ABSTRACT
Autophagy is initiated by the formation of a phagophore assembly site (PAS), the precursor of autophagosomes. In mammals, autophagosome formation sites form throughout the cytosol in specialized subdomains of the endoplasmic reticulum (ER). In yeast, the PAS is also generated close to the ER, but always in the vicinity of the vacuole. How the PAS is anchored to the vacuole and the functional significance of this localization are unknown. Here, we investigated the role of the PAS–vacuole connection for bulk autophagy in the yeast Saccharomyces cerevisiae. We show that Vac8 constitutes a vacuolar tether that stably anchors the PAS to the vacuole throughout autophagosome biogenesis via the PAS component Atg13. S. cerevisiae lacking Vac8 show inefficient autophagosome–vacuole fusion, and form fewer and smaller autophagosomes that often localize away from the vacuole. Thus, the stable PAS–vacuole connection established by Vac8 creates a confined space for autophagosome biogenesis between the ER and the vacuole, and allows spatial coordination of autophagosome formation and autophagosome–vacuole fusion. These findings reveal that the spatial regulation of autophagosome formation at the vacuole is required for efficient bulk autophagy.
KEY WORDS: Autophagy, Vac8, Autophagosome, Phagophore assembly site, PAS
Highlighted Article: Vac8 tethers the phagophore assembly site to the vacuole, which creates a confined space for autophagosome biogenesis and coordinates autophagosome formation with vacuolar fusion.
INTRODUCTION
Macroautophagy, hereafter referred to as autophagy, is an evolutionarily conserved process in eukaryotic cells that enables degradation and recycling of bulk cytoplasmic components. It is the main pathway for the turnover of aggregated proteins, as well as damaged and superfluous organelles, and for providing nutrients during starvation conditions. During autophagy, cargo destined for degradation becomes enwrapped in a double-membrane called the phagophore (also known as the isolation membrane). The phagophore expands around the cargo until its edges close to form a sealed, double-membrane vesicle, the autophagosome. Ultimately, the completed autophagosome fuses with the lytic compartment (the vacuole in yeast and plants, the lysosome in mammals), resulting in degradation of the cargo. In mammalian cells, autophagosomes form in phosphatidylinositol 3-phosphate (PIP3)-rich subdomains of the ER, called omegasomes (Mercer et al., 2018). Similarly, in budding yeast, a connection between the phagophore and the ER is required for autophagosome biogenesis (Gómez-Sánchez et al., 2018; Graef et al., 2013). Moreover, autophagosomes in yeast form in close vicinity to the vacuole (Graef et al., 2013; Suzuki et al., 2013).
Autophagy can operate in a selective and non-selective manner. Selective autophagy involves the specific recognition of a cargo, for example, damaged mitochondria, by dedicated receptors. These receptors subsequently recruit the core autophagy machinery and promote the in situ formation and expansion of the growing phagophore around the cargo (Kraft et al., 2009). Conversely, cytoplasmic material is randomly sequestered by autophagosomes during non-selective bulk autophagy. Bulk autophagy is strongly induced upon starvation conditions to provide amino acids and other nutrients required for cellular survival. Therefore, autophagy constitutes a critical mechanism to maintain cellular homeostasis.
The initial step in autophagy is the formation of the phagophore assembly site (PAS, also called the pre-autophagosomal structure), which defines where the phagophore and, ultimately, the autophagosome form. The assembly of the PAS is hierarchical and involves the recruitment of several autophagy-related (Atg) proteins (Suzuki et al., 2007). During selective autophagy in budding yeast, the PAS assembles on the cargo at the vacuole, resulting in local activation of the serine-threonine protein kinase Atg1 (Torggler et al., 2016). In bulk autophagy, however, a specific cargo is not available to serve as a PAS assembly platform. Instead, Atg1 assembles into a pentameric complex with Atg13, Atg17, Atg29 and Atg31. These pentameric complexes further interact with each other resulting in a higher-order oligomeric structure that constitutes the early PAS for bulk autophagy (Yamamoto et al., 2016). Clustering of the Atg1 complex leads to the activation of Atg1 kinase and recruitment of further Atg proteins. Thereby the PAS matures to a site where the phagophore can form. Initially, Atg9 vesicles and the autophagy-specific phosphoinositide 3-kinase (PI3K) complex containing Atg14 are recruited. Subsequently, the Atg2–Atg18 module and the Atg8 lipidation machinery, which consists of the Atg5–Atg12 conjugate and Atg16, are recruited independently (Suzuki et al., 2007). Atg2 appears to be important for establishing the connection between the phagophore and the ER, both during selective and bulk autophagy (Gómez-Sánchez et al., 2018; Kotani et al., 2018). In contrast, however, it remains unclear how the PAS and growing autophagosomes are anchored to the vacuole, and whether this connection fulfills a functional role during autophagosome formation (Suzuki and Ohsumi, 2010).
Vac8 is a vacuolar membrane protein, anchored to lipid bilayers via myristoylation of a glycine residue and palmitoylation of three cysteine residues in its N-terminus (Wang et al., 1998). Vac8 plays a crucial role in vacuole inheritance (Wang et al., 1998), homotypic vacuole fusion (Veit et al., 2001) and establishment of nucleus–vacuole junctions (Pan et al., 2000). Deletion of VAC8 therefore results in an altered vacuolar morphology, visible as multi-lobed vacuoles.
The crystal structure of Vac8 bound to Nvj1 revealed that Vac8 comprises 12 armadillo repeat domains, organized into a superhelical structure that serves as a protein binding platform (Jeong et al., 2017). Vac8 is known to associate with the Atg1 complex via Atg13 and it has been reported to be involved in bulk autophagy (Scott et al., 2000). However, Vac8 has mainly been associated with selective autophagy, such as the cytoplasm-to-vacuole targeting (Cvt) pathway and piecemeal autophagy of the nucleus (Cheong et al., 2005; Roberts et al., 2002). Despite its characterized roles in vacuolar functions, the function of Vac8 in autophagy is largely unknown.
In this study, we show that Vac8 plays a direct and important role in bulk autophagy. It acts early in the pathway by regulating PAS assembly, as well as during later steps of autophagosome formation and fusion with the vacuole. In the absence of Vac8, autophagosome formation takes place in vicinity to the ER, but a stable vacuolar connection is lost, suggesting that Vac8 is required for tethering the PAS and forming autophagosomes to the vacuole. Moreover, we show that Vac8 tethering of the PAS is mediated by Atg13. Together, our findings show that Vac8 helps to confine and coordinate autophagosome formation between the ER and the vacuole.
RESULTS
Vac8 plays a direct and essential role during bulk autophagy
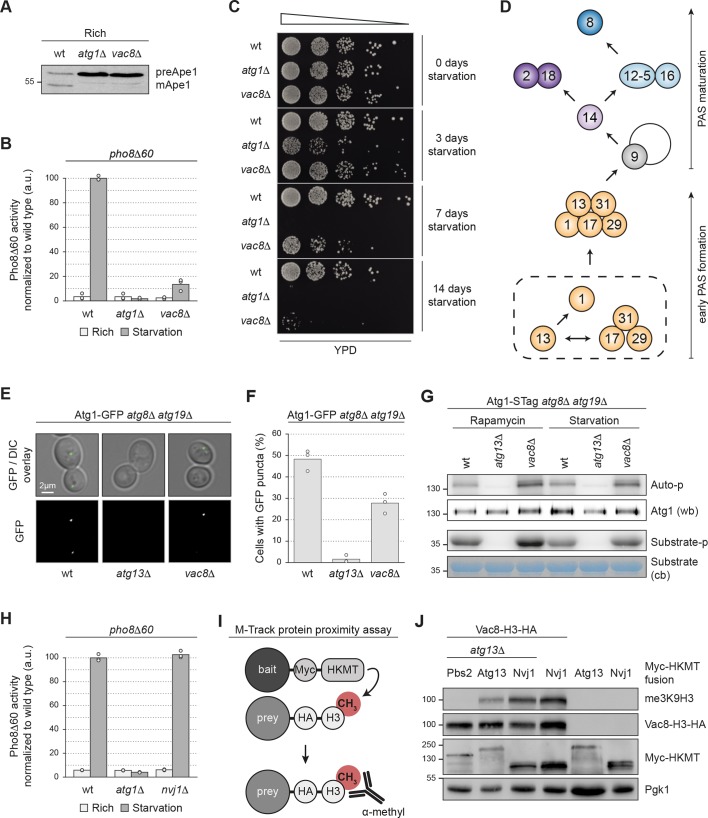
Previous reports have described Vac8 as essential for the selective Cvt pathway, but less important for bulk autophagy, although conflicting conclusions exist (Scott et al., 2000). Its mechanistic role in autophagy, however, remains unknown. To address this question, we first revisited the involvement of Vac8 in the Cvt pathway. As expected, Ape1 processing via the Cvt pathway was strongly impaired in vac8Δ and in atg1Δ mutant cells compared to that in wild-type cells (Scott et al., 2000; Fig. 1A). To assess bulk autophagy, we monitored the activity of Pho8Δ60 reporter phosphatase, which requires bulk autophagy for delivery to and enzymatic activation in the vacuole (Noda et al., 1995). Pho8Δ60 activity was substantially lower in both vac8Δ and atg1Δ mutant cells compared to wild-type cells, as previously shown (Fig. 1B) (Cheong et al., 2005; Scott et al., 2000). Bulk autophagy is required for cell survival during starvation (Tsukada and Ohsumi, 1993), vac8Δ mutants should therefore be starvation sensitive. Wild-type, atg1Δ and vac8Δ cells were nitrogen-starved for several days followed by serial dilution spotting onto nutrient rich agar plates to test cell viability. Whereas wild-type cells were able to cope with nitrogen starvation, atg1Δ control cells displayed a severe starvation sensitivity (Fig. 1C). vac8Δ cells were also starvation sensitive, especially after prolonged starvation, which is in line with the severe bulk autophagy defect of these mutants.
Fig. 1.
