. EHD1 depletion from these cells did not impact the MICAL-L1 recruitment to the basal body (Fig. S4A; graph shows the level of MICAL-L1 at the basal body). Surprisingly, it is possible to deplete γ-tubulin from cells while maintaining them in culture (Shi et al., 2011); accordingly we used siRNA to decrease γ-tubulin levels. As expected, this caused a dramatic decrease in the number of ciliated cells observed upon serum starvation (Fig. S4B). However, the γ-tubulin decrease significantly reduced MICAL-L1 localized to the basal body (Fig. S4C).
Overall, we identify MICAL-L1 as a novel endocytic protein that regulates ciliogenesis. Our data suggest a model whereby γ-tubulin and/or α/β-tubulins serve as anchors to recruit MICAL-L1 to the centrosome, which in turn recruits EHD1 (Fig. 4M). EHD1 is required for recruitment of the SNARE protein, SNAP29 and release of CP110 from the m-centriole, thus facilitating ciliary vesicle formation and generation of a cilium (Lu et al., 2015). Our findings provide new insight into the complexities of ciliogenesis by identifying a new endocytic regulator in this process.
MATERIALS AND METHODS
Reagents and antibodies
Antibodies were purchased from suppliers as indicated: mouse monoclonal anti-acetylated tubulin antibodies (Sigma-Aldrich, St. Louis, MO), rabbit polyclonal antibodies to anti-acetylated tubulin (Cell Signaling, Danvers, MA), rabbit polyclonal anti-centrin 1 antibodies (ProteinTech, Rosemont, IL), rabbit polyclonal anti-CP110 antibodies (ProteinTech), mouse monoclonal anti-γ-tubulin antibodies (Sigma-Aldrich), rabbit polyclonal anti-γ-tubulin antibodies (Abcam), goat polyclonal anti-GST antibodies conjugated with Horse Radish Protein (HRP) (GenScript, Piscataway, NJ), rabbit polyclonal anti-myosin Va antibodies (Novus Biologicals, Littleton, CO), rabbit polyclonal anti-pericentrin antibodies (Abcam, San Francisco, CA), mouse monoclonal anti-GFP antibodies (Sigma-Aldrich), mouse monoclonal anti-MICAL-L1 antibodies (Novus Biologicals), rabbit polyclonal anti-MICAL-L1 antibodies (Abcam), rabbit anti-syndapin 2 antibody (Sigma) and mouse monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies conjugated with HRP (Protein Tech), mouse monoclonal anti-actin antibodies (Novus Biologicals). Alexa 488 fluorophore was attached to anti-γ-tubulin antibodies using APEX™ Alexa Fluor 488 Antibody Labeling Kit (Invitrogen, Eugene, OR). All secondary antibodies used for immunofluorescence were purchased from Molecular Probes (Eugene, OR). For co-immunoprecipitation assays, rabbit polyclonal anti-GFP antibodies (Pearson, London, UK) were used as a negative control. Purified porcine α-tubulin–β-tubulin heterodimers were obtained from Cytoskeleton, Inc. (cat. # T240). Purified γ-tubulin was obtained from Novus (catalog #NBP1-72534). See Table S1 information for additional antibody details.
Cell culture and treatments
The human epithelial cell line hTERT RPE-1 (ATCC-CRL4000) was grown at 37°C in 5% CO2 in DMEM/F12 (Thermo Fisher Scientific, Carlsbad, CA) containing 10% fetal bovine serum (FBS; Sigma-Aldrich), 2 mM L-glutamine, 100 U/ml penicillin/streptomycin and 1× non-essential amino acids (Thermo Fisher Scientific). The CRISPR/Cas9 gene-edited NIH3T3 cell line expressing endogenous levels of EHD1 with GFP attached to its C-terminus, as well as the EHD1-knockout cells were generated as described previously (Yeow et al., 2017). Parental and gene-edited NIH3T3 cells were cultured at 37°C in 5% CO2 in DMEM (GE Healthcare, Chicago, IL) containing 10% FBS, with 2 mM L-glutamine and 100 U/ml penicillin/streptomycin. To induce ciliogenesis, RPE-1 cells were incubated in DMEM/F12 with 2 mM L-glutamine, 1× non-essential amino acids and 0.2% FBS, NIH3T3 was treated with serum-free DMEM with 2 mM L-glutamine for the indicated times. HeLa cells (ATCC-CCL-2) were obtained from the ATCC. All cell cultures were routinely tested for Mycoplasma contamination.
