ABSTRACT
During the extended prophase of Drosophila gametogenesis, spermatocytes undergo robust gene transcription and store many transcripts in the cytoplasm in a repressed state, until translational activation of select mRNAs in later steps of spermatogenesis. Here, we characterize the Drosophila Doublefault (Dbf) protein as a C2H2 zinc-finger protein, primarily expressed in testes, that is required for normal meiotic division and spermiogenesis. Loss of Dbf causes premature centriole disengagement and affects spindle structure, chromosome segregation and cytokinesis. We show that Dbf interacts with the RNA-binding protein Syncrip/hnRNPQ, a key regulator of localized translation in Drosophila. We propose that the pleiotropic effects of dbf loss-of-function mutants are associated with the requirement of dbf function for translation of specific transcripts in spermatocytes. In agreement with this hypothesis, Dbf protein binds cyclin B mRNA and is essential for translation of cyclin B in mature spermatocytes.
KEY WORDS: Drosophila, Male meiosis, Centrosome, Cytokinesis, Spermatogenesis
Summary: The Drosophila C2H2 zinc-finger protein Doublefault controls meiotic chromosome segregation and cytokinesis, and is required for cyclin B translation in mature spermatocytes.
INTRODUCTION
Spermatogenesis in Drosophila melanogaster offers a well-suited model system in which to investigate how transcriptional and post-transcriptional mechanisms of gene regulation coordinate cell cycle progression with cell differentiation programs (Lin et al., 1996; White-Cooper et al., 1998; Fuller and Spradling, 2007). In Drosophila adult testes, the successive stages of spermatogenesis are displayed in a linear way, with the spermatogonia and spermatocytes in meiosis occupying the first one-third of the testis. At the apical tip of the testis, asymmetric division of a male germline stem cell generates a new germline stem cell and a daughter cell, which initiates differentiation as a gonialblast (Fuller and Spradling, 2007). Each gonialblast undergoes four rounds of synchronous mitotic spermatogonial divisions with incomplete cytokinesis to produce a cyst of 16 interconnected germ cells. After the pre-meiotic S phase, the resulting 16 spermatocytes enter an extended meiotic prophase, characterized by an impressive growth phase leading to a 25-fold increase in the cell volume (Fig. S1). During the growth phase, which lasts ∼90 h, spermatocytes undergo robust gene expression. Because gene transcription is mostly shut down upon entry into the first meiotic division, the transcripts are stored in the cytoplasm of male meiotic cells until translation in later stages of spermatogenesis (Olivieri and Olivieri, 1965; Gould-Somero and Holland, 1974; Fuller, 1993). At the completion of meiotic prophase, the spermatocytes undergo two rapid meiotic divisions that generate 64 haploid round spermatids (Fuller, 1993; Giansanti and Fuller, 2012; Giansanti et al., 2012). The first cellular event of spermiogenesis occurs soon after meiosis when all the mitochondria contained in each spermatid, aggregate around the basal body at one side of the nucleus and fuse to form a complex interlaced structure dubbed the nebenkern (Fabian and Brill, 2012). Subsequent morphological changes, including organelle remodeling, flagellar elongation and nuclear shaping, result in the formation of motile sperm (Fabian and Brill, 2012).
Phenotypic analysis of mutants in several genes that control G2/M cell cycle program indicates that activation of spermatid differentiation is not dependent on prior completion of meiosis. For example, testes from mutants in the cell cycle regulating phosphatase twine/Cdc25, undergo spermatid differentiation, despite failure of the spermatocytes to undergo the meiotic division (Lin et al., 1996; White-Cooper et al., 1998). The correct timing of the G2/M transition that completes meiotic prophase is regulated by stage-specific repression of translation that restricts the expression of the core cell cycle protein Cyclin B and other meiotic regulators to late G2 phase (White-Cooper et al., 1998; Baker and Fuller, 2007; Baker et al., 2015).
Here, we identify Doublefault (Dbf) as a testis-specific C2H2 zinc-finger protein that binds cyclin B mRNA and is required for translation of cyclin B in mature spermatocytes. Moreover Dbf interacts with the RNA-binding protein Syncrip/hnRNPQ, a key regulator of localized translation in Drosophila. We show that Dbf is required for multiple aspects of meiotic division, including centriole disengagement, centrosome structure, chromosome segregation and cytokinesis. Our results suggest that the pleiotropic phenotype of dbf mutant males could be associated with the requirement for dbf for translation of specific mRNAs in spermatocytes.
RESULTS
Molecular cloning of the Drosophila doublefault gene
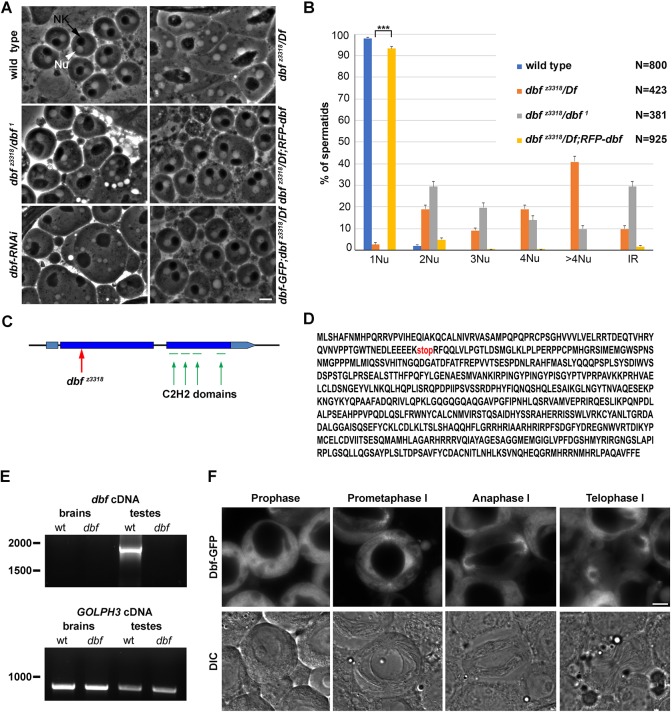
Screening the Zuker collection (Wakimoto et al., 2004) of male sterile mutants, we identified z3318 as a novel mutant allele of the P-element insertion doublefault1 (dbf1), a mutation previously associated with abnormally sized nuclei and nebenkern (Castrillon et al., 1993). When viewed by phase-contrast optics, each onion-stage spermatid from wild-type males contains a phase-dark nebenkern associated with a phase-light nucleus of similar size. Mutants that fail both cytokinesis and chromosome segregation in male meiotic cells can be readily identified by the presence of spermatids containing a large nebenkern associated with multiple nuclei of different size (Giansanti et al., 2004; Giansanti and Fuller, 2012; Giansanti et al., 2012). dbf z3318 and dbf 1/dbf z3318 males displayed aberrant spermatids containing mitochondrial derivatives of variable size associated with nuclei that were also different in size and number, indicating defects of meiotic chromosome segregation and cytokinesis similar to those described previously in dbf 1(Castrillon et al., 1993). dbf 1 and dbf z3318 failed to complement Df(2L)Exel8026 and Df(2L)BSC211 for both male sterility and the male meiotic defects (Fig. 1A,B). DNA sequencing revealed that the dbf z3318 mutant allele carried a nonsense mutation in the annotated CG17098 gene, predicted to encode a truncated protein containing 84 out of 652 amino acids (Fig. 1C,D). In addition, testes from males expressing double-stranded RNA against CG17098 in spermatocytes (Fig. 1B and Fig. S2A), contained frequent irregular spermatids, indicating male meiotic defects comparable with those observed in dbf z3318 and in dbf 1 mutants. Data reported in FlyAtlas indicated robust expression of the CG17098 transcript only in testes (Chintapalli et al., 2007). Consistent with these data, RT-PCR detected dbf cDNA in wild-type testes but not in wild-type larval brains. The dbf cDNA was not detected in dbf mutant testes, indicating that dbf mRNA may be degraded through the nonsense-mediated mRNA-decay pathway (Fig. 1E). Based on these results, dbf encodes a 73 kDa polypeptide, containing four C2H2-like zinc fingers. Searches for homologies via DIOPT (www.flyrnai.org/diopt) identified the RNA-binding, zinc-finger protein ZNF346 as the human ortholog of Dbf (Burge et al., 2014). Transgenes expressing either RFP-Dbf or Dbf-GFP fusion proteins expressed under the α tubulin promoter fully rescued both the phenotypic defects and the male sterility associated with dbf z3318/Df(2L)Exel8026, indicating that the encoded fusion proteins are fully functional (Fig. 1A,B). Analysis of testes from Dbf-GFP or RFP-Dbf flies showed the Dbf protein in the cytoplasm of immature spermatocytes, and becoming enriched around the astral membranes and spindle poles during meiotic divisions (Fig. 1F and Fig. S3).
Fig. 1.
Doublefault is required for chromosome segregation and cytokinesis in dividing spermatocytes. (A) Phase-contrast micrographs of onion-stage spermatids. Each wild-type spermatid displays a single nucleus (Nu, white arrowhead) associated with a single mitochondrial derivative (nebenkern, NK) of similar size (black arrow). Multiple nuclei of varying size associated with one enlarged dark nebenkern were observed in testes from dbf z3318/Df(2L)Exel8026 (dbf z3318/Df), dbf z3318/dbf 1 males and in testes from individuals expressing UAS::dbfRNAi under the control of bam-GAL4 (dbf-RNAi). RFP-Dbf or Dbf-GFP proteins, expressed by a single transgene copy, rescue the cytokinesis defects associated with dbf z3318/Df mutation. (B) Quantification of the percentage of multinucleate spermatids in testes from wild type, dbf z3318/Df(2L)Exel8026 (dbf z3318/Df), dbf z3318/dbf 1 and dbf z3318/Df(2L)Exel8026; RFP-dbf (dbf z3318/Df; dbf) with 1, 2, 3, 4 and >4 nuclei per nebenkern. IR indicates the frequency of spermatids containing a single large nucleus associated with a large nebenkern. Data are mean±s.e.m. Spermatids were examined from testes of at least 10 males per genotype. ***P<0.0001 (unpaired Student's t-test). (C) Schematic of the dbf locus, showing intron/exon organization, localization of the stop codon in the dbf z3318 allele. (red arrow) and the predicted zinc-finger motifs (green). (D) Amino acid sequence of the Dbf protein with the truncation position corresponding to the dbf z3318 mutation (stop). (E) RT-PCR from control (wt) and dbf mutants [dbf z3318/Df(2L)Exel8026]. Lower panel: GOLPH3 loading control in the experiment. (F) Fluorescence (upper panels) and corresponding DIC micrographs (lower panels) of live wild-type spermatocytes expressing Dbf-GFP at the indicated meiotic stages. Scale bars: 10 µm.