Vac8 has a role in early bulk PAS formation. (A) The indicated strains were grown to mid-log phase in YPD medium and cell extracts were prepared by TCA precipitation. Ape1 processing was analysed by anti-Ape1 western blotting. One representative experiment out of three is shown. (B) The indicated strains were grown to mid-log phase in YPD medium and subsequently starved for 4 h in SD-N medium where indicated. Pho8Δ60 alkaline phosphatase activity was measured in three independent experiments. The values of each replicate (circles) and the mean (bars) were plotted. All values were normalized to the mean Pho8Δ60 alkaline phosphatase activity of the wild type. (C) Wild-type, atg1Δ and vac8Δ cells were grown to mid-log phase in YPD medium and shifted to SD-N medium to induce starvation. After 0, 3, 7 and 14 days, cells were spotted in serial dilutions onto YPD plates and incubated for 48 h at 30°C. One representative experiment out of four is shown. (D) Model of hierarchical recruitment of bulk PAS factors, originally described by Suzuki et al. (2007). (E,F) Strains containing endogenously expressed Atg1–GFP as indicated were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. The number of cells with Atg1–GFP puncta was counted in three independent experiments. For each strain and replicate, at least 100 cells were analysed. (E) Representative microscopy images. (F) Quantification of the percentage of cells with at least one Atg1–GFP punctum. The values of each individual experiment (circles) and the mean (bars) were plotted. DIC, differential interference contrast. (G) The indicated strains were grown to mid-log phase in YPD medium and treated for 1 h with 220 nM rapamycin or starved for 1 h in SD-N medium. Endogenously expressed Atg1-STag (Atg1-S-peptide-2×ProteinA) was immunoprecipitated and incubated with γ-[32P]ATP and substrate (Atg19 C-terminus; Pfaffenwimmer et al., 2014). Kinase activity was measured through autoradiography detecting Atg1 auto-phosphorylation and substrate phosphorylation. Atg1–STag levels were assessed by anti-ProteinA (PAP) western blotting (wb) and substrate levels were analysed through Coomassie Blue staining (cb). One representative experiment out of three is shown. (H) Indicated strains were grown to mid-log phase in YPD medium and subsequently starved for 4 h in SD-N medium where indicated. Pho8Δ60 alkaline phosphatase activity was measured in three independent experiments. The values of each replicate (circles) and the mean (bars) were plotted. All values were normalized to the mean Pho8Δ60 alkaline phosphatase activity of the wild type. (I) Schematic illustration of the M-track protein proximity assay. The bait protein is tagged with a histone lysine methyltransferase (HKMT) and the prey protein with a histone 3 (H3) domain. Upon close proximity of bait and prey, the H3 domain becomes stably methylated by the HKMT, which can be detected by using an antibody specific for methylated H3. (J) Indicated strains carrying a plasmid expressing Myc–HKMT-tagged Atg13, Nvj1 or, as a control, Pbs2, and either endogenously expressed H3–HA-tagged Vac8 or untagged Vac8 were grown to mid-log phase in SD medium and treated for 4 h with 220 nM rapamycin. Cell extracts were prepared by TCA precipitation. The Vac8–H3–HA tri-methylation signal was assessed by anti-me3K9H3 western blotting. Vac8–H3–HA expression levels, Myc–HKMT-tagged Atg13, Pbs2 or Nvj1 expression levels and loading were monitored by anti-HA, anti-Myc and anti-Pgk1 western blotting, respectively. One representative experiment out of three is shown. a.u., arbitrary units; wt, wild type.
Vacuolar integrity is important for late steps of autophagy, such as the fusion of autophagosomes with the vacuole and the breakdown of the autophagic cargo in the vacuolar lumen. Therefore, a defect in vacuolar function or integrity could indirectly affect autophagy. Indeed, vac8Δ cells show a severe defect in vacuole–vacuole fusion and contain fragmented vacuoles (Pan and Goldfarb, 1998; Veit et al., 2001).
We reasoned that if the bulk autophagy defect in vac8Δ mutants arises from abnormal vacuolar morphology only, then early steps of autophagy should be unaffected. To test this hypothesis, we determined whether the initiation of autophagy was affected in vac8Δ mutants. Specifically, we used a fluorescent Atg1–GFP reporter to detect the formation of the Atg1 complex within the early PAS (Fig. 1D) (Suzuki et al., 2007). Importantly, to monitor bulk autophagy only, we used cells lacking the receptor Atg19 or the cargo Ape1, which thus prevents Cvt pathway-induced PAS formation. Furthermore, we used an atg8Δ background, which means cells are unable to form autophagic membranes; this results in stalling the PAS and prevents its turnover. This experimental setup allows the specific analysis of early PAS formation during bulk autophagy.
We observed Atg1–GFP puncta in 50% of atg8Δatg19Δ control cells after 1 h of starvation, but in less than 2% of negative control cells that lack the Atg1 complex component Atg13 (atg13Δatg8Δatg19Δ) (Fig. 1E,F). Importantly, only 25% of vac8Δatg8Δatg19Δ cells displayed Atg1–GFP puncta, suggesting that Vac8 is required for an early step of autophagy, namely during PAS formation.
Because Vac8 associates with the Atg1 complex via Atg13, we considered that Vac8 influences Atg1 kinase activation during bulk autophagy. To test this possibility, we immunopurified Atg1–STag and performed an in vitro kinase assay. As expected, Atg13 was required for Atg1 kinase activation. However, the deletion of Vac8 did not diminish Atg1 kinase activity during bulk autophagy-inducing conditions, suggesting that the bulk autophagy defect in vac8Δ mutants is caused by other means than reduced Atg1 kinase activity (Fig. 1G).
Another interaction partner of Vac8 is Nvj1. Binding of Nvj1 to Vac8 is required for establishing nucleus–vacuole junctions (NVJ), which generates a connection between the vacuole and nuclear ER (Pan et al., 2000). To test whether the defect in PAS formation of vac8Δ mutants is caused by impaired NVJ formation, we tested whether nvj1Δ mutants are defective in bulk autophagy. In a Pho8Δ60 assay, nvj1Δ cells showed no defect in bulk autophagy (Fig. 1H). Similarly, when monitoring the cleavage of Pgk1–GFP, an assay that also is used to monitor bulk autophagy flux (Welter et al., 2010), both wild-type and nvj1Δ cells showed similar levels of cleaved GFP after 4 h of nitrogen starvation (Fig. S1), suggesting that Nvj1 is dispensable for bulk autophagy function and therefore does not influence PAS formation.
In vitro analysis suggests that Nvj1 and Atg13 bind to the same region on Vac8 in an exclusive and competitive manner (Jeong et al., 2017), suggesting that Nvj1 and Atg13 are unlikely to assemble into the same holo complex with Vac8. To test whether Nvj1 also interacts with Vac8 in the absence of Atg13 in vivo, we evaluated this interaction using the methylation tracking (M-track) proximity assay. The approach employs a histone lysine methyltransferase (HKMT) domain fused to a bait protein, and the methylation acceptor histone 3 (H3) fused to a prey protein. Upon binding to the bait, the prey is stably methylated in vivo, and this modification is detected using anti-methylation-specific antibodies (Fig. 1I) (Brezovich et al., 2015; Zuzuarregui et al., 2012). We fused Atg13 and Nvj1 to the HKMT domain, and Vac8 to the H3 tag. As expected, we observed methylation of Vac8–H3 with Atg13–HKMT. In addition, Nvj1–HKMT interacted with Vac8–H3 in atg13Δ cells, confirming that Nvj1 interacts with Vac8 independently of Atg13 in vivo (Fig. 1J).
Together, these results suggest that Vac8, independently of its function in NVJ formation, plays a direct and important role in bulk autophagy that involves the formation of the early PAS.
Vac8 regulates early PAS formation but not PAS maturation
Although vac8Δ mutants showed a 50% reduction in early PAS formation, the Pho8Δ60 assay suggests that bulk autophagic flux is almost completely impaired in vac8Δ cells (Fig. 1B). This apparent discrepancy suggests that Vac8 might also regulate later steps in autophagy, after establishing the early PAS.
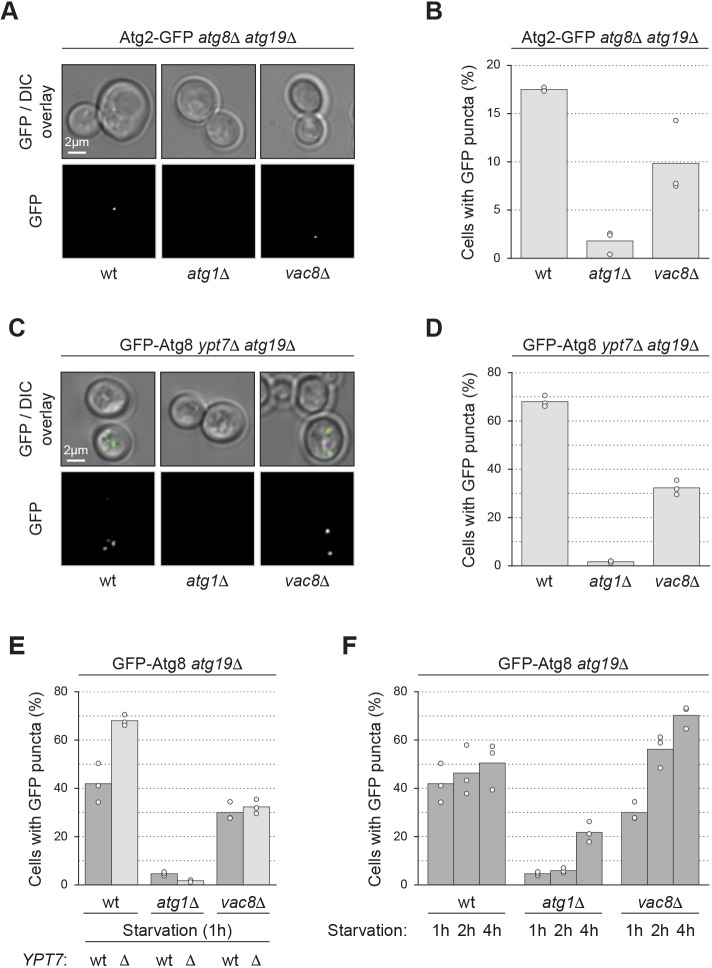
The early PAS formed by the Atg1 complex subsequently matures via the recruitment of downstream Atg proteins. First, Atg9 and Atg14 join the PAS, followed by the independent recruitment of the Atg2–Atg18 module as well as the Atg12 and Atg8 conjugation systems (Suzuki et al., 2007). We assessed the recruitment of Atg2 to the PAS by monitoring Atg2–GFP localization in atg8Δatg19Δ cells. The frequency of cells with Atg2–GFP puncta was reduced by 50% in vac8Δ mutants compared to Vac8-containing cells (Fig. 2A,B), similar to the observations with Atg1–GFP.
Fig. 2.
Vac8 has an additional function post bulk PAS assembly. (A,B) The indicated strains containing endogenously expressed Atg2–GFP were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. The number of cells with Atg2–GFP puncta was counted in three independent experiments. For each strain and replicate, at least 100 cells were analysed. (A) Representative microscopy images. (B) Quantification of the percentage of cells with at least one Atg2–GFP punctum. The values of each replicate (circles) and the mean (bars) were plotted. (C,D) Analysis of fluorescent GFP–Atg8 punctum formation in vac8Δ cells after starvation. GFP–Atg8 was expressed endogenously. (C) Representative microscopy images and (D) quantification. Experiments were performed and presented as described in B. (E,F) Analysis of GFP–Atg8 punctum turnover in vac8Δ cells during starvation. Experiments were performed and presented as described in B; strains were starved for 1, 2 or 4 h in SD-N medium. Data from ypt7Δ mutants (light grey bars) shown in E correspond to the experiments shown in D; data from YPT7 wild type (dark grey bars) shown in E correspond to the experiments shown in F. DIC, differential interference contrast; wt, wild type.
Additionally, to assess the recruitment of Atg8 to the PAS, we monitored GFP–Atg8 in cells lacking Ypt7 and Atg19. ypt7Δ cells are deficient in autophagosome–vacuole fusion and thus accumulate mature autophagosomes (Kirisako et al., 1999). As a result, GFP–Atg8 puncta represent either the PAS or forming autophagosomes. In both cases, GFP–Atg8 puncta indicate that a PAS had been initiated. Similar to what is seen for Atg1–GFP and Atg2–GFP, the frequency of cells with GFP–Atg8 puncta was reduced by 50% in the vac8Δ mutant compared to the corresponding wild type (Fig. 2C,D).
We infer that the reduced recruitment of Atg2–GFP and GFP–Atg8 to the PAS reflects the defect in early PAS formation in the vac8Δ mutant. Vac8 therefore regulates early PAS formation, but does not further affect PAS maturation, that is recruitment of downstream Atg proteins to the PAS.