Co-immunoprecipitation
Mock- or γ-tubulin siRNA-treated HeLa cells growing in 100 mm dishes were lysed in lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% NP-40 and freshly added protease inhibitor cocktail. Cell debris was eliminated by centrifugation at 1889 g at 4°C for 10 min. The cleared lysate was collected and incubated with anti-γ-tubulin antibodies or anti-GFP antibodies (negative control) overnight at 4°C. Protein G–agarose beads (GE healthcare) were added to the lysate–antibody mix and left to rock at 4°C for 4 h. Samples were then washed three times with lysis buffer. Protein complexes were eluted from the beads by boiling the sample for 10 min in the presence of 4× loading buffer containing 250 mM Tris-HCl pH 6.8, 8% SDS, 40% glycerol, 5% β-mercaptoethanol and 0.2% (w/v) Bromophenol Blue. Eluted proteins were detected by immunoblotting.
Immunoblotting
For examination of protein knockdown efficiency, siRNA-treated cells were washed twice with pre-chilled PBS and harvested with a rubber cell scraper. Cell pellets were lysed by resuspending in lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and freshly added protease inhibitor cocktail (Roche) for 30 min on ice. The cell lysates were then centrifuged at 1889 g at 4°C for 10 min. The amounts of protein from each sample were measured and equalized by Bio-Rad protein assay (Bio-Rad, Hercules, CA), and eluted by boiling with 4× loading buffer. Proteins from either cell lysates or immunoprecipitations were separated by SDS-PAGE on 10% gels. Proteins were transferred onto nitrocellulose membranes (Midsci, St Louis, MO). Membranes were blocked for 30 min at room temperature in PBS plus 0.3% (v/v) Tween-20 (PBST) containing 5% dried milk, and then incubated overnight at 4°C with diluted primary antibodies. Protein–antibody complexes were detected with HRP-conjugated goat anti-mouse-IgG (Jackson Research Laboratories, Bar Harbor, ME) or donkey anti-rabbit-IgG (GE Healthcare) secondary antibodies for 1 h at room temperature, followed by enhanced chemiluminescence substrate (Thermo Fisher Scientific).
DNA constructs, transfection and siRNA treatment
MICAL-L1-Calponin homology domain (CH domain) and MICAL-L1-CH+Lin11, Isl-1, Mec-3 (LIM) domain (CH+LIM domain) constructs were cloned into pGEX-6p-2 generated as described previously (Sharma et al., 2009). A MICAL-L1 cDNA with silent point mutations rendering it insensitive to the MICAL-L1 siRNA oligonucleotides was cloned into pEGFP-C3 as described previously (Sharma et al., 2009). 1.5 μg DNA was transfected into RPE cells for 48 h at 37°C using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Four specific siRNA oligonucleotides (On-Target SMART pool, Dharmacon, Lafayette, CO) were simultaneously directed at human MICAL-L1 (5′-CCGGGUUCCUGGCAAACUA-3′, 5′-GCUAGGAAACAAACGUGAU-3′, 5′-GGUUCAAGCUCAUCCACGA-3′, 5′-GAAUGGGCCUGACGGGCAA-3′) or Syndapin2 (5′-CAAAUUAUGUGGAGGCGAU-3′, 5′-CCCUUAAUGUCCCGAGCAA-3′, 5′-CCUCACUGAUGAACGAUGA-3′, 5′-CUGAGGUGGUUCCGAGCCA-3′). On-Target SMARTpool siRNA targeting human γ-tubulin (5′-UCUUAGAACGGCUGAAUGA-3′, 5′-CUAGAGGGCUUUGUGCUGU-3′, 5′-GAACCUGUCGCCAGUAUGA-3′, 5′-AAAGAUCCAUGAGGACAUU-3′) was purchased from Dharmacon. A custom EHD1 siRNA oligonucleotide (5′-AAGAAAGAGATGCCCAATGTC-3′) was also obtained from Dharmacon. For knockdown experiments, ∼5.0×104 RPE-1 or NIH3T3 cells were plated on coverslips in a 35 mm culture dish for 24 h, and then transfected with 50 nM of On-Target SMARTpool oligonucleotides targeting MICAL-L1 using Lipofectamine RNAi/MAX (Invitrogen) for 72 h in the absence of antibiotics, as per the manufacturer's protocol. At 24 h after transfection, the culture medium was changed for fresh medium containing 0.2% serum for ciliogenesis induction. The efficiency of knockdown was determined by immunoblotting.