Doublefault is required for sex chromosome condensation and centrosomal structure during male meiosis
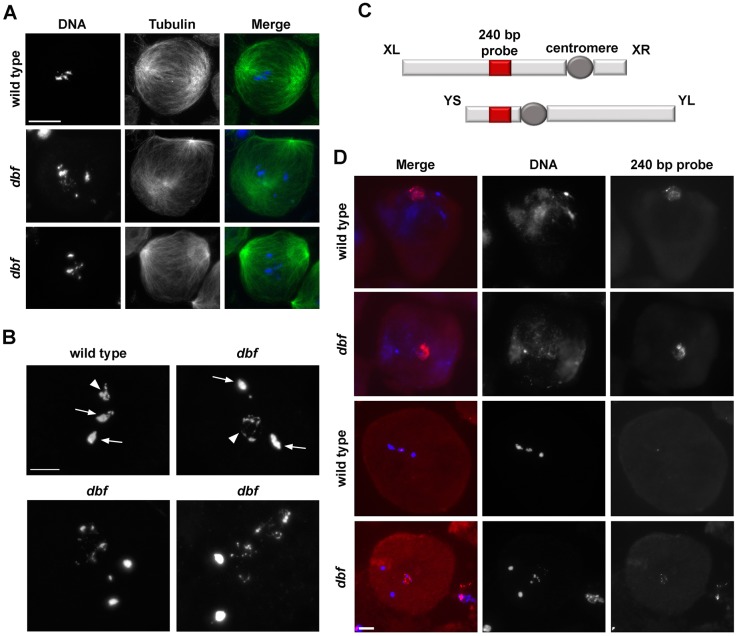
Examination of chromatin condensation during meiosis I revealed that dbf function is required for sex chromosome condensation. Male meiotic cells were staged according to the nomenclature introduced by Cenci et al. (1994), as described in Fig. S1. In wild-type spermatocytes during prometaphase/metaphase (M1-M3, Fig. 2A,B), chromatin compacted into three clumps, corresponding to the two autosomal bivalents and the sex chromosome bivalent, while two prominent asters defined the two poles of a bipolar spindle (Fig. 2A). In contrast, in dividing spermatocytes from dbf/Df mutant males at the same stage, one chromosome pair failed to condense properly (Fig. 2A,B). Fluorescent in situ hybridization using a 240 bp probe (Thomas et al., 2005), located in the intergenic spacers of the rRNA genes (Fig. 2C,D), revealed it was specifically the X-Y bivalent chromatin that failed to condense in all the dbf metaphase spermatocytes (Fig. 2C,D).
Fig. 2.
Doublefault is required for sex chromosome condensation in male meiotic cells. (A) Wild-type and dbf z3318/Df(2L)Exel802 (dbf) mutant spermatocytes in prometaphase (stage M2) stained for α-tubulin (green) and DNA (blue). (B) Metaphase chromosomes from wild-type and dbf z3318/Df(2L)Exel802 (dbf) mutant spermatocytes. Arrows indicate bivalents of the metacentric autosomes. Arrowheads indicate the X-Y pair. n=60 wild-type metaphase spermatocytes; n=54 dbf mutant metaphase spermatocytes, randomly selected from images taken in five experiments. (C) Diagram of the X and Y chromosomes. Red boxes indicate rRNA loci. The fluorescently labeled probe used in fluorescent in situ hybridization analysis is complementary to the 240 bp repeat in the spacers of the rDNA repeats. XL and XR indicate the left and right arms of the X chromosome; YS and YL indicate the short and long arms of the Y chromosome. (D) Fluorescent in situ hybridization analysis in wild-type and dbf z3318/Df(2L)Exel802 (dbf) mutant spermatocytes at metaphase (stage M2/M3) using a fluorescently labeled 240 bp probe (red). DNA was counterstained with DAPI (blue). n=53 wild-type metaphase spermatocytes; n=20 dbf mutant spermatocytes randomly selected from images taken in five experiments. Scale bars: 10 µm.
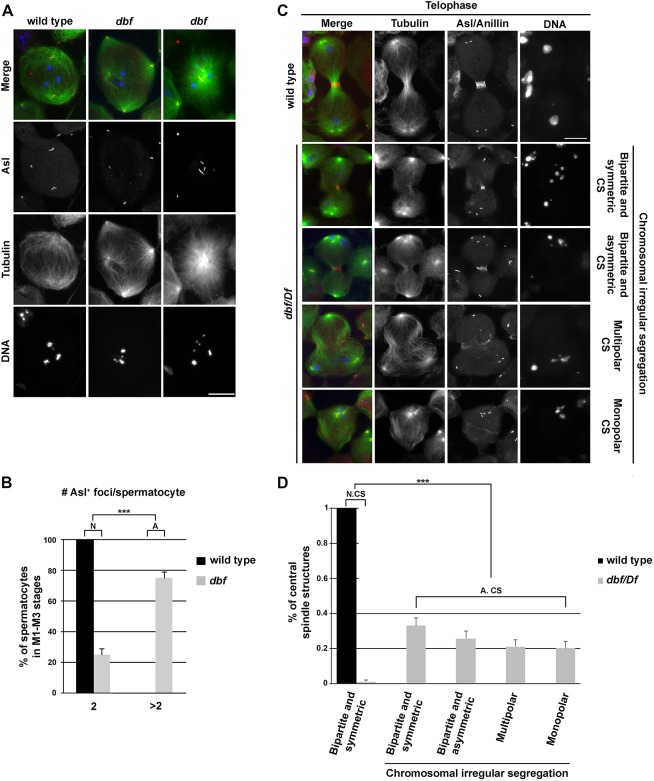
Loss of Dbf activity also affected centrosomes. In wild type, dividing spermatocytes in meiosis I immunolabeled for the centrosomal markers Centrosomin (Li et al., 1998) and Spd2 (Dix and Raff, 2007; Giansanti et al., 2008) displayed two prominent centrosomes (each containing a pair of centrioles) that formed bipolar meiotic spindles (Fig. S4A,B). In contrast, 43% of dividing spermatocytes in meiosis I from dbf/Df mutant males displayed a variable number of centrosomal foci and assembled monopolar or multipolar spindles, suggesting a defect in centriole disengagement, migration and/or centrosome structure (Fig. S4A,B). Analysis of spermatocytes using antibodies directed against the Asterless protein to stain centrioles at stages S5-S6 revealed that, whereas wild-type primary spermatocytes had two pairs of elongated centrioles with a characteristic V-shaped structure (Fig. 3A,B; Fig. S5A,B), most S5-S6 dbf primary spermatocytes displayed more than two Asl-labeled foci, indicating premature centriole disengagement (Fig. 3A,B; Fig. S5A,B). Taken together, these results suggest that function of Dbf is required to maintain attachment of mother and daughter centrioles in each centriole pair during male meiotic prophase (G2).
Fig. 3.
Mutant spermatocytes in dbf display premature centriole disengagement and irregular ana-telophase. (A) Wild-type and dbf z3318/Df(2L)Exel802 (dbf) mutant spermatocytes during prometaphase stained for the centriolar protein Asterless (Asl, red), α-tubulin (green) and DNA (blue). Scale bar: 10 µm. (B) Graph of the percentage (±s.e.m.) of stage M1-M3 spermatocytes containing the indicated number of Asl-labeled foci. n=112 mutant spermatocytes and n=120 wild-type spermatocytes at stage M1-M3 randomly selected from images taken in six independent experiments. ***P<0.0001 (Fisher's exact test). N, normal; A, abnormal foci number. (C) Wild-type and dbf z3318/Df(2L)Exel802 (dbf/Df) mutant spermatocytes during ana-telophase, stained for Anillin (red), Asl (red), α-Tubulin (green) and DNA (blue). CS, central spindle. Scale bar: 10 µm. (D) Percentages (±s.e.m.) of central spindle types observed in dbf z3318/Df(2L)Exel802 (dbf/Df) mutant spermatocytes compared with wild type. n=110 wild-type ana-telophase spermatocytes and n=110 dbf z3318/Df(2L)Exel802 mutant anatelophase spermatocytes randomly selected from images taken in six independent experiments. ***P<0.0001 (Fisher's exact test). N, normal; A, abnormal; CS, central spindle.
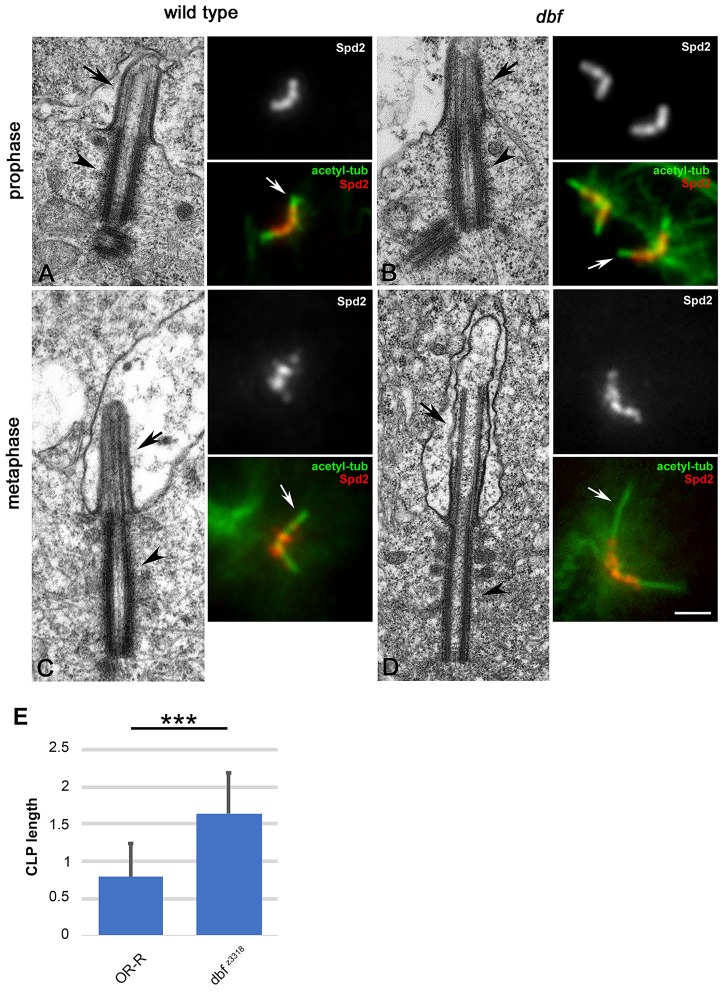
Loss of Doublefault results in aberrant centriole and axoneme structures during spermatogenesis
Transmission electron microscopy (EM) showed that centrioles and cilium-like projections (CLPs) in primary prophase I dbf spermatocytes at stage S5 were very similar in size to wild type (Fig. 4A-E). However, as meiosis progressed, the centrioles and the CLPs of dbf spermatocytes appeared more extended than those of control spermatocytes at the same stage. Immunostaining for the CLP marker acetylated tubulin confirmed the increase in CLP size in dbf mutant spermatocytes at stages M2-M3 compared with control (Fig. 4A-E). Immunofluorescence analysis revealed that localization of the centriole protein Spd2 at the dbf centrioles in S5-S6 spermatocytes did not appear different when compared with wild type at the same stage (Fig. 4A,B). However, Spd2 localization changes from ring-like in wild type to more diffuse and extended onto elongated tubulin structures in dbf mutants during later stages of meiosis (Fig. 4C,D).
Fig. 4.