Vac8 regulates autophagosome turnover
Next, we asked whether autophagosome turnover is affected by Vac8. To evaluate autophagosome turnover, we compared the frequency of cells with GFP–Atg8 puncta in Ypt7-containing cells versus fusion-deficient ypt7Δ mutants after 1 h of starvation. As expected, the frequency of cells with GFP–Atg8 puncta increased from 40% in Ypt7-containing atg19Δ cells to almost 70% in fusion-defective atg19Δypt7Δ cells, indicating that in Ypt7-containing cells autophagosomes are turned over by fusion with the vacuole after 1 h of starvation. In contrast, vac8Δatg19Δ and vac8Δatg19Δypt7Δ cells displayed a similar frequency of GFP–Atg8 punctum-containing cells after 1 h of starvation (Fig. 2E), suggesting that vac8Δ cells are deficient for autophagosome turnover.
To confirm the defect of vac8Δ cells in autophagosome turnover, we monitored whether atg19Δ and vac8Δatg19Δ cells accumulate GFP–Atg8 puncta over a prolonged period of starvation. In fact, the number of cells containing GFP–Atg8 puncta remained almost constant in atg19Δ cells, whereas a substantial increase in the vac8Δatg19Δ mutant was observed during 4 h of starvation (Fig. 2F). This supports the hypothesis that vac8Δ cells are inefficient in turning over GFP–Atg8 structures. This phenotype could be caused by a kinetic defect in phagophore formation and autophagosome completion, or be due to inefficient fusion of autophagosomes with the vacuole.
Together, these findings demonstrate that in addition to early PAS formation, Vac8 also regulates a later step in autophagy, the formation or fusion of autophagosomes.
Vac8 mutants display smaller autophagosomes and reduced autophagosome-vacuole fusion
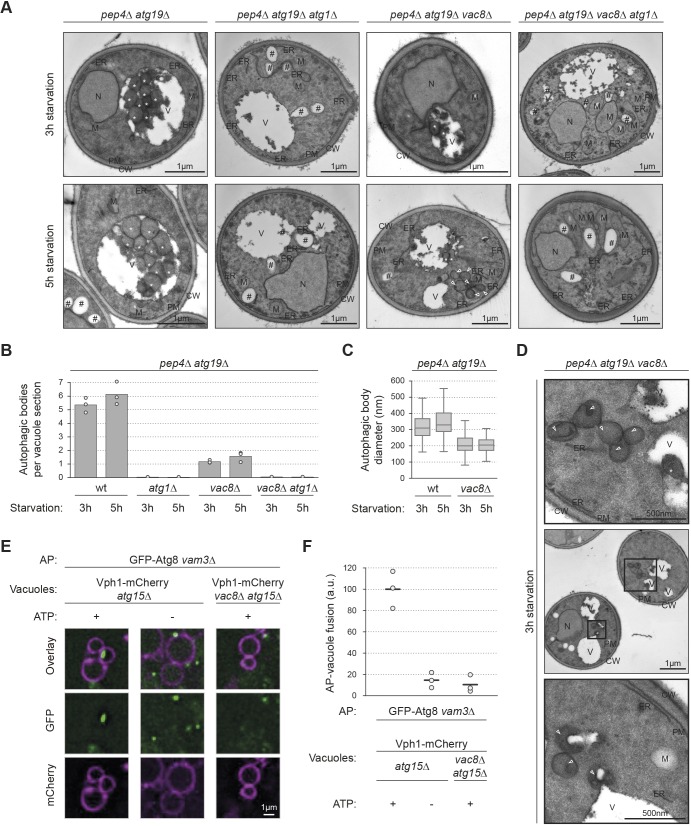
To further assess the role of Vac8 in later steps of autophagy, we analysed vac8Δ mutants by transmission electron microscopy. We visualized the accumulation of autophagic bodies in the vacuolar lumen of vacuolar protease mutant pep4Δ cells (Guimaraes et al., 2015). As expected, autophagic bodies accumulated in the vacuole of pep4Δatg19Δ cells. However, autophagic bodies were not observed in pep4Δatg19Δatg1Δ cells, owing to their inability to form autophagosomes (Cheong et al., 2007). The pep4Δatg19Δvac8Δ mutant accumulated very few autophagic bodies in the vacuole compared to pep4Δatg19Δ cells (Fig. 3A,B), consistent with a highly reduced bulk autophagic flux in the absence of Vac8 (Fig. 1B). Furthermore, autophagic bodies in pep4Δatg19Δvac8Δ cells were only 200 nm in diameter, whereas the average diameter of autophagic bodies in VAC8 wild-type cells was 300 nm, suggesting that deletion of Vac8 impairs the formation of fully sized autophagosomes (Fig. 3A,C).
Fig. 3.
Deletion of Vac8 results in the delivery of fewer and smaller autophagosomes to the vacuole. (A–D) The indicated strains were grown to mid-log phase, starved for 3 or 5 h in SD-N medium, fixed in potassium permanganate and analysed by transmission electron microscopy. (A,D) Representative electron micrographs. CW, cell wall; ER, endoplasmic reticulum; M, mitochondria; N, nucleus; PM, plasma membrane; V, vacuole; asterisks, autophagic bodies; #, lipid droplets; arrowheads, autophagosomes. In the pep4Δatg19Δvac8Δ mutant, after 3 h of starvation, 85% of cytoplasmic autophagosomes were not connected with the vacuole, whereas 15% of autophagosomes were adjacent to vacuoles (60 cell profiles containing autophagosomes were quantified, the s.e.m. was 1.92). (B) The number of autophagic bodies per vacuole section was quantified in three independent technical replicates. For each condition and replicate 50 cell sections were analysed. The average of each replicate (circles) and the overall mean (bars) were plotted. (C) Boxplot of autophagic body size. At least 90 autophagic bodies from three independent technical replicates were analysed per strain and condition. Dark horizontal lines represent medians, boxes represent the 25th and 75th percentiles, whiskers expand to the largest value no further than 1.5 times the interquartile range; outliers are not shown. (E) Vacuoles were isolated from Vph1–mCherry atg15Δ or Vph1–mCherry atg15Δvac8Δ cells and incubated with autophagosomal (AP) fractions prepared from GFP–Atg8 vam3Δ cells and an energy regeneration system for 2 h. Apyrase was added to deplete ATP in the negative control. Fusion was monitored by fluorescence microscopy and judged by the appearance of a mobile green dot in the vacuole. Shown is the first time point of a 20 s time lapse (see also Fig. S2). (F) Quantification of successful fusion events in (E). The number of successful fusion events per vacuole was counted in three independent experiments and normalized to the positive control, values of each individual experiment (circles) and the mean (horizontal lines) were plotted. a.u., arbitrary units; wt, wild type.
Unexpectedly, pep4Δatg19Δvac8Δ cells accumulated large vesicles in the cytosol (Fig. 3D, arrowheads). These cytosolic structures were not observed in pep4Δatg19Δ or in pep4Δatg19Δatg1Δ cells. Furthermore, they were not present in pep4Δatg19Δvac8Δatg1Δ cells (Fig. 3A), indicating that they correspond to smaller autophagosomes formed in vac8Δ cells. These findings are consistent with the observed accumulation of GFP–Atg8 puncta in vac8Δ cells after prolonged starvation (Fig. 2F). The autophagosomes that formed in the absence of Vac8 were mostly not connected to the vacuole (85%, Fig. 3A,D), suggesting that Vac8 is involved in targeting or tethering autophagosomes to the vacuolar membrane. Eventually, however, some vacuolar contact must happen, as some fusion also occurred in vac8Δ cells.
Next, we asked whether autophagosome–vacuole fusion is affected by Vac8. To test for autophagosome–vacuole fusion, we took advantage of our recently established in vitro fusion assay (Bas et al., 2018), which recapitulates autophagosome–vacuole fusion in vitro using isolated yeast vacuoles and an autophagosome-enriched fraction. As previously described, autophagosomes were isolated from starved and fusion-deficient GFP–Atg8 vam3Δ cells. Vacuoles were isolated from cells lacking the vacuolar lipase Atg15, which results in the stabilization of autophagic bodies within the vacuole and allows their visualization. The vacuolar membrane protein Vph1 was tagged with mCherry to visualize the vacuole. Vacuoles isolated from Vph1–mCherry atg15Δ cells were proficient in autophagosome–vacuole fusion, as expected. In the absence of ATP, fusion was inhibited, confirming previous findings that the fusion process requires ATP (Bas et al., 2018). Intriguingly, when vacuoles were isolated from Vph1–mCherry vac8Δatg15Δ cells, fusion was blocked, suggesting that Vac8 supports autophagosome–vacuole fusion (Fig. 3E,F; Fig. S2).
Vac8 membrane association is required for bulk autophagy
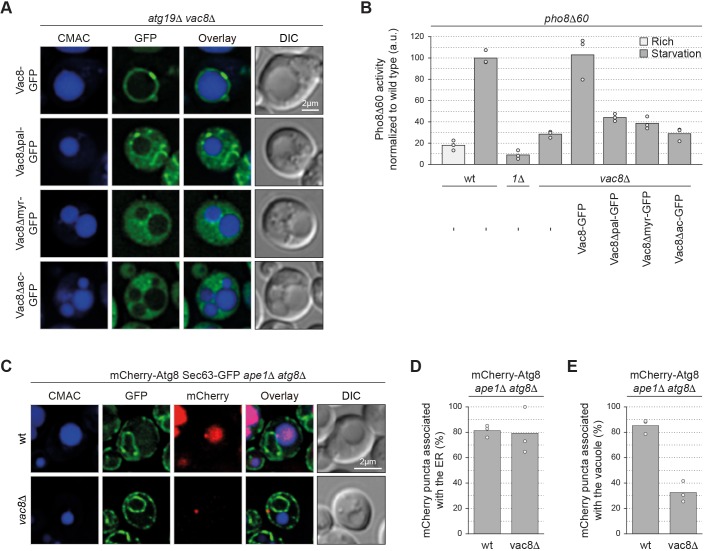
Vacuolar membrane anchoring of Vac8 depends on both palmitoylation and myristoylation in its N-terminus (Wang et al., 1998). To determine whether the vacuolar membrane association of Vac8 is important for bulk autophagy, we assessed whether vacuolar localization-defective Vac8 mutants that disrupt the Vac8 myristoylation site (Vac8Δmyr–GFP), the palmitoylation sites (Vac8Δpal–GFP) or all acylation sites (Vac8Δac–GFP) (Wang et al., 1998) could restore Pho8Δ60 activity in vac8Δ cells. As expected, all three mutant proteins localized primarily to the cytoplasm in atg19Δvac8Δ cells, whereas Vac8–GFP showed vacuolar localization (Fig. 4A). Unlike Vac8–GFP, none of the mutants could substantially restore Pho8Δ60 activity to vac8Δ cells (Fig. 4B), suggesting that both myristoylation and palmitoylation are required for bulk autophagy. In contrast to previous observations (Wang et al., 1998), only Vac8–GFP expression fully rescued the Cvt pathway defect in the vac8Δ knockout, while vac8Δ cells expressing Vac8Δpal–GFP and Vac8Δmyr–GFP displayed mild Cvt pathway defects, and those expressing Vac8Δac–GFP showed a severe Cvt pathway defect (Fig. S3A). These data suggest that vacuolar localization of Vac8 is required for bulk autophagy.
Fig. 4.