GST pulldown
NIH3T3 cells grown on a 100-mm culture dish were collected by Cellstripper (Corning, Corning, NY), and lysed with GST lysis buffer (25 mM HEPES pH 7.4, 100 mM NaCl, 1 mM EDTA and 0.5% Triton-X) with freshly added AEBSF protease inhibitor for 30 min at 4°C. Cell debris was cleared by ultracentrifugation at 1889 g for 10 min. Then, 30 μg of purified GST, GST–MICAL-L1-CH or GST–MICAL-L1-CH+LIM domain was bound to GST beads (Genscript, Nanjing, China) by rocking at 4°C for 4 h. The protein-bound beads were then incubated with NIH3T3 cell lysates at 4°C overnight. The protein complex pulled down with beads was washed three times with GST wash buffer (25 mM HEPES pH 7.4, 100 mM NaCl, 1 mM EDTA and 0.1% Triton-X), eluted by boiling with SDS-containing loading buffer, and then subjected to immunoblotting for protein detection.
Modified protein–protein interaction assay
The protein interaction assay was modified from the protocol described above in the GST pulldown section. The GST beads were washed four times with TGEM buffer (20 mM Tris-HCl pH 7.9, 20% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 0.2 mM PMSF and 0.1 M NaCl), and spun down at 800 g at 4°C for 2 min. Then, 500 ng purified GST, GST-MICAL-L1-CH or GST-MICAL-L1-CH+LIM (bait proteins) were diluted in 40 μl TGEM buffer, and bound to the beads by rocking at 4°C for 2 h. Then the beads were washed two times with 100 μl ice-cold TGEM buffer with 1 M NaCl, and two times with TGMC buffer (20 mM Tris-HCl pH 7.9, 20% glycerol, 5 mM MgCl2, 5 mM CaCl2, 0.1% NP-40, 1 mM DTT, 0.2 mM PMSF, 0.1 M NaCl), and resuspended in 20 μl TGMC. α-tubulin–β-tubulin heterodimers or γ-tubulin (as a target protein) were diluted in 30 μl TGMC buffer. 1 U of micrococcal nuclease was added to both the bait proteins and the target proteins, and then incubated at 30°C for 10 min. The binding reaction was set up by adding target proteins to bait proteins on ice and rocking at 4°C for 2 h. The protein complex were analyzed by immunoblotting.
Protein purification
GST–MICAL-L1-CH and GST–MICAL-L1-CH+LIM were transformed into E.coli bacteria, cultured in Luria broth (Miller's LB Broth, Kappa Chemicals, St Louis, MO) at 37°C until the optical density (OD) at 600 nm reached 0.55. Protein expression was then induced with 0.2 mM IPTG at 30°C for 5 h. Bacteria were then lysed via sonication. Bacterial debris was removed by ultracentrifugation for 20 min as 4°C at 6000 rpm with a SS34 Sorval rotor. Proteins were purified from the lysates using GST beads, eluted and dialyzed into PBS buffer.