Defects in morphology and ultrastructure of centrioles and cilium-like projections in male meiotic cells from dbf mutant males. (A,B) Transmission electron micrographs and immunofluorescence pictures showing centrioles and cilium-like projections in control (A) and dbf z3318/Df(2L)Exel802 (dbf, B) primary spermatocytes at stage S5. Spd2 marks the centrioles; acetylated tubulin (white arrows) marks the axonemal microtubules of the CLPs. (C,D) Electron microscopy and immunofluorescence of wild-type (C) and dbf (D) metaphase I spermatocytes. Black arrowheads indicate centrioles; black arrows indicate CLPs. Scale bars: 0.4 µm (EM panels); 2 µm (IF panels). (E) Average CLP length based on acetylated tubulin staining in dividing spermatocytes at metaphase I (stage M3). n=65 wild-type CLPs and n=72 dbf z3318/Df(2L)Exel802 (dbf z3318) mutant CLPs randomly selected from images taken in five independent experiments. Data are mean±s.d. ***P<0.0001 (unpaired Student's t-test).
Axoneme formation was dramatically affected in early spermatids from dbf mutants. Although basal bodies from spermatids of dbf mutants nucleated axonemal doublets as in wild-type spermatids (Fig. 5A), axonemes quickly lost integrity and the doublets spread in dbf spermatids (Fig. 5B). Canonical 9+2 axonemes (Fig. 5C,D) were not observed in cross-sections of elongating dbf spermatids (Fig. 5E,F). Instead, isolated microtubule doublets or triplets were observed (Fig. 5F). Strikingly Spd2 protein persisted at the basal body of early round spermatids from dbf males (Fig. 5B, inset), whereas this antigen disappeared in control spermatids at the same stage (Fig. 5A, inset).
Fig. 5.
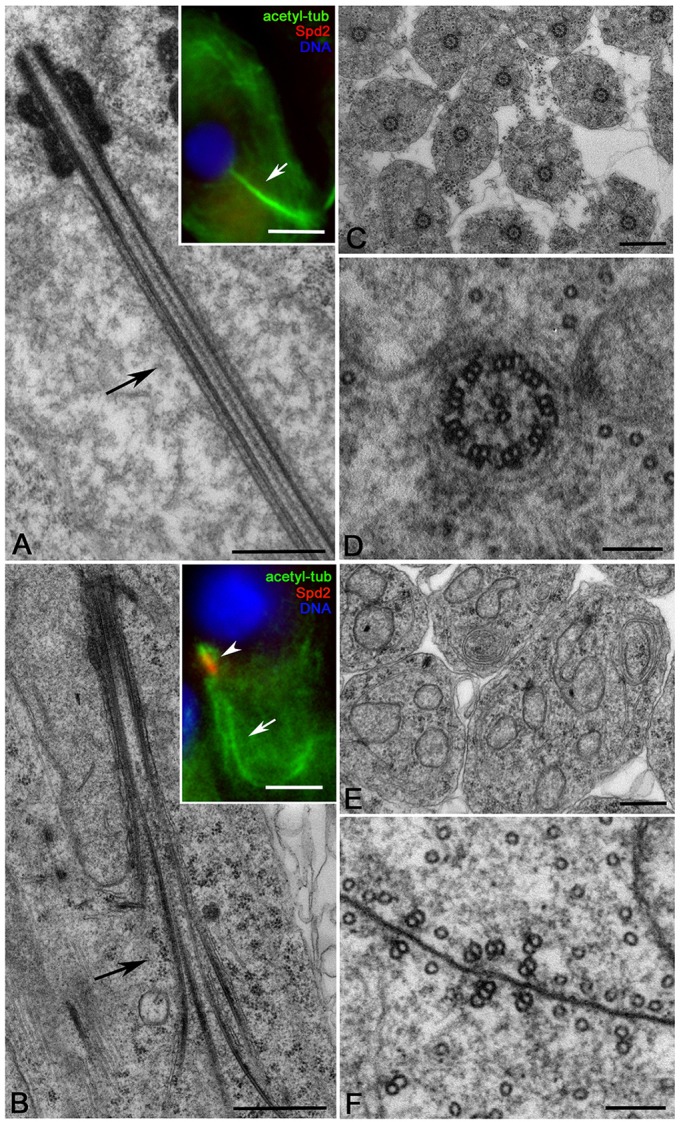
dbf mutant spermatids display defects of axoneme assembly. Longitudinal sections of control (A, inset) and dbf z3318/Df(2L)Exel802 (B, inset) elongating spermatids. Electron microscopy: n=16 wild-type axonemes, n=11 dbf mutant axonemes randomly selected from images taken in five independent experiments. Immunofluorescence (insets): n=71 wild-type and n=52 dbf mutant elongating spermatids; randomly selected from images taken in five independent experiments. (C,E) Cross-sections of control (C; n=173 randomly selected from images taken in five independent experiments) and dbf (E; n=122 randomly selected from images taken in five independent experiments) elongating spermatids. (D,F) High magnifications show that the defect of axoneme assembly in dbf results in isolated doublets or triplets (F), whereas control spermatids display distinct 9+2 axonemes (D). Scale bars: 0.5 µm in A-C,E; 5 µm in A,B (insets); 100 nm in D,F.
doublefault mutant spermatocytes undergo irregular ana-telophase
Staining of dividing spermatocytes for Anillin, tubulin and Asl revealed that dbf mutant spermatocytes undergo highly irregular ana-telophase (Fig. 3C,D). All ana-telophase figures from wild type displayed bipartite and symmetric central spindles, and accumulated Anillin at the cleavage site (Fig. 3C,D). In contrast, most ana-telophases from dbf z3318/Df(2L)Exel8026 males exhibited irregular spindles, with either multipartite or bipartite central spindles asymmetrically located with respect to cell poles (Fig. 3C,D). Moreover all the ana-telophase figures from dbf mutants exhibited irregular chromosome segregation (Fig. 3C,D).
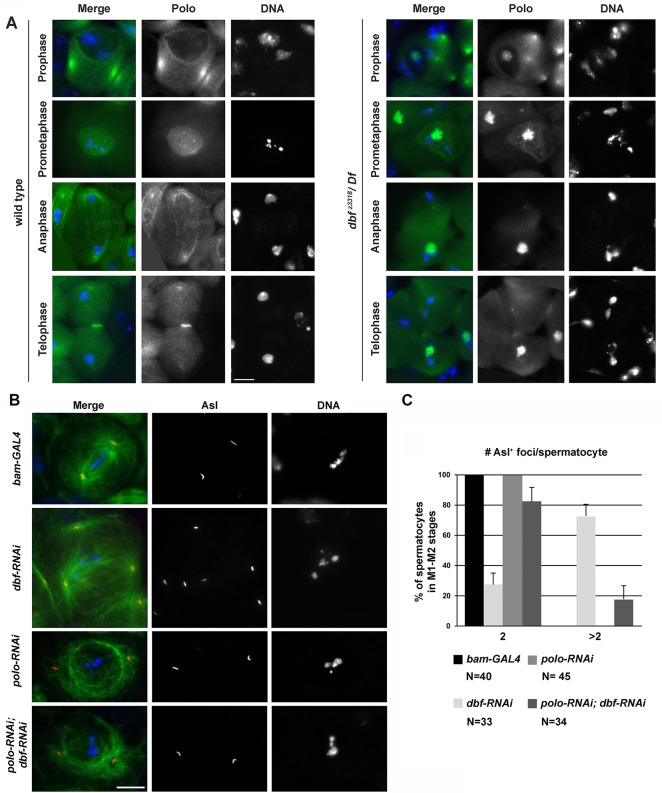
Loss of Dbf also affects subcellular localization of Polo kinase, a key regulator of chromosomal events during mitosis and meiotic divisions (Pintard and Archambault, 2018). Immunofluorescence analysis of wild-type dividing spermatocytes revealed that Polo protein concentrated at the centrosomes and at the nuclear envelope in late prophase (stages S6-M1), accumulated at the kinetochores during prometaphase (stage M2) and was enriched in the midzone by late anaphase (Fig. 6A). In contrast, in dbf z3318/Df(2L)Exel8026 mutant spermatocytes at late prophase, Polo protein, although enriched at the centrosomes, was also visible in a distinct structure inside the nucleus (Fig. 6A). During prometaphase and later stages, the majority of Polo protein in dbf z3318/Df(2L)Exel8026 dividing spermatocytes was associated with the mass of uncondensed sex bivalent chromatin (Fig. 6A). The accumulation of Polo protein inside the nucleus of dbf z3318/Df(2L)Exel8026 was not an artefact of the monoclonal antibody used to detect Polo, as similar results were obtained by analyzing live and fixed spermatocytes expressing Polo-GFP (Fig. S6A,B). Live imaging revealed enrichment of Polo-GFP inside the nucleus of wild-type and dbf-RNAi spermatocytes at stage S5 (Figs S2A and S6A). Co-immunostaining for the nucleolar marker Fibrillarin, revealed that Polo-GFP localization inside the nucleus was coincident with the nucleolus (Fig. S6B). We cannot exclude the possibility that the lack of nucleolar localization of the endogenous Polo in wild-type spermatocytes, immunostained using anti-Polo, may reflect some loss of the protein during fixation. In later stages of wild-type dividing spermatocytes, Polo-GFP accumulated at the nuclear envelope and at kinetochores (Fig. S6A). In contrast, in dbf-RNAi spermatocytes at late prophase (stage S6), most Polo-GFP remained inside the nucleus while a pool of the fluorescent-tagged protein was associated with the centrosomes that failed to contact the nuclear envelope (Fig. S6A). Western blot analysis revealed a significant increase of Polo-GFP protein levels in dbf-RNAi mutant testes compared with control, as well as a significant increase of the endogenous Polo protein in dbf z3318/Df(2L)Exel8026 testes compared with control (Figs S6C,D and S7). Knocking down expression of Polo kinase by RNA interference in spermatocytes under the control of the bam-Gal4 expression driver (Fig. S2A,B) rescued the premature centriole disengagement in S5-to-prometaphase spermatocytes (Fig. 6B,C). Together, these results indicate that the defects in centrosome structure and the consequent meiotic spindle defects are likely due to aberrant Polo localization/expression.
Fig. 6.
Defective localization of Polo protein in dbf mutant male meiotic cells. (A) Wild-type and dbf z3318/Df(2L)Exel802 (dbf z3318/Df) mutant spermatocytes stained to visualize Polo (green) and DNA (blue). (B) Dividing spermatocytes at late prophase/prometaphase (stages M1-M2) stained for Tubulin (green), Asl (red) and DNA (blue). Scale bars: 10 µm. (C) Quantification of centriole disengagement in spermatocytes at stages M1-M2. Graph shows percentage (±s.e.m.). Depletion of Dbf by bam-GAL4>dbfsiRNA (dbf-RNAi) resulted in 27% of spermatocytes displaying two Asl+ foci and 73% of spermatocytes displaying more than two Asl+ foci, indicating premature centriole disengagement. Depletion of Polo by bam-GAL4>polosiRNA (polo-RNAi) resulted in 100% of spermatocytes displaying two Asl+ foci. Depletion of Polo by bam-GAL4>polosiRNA (polo-RNAi) in a dbf-RNAi background (polo-RNAi, dbf-RNAi) suppressed the centriole disengagement defect, resulting in 82% of spermatocytes displaying two Asl+ foci and 18% of spermatocytes with more than two Asl+ foci. (P<0.0001, compared with dbf-RNAi, Fisher's exact test). The cells examined were randomly selected from images taken in six experiments.