Vac8 anchors the bulk PAS and forming autophagosomes to the vacuole. (A) atg19Δvac8Δ cells carrying a plasmid expressing Vac8–GFP, Vac8Δpal–GFP (Vac8-C4G-C5T-C7S-GFP), Vac8Δmyr–GFP (Vac8-G2A-GFP) or Vac8Δac–GFP (Vac8-G2A-C4G-C5T-C7S-GFP) were grown to mid-log phase in SD medium, starved for 1 h in SD-N medium and labelled with CMAC. Representative fluorescence images are shown. (B) The indicated strains transformed with a plasmid expressing Vac8–GFP, Vac8Δpal–GFP, Vac8Δmyr–GFP or Vac8Δac–GFP or an empty vector were grown to mid-log phase in SD medium and starved for 4 h in SD-N medium where indicated. Pho8Δ60 alkaline phosphatase activity was measured in three independent experiments. The values of each replicate (circles) and the mean (bars) were plotted. All values were normalized to the mean Pho8Δ60 alkaline phosphatase activity of the wild type. (C–E) Cells containing endogenously expressed Sec63–GFP and plasmid expressed mCherry-V5-Atg8 were grown to mid-log phase in SD medium, starved for 1 h in SD-N medium, and labelled with CMAC. Three independent experiments were performed. For each strain and replicate at least 100 cells were analysed. (C) Representative fluorescence images. The percentage of mCherry–Atg8 puncta associated with (D) the ER and (E) the vacuole was quantified; the values of each replicate (circles) and the mean (bars) were plotted. DIC, differential interference contrast; a.u., arbitrary units; wt, wild type.
Vac8 stably anchors the PAS and forming autophagosomes at the vacuole
It has been reported that autophagosomes are simultaneously tethered to the vacuole and the ER during their formation (Graef et al., 2013; Suzuki et al., 2013). Atg2 was recently shown to play a role in connecting the autophagosomes to the ER (Gómez-Sánchez et al., 2018), but the factor(s) that connect the autophagosomes to the vacuole are unknown. Because we observed autophagosomes in the cytosol of the vac8Δ mutant (Fig. 3D), we hypothesized that Vac8 could mediate the autophagosome–vacuole connection. To test this possibility, we used mCherry–Atg8 to monitor the localization of the PAS and forming autophagosomes, and quantified their proximity to the vacuole (CMAC) and the ER (Sec63–GFP). mCherry–Atg8-positive structures, which represent both PAS and forming autophagosomes, were associated with the ER in ∼80% of both wild-type and vac8Δ cells (Fig. 4C,D). In addition, mCherry–Atg8-positive structures also localized adjacent to the vacuole in ∼80% of wild-type cells, suggesting that PAS assembly and autophagosome formation takes place in between the ER and the vacuole in 60–80% of cells (Fig. 4E), as previously suggested (Graef et al., 2013). However, the vacuolar proximity of mCherry–Atg8 puncta was substantially reduced to ∼30% in vac8Δ mutants (Fig. 4E). Similarly, mCherry–Atg8 displayed reduced vacuolar localization in the vac8Δ cells expressing cytoplasmic Vac8Δac–GFP (Fig. S3B,C). These findings suggest that Vac8 is required to stably anchor the PAS and/or autophagosomes at the vacuole.
Ectopic Vac8 recruits the PAS protein Atg13
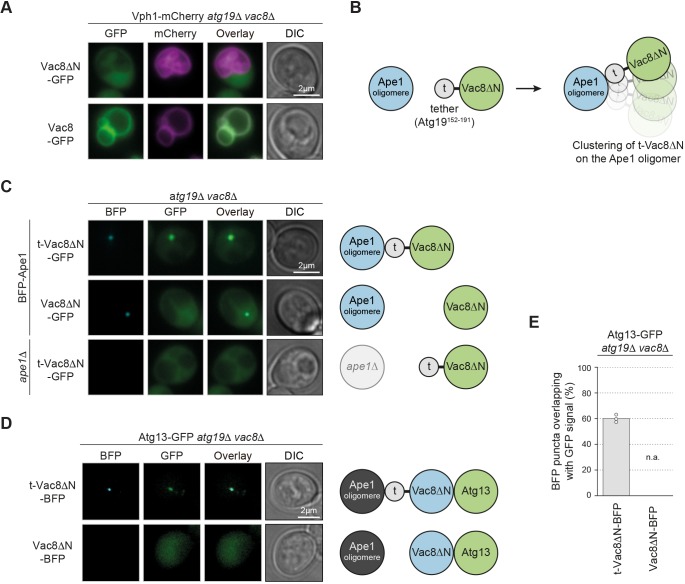
We hypothesized that if endogenous Vac8 tethers the PAS to the vacuole, then ectopic localization of Vac8 should tether the PAS to another site in the cell. To test this hypothesis, we used Ape1 oligomers for ectopic recruitment of Vac8. Ape1 oligomers provide a protein-clustering platform that enables the visualization of recruited proteins by fluorescence microscopy. Importantly, in the absence of the cargo receptor Atg19, the Ape1 oligomer remains cytosolic and does not interact with other Atg proteins or the vacuole (Shintani et al., 2002; Watanabe et al., 2010). To prevent vacuolar recruitment of Vac8, we used a truncation mutant lacking the N-terminal 18 amino acids (Vac8ΔN), which remains cytosolic due to the lack of acylation (Fig. 5A). Vac8ΔN was fused to the minimal reported Ape1-interacting region of Atg19, amino acids 152–191 (t-Vac8ΔN). This 40 amino acid-long peptide of Atg19 is sufficient for Ape1 binding but lacks the Atg11-, Ams1- and Atg8-binding regions (Shintani et al., 2002). Therefore, the fusion construct t-Vac8ΔN should be recruited to cytosolic Ape1 oligomers in atg19Δ cells (Fig. 5B). Indeed, t-Vac8ΔN–GFP colocalized with BFP–Ape1 in atg19Δvac8Δ cells, whereas Vac8ΔN–GFP showed dispersed cytoplasmic signal. t-Vac8ΔN–GFP formed a distinct punctum in the cell, which was lost in the absence of Ape1, validating our experimental setup (Fig. 5C).
Fig. 5.
Vac8 binds to Atg13 in vivo. (A) Vph1–mCherry atg19Δvac8Δ cells carrying a plasmid expressing Vac8ΔN–GFP (Vac819-578-GFP) or Vac8–GFP were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. Representative fluorescence images are shown. (B) Schematic of the experimental setup used in (C). N-terminal truncation of 18 amino acids results in cytosolic distribution of Vac8ΔN (Vac819-578). Fusion of the Ape1-binding region of Atg19152-191 to Vac8ΔN (t-Vac8ΔN) results in ectopic localization and clustering of t-Vac8ΔN on Ape1 oligomers. (C) atg19Δvac8Δ cells carrying a plasmid expressing BFP–Ape1 and either t-Vac8ΔN–GFP or Vac8ΔN–GFP, and atg19Δvac8Δape1Δ cells carrying an empty plasmid and a plasmid expressing t-Vac8ΔN–GFP were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. Representative fluorescence images are shown. (D) Atg13–GFP atg19Δvac8Δ cells carrying a plasmid expressing t-Vac8ΔN–BFP or Vac8ΔN–BFP were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. Representative fluorescence images are shown. (E) Quantification of D. The percentage of BFP puncta overlapping with GFP signal was quantified in three independent experiments; the values of each replicate (circles) and the mean (bars) were plotted. t, tether; DIC, differential interference contrast; n.a., not applicable (none visible).
Vac8 has been reported to directly interact with the early PAS component Atg13 (Scott et al., 2000), therefore we asked whether ectopically localized Vac8 was still able to recruit Atg13. Indeed, t-Vac8ΔN–BFP formed distinct puncta and was able to recruit Atg13–GFP, which was not observed when expressing cytosolic Vac8ΔN–BFP, suggesting that ectopically localized Vac8 is able to recruit the PAS component Atg13 (Fig. 5D,E).
Vac8 mediates PAS anchoring via the C-terminus of Atg13
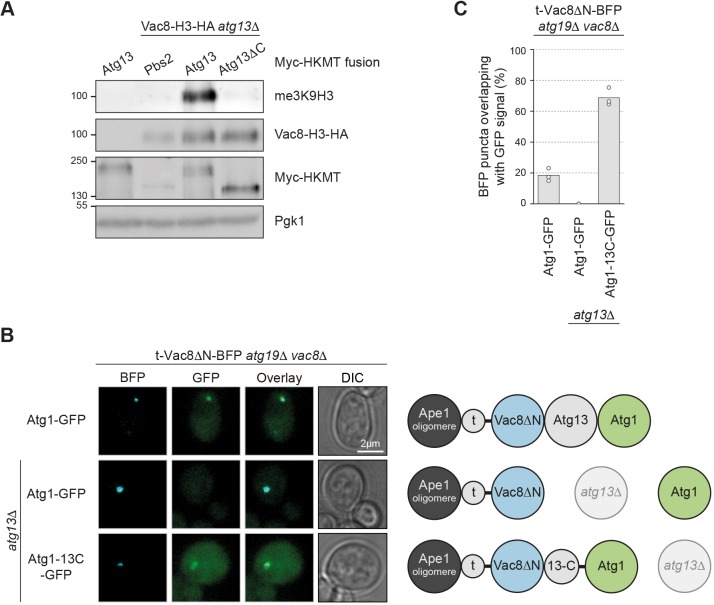
Vac8 interacts with the C-terminus of Atg13 in a yeast two-hybrid assay (Scott et al., 2000). We re-evaluated this interaction in vivo by performing a methylation tracking (M-track) proximity assay (Fig. 1I). We fused Atg13 and Atg13ΔC to the HKMT domain, and Vac8 to the H3 tag. As expected, we observed methylation of Vac8–H3 with Atg13–HKMT, but only background levels of methylation with Atg13ΔC–HKMT (Fig. 6A), confirming that the Atg13 C-terminus is required for the interaction between Vac8 and Atg13 in vivo.
Fig. 6.
Vac8 mediates PAS anchoring via the C-terminus of Atg13. (A) Cells carrying a plasmid expressing Myc–HKMT-tagged Atg13, Atg13ΔC (Atg131-520) or as a control Pbs2, and either endogenously expressed H3–HA-tagged Vac8 or untagged Vac8 were grown to mid-log phase in SD medium and treated for 4 h with 220 nM rapamycin. Cell extracts were prepared by TCA precipitation. The Vac8–H3–HA tri-methylation signal was assessed by anti-me3K9H3 western blotting. Vac8–H3–HA expression levels, Myc–HKMT-tagged Atg13 or Pbs2 expression levels, and loading were monitored by anti-HA, anti-Myc and anti-Pgk1 western blotting, respectively. One representative experiment out of three is shown. (B) atg19Δvac8Δ or atg13Δatg19Δvac8Δ cells containing endogenously expressed Atg1–GFP or Atg1–Atg13C567-738–GFP were transformed with a plasmid expressing t-Vac8ΔN–BFP. Cells were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. Representative fluorescence images are shown. t, tether; DIC, differential interference contrast. (C) Quantification of B. The percentage of BFP puncta overlapping with GFP signal was quantified in three independent experiments; the values of each replicate (circles) and the mean (bars) were plotted.
Next, we asked whether ectopic Vac8 is able to recruit additional PAS proteins. Atg1 is a PAS component and its PAS recruitment depends on Atg13 (Suzuki et al., 2007). We reasoned that if Vac8 acts as a PAS anchor by interacting with Atg13, then Atg1 should also be recruited to ectopic Vac8, but in an Atg13-dependent manner. Indeed, Atg1–GFP colocalized with t-Vac8ΔN–BFP puncta in atg19Δvac8Δ cells, supporting the idea that other PAS components are also recruited to ectopic Vac8. This colocalization and Atg1–GFP punctum formation were lost in the absence of Atg13 (Fig. 6B,C). When fusing the C-terminal 172 amino acid Vac8-binding region of Atg13 to Atg1 (Atg1-13C), the localization of Atg1 to ectopic Vac8 was restored. Overall, these data suggest that Vac8 interacts with the PAS via the C-terminus of Atg13.