Immunofluorescence and microscopy imagining
Cells plated on coverslips were fixed in pre-chilled 100% methanol at −20°C for 5 min, or in 3.7% paraformaldehyde in PBS for 10 min at room temperature, followed by three rinses with PBS buffer. For immunofluorescence staining, cells were first permeabilized in 0.5% Triton-X plus 0.5% BSA in PBS for 30 min, and then stained with appropriate primary antibodies diluted in PBS buffer containing 0.1% Triton-X and 0.5% BSA for 1 h at room temperature. PBS washes were applied to remove unbound primary antibodies. Cells were then incubated with fluorochrome-conjugated secondary antibodies for 1 h at room temperature. Coverslips were mounted in Fluoromount G Mounting medium (SouthernBiotech). Imaging was performed with a Zeiss LSM 800 confocal microscopy (Carl Zeiss, Oberkochen, Germany) using a Plan-Apochromat 63×/1.4 NA oil objective and appropriate filters, as previously described (Xie et al., 2016). Image acquisition was carried out with Zen software (Carl Zeiss). Z-slices were either z-projected with ImageJ (National Institutes of Health, Bethesda, MD), or 3D-rendered with Imaris (Oxford Instruments, Abingdon, UK). Images were cropped, adjusted for brightness (whole-image adjustment) with minimal manipulation, for better presentation. For quantification, three independent experiments were carried out, and the number of samples collected for quantification is described in the text. sr-SIM images were captured with a Zeiss ELYRA PS.1 illumination system (Carl Zeiss) using a Plan-Apochromat 63×/1.4 NA oil objective lens. Three orientation angles of the excitation grid were acquired for z-slices with 110 nm intervals. Images acquired were processed with SIM module of Zen Black (Carl Zeiss), and presented as maximal intensity projection along the z-axis. Surface rendering was performed using Imaris imaging software.
Statistical analysis
Data obtained from ImageJ was exported to Prism 7 (GraphPad). Bar graphs were created representing the mean and standard deviation from data obtained from three independent experiments. Statistical significance was calculated with an unpaired two-tailed t-test.
Supplementary Material
Acknowledgements
The University of Nebraska Medical Center Advanced Microscopy Core Facility receives partial support from the National Institute for General Medical Science (NIGMS) INBRE - P20 GM103427 and COBRE - P30 GM106397 grants, as well as support from the National Cancer Institute (NCI) for The Fred & Pamela Buffett Cancer Center Support Grant- P30 CA036727, and the Nebraska Research Initiative. This publication’s contents and interpretations are the sole responsibility of the authors.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.X., N.N., S.C.; Methodology: T.F., N.N., S.C.; Validation: S.X., T.F., N.N., S.C.; Formal analysis: S.X., N.N., S.C.; Investigation: S.X., T.F., N.N.; Resources: S.C.; Data curation: S.X., T.F., N.N.; Writing - original draft: S.X., S.C.; Writing - review & editing: S.X., N.N., S.C.; Visualization: S.X., T.F., N.N., S.C.; Supervision: N.N., S.C.; Project administration: N.N., S.C.; Funding acquisition: S.C.
Funding
The authors acknowledge support from the National Institutes of General Medical Sciences (1R01GM123557 and P30GM106397). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.233973.supplemental
References
- Brancati F., Dallapiccola B. and Valente E. M. (2010). Joubert syndrome and related disorders. Orphanet J. Rare Dis. 5, 20 10.1186/1750-1172-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K. and Holland A. J. (2019). Mechanism and regulation of centriole and cilium biogenesis. Annu. Rev. Biochem. 88, 691-724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C. E. and Anderson K. V. (2007). The graded response to sonic hedgehog depends on cilia architecture. Dev. Cell 12, 767-778. 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Fabbri L., Bost F. and Mazure N. M. (2019). Primary cilium in cancer hallmarks. Int. J. Mol. Sci. 20, 1336 10.3390/ijms20061336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S. D., Gowen B. E., Reinsch S., Sawyer A., Buendia B., Wepf R. and Karsenti E. (1995). The core of the mammalian centriole contains gamma-tubulin. Curr. Biol. 5, 1384-1393. 10.1016/S0960-9822(95)00276-4 [DOI] [PubMed] [Google Scholar]