Doublefault interacts with the RNA-binding protein Syncrip and regulates Cyclin B expression
Using RFP trap-affinity purification coupled with mass spectrometry (AP-MS), we identified the RNA-binding protein Syncrip (McDermott et al., 2012), the ribosomal protein S13 (RpS13, Alonso and Santarén, 2006) and the translational initiation factor eIF4E-7 (Hernández et al., 2005) as putative molecular partners of RFP-Dbf, suggesting involvement of Dbf in translational control in premeiotic spermatocytes (Fig. 7A). Co-immunoprecipitation from extracts of testes expressing RFP-Dbf confirmed that the Dbf and Syncrip proteins interacted and suggested that the interaction was dependent on RNA targets (Fig. 7B).
Fig. 7.
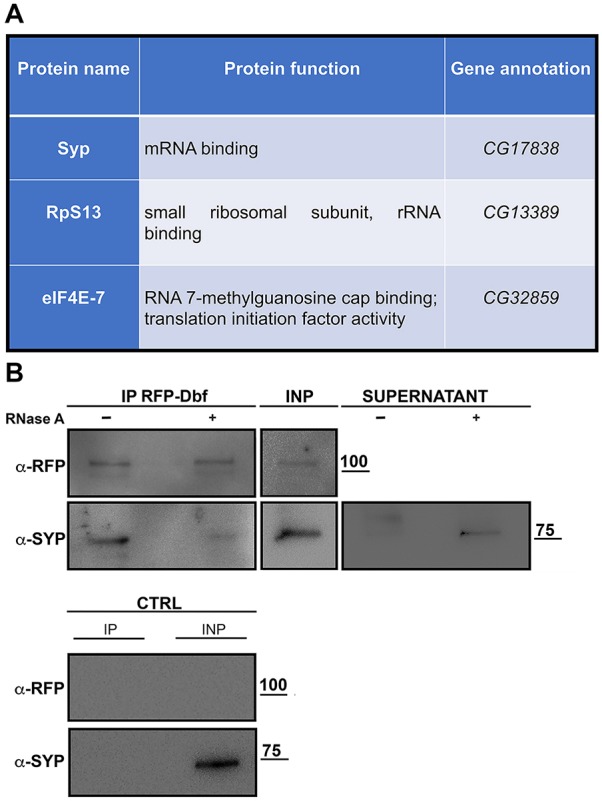
Dbf protein interacts with Syncrip. (A) Proteins identified via mass spectrometry as immunoprecipitations from testis extracts expressing Drosophila RFP-Dbf compared with control testes from males expressing RFP under the same promoter. (B) Western blot of co-immunoprecipitation of Syncrip with RFP-Dbf. RFP-Dbf was isolated with RFP-trap beads with (+) or without (−) RNase from testis extracts of males expressing RFP-Dbf. Two percent of the total lysates (INP) and one-third of the immunoprecipitates were probed using anti-RFP and anti-Syncrip antibodies. Control is testis extracts from Oregon-R. Molecular masses are in kDa.
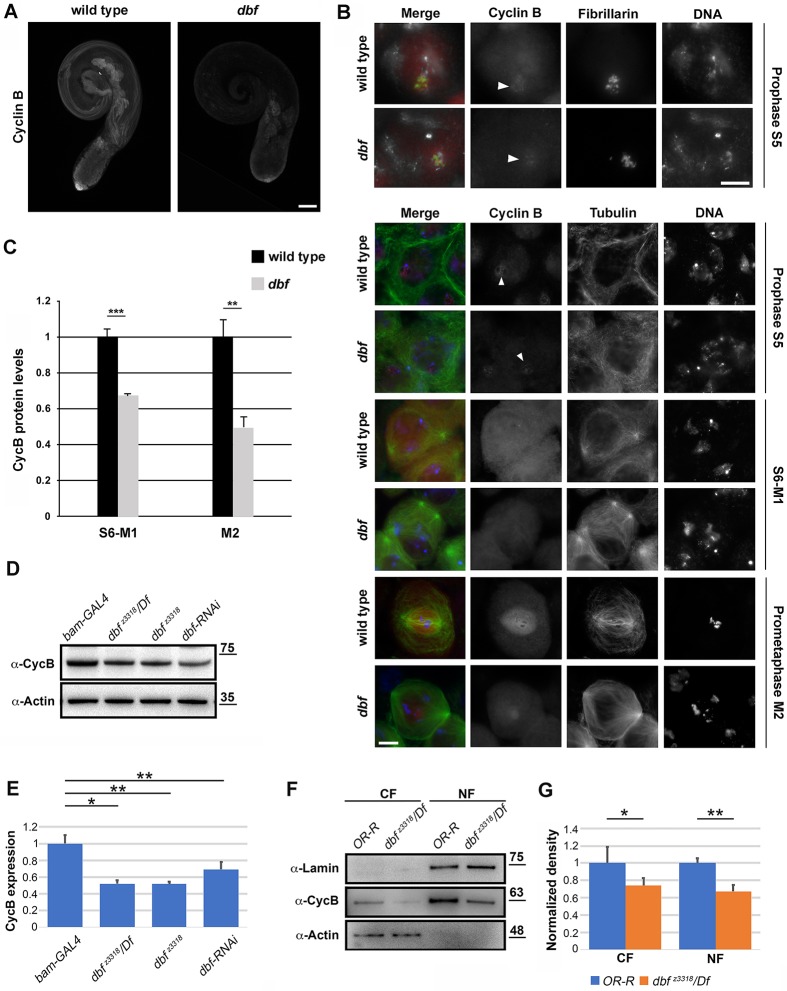
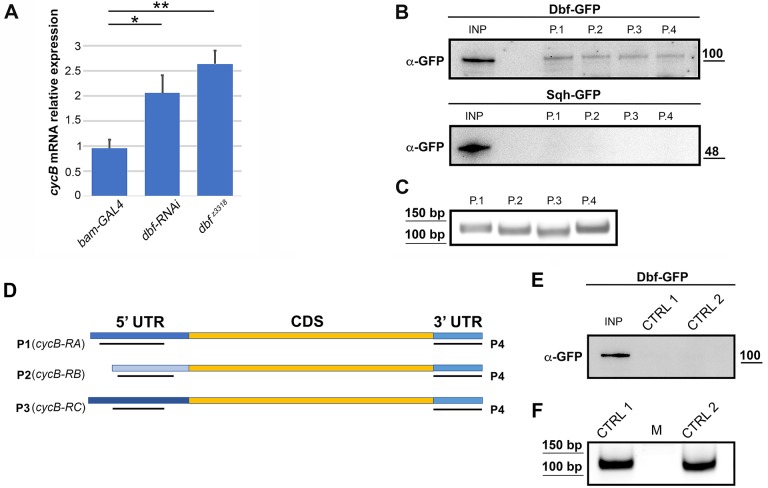
Immunofluorescence staining of whole-mount adult testes revealed that the function of Dbf is required for normal accumulation of CycB protein in mature spermatocytes, in preparation for entry into the first meiotic division (Fig. 8A). As expected, immunofluorescence analysis of squashed preparations of fixed testes revealed only low levels of CycB in immature spermatocytes of wild-type testes (Fig. 8B,C). We detected enrichment of CycB in the nucleolus of S5 spermatocytes under our fixation and staining conditions that was not reported in previous studies. As described by White-Cooper et al. (1998), in wild type, CycB started to accumulate in the cytoplasm of premeiotic spermatocytes at S6 as chromosome condensation initiated, with CycB accumulating to high levels in the nuclei of M2 spermatocytes during meiotic entry (Fig. 8B,C). Immunofluorescence of dbf spermatocytes revealed a significant decrease in the intensity of CycB protein in S6-M2 spermatocytes compared with control cells (Fig. 8B,C). Consistent with these results, western blot analysis revealed significant reduction of CycB protein levels in dbf mutant testes compared with control (Fig. 8D,E). In addition, protocols aimed at separating nuclear and cytoplasmic fractions showed that loss of Dbf protein diminished CycB levels in both nuclear and cytoplasmic protein extracts (Fig. 8F,G). Analysis of mRNA levels by quantitative real-time PCR (RT-qPCR) revealed that cycB transcript levels were significantly increased in testes from dbf z3318 males and in testes from males expressing double-stranded RNA against dbf (Fig. 9A). Taken together, these results suggest that function of dbf might be required for effective cycB mRNA translation in mature spermatocytes. Consistent with these results, biotin RNA pull-down assays from dbf-GFP testis extracts revealed that Dbf protein associated with the cyclin B 5′ UTR and 3′ UTR (Fig. 9B-D). When biotinylated RNA probes corresponding to 5′-UTR regions or to the short cycB 3′ UTR expressed in spermatocytes (Baker et al., 2015) were synthesized in vitro (Fig. 9D) and incubated with testis extracts from Dbf-GFP flies, Dbf-GFP was pulled down with the RNA probes (Fig. 9B). In contrast, Spaghetti Squash-GFP (Sqh-GFP) protein, used as a control, did not bind any of the biotinylated RNA probes (Fig. 9B) and Dbf-GFP was not pulled down with the biotinylated RNA probes (CTRL1 and CTRL2) corresponding to sequences within the GFP transcript (Fig. 9E,F).
Fig. 8.
Loss of Dbf affects CycB accumulation in male meiotic cells (A) Whole-mounts of adult testes immunostained for CycB. n=30 testes per genotype over five independent experiments. Scale bar: 100 µm. (B) Wild-type and dbf z3318/Df(2L)Exel802 (dbf) mutant spermatocytes stained to visualize CycB (red), DNA (blue) and either Fibrillarin (green, upper panels) or Tubulin (green, lower panels). Arrowheads indicate CycB enrichment in the nucleolus. Scale bars: 10 µm. (C) CycB levels per cell, quantified as mean fluorescence intensity using ImageJ. n=40 spermatocytes at stages S6-M1 per each genotype; n=30 spermatocytes at stage M2 per each genotype. The cells examined for CycB analysis were randomly selected from five independent experiments. Data are mean±s.e.m. **P<0.01; ***P<0.0001 (unpaired Student's t-test). (D) Western blot from adult testis extracts showing decreased CycB protein levels in testes from dbf mutant males and in testes depleted of Dbf (dbf-RNAi). Testis extracts from males carrying bam-GAL4 were used as a control. Molecular masses are in kDa. Actin was used as a loading control. (E) Quantification of the expression levels of CycB protein in western blots from adult testis extracts using ImageJ software. n=3 independent experiments. The intensity of each band relative to the intensity of the loading control (Actin) was normalized to the control (bam-GAL4). Data are mean±s.d. *P<0.05; **P<0.01. (F) Testis proteins from wild-type (Oregon-R, OR-R) and dbf mutant males were separated into cytoplasmic (CF) and nuclear pools (NF) by fractionation methods, and examined by western blot using anti-CycB antibodies. Cleanliness of fractionation was determined by probing for Lamin (nuclear) and Actin (cytoplasmic) proteins. Molecular mass is in kDa. (G). Quantification of the expression levels of CycB proteins in western blots from cytoplasmic (CF) and nuclear (NF) testis fractions using ImageJ. n=3 independent experiments. The intensity of each band relative to the intensity of the loading control (Actin for CF and lamin for NF) was normalized to the control (Oregon-R). Data are mean±s.d. *P<0.05; **P<0.01 (unpaired Student's t-test).