The C-terminus of Atg13 links the PAS to the vacuolar tether Vac8
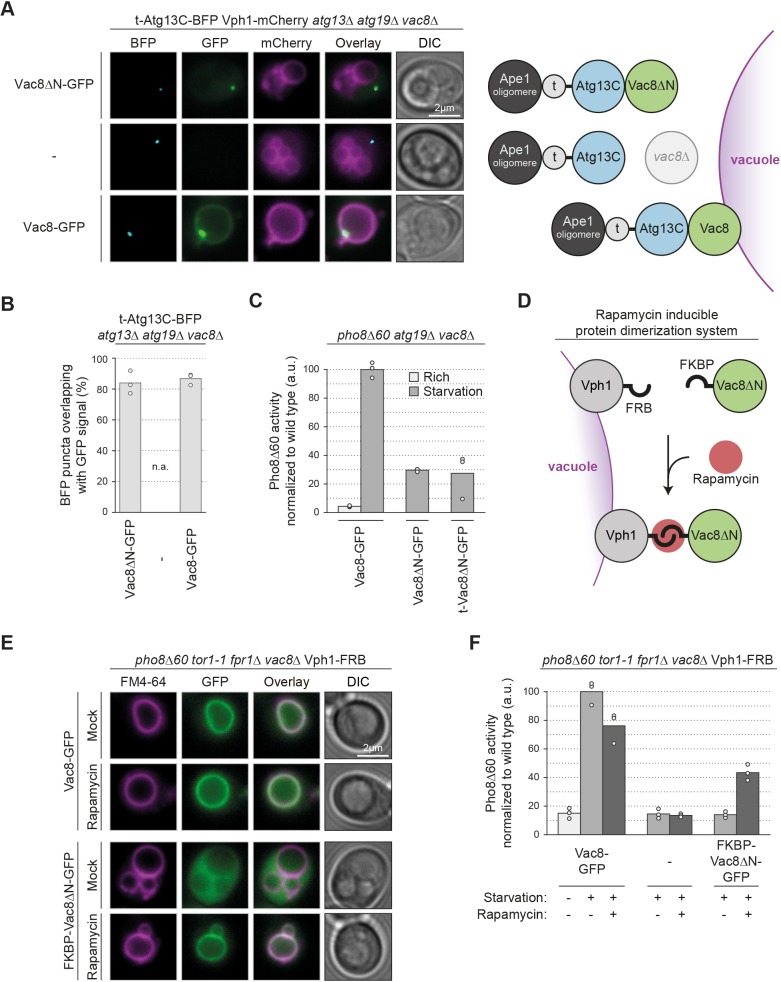
If the C-terminus of Atg13 links the PAS to Vac8 on the vacuole, then ectopically localized Atg13C should also be sufficient to recruit Vac8ΔN. To test this scenario, we fused the Ape1-interacting region of Atg19 to the C-terminus of Atg13 (t-Atg13C). Indeed, t-Atg13C–BFP was sufficient to recruit Vac8ΔN–GFP to cytosolic Ape1 structures, visible as colocalizing fluorescence puncta in the cell. As expected, these puncta remained largely cytosolic in atg13Δatg19Δvac8Δ cells or in cells containing cytosolic Vac8ΔN–GFP (Fig. 7A,B; Fig. S4A). However, in the presence of wild-type vacuole-localized Vac8–GFP, the t-Atg13C fluorescent puncta were efficiently recruited to the vacuole. Interestingly, this also resulted in an enrichment of Vac8 at the vacuolar contact site of the t-Atg13C–BFP punctum (Fig. S4A,B). Together, these findings further substantiate that Vac8 anchors the PAS via its interaction with the C-terminus of Atg13, and suggest that Atg13 provides a link between the PAS and the vacuolar tether Vac8.
Fig. 7.
Vac8-mediated vacuolar localization of the PAS is required for bulk autophagy. (A) Vph1–mCherry atg13Δatg19Δvac8Δ cells carrying a plasmid expressing t-Atg13C–BFP and either Vac8ΔN–GFP, Vac8–GFP or an empty plasmid were grown to mid-log phase in SD medium and starved for 1 h in SD-N medium. Representative fluorescence images are shown (see also Fig. S4A). (B) Quantification of A. The percentage of BFP puncta overlapping with GFP signal was quantified in three independent experiments; the values of each replicate (circles) and the mean (bars) were plotted. (C) pho8Δ60 atg19Δ vac8Δ cells transformed with a plasmid expressing Vac8–GFP, Vac8ΔN–GFP or t-Vac8ΔN–GFP were grown to mid-log phase in SD medium and starved for 4 h in SD-N medium where indicated. Pho8Δ60 alkaline phosphatase activity was measured in three independent experiments. The values of each replicate (circles) and the mean (bars) were plotted. All values were normalized to the mean Pho8Δ60 alkaline phosphatase activity of cells expressing Vac8–GFP. (D) Schematic of experimental setup to inducibly tether Vac8 to the vacuole. The vacuolar transmembrane protein Vph1 is fused to an FRB domain and cytosolic Vac8ΔN to an FKBP domain. Upon addition of rapamycin the FRB and FKBP domains dimerize, which results in rapid and stable recruitment of Vac8ΔN to the vacuole. (E) pho8Δ60 tor1-1 fpr1Δ vac8Δ Vph1–FRB cells transformed with a plasmid expressing Vac8–GFP or FKBP–Vac8ΔN–GFP were grown to mid-log phase in SD medium, labelled with FM4-64 and starved for 1 h in SD-N medium with or without 1.5 µM rapamycin where indicated. Representative fluorescence images are shown. (F) pho8Δ60 tor1-1 fpr1Δ vac8Δ Vph1–FRB cells transformed with a plasmid expressing Vac8–GFP or FKBP–Vac8ΔN-GFP or an empty vector were grown to mid-log phase in SD medium, starved for 4 h in SD-N medium with or without 1.5 µM rapamycin where indicated. Pho8Δ60 alkaline phosphatase activity was measured in three independent experiments. The values of each replicate (circles) and the mean (bars) were plotted. All values were normalized to the mean Pho8Δ60 alkaline phosphatase activity of cells expressing Vac8–GFP. t, tether; DIC, differential interference contrast; a.u., arbitrary units.
Vac8-mediated PAS localization to the vacuole is required for autophagy function
Ectopic t-Vac8ΔN was able to recruit PAS components (Fig. 6C). To test whether this ectopic PAS localization away from the vacuole is sufficient to rescue the vac8Δ bulk autophagy defect, we monitored Pho8Δ60 activity. Both cells expressing Vac8ΔN and t-Vac8ΔN were strongly impaired in Pho8Δ60 activity, suggesting that ectopic PAS localization is not sufficient to drive autophagy (Fig. 7C).
To investigate whether vacuolar localization of Vac8, and thus PAS–vacuole contacts, are required for autophagosome formation, we artificially recruited cytosolic Vac8ΔN to vacuoles and asked whether this restores bulk autophagy function. We employed the inducible FKBP/FRB/rapamycin tethering system (FKBP, 12-kDa FK506 binding protein; FRB, FKBP-rapamycin binding domain). The method is based on rapamycin-induced dimerization of two proteins of interest tagged with FKBP and FRB, respectively (Choi et al., 1996). These experiments were performed in a tor1-1 mutant background, in which a mutation in the FRB region of Tor1 prevents FKBP–rapamycin binding, thereby rendering tor1-1 mutant cells resistant to rapamycin. In addition, FPR1 (FK506-sensitive proline rotamase, the yeast homolog of FKBP) was deleted to prevent competition for rapamycin binding (Heitman et al., 1991).
We used pho8Δ60 tor1-1 fpr1Δ vac8Δ Vph1-FRB cells expressing FKBP–Vac8ΔN. Rapamycin addition resulted in the tethering of FKBP–Vac8ΔN to Vph1–FRB (Fig. 7D,E). To test whether this is sufficient to restore the function of cytosolic Vac8ΔN, we monitored Pho8Δ60 activity. In the absence of rapamycin, cells expressing FKBP–Vac8ΔN were impaired in Pho8Δ60 activity. However, upon rapamycin treatment Pho8Δ60 activity was restored to 50% of wild-type levels (Fig. 7F). Similar results were obtained for Vac8ΔN tethered constitutively to the vacuole by fusing it directly to Vph1 (Fig. S4C). These findings further support the hypothesis that that PAS–vacuole contacts are required for efficient autophagosome formation.
The middle domain of Atg13 (amino acids 269–520, Atg13M) contains binding regions for Atg1 and Atg17 (Fujioka et al., 2014); thus, we speculated that vacuolar localized Atg13M might be sufficient to recruit the PAS to the vacuole and to promote bulk autophagy independently of Vac8. To test this possibility, we fused Atg13M to the vacuolar protein Vph1 and expressed it in addition to endogenous Atg13 in wild-type and vac8Δ cells (Fig. S4D). However, in a Pho8Δ60 assay, the Vph1–Atg13M fusion protein failed to bypass the requirement for Vac8 in bulk autophagy (Fig. S4C). This could be due to technical reasons or it could indicate that Vac8 not only anchors the PAS at the vacuole but also serves additional functions in bulk autophagy.
In summary, our findings suggest that Vac8 promotes the formation of autophagosomes and autophagosome–vacuole fusion by tethering the PAS and the different autophagosomal intermediates via the C-terminus of Atg13 to the vacuole.
DISCUSSION
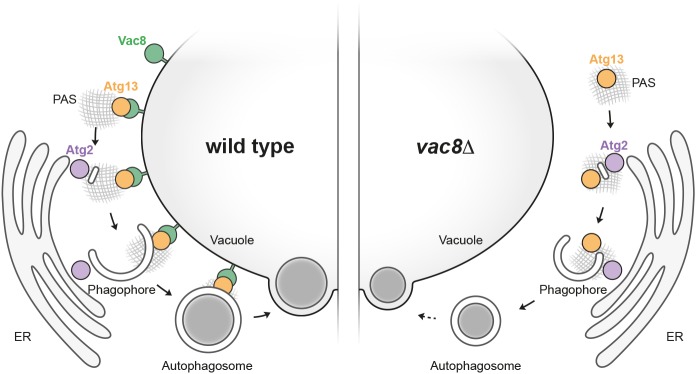
Our work shows that Vac8 plays a direct and important role in bulk autophagy. Vac8 is not only required for efficient initiation of PAS formation but also for formation and subsequent fusion of autophagosomes with the vacuole. Based on the Vac8-dependent association of Atg8-positive structures with the vacuole, we propose that Vac8 anchors the early PAS to the vacuole and maintains this connection throughout the biogenesis of the autophagosome. In this way, Vac8 coordinates the site of autophagosome formation with vacuole fusion and improves the efficiency of bulk autophagy by localizing autophagosome formation within the cell (Fig. 8).
Fig. 8.
Vac8 confines autophagosome formation between the vacuole and the ER by tethering the PAS to the vacuole. Vac8 tethers the early PAS to the vacuole via interaction with Atg13. Maturation of the PAS by recruitment of downstream Atg factors, including Atg2, results in establishment of an ER–PAS–vacuole connection. Consequently, autophagosome formation takes place in a confined space between the ER and the vacuole. This allows autophagosome formation to be spatially coordinated with autophagosome–vacuole fusion. In a VAC8 deletion mutant, the connection between PAS and vacuole is lost and the site of autophagosome formation remains only at the ER. As a consequence, PAS initiation is decreased, autophagosomes form with a reduced size and are ultimately released into the cytosol, which makes the autophagosome–vacuole fusion process inefficient.