- Garcia G. III, Raleigh D. R. and Reiter J. F. (2018). How the ciliary membrane is organized inside-out to communicate outside-in. Curr. Biol. 28, R421-R434. 10.1016/j.cub.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghata J. and Cowley B. D. Jr (2017). Polycystic kidney disease. Compr. Physiol. 7, 945-975. 10.1002/cphy.c160018 [DOI] [PubMed] [Google Scholar]
- Giridharan S. S. P., Cai B., Naslavsky N. and Caplan S. (2012). Trafficking cascades mediated by Rab35 and its membrane hub effector, MICAL-L1. Commun. Integr. Biol. 5, 384-387. 10.4161/cib.20064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan S. S. P., Cai B., Vitale N., Naslavsky N. and Caplan S. (2013). Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol. Biol. Cell 24, 1776-1790, S1-15 10.1091/mbc.e13-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y., Kobayashi T., Kikko Y., Kontani K. and Katada T. (2008). Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem. Biophys. Res. Commun. 373, 119-124. 10.1016/j.bbrc.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Insinna C., Lu Q., Teixeira I., Harned A., Semler E. M., Stauffer J., Magidson V., Tiwari A., Kenworthy A. K., Narayan K. et al. (2019). Investigation of F-BAR domain PACSIN proteins uncovers membrane tubulation function in cilia assembly and transport. Nat. Commun. 10, 428 10.1038/s41467-018-08192-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J. and Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA 107, 6346-6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P. A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.-S. et al. (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17, 228-240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megaw R. and Hurd T. W. (2018). Photoreceptor actin dysregulation in syndromic and non-syndromic retinitis pigmentosa. Biochem. Soc. Trans. 46, 1463-1473. 10.1042/BST20180138 [DOI] [PubMed] [Google Scholar]
- Novas R., Cardenas-Rodriguez M., Irigoín F. and Badano J. L. (2015). Bardet-Biedl syndrome: is it only cilia dysfunction? FEBS Lett. 589, 3479-3491. 10.1016/j.febslet.2015.07.031 [DOI] [PubMed] [Google Scholar]
- Rahajeng J., Giridharan S. S. P., Cai B., Naslavsky N. and Caplan S. (2012). MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic 13, 82-93. 10.1111/j.1600-0854.2011.01294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Giridharan S. S. P., Rahajeng J., Naslavsky N. and Caplan S. (2009). MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol. Biol. Cell 20, 5181-5194. 10.1091/mbc.e09-06-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Giridharan S. S. P., Rahajeng J., Caplan S. and Naslavsky N. (2010). MICAL-L1: an unusual Rab effector that links EHD1 to tubular recycling endosomes. Commun. Integr. Biol. 3, 181-183. 10.4161/cib.3.2.10845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Sun X., Liu M., Li D., Aneja R. and Zhou J. (2011). CEP70 protein interacts with gamma-tubulin to localize at the centrosome and is critical for mitotic spindle assembly. J. Biol. Chem. 286, 33401-33408. 10.1074/jbc.M111.252262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K. F., Schermelleh L., Leonhardt H. and Nigg E. A. (2012). 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open 1, 965-976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15, 363-377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. P. (1968). Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3, 207-230. [DOI] [PubMed] [Google Scholar]
- Wang L. and Dynlacht B. D. (2018). The regulation of cilium assembly and disassembly in development and disease. Development 145, dev151407 10.1242/dev.151407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Bahl K., Reinecke J. B., Hammond G. R. V., Naslavsky N. and Caplan S. (2016). The endocytic recycling compartment maintains cargo segregation acquired upon exit from the sorting endosome. Mol. Biol. Cell 27, 108-126. 10.1091/mbc.E15-07-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Reinecke J. B., Farmer T., Bahl K., Yeow I., Nichols B. J., McLamarrah T. A., Naslavsky N., Rogers G. C. and Caplan S. (2018). Vesicular trafficking plays a role in centriole disengagement and duplication. Mol. Biol. Cell 29, 2622-2631. 10.1091/mbc.E18-04-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow I., Howard G., Chadwick J., Mendoza-Topaz C., Hansen C. G., Nichols B. J. and Shvets E. (2017). EHD proteins cooperate to generate caveolar clusters and to maintain Caveolae during repeated mechanical stress. Curr. Biol. 27, 2951-2962.e5. 10.1016/j.cub.2017.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.