Fig. 9.
Dbf protein is required for normal cycB translation. (A) RT-qPCR expression quantification of cycB mRNA in testes of males of the indicated genotypes. Relative expression levels were normalized to actin and to control siblings (bamGAL4). Data are mean±s.d. *P<0.05, **P<0.01 (unpaired Student's t-test). (B) Anti-GFP western blot of a biotin RNA pull-down from testes expressing either Dbf-GFP or Sqh-GFP (used as a control). Molecular mass is in kDa. P1, P2 and P3 RNA probes were designed for three distinct cycB 5′ UTRs; P4 RNA probe was designed for the short 3′ UTR expressed in spermatocytes (see Materials and Methods). (C) Agarose gel analysis of the integrity of each probe used in B. (D) Schematic showing where the P1-P4 RNA probes are located with respect to the cycB 5′ UTRs and the short 3′ UTR expressed in spermatocytes (see Materials and Methods for details). (E) Anti-GFP western blot of a biotin RNA pull-down from testes expressing Dbf-GFP. Molecular mass is in kDa. CTRL1 and CTRL2 RNA probes were specific for sequences within the GFP transcript. (F) Agarose gel analysis of the integrity of each probe used in E.
DISCUSSION
Our findings demonstrate that the Dbf protein, which is primarily expressed in testes, regulates multiple events of male meiotic division and spermiogenesis, ranging from centrosome structure and spindle assembly to sex chromosome condensation, cytokinesis and axoneme/cilium structure. We suggest that the pleiotropic effects of dbf loss of function in males could be due to a requirement for dbf function for translation of specific mRNAs in spermatocytes. Dbf protein forms a complex with the RNA-binding protein Syncrip/hnRNPQ, a key regulator of localized translation in Drosophila (McDermott et al., 2012; McDermott et al., 2014). Syncrip is the fly homolog of mammalian SYNaptotagmin-binding Cytoplasmic RNA-Interacting Protein (SYNCRIP)/hnRNPQ, a component of RNA transport granules associated with localized dendritic mRNAs of hippocampal neurons (Bannai et al., 2004; Kanai et al., 2004; Elvira et al., 2006). Previous work has demonstrated that Drosophila Syncrip protein binds specific mRNAs and plays an important function in regulating their localization and translation in oocytes and neuromuscular junctions (McDermott et al., 2012; McDermott et al., 2014).
The closest human homolog of Dbf is a ZNF346/JAZ, which has been shown to bind to RNA with high affinity through its C2H2 zinc fingers (Yang et al., 1999; Burge et al., 2014). Human JAZ protein is a nucleocytoplasmic shuttling protein that associates with exportin 5 in the presence of RanGTP and a hairpin RNA, suggesting involvement in regulating the transport of certain classes of RNA (Chen et al., 2004). Our analysis, however, did not reveal accumulation of Drosophila Dbf protein in the nucleus. Rather, imaging of Dbf-GFP and RFP-Dbf revealed localization to the cytoplasm of premeiotic spermatocytes, with the protein relocalizing to the spindle poles during meiotic divisions.
Our data suggest that translation of cycB mRNA in mature spermatocytes depends on Dbf protein. Translation of Cyclin B1 (CycB) is normally repressed in immature spermatocytes, then activated in mature spermatocytes to prepare for entry into the first meiotic division (White-Cooper et al., 1998; Baker et al., 2015). The levels of CycB protein expression in late prophase/prometaphase were substantially decreased in dbf mutant spermatocytes compared with wild type. Dbf protein was pulled down by cycB mRNA, suggesting dbf may play a direct role in controlling translation of cycB in mature spermatocytes. Rising levels of CycB and mechanisms that control its translocation into the nucleus have a key role in regulating the timing of different events of G2/M transition during both mitosis and meiosis (Malumbres and Barbacid, 2009; Gavet and Pines, 2010a,b). Despite the defective CycB expression, dbf mutant spermatocytes embark on highly irregular meiosis. The work of Gavet and Pines (2010b) in HeLa cells demonstrated that different thresholds of CyclinB-Cdk1 activity are required to trigger specific mitotic events during prophase, with the higher levels required for disassembly of the nucleolus and nuclear envelope breakdown (Gavet and Pines, 2010b). Thus, it is possible that the residual amount of CycB protein in dbf mutants is not sufficient to coordinate all the changes in the cell architecture during meiotic divisions.
A key point for translational control is the recruitment of mRNAs to the ribosome, a step that for most mRNAs requires the recognition of the mRNA 5′ cap structure by the eukaryotic binding complex eIF4F (Kong and Lasko, 2012). This complex consists of the cap-binding protein eIF4E, in association with the RNA Helicase eIF4A and the scaffolding protein eIF4G, which also binds the poly (A)-binding protein, leading to recruitment of the 40S ribosomal subunit and circularization of mRNA (Sonenberg and Hinnebusch, 2009). Seven isoforms of eIF4E proteins and two isoforms of eIF4G have been identified in Drosophila melanogaster (Hernández et al., 2005; Baker and Fuller, 2007; Franklin-Dumont et al., 2007; Hernández et al., 2012; Ghosh and Lasko, 2015). Several of these isoforms are expressed in testes, including eIF4E-7, one of the proteins that we identified as a molecular partner of Dbf protein through mass spectrometry. Previous characterization of the male meiotic phenotype associated with mutations in testes-specific translation factors, showed that eIF4E-1 and eIF4E-3 are required for chromosome condensation and segregation, and cytokinesis (Hernández et al., 2012; Ghosh and Lasko, 2015). eIF4G2 is essential for normal meiotic chromosome condensation, transition to meiotic division and spermatid differentiation (Baker and Fuller, 2007; Franklin-Dumont et al., 2007; Ghosh and Lasko, 2015). Interestingly, analysis of protein expression in eIF4E-3 mutant testes revealed that the protein that was reduced the most (Hernández et al., 2012) was a microtubule-stabilizing component of centrioles and of ciliary and flagellar axonemes (Amos, 2008), suggesting a possible link with meiotic chromosome segregation defects.
Phenotypic defects similar to dbf mutants, including multipolar spindles and cytokinesis failure, were also reported in testes of mutants in the Drosophila gene for Larp (La-related-protein), a protein required for mRNA translation (Blagden et al., 2009). Moreover Larp physically interacts with PABP and hypomorphic mutant alleles are associated with centrosome and cytokinesis defects during male meiosis, similar to larp mutants (Blagden et al., 2009).
Polo kinase controls all the events during cell division, including chromosome condensation and segregation, spindle formation and cytokinesis (Pintard and Archambault, 2018), which were affected in dbf dividing spermatocytes. Our findings demonstrate that the premature centriole disengagement in premeiotic dbf mutant spermatocytes could be suppressed by depletion of Polo, indicating the involvement of this kinase in the centrosome dynamics defects and the consequent multipolar spindles. Polo-like kinase I regulates mitotic centriole disengagement in several cell systems, including Drosophila melanogaster (Tsou et al., 2009; Wang et al., 2008; Cabral et al., 2013; Varadarajan et al., 2016). Moreover, pharmacological inhibition of Polo kinase by treatment with the ATP-competitive kinase inhibitor BI2536 blocks centriole disengagement in Drosophila dividing spermatocytes (Riparbelli et al., 2014). Consistent with these data, dbf loss of function and bam-driven knockdown of Dbf protein affect Polo localization in dividing spermatocytes and result in increased Polo levels in testis protein extracts.
Dbf protein is also required to regulate centriole and axoneme structures. Loss of Dbf results in increased length of centrioles and CLPs in dividing spermatocytes, and impairs the assembly of canonical 9+2 axonemes in elongating spermatids. These phenotypic defects are likely to result from the effects dbf mutation on expression of specific proteins required for centriole/CLP structure. In addition, the enrichment of Dbf protein at the polar regions of meiotic spindles suggests that it might also provide a structural role at the centrosome and at the CLP. Several studies have also revealed that ciliary basal bodies and centrosomes are enriched in RNA-processing proteins, and share molecular components with stress granules and P-bodies (Aizer et al., 2008; Moser et al., 2011; Youn et al., 2018; Johnson and Malicki, 2019). Moreover, recent work has shown the localization of several translation factors at the centrosomes (Iaconis et al., 2017) leading to speculation about the existence of localized centrosomal translation that would regulate translation of ciliary proteins during ciliogenesis.
MATERIALS AND METHODS
Fly stocks and transgenes
Flies were reared according to standard procedures at 25°C unless otherwise specified. Oregon-R flies were used as wild-type controls unless otherwise noted. The dbf z3318 mutant strain was identified by a cytological screen of the Zuker's collection of male sterile mutants (Wakimoto et al., 2004). The chromosomal deficiencies Df(2L)Exel8026 and Df(2L)BSC211, the P element dbf 1 and P(sqh-GFP.RLC) (Royou et al., 2002) were obtained from the Bloomington Drosophila Stock Center (Indiana University). Transgenic bam-GAL4 (Chen and McKearin, 2003) was used to deplete Dbf and Polo proteins in spermatocytes. The following fly stocks, used for RNAi in testes, were from the Vienna Drosophila Resource Center (VDRC): UAS::dbf-RNAi (ID46309), UAS::polo-RNAi (ID20177) and UAS::cycB-RNAi (ID109611).
Transmission electron microscopy
Testes of control (Oregon-R) and mutant dbf pupae were dissected in phosphate-buffered saline (PBS), and fixed in 2.5% glutaraldehyde in PBS for 2 h at 4°C. Samples were washed in PBS and post-fixed in 1% osmium tetroxide in PBS for 1 h at 4°C. The material was then dehydrated through a graded series of ethanol, infiltrated with a mixture of Epon-Araldite resin and polymerized at 60°C for 48 h. Ultrathin sections (40-50 nm) were obtained using a Reichert ultramicrotome, collected with formvar-coated copper grids, and stained with uranyl acetate and lead citrate. TEM preparations were observed with a Tecnai G2 Spirit EM (FEI) operating at 100 kV and equipped with a Morada CCD camera (Olympus).