Although decreased autophagic function had been observed in vac8Δ mutants (Cheong et al., 2005; Scott et al., 2000), the molecular function of Vac8 in autophagy had remained unknown. Deletion of VAC8 was found to reduce the degradation of autophagic cargo in the vacuole, the end point of the pathway, and this was interpreted as a severe bulk autophagy flux defect (Cheong et al., 2005; Scott et al., 2000). However, the assay used relies on vacuolar function. Since Vac8 is also required for efficient homotypic vacuole fusion, it had remained unclear whether Vac8 actually plays a direct role in autophagy or whether the observed autophagy defect in vac8Δ mutants rather stemmed from a general defect in vacuolar function. Our work reveals a direct function for Vac8 in autophagy. It acts early in autophagy by tethering the PAS and forming autophagosomes to the vacuole. Vac8 itself is stably anchored to the vacuole by N-terminal acylation and its vacuolar localization has been reported to be important for establishing nucleus–vacuole, vacuole–vacuole and vacuole–cytoskeleton connections. We show that N-terminal acylation of Vac8 is also essential for its function in autophagy, which is in line with Vac8 serving as a vacuolar tether of the PAS.
PAS anchoring is mediated via Vac8 binding to the C-terminus of Atg13. Atg13 in turn then recruits further Atg proteins to assemble the PAS at the vacuole. Interestingly, clustering of Atg13 C-terminus on ectopic cytosolic Ape1 results in the recruitment of this structure to the vacuole, but also the enrichment of vacuolar Vac8 at the Ape1–Atg13C–vacuole contact site (Fig. 7A; Fig. S4A,B). This enrichment of Vac8 might be caused by a high local concentration of t-Atg13C on Ape1. Similarly, Vac8 has been observed to be enriched at nucleus–vacuole junctions, suggesting that it has the ability to locally cluster on the vacuolar surface where needed. The role of this local clustering of Vac8 and whether this also happens during native PAS assembly is an interesting subject for future studies.
Ectopic Vac8ΔN clustered on a cytosolic oligomer is able to assemble PAS components, but it is not sufficient to restore bulk autophagy function (Figs 6B and 7C). However, vacuolar tethering of Vac8ΔN partially rescued the bulk autophagy defect of vac8Δ mutants (Fig. 7F; Fig. S4C), underlining the necessity for the vacuolar localization of the PAS. Vac8ΔN artificially tethered to the vacuole lacks N-terminal myristoylation and palmitoylation. Interestingly, it was previously reported that palmitoylation of Vac8 might have a direct function in mediating vacuole–vacuole fusion and in vacuolar inheritance (Subramanian et al., 2006). Thus, it could be speculated that palmitoylation of Vac8 also affects its function in bulk autophagy. Attempts to fully bypass Vac8 function by tethering the PAS to the vacuole via a vacuolar localized middle domain of Atg13 failed (Fig. S4C). This could be due to technical reasons, for example, that the bypass system fails to appropriately recruit PAS factors to the vacuole. Alternatively, this result could indicate that Vac8 not only anchors the PAS to the vacuole via interaction with C-terminus of Atg13 but that it is also required for additional functions in bulk autophagy, such as fusion of autophagosomes with the vacuole. Further studies will be required to fully understand the mechanistic details of the PAS–autophagosome–vacuole connection.
The growing phagophore has been observed to be positioned adjacent to the vacuole and, concurrently, also have one edge connected to the ER, with Atg2 being required for maintaining the ER connection (Gómez-Sánchez et al., 2018; Graef et al., 2013; Suzuki et al., 2013). Our results therefore suggest that Atg2 and Vac8 are key determinants in establishing an ER–PAS-vacuole connection, thereby localizing the formation of autophagosomes between the ER and the vacuole. This organization principle allows autophagosome formation to be spatially coordinated with vacuole fusion. The endosomal Rab family protein Vps21 is required for sealing autophagosomes, and indeed a VPS21 deletion mutant displays accumulation of unsealed autophagosomes that remain attached to the vacuole (Zhou et al., 2017). This further supports our model where autophagosomes are anchored to the vacuole by Vac8 throughout the whole autophagosome formation process until they fuse with the vacuole (Fig. 8).
In VAC8 deletion mutants, autophagosomes form adjacent to the ER without being stably tethered to the vacuole. Loss of this vacuolar connection results in autophagosomes accumulating in the cytosol of vac8Δ cells (Figs 2F and 3D), suggesting that complete autophagosomes are released from the ER but fail to establish a connection to the vacuole. Nevertheless, autophagosomes ultimately can fuse with the vacuole, although with a substantial temporal delay. A correct topological orientation of the phagophore relative to the ER is important for autophagosome formation (Gómez-Sánchez et al., 2018). So far, however, the relevance of the vacuolar connection for phagophore expansion has remained unclear. Bulk autophagosomes formed in vac8Δ mutants are smaller than in wild-type cells, indicating that the PAS–vacuole connection also plays an important structural role for normal autophagosome formation. In mammals, the omegasome, a specialized subdomain of the ER, has been reported to serve as a cradle for the formation of the phagophore (Hayashi-Nishino et al., 2009; Kast and Dominguez, 2017). It is conceivable that the ER–vacuole connection might serve an analogous role in yeast. The precise molecular reason for the bulk autophagosome size defect caused by deletion of VAC8 remains to be determined.
In mammalian cells, autophagosomes are made throughout the cytosol, forming in specialized subdomains of the ER (Karanasios et al., 2016; Nakamura and Yoshimori, 2017). Endosomes and lysosomes, by contrast, are predominantly found in the perinuclear region. As a result, completed autophagosomes need to be actively transported into the perinuclear region to allow effective fusion with lytic organelles. This transport is mediated by the microtubular network (Mackeh et al., 2013). In yeast, however, the cytoskeleton is dispensable for bulk autophagy (Kirisako et al., 1999; Reggiori et al., 2005). Our study sheds light on the reasons for this fundamental difference among eukaryotic species. Bulk autophagosome formation in yeast is initiated at the vacuole and apparently this connection is maintained while the autophagosome is forming and, most likely, until the point of fusion with the vacuole. Hence, active transport of mature autophagosomes to the lytic organelle is superfluous. The observation that vac8Δ cells accumulate cytosolic autophagosomes underpins the significance of a stable autophagosome–vacuole connection in yeast. This difference in autophagosome formation might stem from the apparent difference in the size of the lytic compartment and its distribution in the cell. Whereas yeast cells contain one to only a few large vacuoles that take up a substantial part of the cell, mammalian cells contain many small lysosomes that are enriched in the perinuclear region.
In contrast to bulk autophagy, selective autophagy requires the actin cytoskeleton in yeast (Reggiori et al., 2005). Future studies are needed to better understand the functional relevance of the cytoskeleton and the vacuolar connection of the PAS during selective autophagy in yeast.
MATERIALS AND METHODS
Yeast strains
Yeast strains are listed in Table S1 (Bas et al., 2018; Kijanska et al., 2010; Robinson et al., 1988; Torggler et al., 2016). Yeast strains and plasmids used in each figure are listed in Table S3. Genomic insertions (tagging) were performed according to Janke et al. (2004) or Longtine et al. (1998); multiple deletions or mutations were generated by PCR knockout or mating and dissection.
Plasmid construction
Plasmids are listed in Table S2 (Mari et al., 2010; Pfaffenwimmer et al., 2014; Sikorski and Hieter, 1989; Torggler et al., 2016; Welter et al., 2010). For pMS5 (Vac8–GFP), Vac8 with its endogenous promoter (626 bases) was amplified from genomic DNA and ligated into pRS415 vector using NotI/PstI restriction sites; GFP was PCR amplified and subcloned via PstI/SalI. Vac8–GFP mutants (pMS13, pMS130, pMS131 and pDH1) were generated by ‘round the horn’ PCR using pMS5 (Vac8–GFP) as a template and primers containing the respective mutations. To create pDH2 [FKBP-Vac8(19-578)–GFP], FKBP was PCR amplified and subcloned into pDH1 via SpeI/SbfI. For pDH8 [Atg19(152-191)–Vac8(19-578)–GFP], the Ape1-binding region of Atg19 (amino acids 152–191) was PCR amplified from genomic DNA and subcloned into pDH2 via SpeI/BamHI. pDH10 [Atg19(152-191)–Vac8(19-578)–GFP], pDH11 [Vac8(19-578)–GFP] and pDH12 [Vac8–GFP] were generated by exchanging the pRS415 vector to pRS413 via SacI/SalI restriction cloning. For pDH9 [Atg19(152-191)–Vac8(19-578)–mTagBFP2] and pDH13 [Vac8(19-578)–mTagBFP2], the GFP tag of pDH8 and pDH1, respectively, was replaced with mTagBFP2 via PstI/SalI; mTagBFP2 was PCR amplified. For pDH14 [Atg19(152-191)-Atg13(567-738)-mTagBFP2], the C-terminal region of Atg13 (amino acids 567–738) was PCR amplified from genomic DNA and ligated into pDH9 via BamHI/PstI. For pDH33, the VAC8 promoter was cut out from pDH1 using NotI/SpeI, Vph1 was PCR amplified from genomic DNA and cut with SpeI/BamHI, mScarlet was PCR amplified with an N-terminal and C-terminal AGSAAGS linker and cut with BamHI/SalI, and all were ligated into a pRS416 vector. For pDH32, the middle region of Atg13 (amino acids 269–520) was PCR amplified from genomic DNA and ligated into pDH33 using SbfI/SalI restriction sites. For pDH39, Vac8 (amino acids 19–578) was PCR amplified from genomic DNA and ligated into pDH33 using the SbfI/SalI restriction sites. For pDH41, the ATG13 endogenous promoter (698 bases) was PCR amplified from genomic DNA and ligated into pDH32 using NotI/SpeI restriction sites. For pAB19, Pbs2 with its endogenous promoter (296 bases) was amplified from genomic DNA and ligated into pCK903 vector (Brezovich et al., 2015) using the NotI/SbfI restriction sites. For pRT118, Atg13 with its endogenous promoter (698 bases) was amplified from genomic DNA and ligated into pCK903 vector using the NotI/SbfI restriction sites. For pRT119, Atg13 (amino acids 1–520) with its endogenous promoter (698 bases) was amplified from genomic DNA and ligated into pCK903 vector using NotI/SbfI restriction sites. For pAC160, Nvj1 with its endogenous promoter (700 bases) was amplified from genomic DNA and ligated into pCK903 vector using NotI/SbfI restriction sites.
Antibodies
The following primary antibodies were used in this study: rabbit polyclonal PAP antibody (1:5000, P1291, Sigma-Aldrich), polyclonal rabbit anti-Ape1 antibody (1:20,000; Torggler et al., 2016), mouse monoclonal anti-Pgk1 antibody (1:10,000; 22C5D8, Invitrogen), mouse monoclonal anti-GFP antibody (1:100; 2B6, Max F. Perutz Laboratories, Monoclonal Antibody Facility), rabbit monoclonal anti-HA antibody (1:1000; EPR4095, Abcam), mouse monoclonal anti-me3K9H3 antibody (1:2000; 6F12-H4, Novus Biochemicals) and mouse monoclonal anti-Myc antibody (used 1:5000) (4A6, Millipore).