Microscopy and histology
Images of living spermatocytes expressing fluorescence-tagged-Dbf and Polo-GFP were captured using a charged-coupled device (Axiocam 503 mono CCD camera) and ZEN2 software connected to a Zeiss Cell Observer Z1 microscope equipped with an HXP 120 V inclusive built-in power supply, lamp module and a 63×/1.4 objective. To quantify multinucleate spermatids, squashed live testis preparations were observed on a Nikon Axioplan epifluorescence microscope equipped with a 40× phase-contrast objective. Metaphase chromosome preparations were obtained from larval testes dissected in saline buffer (0.7% NaCl), transferred to a drop of 45% acetic acid on a siliconized coverslip. After 2 min, preparations were gently squashed and immersed into liquid nitrogen. After coverslip removal with a razor blade, preparations were mounted in Vectashield mounting medium with DAPI (H-1200, Vector Laboratories). Cytological preparations for immunofluorescence analysis were made with testes from third instar larvae unless otherwise indicated. To visualize Polo or α-Tubulin together with one of the following antigens, Cnn, Spd2, Asl, CycB or Fibrillarin, testes were fixed with methanol and formaldehyde according to Frappaolo et al. (2017). To visualize Polo-GFP and Fibrillarin, testes were fixed in 4% formaldehyde as described by Belloni et al. (2012). For immunostaining of larval testes for α-Tubulin, Asl and Anillin, preparations were fixed using 3.7% formaldehyde in PBS and then squashed in 60% acetic acid, as previously described by Szafer-Glusman et al. (2011). For immunostaining for Spd2 and acetylated tubulin, testes were dissected in phosphate-buffered saline (PBS), squashed under a coverslip and frozen in liquid nitrogen. After removal of the coverslip, the samples were fixed in methanol for 10 min at −20°C. For CycB whole-mount immunostaining, we used the protocol described by Baker et al. (2015). Briefly, adult testes were dissected in PBS, collected in 1.5 ml tubes and fixed in ice-cold methanol (5 min) and ice-cold acetone (2 min). All samples were permeabilized and blocked in PBS (2×5 min) with 0.1% Triton X-100 and 3% BSA before immunostaining.
Monoclonal antibodies were used to stain α-Tubulin (1:300; Sigma-Aldrich, T6199), Polo (M294; 1:20, a gift from D. M. Glover, University of Cambridge, UK; Carmena et al., 1998), mouse anti-Fibrillarin (1:200; 38F3, Abcam 4566) and acetylated tubulin (1:100; Sigma-Aldrich, T7451). Polyclonal antibodies were as follows: rabbit anti-Spd2 (1:500; Rodrigues-Martins et al., 2007; Giansanti et al., 2008); rabbit anti-Anillin (1:1000; Giansanti et al., 2015; Sechi et al., 2017); rabbit anti-Cnn (1:300; a gift from T. Megraw, Florida State University, Tallahassee, FL, USA; Li et al., 1998), rabbit anti-Asl (1:500; a gift from G. Gonzalez, IRB-Barcelona, Spain; Varmark et al., 2007) and rabbit anti-CycB (Rb271, 1:600; a gift from D. M. Glover; Whitfield et al., 1990). Secondary antibodies were: Alexa 555-conjugated anti-rabbit IgG (1:300; Life Technology), FITC-conjugated anti-mouse IgG (1:30; Jackson ImmunoResearch) and Alexa Fluor-488 anti-mouse-IgG (1:800; Invitrogen). All incubations with primary antibodies (diluted in PBT containing 3% BSA) were performed overnight at 4°C. Incubations with secondary antibodies were performed at room temperature for 1 h. After immunostaining, samples were rinsed in PBS and mounted in Vectashield mounting medium with DAPI (H-1200, Vector Laboratories).
Images of testes stained for Spd2/acetylated tubulin were taken using an Axio Imager Z1 microscope (Zeiss) equipped with an AxioCam HR cooled charge-coupled camera (Zeiss). All the other images were captured with a charged-coupled device (CCD camera, Qimaging QICAM Mono Fast 1394 Cooled) connected to a Nikon Axioplan epifluorescence microscope equipped with an HBO 100-W mercury lamp and 4×, 40× and 100× objectives.
Molecular cloning
To identify the mutation in the dbf z3318 allele, the genomic DNA corresponding to CG17098 was amplified by PCR and sequenced on both strands (BMR research service). DNA sequences from dbf z3318 individuals were compared with sequences of the original Zuker-background chromosome. To generate the RFP-dbf fusion construct, the cDNA of the CG17098 gene was fused in frame with the 3′ end of mRFP sequence and cloned into pCasper4-α tubulin (see Sechi et al., 2014). The mRFP sequence (used as a control) was cloned into pCasper4-α tubulin. To construct the dbf-GFP fusion, the EGFP CDS was fused in frame to the 3′ end of CG17098 cDNA and cloned into pCaSper4-tubulin. The polo-GFP fusion construct was cloned into pCaSper4. Transgenic flies were generated by P-element-mediated germline transformation (Bestgene). dbf-GFP and mRFP-dbf were crossed into the dbf z3318 mutant background to test for phenotypic rescue of male sterility and meiotic defects.
Nuclear and cytoplasmic protein extracts from Drosophila testes
Two-hundred and fifty testes from each genotype were homogenized in 300 µl of buffer 1 [30 mM HEPES-KOH (pH 7.6), 20 mM KCl, 10 mM MgCl2, 0.2 mM EDTA (pH 8), 20% glycerol, 0.5 mM DTT, 0.5% Triton X-100 and 1× protease inhibitor cocktail] using a glass Dounce homogenizer for 30 min on ice. The lysate was incubated for 15 min on the wheel at 4°C, and centrifuged for 2 min at 1000 rpm (67 g). The supernatant was collected as the cytosolic fraction. The remaining pellet was gently washed with buffer 1 and centrifuged for 1 min at 1000 rpm (67 g). The supernatant was discarded and the pellet was resuspended in 150 µl of buffer 2 [350 mM sucrose, 15 mM, HEPES-KOH (pH 7.6), 385 mM KCl, 0.2 mM EDTA (pH 8), 0.5 mM DTT, 5 μl/ml MNase, 0.1% Triton X-100 and 1× protease inhibitor cocktail] by vortexing and incubated on the wheel at 4°C for 30 min (nuclear fraction). Each protein fraction was then boiled for 10 min in sample buffer [glycerol 20%, SDS 4%, BBF 0.2%, Tris-HCl 100 mM (pH 6.8) and DTT 200 mM].
Co-immunoprecipitation
For the co-immunoprecipitation experiment, 400 adult testes expressing either RFP-Dbf or mRFP were homogenized in 500 µl of Lysis buffer [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× protease inhibitor cocktail] for 40 min on ice using a Dounce homogenizer. Lysates were clarified by centrifugation and protein concentration was quantified using NanoDrop 2000c Spectrophotometer (Thermo Scientific). Four percent of each lysate was retained as the ‘input’, the remainder was precleared with control agarose beads (ChromoTek, Bab-20). Co-IP was performed using the RFP trap-A or control binding beads purchased from ChromoTek (ChromoTek, rta-20), following the protocol that was previously described (Belloni et al., 2012). For RNase treatment, 0.1 U of RNase A (Qiagen, 19101) was added to one of the samples for 45 min. The beads were rinsed once with ice-cold IP buffer [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× protease inhibitor cocktail] and washed extensively (4×5 min) on the wheel at 4°C. After the final wash, the beads were resuspended in 30 µl of protein sample buffer [20% glycerol, 4% SDS, 0.2% BBF, 100 mM Tris-HCl (pH 6.8), 200 mM DTT], boiled for 10 min and the supernatant loaded onto Mini-protean TGX precast gels (Bio-Rad) for SDS-PAGE.
Proteomics and data analysis
Visualization of protein bands was obtained using a colloidal Coomassie staining. From each SDS-PAGE lane, ten slices were excited and submitted to a trypsin proteolysis (Di Francesco et al., 2012). Peptides mixtures were then extracted from the gel matrix and submitted to a desalting step by solid phase extraction before mass spectrometric analyses (Rappsilber et al., 2007). Nano-liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analyses were performed using an Ultimate3000 system (Dionex) equipped with a splitting cartridge for nanoflows and connected on-line via a nanoelectrospray ion source (Thermo-Fisher Scientific) to an LTQ-Orbitrap XL mass spectrometer (Thermo-Fisher Scientific). Each sample was automatically loaded from the autosampler module of the Ultimate 3000 system at a flow rate of 20 µl/min onto a trap column (AcclaimPepMap µ-Precolumn, 300 µm×1 mm, Dionex) in 4% ACN containing 0.1% FA. After 4 min, peptides were eluted at 300 nl/min onto a 15 cm column (360 µm OD×75 μmID, 15 µm Tip ID; PicoFrit, New Objective) and custom packed by reverse phase (C18.5 µm particle size, 200 Å pore size; Magic C18AQ, Michrom) using a 90 min two-step gradient of solvent B (from 5% to 40% in 120 min and from 40% to 85% in 15 min). Data-dependent tandem mass spectroscopy (MS) was performed using full precursor ion scans (MS1) collected at 30,000 resolution, with an automatic gain control (AGC) of 1×106 ions and maximal injection time of 1000 ms. The five most intense (>200 counts) ions with charge states of at least +2 were selected for collision-induced dissociation (CID). Dynamic exclusion was active, with 90 ms exclusion for ions selected twice within a 30 ms window. For MS/MS scanning, the minimum MS signal was set to 500, activation time to 30 ms, target value to 10,000 ions and injection time to 100 ms. All MS/MS spectra were collected using a normalized collision energy of 35% and an isolation window of 2 Th. All MS/MS samples were analyzed using Sequest algorithm in Thermo Proteome Discoverer (Thermo Fisher Scientific version 1.4.0.288). Peptides sequences were searched against the Drosophila melanogaster Uniprot proteome database.
Western blotting
Aliquots of co-immunoprecipitates or testis protein extracts, separated on Mini-protean TGX precast gels (Bio-Rad) were blotted to PVDF membranes (Bio-Rad, 162-0177). Membranes were blocked in Tris-buffered saline (Sigma-Aldrich) with 0.05% Tween-20 (TBS-T) containing 5% non-fat dry milk (Bio-Rad; Blotting Grade Blocker) for 1 h at room temperature followed by incubation with primary and secondary antibodies diluted in TBS-T. Primary antibodies used for immunoblotting, were as follows: mouse α-Tubulin (1:5000; Sigma-Aldrich, T6199), mouse anti-RFP (1:1000; Chromotek, 6G6), monoclonal HRP-conjugated anti-actin (1:1000; sc-1615, Santa Cruz Biotechnology), guinea pig anti-Syncrip (1:2500; a gift from I. Davis, University of Oxford, UK; McDermott et al., 2012), rabbit anti-CycB (Rb271, 1:2500) provided by D. M. Glover, rabbit anti-GAPDH (1:5000; GTX100118, Genetex) and rabbit anti-GFP (1:1000; TP401, Torrey Pines). HRP-conjugated secondary antibodies were as follows: goat anti guinea-pig IgG (whole molecule)-peroxidase (Sigma-Aldrich, A-7289), goat anti-mouse IgG (H+L) (Pierce, N.31431) and goat anti-rabbit IgG (H+L) (Pierce, N.31466). All secondary antibodies were used at 1:5000. After incubation with the antibodies, blots were washed (3×5 min) in TBS-T [20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.05% Tween 20]. Blots were imaged using ECL (Cyanagen, XLS100) and signals revealed with the ChemiDoc XRS imager (Bio-Rad).