Growth conditions
Yeast cells were grown in synthetic medium (SD, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose and amino acids as required) or rich medium (YPD, 1% yeast extract, 2% peptone and 2% glucose) to mid-log phase. To induce bulk autophagy, cells were washed and resuspended in nitrogen starvation medium (SD-N: 0.17% yeast nitrogen base without amino acids, 2% glucose) or treated with 220 nM rapamycin for the indicated time to induce autophagy. Yeast liquid cultures were incubated with shaking (200 rpm) at 30°C.
Serial dilution spot assay
Yeast cell cultures were grown to mid-log phase in YPD medium, shifted to SD-N medium containing 10 µg/ml tetracycline, diluted to an optical density at 600 nm (OD600) of 0.1 and incubated at 30°C. For analysing cell viability, a 7-fold serial dilution series was prepared by diluting cell cultures in SD-N medium, and 5 µl of each dilution was spotted onto YPD plates. Pictures were taken after 48 h incubation at 30°C.
Standard biochemical assays
For trichloroacetic acid (TCA) extract preparation, 1.5 ml of yeast cell culture was precipitated with 7% TCA and incubated for 10 min on ice. Precipitated proteins were pelleted at 13,000 g for 5 min at 4°C, washed with 1 ml acetone, air-dried, resuspended in urea loading buffer (120 mM Tris-HCl pH 6.8, 5% glycerol, 8 M urea, 143 mM β-mercaptoethanol, 8% SDS) and boiled before being loaded onto SDS-PAGE gels. Protein extracts were transferred onto nitrocellulose membranes and proteins were detected by immunoblotting, using the ECL detection system.
For preparation of freezer milled yeast powder, cells were harvested by centrifugation (3000 g, 10 min, room temperature), washed with 1× PBS containing 2% glucose and resuspended in 1 µl/OD600 unit (one OD600 unit corresponds to 1 ml yeast culture with an OD600 of 1) IP buffer [1× PBS, 10% glycerol, 0.5% Tween-20, 1 mM NaF, 1 mM PMSF, 20 mM β-glycerophosphate, 1 mM Na3VO4 and cOmplete™ protease inhibitor cocktail (Roche)]. Cells were milled in a cryogenic grinder (SPEX Freezer Mill 6875, SPEX SamplePrep), using five rounds of 3 min breakage at 15 cycles per second and 2 min cooling. Yeast powder was stored at −80°C.
Atg1 kinase assay
0.8 g freezer milled yeast powder was thawed on ice and 500 µl IP buffer was added (see section ‘Standard biochemical assays’). Extract was cleared by centrifugation (twice for 10 min, 5000 g, 4°C) and protein concentration was adjusted to 20–25 µg/µl in 700 µl IP buffer. Extract was incubated with 10 µl IgG-coupled Dynabeads (Thermo Fisher Scientific) on an end-over-end rotating wheel for 1 h at 4°C. Beads were washed three times for 5 min in IP buffer and afterwards split for western blotting (5 µl beads) and the kinase assay (5 µl beads). Beads for western blotting were eluted in 10 µl urea loading buffer and blotted for the ProteinA tag. Beads for the kinase assay were washed in 1× kinase buffer (20 mM HEPES pH 7.4, 150 mM potassium acetate, 10 mM magnesium acetate, 0.5 mM EGTA, 5 mM NaCl) and resuspended in 11 µl kinase mix {1× kinase buffer, 10 mM Na3VO4, 3 µg GST-tagged Atg19 C-terminal fragment used as substrate (Pfaffenwimmer et al., 2014), 2.5 µCi γ-[32P]ATP}. After 20 min at 30°C, beads and supernatant were separated and mixed with urea loading buffer. Samples were analysed by SDS-PAGE and phosphor-imaging.
Pho8Δ60 assay
25 OD600 units of yeast culture were harvested by centrifugation (3000 g, 5 min, room temperature). Pellets were washed in 1 ml distilled H2O, followed by centrifugation (2000 g, 5 min, 4°C) and resuspension of the pellet in 2 ml ice-cold 0.85% NaCl containing 1 mM PMSF. After another centrifugation step (2000 g, 5 min, 4°C), pellets were resuspended in 16 µl/OD600 unit lysis buffer [20 mM PIPES pH 6.8, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 µM ZnSO4, 1 mM PMSF, cOmplete™ protease inhibitor cocktail (Roche)]. Cells were lysed by bead beating, and extracts were cleared by centrifugation (16,000 g, 5 min, 4°C). Protein concentration of the supernatant was adjusted to 50 µg in 100 µl lysis buffer. 400 µl reaction buffer (0.4% Triton X-100, 10 mM MgSO4, 10 µM ZnSO4, and 250 mM Tris-HCl pH 8.5) containing 6.25 mM α-naphthylphosphate (Sigma-Aldrich) was added to enzymatic reactions, or only reaction buffer was added to control reactions. Reactions were incubated at 37°C for 10 min and stopped by adding 500 µl stop buffer (1 M glycine pH 11). Fluorescence was measured using 345 nm for excitation and 472 nm for emission.
For each experiment, three independent replicates were performed. Normalized Pho8Δ60 activity was calculated as follows: a standard curve was generated by either using a dilution series of the product (1-naphtol, Sigma-Aldrich) or a reaction time course series of the starvation-induced wild-type sample (20, 10, 5, 2.5 and 0 min). Least squares linear regression was performed using the standard curve, which was then used to calculate the relative abundance values of the samples. Replicates were normalized to each other by subtracting the averaged differences of the sample means. The replicate mean of the starvation-induced wild-type sample was set to 100.
M-track in vivo protein–protein proximity assay
M-track assays were performed as described previously (Brezovich et al., 2015; Zuzuarregui et al., 2012). Cell extracts were prepared by TCA precipitation and analysed by western blotting.
In vitro fusion assay
Fusion assays were performed according to Bas et al. (2018) with some adaptations. Vacuoles were isolated from Vph1–mCherry atg15Δ pep4Δ cells. Cells were lysed with DEAE-dextran according to Gao et al. (2018) and Haas (1995) and vacuoles purified on a Ficoll gradient at the 0–4% interface. Vacuoles were incubated with autophagosomal fractions prepared from GFP–Atg8 pep4Δvam3Δ cells and successful fusion was monitored by fluorescence microscopy. ATP was depleted by apyrase treatment, which converts ATP into AMP.
Quantitative live-cell imaging
Live-cell imaging was performed at room temperature using an AxioObserver Z1 inverted microscope (ZEISS) equipped with an α-Plan Apochromat 100× oil/1.46 NA differential interference contrast RMS objective, a PCO 1600 camera, and a Lumencor SOLA 6-LCR-SB light source with VisiView software (Visitron Systems) (Figs 1E,F, 2A–F, 5A,C–E, 6B,C and 7A,B,E; Fig. S4A,B,D) or with a DeltaVision Elite RT microscope system (GE Healthcare, Applied Precision), equipped with a UPLASPO 100× oil/1.40 NA objective, a pco.edge 5.5 sCMOS camera (PCO) and a seven-colour InsightSSI solid-state illumination system (GE Healthcare, Applied Precision) (Fig. 4A,C–E; Fig. S3B,C).
Quantitative analysis of PASs, forming autophagosomes or completed autophagosomes was performed by counting the number of GFP–Atg8, Atg1–GFP or Atg2–GFP puncta per cell. Images were generated by collecting a z-stack of 21 pictures with focal planes 0.20 μm or 0.25 µm apart, in order to cover the entire volume of a yeast cell. Background subtraction was performed using the rolling ball algorithm with a 7-pixel radius for the green channel (PAS or autophagosomes). The number of PASs, forming autophagosomes or completed autophagosomes per cell was counted blindly after randomizing image names. Image analysis was performed using FIJI (Schindelin et al., 2012) (Figs 1E,F and 2A–F).
Subcellular positioning of PASs, forming autophagosomes or complete autophagosomes was investigated by analysing the localization of mCherryV5–Atg8 puncta in regard to the ER, using the genomically tagged ER marker protein Sec63–GFP, and vacuoles labelled with CellTracker™ Blue 7-amino-4-chloromethylcoumarin (CMAC) dye (Thermo Fisher Scientific). Distribution of Vac8–GFP variants was analysed in regard to CMAC-stained vacuoles. Images were generated by collecting a z-stack of 22 pictures with focal planes 0.20 μm apart. Image deconvolution and analysis were performed using SoftWoRx (Applied Precision) and ImageJ software (Schneider et al., 2012) (Fig. 4A,C–E, Fig. S3B,C).
Recruitment of Vac8 (t-Vac8ΔN) artificially tethered to Ape1 was investigated by analysing the localization of Vac8–GFP signal in regard to mTagBFP2–Ape1 puncta. Colocalization of GFP-tagged proteins to Ape1 tethered t-Vac8ΔN–BFP was investigated by analysing the localization of GFP signal in regard to t-Vac8ΔN–BFP puncta. Images were generated on one focal plane. For quantification of BFP puncta overlapping with GFP signal, background subtraction was performed using the rolling ball algorithm with a 5-pixel radius for the green and blue channel. Quantification was performed blindly after randomizing image names. Image analysis was performed using FIJI (Schindelin et al., 2012) (Figs 5A,C–E and 6B,C).
Recruitment of Vac8 or Vac8ΔN to Ape1-tethered Atg13C (t-Atg13C) was investigated by analysing the localization of Vac8–GFP or Vac8ΔN–GFP signal in regard to t-Atg13C–BFP puncta. Subcellular positioning of Ape1-tethered Atg13C (t-Atg13C) was investigated by analysing the localization of t-Atg13C–BFP puncta in regard to vacuoles, using the genomically tagged vacuolar marker protein Vph1–4×mCherry. Images were generated by collecting a z-stack of 11 pictures with focal planes 0.25 μm apart. Image analysis was performed using FIJI (Schindelin et al., 2012) (Fig. 7A,B, Fig. S4A,B).
Distribution of FKBP–Vac8ΔN–GFP was analysed in regard to vacuoles stained with the FM™ 4-64 dye [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide] dye (Thermo Fisher Scientific). Images were generated by collecting a z-stack of 11 pictures with focal planes 0.25 µm apart. Image analysis was performed using FIJI (Schindelin et al., 2012) (Fig. 7E).
Localization of Vph1–mScarlet fusion proteins was investigated by analysing the mScarlet signal. Images were generated by collecting a z-stack of 11 pictures with focal planes 0.25 µm apart. Image analysis was performed using FIJI (Schindelin et al., 2012) (Fig. S4D).
Subcellular positioning of the PASs or autophagosomes, or the number of PASs or autophagosomes per cell was determined by analysing ≥100 cells from three independent experiments. The number of Ape1-tethered t-Vac8ΔN–BFP or t-Atg13C–BFP puncta overlapping with the signal of GFP-tagged proteins was determined by analysing ≥75 BFP puncta from three independent experiments.
Images from each figure panel were taken with the same imaging setup and are shown with the same contrast settings. Single focal planes of representative images are shown.
Electron microscopy
Fifteen OD600 units of cells were harvested by centrifugation (1800 g, 5 min, room temperature). Cells were washed in distilled H2O and pelleted by centrifugation (1800 g, 5 min, room temperature). Cells were resuspended in 3 ml of freshly prepared ice-cold 1.5% KMnO4 (Sigma) and transferred into two 1.5 ml microfuge tubes. After topping up the tube with the same solution to exclude air, samples were mixed on a rotatory wheel for 30 min at 4°C. After centrifugation (1400 g, 3 min, 4°C), the 1.5% KMnO4 incubation was repeated once more before washing the pellets five times with 1 ml of H2O.