RNA extraction and analysis by RT PCR and real time PCR
Total RNA extraction was performed from samples of 250 adult testes or 200 larval brains of Drosophila melanogaster, using a RNeasy-Protect Mini Kit (Qiagen, 74124). The quality of the RNA obtained was controlled by 2% agarose gel electrophoresis and quantified using a NanoDrop 2000c Spectrophotometer (Thermo Scientific). For each sample, 1 μg of total RNA was transcribed into cDNA using the SuperScript III First-strand kit (Invitrogen, 18080-051) and diluted to obtain the final concentration of 15 ng/μl for each sample by addition of DEPC water. The following primers were used for RT-PCR on dbf and GOLPH3 transcripts: dbf forward, ATGTTGTCCCACGCCTTCAAC; dbf reverse, TCACTCGAAGAAGACCGCTTG; GOLPH3 forward, ATGAATCGCTCCGACGGATTG; GOLPH3 reverse, CTATTTCGTGAACGCCATGAAC. The PCR amplicons were separated by 1.2% agarose gel electrophoresis in TAE buffer [40 mM Tris, 1 mM EDTA, 20 mM CH3COOH (pH 8.3) (Bio-Rad)]. q-RTPCRs were performed as described previously (Burla et al., 2015) using the following primers: polo (CG12306) forward, ACTCGTCTGGAGTCGACCTT; polo reverse, CTGCGATAGTCCTCGCTGTGT; dbf (CG17098) forward, AATAAGGCCCTTGGGCTCAC; dbf reverse, TTGCAGGCATCGCAGTAGAA; cyc B (CG3510) forward, ACCTGGTACTGGTCTCCGAA; cyc B reverse, GGCCAGGTGATCCTTGTGAA; actin (CG12051) (used as normalizer) forward, CCGCGTGCAGTTTTTCCTTC; actin reverse, GTGCCTCATCGCCGACATAA.
Fluorescent in situ hybridization experiments
To obtain probes used in the fluorescent in situ hybridization experiments, a PCR reaction was performed to amplify the 240 bp fragment. The oligos used for the PCR reaction (rDNA forward, AGCTGTTCTACGACAGAGGGTTC, rDNA reverse, AGCTTACACTACTATATCCATTC) were designed using the nucleotide sequences provided by FlyBase (flybase.org). The PCR amplicon was separated by 2% agarose gel electrophoresis in TAE buffer [Bio-Rad; 40 mM Tris, 1 mM EDTA, 20 mM CH3COOH (pH 8.3)] and purified using the NucleoSpin Gel and PCR clean-up kit (grisp, gk01.0100). The amplicon was then quantified with the NanoDrop2000c Spectrophotometer (Thermo Scientific) and labeled with the Atto550 NT Labeling Kit (Jena Bioscience, PP-305S-550), following the protocol recommended by Jena Bioscience. In situ hybridization was carried out as described by Balicky et al. (2002). Briefly, larval testes were dissected in 0.7% NaCl, then transferred to 5 μl of 45% acetic acid/2% formaldehyde on a siliconized 18 mm coverslip (3 min) and gently squashed. The slides were immersed in liquid nitrogen for at least 2 h. After removal of coverslips with a razor blade, samples were rinsed in PBS. Before hybridization, slides were incubated for 2×10 min in 70% ethanol, 1×10 min in 100% ethanol, and air-dried at room temperature. The preparations were then incubated for 3×10 min in 2× SSC/0.1% Tween 20 (SSCT), in 25% formamide/2× SSCT for 10 min, and rinsed in 50% formamide/2× SSCT for 10 min. Each sample was covered with 500 μl of 50% formamide/2× SSCT and allowed to prehybridize for at least 3 h at 37°C in a humid chamber. The probe was diluted to 10 ng/μl in hybridization buffer (3× SSC, 50% formamide, 10% dextran sulfate). Probe and chromosomal DNA were denatured at 94°C for 2 min. After denaturation, samples were hybridized overnight in a humid chamber at 37°C. After hybridization, coverslips were removed and samples were incubated (twice for 1 h) in 50% formamide/2× SSCT at 37°C, rinsed for 10 min in 25% formamide/2× SSCT and for 3×10 min in 2× SSCT without formamide. Samples were then mounted in Vectashield mounting medium with DAPI (H-1200, Vector Laboratories).
Biotin pull-down
PCR was used to amplify specific sequences within three different 5′ UTRs (probes 1-3) of cycB and the short 3′ UTR of cycB (Probe 4) using the following primers: probe 1 (flybase ID FBtr0071911) forward primer, TAATACGACTCACTATAGGGAGAGGCAGCTTGGCCTCGCTTCG; reverse primer, GCGTTTCTGTGACCGAACG; probe 2 (flybase ID FBtr0071913) forward primer, TAATACGACTCACTATAGGGAGAGAAAGAGTGCCGTTTGTCC; reverse primer, CTCCTCTGATTTGGTGCG; probe 3 (flybase ID FBtr0071914), forward primer, TAATACGACTCACTATAGGGAGAGGTTCAAAGTGCAGATATGC; reverse primer, GGAAGGAACCGTCAATTTCTTC; probe 4 (flybase ID FBtr0071913) forward primer, TAATACGACTCACTATAGGGAGAGCGGTCCAAGGCGGACTGG; reverse primer, CGGTAACTATACGCATTTTTTAAC. PCR was used to amplify specific sequences within GFP from the pCasper-dbf-GFP construct, using the following primers: ctrl 1 forward primer, TAATACGACTCACTATAGGGAGAGATGCCACCTACGGCAAGCTG; reverse primer, GTCGTGCTGCTTCATGTGGTC; ctrl 2 forward primer, TAATACGACTCACTATAGGGAGACACAAGCTGGAGTACAACTAC; reverse primer, GTGTTCTGCTGGTAGTGGTC. The PCR products were quantified with a NanoDrop a2000c spectrophotometer (Thermo Scientific). Biotin-labeled probes were generated using T7 RNA polymerase (Roche) and Biotin RNA Labeling Mix (Roche, 11 685 597 910) according to the protocol recommended by Roche. Probe concentration was determined using a Nanodrop 2000c Spectrophotometer (Thermo Scientific) and probe integrity was verified by agarose gel. Six-hundred testes expressing Dbf-GFP or 600 testes expressing Sqh-GFP (control) were dissected and homogenized in 800 µl of lysis buffer [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× protease inhibitor cocktail, 1 µl/ml RNasin Plus (Promega)] using a glass Dounce homogenizer, and lysed at 4°C for 30 min. Each lysate was centrifuged for 7 min at 13,000 rpm (10,000 g), and 20 µl supernatant was set aside as the input. The remaining supernatant was pre-cleared with streptavidin agarose beads (Millipore, 16-126) for 30 min at 4°C, then split into four tubes and incubated with 100 ng of biotin-labeled probe for 30 min at 4°C. Each sample was then incubated with fresh streptavidin beads (30 min, 4°C) and washed five times for 5 min. Laemmli sample buffer [20% glycerol, 4% SDS, 0.2% BBF, 100 mM Tris-HCl (pH 6.8) and 200 mM DTT] was added to each sample and samples were boiled for 10 min and analyzed by western blot.
Statistical analysis
For all the immunofluorescence, differences between wild-type and mutant cells were examined for statistical significance using an unpaired Student's t-test or a Fisher's exact test with Prism 8 (Graphpad). Quantification of the density of western blot bands was performed using ImageJ. Data are expressed as fold change compared with control. All data represent the mean±s.d. from three independent experiments. Significance is indicated as follows: *P<0.05, **P<0.01, ***P<0.0001; ns, not significant. The representative results, from at least three independent experiments were analyzed using unpaired Student's t-test with Prism 8 (Graphpad). For the statistical analysis of RT-qPCR, all reactions were performed in technical triplicate with at least three biological replicates. P-values were calculated using a using unpaired Student's t-test with Prism 8 (Graphpad), performed on untransformed average ddct values.
Supplementary Material
Acknowledgements
We thank the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for fly stocks; and D. M. Glover, J. Raff, T. Megraw, C. Gonzalez and I. Davis for antibodies. We thank R. Piergentili, A. Berducci and F. R. Liberati for their assistance in the early stages of this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., A.F., E.S., M.G.R., G.C., M.G.G.; Methodology: S.S., A.F., A.K.-G., R.B., L.D.F., E.S.-G., E.S., M.G.R., G.C., M.G.G.; Software: R.B., L.D.F.; Validation: S.S., A.F., A.K.-G., M.G., R.B., L.D.F., E.S., M.T.F., M.G.R., G.C., M.G.G.; Formal analysis: S.S., A.F., A.K.-G., M.G., R.B., L.D.F., M.G.R., G.C., M.G.G.; Investigation: S.S., A.F., A.K.-G., M.G., R.B., L.D.F., E.S.-G., G.C., M.G.G.; Resources: M.G.G.; Data curation: S.S., A.F., M.G.G.; Writing - original draft: M.G.G.; Writing - review & editing: S.S., A.F., M.T.F., M.G.G.; Visualization: S.S., A.F., A.K.-G., E.S.-G., E.S., M.T.F., I.S., G.C., M.G.G.; Supervision: M.T.F., I.S., G.C., M.G.G.; Project administration: M.G.G.; Funding acquisition: M.G.G..