Permanganate-fixed cells were dehydrated stepwise with increasing concentrations of acetone (10%, 30%, 50%, 70%, 90%, 95% and three times 100%). Each incubation step was performed for 20 min at room temperature on a rotating wheel, and in between each step, cells were pelleted by centrifugation (1400 g, 3 min, room temperature). After the last centrifugation, pellets were resuspended in 33% Spurr's resin in acetone and mixed on the same device for 1 h at room temperature. Cells were then harvested (7600 g, 3 min, room temperature) and incubated in 100% freshly made Spurr's resin overnight on a rotating wheel at room temperature. This operation was repeated the following day, over the day, after centrifugation of the overnight incubation (9000 g, 5 min, room temperature). The Spurr's resin mixture was prepared by mixing 10 g of 4-vinylcyclohexene dioxide (or ERL4206), 4 g of epichlorohydrin-polyglycol epoxy (DER) resin 736, 26 g of (2-nonen-1-yl)succinic anhydride (NSA) and 0.4 g of N,N-diethylethanolamine (all from Sigma). Afterwards, the cell and Spurr's mixture was transferred in size 00 embedding capsules (Electron Microscopy Science), and cells were pelleted by centrifugation (9000 g, 5 min, room temperature). Embedding capsules were topped up with 100% Spurr's and were baked for a minimum of 3 days at 60°C.
Thin sections of ∼55 nm were cut using an ultramicrotome (Leica Microsystems). Cell sections were collected on formvar carbon-coated 100 meshes copper grids (EMS). Cell sections were stained with a filtered lead-citrate solution (80 mM lead nitrate, 120 mM sodium citrate pH 12) for 2 min at room temperature. Sections were viewed in a CM100bio TEM (FEI, Eindhoven, Netherlands).
Supplementary Material
Acknowledgements
We would like to thank Irmgard Fischer and the MFPL biooptics facility for help with microscopy, Martina Schuschnig for help with strain crossing and testing and Daniel Papinski, Levent Bas, Riccardo Trapannone and Sascha Martens for discussions on experiments and the manuscript. We thank Serge Pelet, Roswitha Krick and Michael Thumm for plasmids. We would like to thank Life Science Editors for editorial assistance. This work reflects only the authors' view and the European Union's Horizon 2020 research and innovation programme is not responsible for any use that may be made of the information it contains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.M.H., R.G.-S., R.T., F.R., C.K.; Methodology: D.M.H., R.G.-S., F.R., C.K.; Validation: D.M.H., R.G.-S., A.C. F.K., M.M., R.T., M.L., F.R., C.K.; Investigation: D.M.H., R.G.-S., A.C. F.K., M.M., R.T., M.L.; Resources: M.M., F.R., C.K.; Data curation: D.M.H., R.G.-S., A.C. F.K., M.M., R.T., M.L., F.R., C.K.; Writing - original draft: D.M.H., C.K.; Writing - review & editing: D.M.H., R.G.-S., A.C. F.K., M.M., R.T., M.L., F.R., C.K.; Supervision: F.R., C.K.; Project administration: F.R., C.K.; Funding acquisition: F.R., C.K.
Funding
The Kraft laboratory has received funding from the Vienna Science and Technology Fund (WWTF, VRG10-001) from the FWF Austrian Science Fund (grant numbers P25522-B20, P28113-B28 and W1261), from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme under grant agreement No 769065, from the European Union's Horizon 2020 research and innovation programme under grant agreement No 765912, from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 403222702 – SFB 1381, and from the European Molecular Biology Organization (EMBO) Young Investigator Program. F.R. is supported by ZonMW VICI (016.130.606), ZonMW TOP (91217002), ALW Open Programme (ALWOP.310) and H2020 Marie Skłodowska-Curie Actions Cofund (713660) and Marie Skłodowska Curie ETN (765912) grants. M.M. is supported by an ALW Open Programme (ALWOP.355). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.235002.supplemental
References
- Bas L., Papinski D., Licheva M., Torggler R., Rohringer S., Schuschnig M. and Kraft C (2018). Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome–vacuole fusion. J. Cell Biol. 217, 3656-3669. 10.1083/jcb.201804028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezovich A., Schuschnig M., Ammerer G. and Kraft C (2015). An in vivo detection system for transient and low-abundant protein interactions and their kinetics in budding yeast. Yeast 32, 355-365. 10.1002/yea.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W. and Klionsky D. J. (2005). Atg17 regulates the magnitude of the autophagic response. MBoC 16, 3438-3453. 10.1091/mbc.e04-10-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J. and Klionsky D. J. (2008). The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in saccharomyces cerevisiae. MBoC 19, 668-681. 10.1091/mbc.e07-08-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S. L. and Clardy J. (1996). Structure of the FKBP12-rapamycin complex interacting with binding domain of human FRAP. Science 273, 239-242. 10.1126/science.273.5272.239 [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Suzuki S. W., Yamamoto H., Kondo-Kakuta C., Kimura Y., Hirano H., Akada R., Inagaki F., Ohsumi Y. and Noda N. N. (2014). Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 21, 513-521. 10.1038/nsmb.2822 [DOI] [PubMed] [Google Scholar]
- Gao J., Reggiori F. and Ungermann C. (2018). A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J. Cell Biol. 217, 3670-3682. 10.1083/jcb.201804039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez R., Rose J., Guimarães R., Mari M., Papinski D., Rieter E., Geerts W. J., Hardenberg R., Kraft C., Ungermann C. et al. (2018). Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217, 2743-2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., Friedman J. R., Graham C., Babu M. and Nunnari J. (2013). ER exit sites are physical and functional core autophagosome biogenesis components. MBoC 24, 2918-2931. 10.1091/mbc.e13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes R. S., Delorme-Axford E., Klionsky D. J. and Reggiori F. (2015). Assays for the biochemical and ultrastructural measurement of selective and nonselective types of autophagy in the yeast Saccharomyces cerevisiae. Methods Autophagy 75, 141-150. 10.1016/j.ymeth.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Haas A. (1995). A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 17, 283-294. 10.1007/BF00986234 [DOI] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T. and Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433-1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva N. R. and Hall M. N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905-909. 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E et al. et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947-962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jeong H., Park J., Kim H.-I., Lee M., Ko Y.-J., Lee S., Jun Y. and Lee C (2017). Mechanistic insight into the nucleus–vacuole junction based on the Vac8p–Nvj1p crystal structure. PNAS 114, E4539-E4548. 10.1073/pnas.1701030114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Walker S. A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H., Ahmed Q., Domart M.-C., Collinson L. and Ktistakis N. T. (2016). Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7, 12420 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast D. J. and Dominguez R. (2017). The Cytoskeleton–Autophagy Connection. Curr. Biol. 27, R318-R326. 10.1016/j.cub.2017.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M., Dohnal I., Reiter W., Kaspar S., Stoffel I., Ammerer G., Kraft C. and Peter M. (2010). Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6, 1168-1178. 10.4161/auto.6.8.13849 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T. and Ohsumi Y. (1999). Formation Process of Autophagosome Is Traced with Apg8/Aut7p in Yeast. J. Cell Biol. 147, 435-446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T., Kirisako H., Koizumi M., Ohsumi Y. and Nakatogawa H. (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 115, 10363-10368. 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Reggiori F. and Peter M. (2009). Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793, 1404-1412. 10.1016/j.bbamcr.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A. M. III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P. and Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Mackeh R., Perdiz D., Lorin S., Codogno P. and Poüs C. (2013). Autophagy and microtubules – new story, old players. J. Cell Sci. 126, 1071-1080. 10.1242/jcs.115626 [DOI] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D. J. and Reggiori F. (2010). An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005-1022. 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. J., Gubas A. and Tooze S. A. (2018). A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 293, 5386-5395. 10.1074/jbc.R117.810366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S. and Yoshimori T. (2017). New insights into autophagosome–lysosome fusion. J. Cell Sci. 130, 1209-1216. 10.1242/jcs.196352 [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y. and Ohsumi Y. (1995). Novel System for Monitoring Autophagy in the Yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126-132. 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- Pan X. and Goldfarb D. S. (1998). YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J. Cell Sci. 111, 2137-2147. [DOI] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S. and Goldfarb D. S. (2000). Nucleus–vacuole junctions in saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. MBoC 11, 2445-2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenwimmer T., Reiter W., Brach T., Nogellova V., Papinski D., Schuschnig M., Abert C., Ammerer G., Martens S. and Kraft C. (2014). Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 15, 862-870. 10.15252/embr.201438932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T. and Klionsky D. J. (2005). The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast saccharomyces cerevisiae. MBoC 16, 5843-5856. 10.1091/mbc.e05-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M. and Goldfarb D. S. (2003). Piecemeal Microautophagy of Nucleus in Saccharomyces cerevisiae. MBoC 14, 129-141. 10.1091/mbc.e02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M. and Emr S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936-4948. 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Nice D. C., Nau J. J., Weisman L. S., Kamada Y., Keizer-Gunnink I., Funakoshi T., Veenhuis M., Ohsumi Y. and Klionsky D. J. (2000). Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 275, 25840-25849. 10.1074/jbc.M002813200 [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E. and Klionsky D. J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825-837. 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S. and Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K., Dietrich L. E. P., Hou H., LaGrassa T. J., Meiringer C. T. A. and Ungermann C (2006). Palmitoylation determines the function of Vac8 at the yeast vacuole. J. Cell Sci. 119, 2477-2485. 10.1242/jcs.02972 [DOI] [PubMed] [Google Scholar]
- Suzuki K. and Ohsumi Y. (2010). Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 584, 1280-1286. 10.1016/j.febslet.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T. and Ohsumi Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209-218. 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H. and Ohsumi Y. (2013). Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126, 2534-2544. 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Torggler R., Papinski D., Brach T., Bas L., Schuschnig M., Pfaffenwimmer T., Rohringer S., Matzhold T., Schweida D., Brezovich A. et al. (2016). Two Independent pathways within selective autophagy converge to activate Atg1 kinase at the vacuole. Mol. Cell 64, 221-235. 10.1016/j.molcel.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Tsukada M. and Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169-174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Veit M., Laage R., Dietrich L., Wang L. and Ungermann C. (2001). Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 20, 3145-3155. 10.1093/emboj/20.12.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-X., Catlett N. L. and Weisman L. S. (1998). Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 140, 1063-1074. 10.1083/jcb.140.5.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Noda N. N., Kumeta H., Suzuki K., Ohsumi Y. and Inagaki F. (2010). Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J. Biol. Chem. 285, 30026-30033. 10.1074/jbc.M110.143545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter E., Thumm M. and Krick R. (2010). Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy 6, 794-797. 10.4161/auto.6.6.12348 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Fujioka Y., Suzuki S. W., Noshiro D., Suzuki H., Kondo-Kakuta C., Kimura Y., Hirano H., Ando T., Noda N. N. et al. (2016). The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev. Cell 38, 86-99. 10.1016/j.devcel.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Zhou F., Zou S., Chen Y., Lipatova Z., Sun D., Zhu X., Li R., Wu Z., You W., Cong X. et al. (2017). A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 13, e1007020 10.1371/journal.pgen.1007020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuzuarregui A., Kupka T., Bhatt B., Dohnal I., Mudrak I., Friedmann C., Schüchner S., Frohner I. E., Ammerer G. and Ogris E. (2012). M-Track: detecting short-lived protein-protein interactions in vivo. Nat. Methods 9, 594-596. 10.1038/nmeth.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.