Funding
This work was supported by a grant from Associazione Italiana per la Ricerca sul Cancro (IG2017 20779) to M.G.G., and by National Institutes of Health grants (R01GM124054 and R01GM62276 to M.T.F.). A.F. was supported by a fellowship from the Federazione Italiana per la Ricerca sul Cancro-Associazione Italiana per la Ricerca sul Cancro (19686). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.183053.supplemental
References
- Aizer A., Brody Y., Ler L. W., Sonenberg N., Singer R. H. and Shav-Tal Y. (2008). The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19, 4154-4166. 10.1091/mbc.e08-05-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. and Santarén J. F. (2006). Characterization of the Drosophila melanogaster ribosomal proteome. J. Proteome Res. 5, 2025-2032. 10.1021/pr0601483 [DOI] [PubMed] [Google Scholar]
- Amos L. A. (2008). The tektin family of microtubule-stabilizing proteins. Genome Biol. 9, 229 10.1186/gb-2008-9-7-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C. and Fuller M. T. (2007). Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 134, 2863-2869. 10.1242/dev.003764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Gim B. S. and Fuller M. T. (2015). Cell type-specific translational repression of Cyclin B during meiosis in males. Development 142, 3394-3402. 10.1242/dev.122341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balicky E. M., Endres M. W., Lai C. and Bickel S. E. (2002). Meiotic cohesion requires accumulation of ORD on chromosomes before condensation. Mol. Biol. Cell 13, 3890-3900. 10.1091/mbc.e02-06-0332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H., Fukatsu K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T. and Mikoshiba K. (2004). An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279, 53427-53434. 10.1074/jbc.M409732200 [DOI] [PubMed] [Google Scholar]
- Belloni G., Sechi S., Riparbelli M. G., Fuller M. T., Callaini G. and Giansanti M. G. (2012). Mutations in Cog7 affect Golgi structure, meiotic cytokinesis and sperm development during Drosophila spermatogenesis. J. Cell Sci. 125, 5441-5452. 10.1242/jcs.108878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden S. P., Gatt M. K., Archambault V., Lada K., Ichihara K., Lilley K. S., Inoue Y. H. and Glover D. M. (2009). Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 334, 186-197. 10.1016/j.ydbio.2009.07.016 [DOI] [PubMed] [Google Scholar]
- Burge R. G., Martinez-Yamout M. A., Dyson H. J. and Wright P. E. (2014). Structural characterization of interactions between the double-stranded RNA-binding zinc finger protein JAZ and nucleic acids. Biochemistry 53, 1495-1510. 10.1021/bi401675h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla R., Carcuro M., Raffa G. D., Galati A., Raimondo D., Rizzo A., La Torre M., Micheli E., Ciapponi L., Cenci G. et al. (2015). AKTIP/Ft1, a new shelterin-interacting factor required for telomere maintenance. PLoS Genet. 11, e1005167 10.1371/journal.pgen.1005167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G., Sans S. S., Cowan C. R. and Dammermann A. (2013). Multiple mechanisms contribute to centriole separation in C. elegans. Curr. Biol. 23, 1380-1387. 10.1016/j.cub.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Riparbelli M. G., Minestrini G., Tavares A. M., Adams R., Callaini G. and Glover D. M. (1998). Drosophila polo kinase is required for cytokinesis. J. Cell Biol. 143, 659-671. 10.1083/jcb.143.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H., Gonczy P., Alexander S., Rawson R., Eberhart C. G., Viswanathan S., DiNardo S. E. and Wasserman S. A. (1993). Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135, 489-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Bonaccorsi S., Pisano C., Verni F. and Gatti M. (1994). Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107, 3521-3534. [DOI] [PubMed] [Google Scholar]
- Chen D. and McKearin D. (2003). A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159-1170. 10.1242/dev.00325 [DOI] [PubMed] [Google Scholar]
- Chen T., Brownawell A. M. and Macara I. G. (2004). Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol. Cell Biol. 24, 6608-6619. 10.1128/MCB.24.15.6608-6619.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J. and Dow J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715-720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Di Francesco L., Correani V., Fabrizi C., Fumagalli L., Mazzanti M., Maras B. and Schininà M. E. (2012). 14-3-3ε marks the amyloid-stimulated microglia long-term activation. Proteomics 12, 124-134. 10.1002/pmic.201100113 [DOI] [PubMed] [Google Scholar]
- Dix C. I. and Raff J. W. (2007). Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759-1764. 10.1016/j.cub.2007.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G., Wasiak S., Blandford V., Tong X.-K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A. W., Boismenu D. et al. (2006). Characterization of an RNA granule from developing brain. Mol. Cell Proteomics 5, 635-651. 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- Fabian L. and Brill J. A. (2012). Drosophila spermiogenesis: big things come from little packages. Spermatogenesis 2, 197-212. 10.4161/spmg.21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Dumont T. M., Chatterjee C., Wasserman S. A. and Dinardo S. (2007). A novel eIF4G homolog, Off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development 134, 2851-2861. 10.1242/dev.003517 [DOI] [PubMed] [Google Scholar]
- Frappaolo A., Sechi S., Belloni G., Piergentili R. and Giansanti M. G. (2017). Visualization of cleavage furrow proteins in fixed dividing spermatocytes. Methods Cell Biol. 137, 85-103. 10.1016/bs.mcb.2016.03.035 [DOI] [PubMed] [Google Scholar]
- Fuller M. T. (1993). Spermatogenesis. In The Development of Drosophila melanogaster (ed. Bate M. and Martinez-Arias A.), pp. 71-147. Cold Spring Harbor, New York: Cold Spring Harbor Lababoratory Press. [Google Scholar]
- Fuller M. T. and Spradling A. C. (2007). Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402-404. 10.1126/science.1140861 [DOI] [PubMed] [Google Scholar]
- Gavet O. and Pines J. (2010a). Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 18, 533-543. 10.1016/j.devcel.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O. and Pines J. (2010b). Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189, 247-259. 10.1083/jcb.200909144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. and Lasko P. (2015). Loss-of-function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E-3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 10, e0122519 10.1371/journal.pone.0122519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G. and Fuller M. T. (2012). What Drosophila spermatocytes tell us about the mechanisms underlying cytokinesis. Cytoskeleton 69, 869-881. 10.1002/cm.21063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Farkas R. M., Bonaccorsi S., Lindsley D. L., Wakimoto B. T., Fuller M. T. and Gatti M. (2004). Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell. 15, 2509-2522. 10.1091/mbc.e03-08-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Bucciarelli E., Bonaccorsi S. and Gatti M. (2008). Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303-309. 10.1016/j.cub.2008.01.058 [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Sechi S., Frappaolo A., Belloni G. and Piergentili R. (2012). Cytokinesis in Drosophila male meiosis. Spermatogenesis 2, 185-196. 10.4161/spmg.21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Vanderleest T. E., Jewett C. E., Sechi S., Frappaolo A., Fabian L., Robinett C. C., Brill J. A., Loerke D., Fuller M. T. et al. (2015). Exocyst-dependent membrane addition is required for anaphase cell elongation and cytokinesis in Drosophila. PLoS Genet. 11, e1005632 10.1371/journal.pgen.1005632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould-Somero M. and Holland L. (1974). The timing of RNA synthesis for spermiogenesis in organ cultures of Drosophila melanogaster testes. Wilhelm Roux Arch. Entwickl. Mech. Org. 174, 133-148. 10.1007/BF00573626 [DOI] [PubMed] [Google Scholar]
- Hernández G., Altmann M., Sierra J. M., Urlaub H., Diez del Corral R., Schwartz P. and Rivera-Pomar R. (2005). Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 122, 529-543. 10.1016/j.mod.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Hernández G., Han H., Gandin V., Fabian L., Ferreira T., Zuberek J., Sonenberg N., Brill J. A. and Lasko P. (2012). Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila. Development 139, 3211-3220. 10.1242/dev.073122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis D., Monti M., Renda M., van Koppen A., Tammaro R., Chiaravalli M., Cozzolino F., Pignata P., Crina C., Pucci P. et al. (2017). The centrosomal OFD1 protein interacts with the translation machinery and regulates the synthesis of specific targets. Sci. Rep. 7, 1224 10.1038/s41598-017-01156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. A. and Malicki J. J. (2019). The nuclear arsenal of cilia. Dev. Cell 49, 161-170. 10.1016/j.devcel.2019.03.009 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N. and Hirokawa N. (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 43, 513-525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Kong J. and Lasko P. (2012). Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383-394. 10.1038/nrg3184 [DOI] [PubMed] [Google Scholar]
- Li K., Xu E. Y., Cecil J. K., Turner F. R., Megraw T. L. and Kaufman T. C. (1998). Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J. Cell Biol. 141, 455-467. 10.1083/jcb.141.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. Y., Viswanathan S., Wood C., Wilson P. G., Wolf N. and Fuller M. T. (1996). Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development 122, 1331-1341. [DOI] [PubMed] [Google Scholar]
- Malumbres M. and Barbacid M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153-166. 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- McDermott S. M., Meignin C., Rappsilber J. and Davis I. (2012). Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol. Open 1, 488-497. 10.1242/bio.2012885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott S. M., Yang L., Halstead J. M., Hamilton R. S., Meignin C. and Davis I. (2014). Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. RNA 20, 1593-1606. 10.1261/rna.045849.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J. J., Fritzler M. J. and Rattner J. B. (2011). Repression of GW/P body components and the RNAi microprocessor impacts primaryciliogenesis in human astrocytes. BMC Cell Biol. 12, 37 10.1186/1471-2121-12-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri G. and Olivieri A. (1965). Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat. Res. 2, 366-380. 10.1016/0027-5107(65)90072-2 [DOI] [PubMed] [Google Scholar]
- Pintard L. and Archambault V. (2018). A unified view of spatio-temporal control of mitotic entry: polo kinase as the key. Open Biol. 8, 180114 10.1098/rsob.180114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M. and Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896-1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Riparbelli M. G., Gottardo M., Glover D. M. and Callaini G. (2014). Inhibition of Polo kinase by BI2536 affects centriole separation during Drosophila male meiosis. Cell Cycle 13, 2064-2263. 10.4161/cc.29083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G. and Glover D. M. (2007). DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17, 1465-1472. 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Royou A., Sullivan W. and Karess R. (2002). Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J. Cell Biol. 158, 127-137. 10.1083/jcb.200203148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi S., Colotti G., Belloni G., Mattei V., Frappaolo A., Raffa G. D., Fuller M. T. and Giansanti M. G. (2014). GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster. PLoS Genet. 10, e1004305 10.1371/journal.pgen.1004305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi S., Frappaolo A., Fraschini R., Capalbo L., Gottardo M., Belloni G., Glover D. M., Wainman A. and Giansanti M. G. (2017). Rab1 interacts with GOLPH3 and controls Golgi structure and contractile ring constriction during cytokinesis in Drosophila melanogaster. Open Biol. 7, 160257 10.1098/rsob.160257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. and Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731-745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafer-Glusman E., Fuller M. T. and Giansanti M. G. (2011). Role of Survivin in cytokinesis revealed by a separation-of-function allele. Mol. Biol. Cell 22, 3779-3790. 10.1091/mbc.e11-06-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. E., Soltani-Bejnood M., Roth P., Dorn R., Logsdon J. M. Jr and McKee B. D. (2005). Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123, 555-568. 10.1016/j.cell.2005.08.043 [DOI] [PubMed] [Google Scholar]
- Tsou M.-F. B., Wang W.-J., George K. A., Uryu K., Stearns T. and Jallepalli P. V. (2009). Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344-354. 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan R., Ayeni J., Jin Z., Homola E. and Campbell S. D. (2016). Myt1 inhibition of Cyclin A/Cdk1 is essential for fusome integrity and premeiotic centriole engagement in Drosophila spermatocytes. Mol. Biol. Cell 27, 2051-2063. 10.1091/mbc.E16-02-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H. and Gonzalez C. (2007). Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17, 1735-1745. 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Wakimoto B. T., Lindsley D. L. and Herrera C. (2004). Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167, 207-216. 10.1534/genetics.167.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Y., Duan Q., Jiang N., Huang Y., Darzynkiewicz Z. and Dai W. (2008). sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell 14, 331-341. 10.1016/j.devcel.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H., Schäfer M. A., Alphey L. S. and Fuller M. T. (1998). Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125, 125-134. [DOI] [PubMed] [Google Scholar]
- Whitfield W. G., Gonzalez C., Maldonado-Codina G. and Glover D. M. (1990). The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO J. 9, 2563-2572. 10.1002/j.1460-2075.1990.tb07437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., May W. S. and Ito T. (1999). JAZ requires the double-stranded RNA-binding zinc finger motifs for nuclear localization. J. Biol. Chem. 274, 27399-27406. 10.1074/jbc.274.39.27399 [DOI] [PubMed] [Google Scholar]
- Youn J.-Y., Dunham W. H., Hong S. J., Knight J. D. R., Bashkurov M., Chen G. I., Bagci H., Rathod B., MacLeod G., Eng S. W. M. et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517-532.e11. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.