Abstract
Introduction
Mantle cell lymphoma (MCL) is a rare subtype of B-cell lymphoma. Circular (circ) RNA is a member of the non-coding RNA family. However, clinical references to circRNAs in MCL are not clear.
Methods
In this study, we detected the expression level of circCDYL in the plasma of MCL patients compared to healthy donors by the quantitative reverse transcription polymerase chain reaction. The diagnostic value of circCDYL was determined using a receiver operating characteristic (ROC) curve. We constructed a circCDYL short hairpin RNA plasmid and infected the MCL cell line, Z138, to detect its effect on cell proliferation.
Results
CircCDYL was high expressed in the plasma of MCL patients. The ROC curve showed that circCDYL had diagnostic value (area under the curve (AUC) = 0.856). Functionally, circCDYL knockdown inhibited MCL cell proliferation. We conducted bioinformatics analyses and identified a circCDYL-micro (mi)RNA–mRNA/long non-coding (lnc)RNA network, highlighted by five miRNAs (hsa-miR-129-5p, hsa-miR-3163, hsa-miR-4662a-5p, hsa-miR-101-3p, and hsa-miR-186-5p), three lncRNAs (MALAT1, NEAT1, and XIST), and five mRNAs (NOTCH1, FMR1, ABCB1, TWIST1, and VEGFA).
Conclusion
These findings indicate that circ-CDYL might serve as a potential diagnostic biomarker in clinical practice.
Keywords: circular RNA, biomarker, AUC, diagnosis, non-coding RNA
Introduction
Mantle cell lymphoma (MCL) is a rare subtype of non-Hodgkin lymphoma.1 It is classified as an aggressive B-cell malignancy,2 accounting for less than 10% of the non-Hodgkin lymphoma patients. The median age at onset is 60 years with an overall survival of 4 to 5 years.3 The diagnosis of MCL is based on the chromosomal translocation (11;14) (q13;q32), and positive immunophenotyping for cyclin D1.3,4
Non-coding RNAs account for 98% of RNA functions in regulating gene expression.5,6 Circular (circ)RNA is a member of the non-coding RNA family. It specializes in forming covalently closed continuous loops with back-splicing events.7 CircRNA has shown great potential to become a biomarker because of its conserved, stable, and specific characteristics for developmental stages and tissues.8,9 In addition, circRNAs exist widely in the blood where they can be measured easily.10 CircRNAs were initially considered as RNA by-products. However, they can compete with endogenous RNAs and interact with proteins.10,11 Clinical references to circRNAs in MCL are not clear. In the previous research, several circRNAs in MCL tissues and cell lines were reported to be clinically significant, including circMAN1A2, circSMARCA5, and circ-chromodomain Y-like (circCDYL).12 In this study, we focused on circCDYL for its diagnostic and prognostic value and assessed its biological function in MCL.
Materials and Methods
Patients and Samples
This study involved 18 patients diagnosed with MCL and 17 healthy donors as a control group. All individuals were recruited from the First Affiliated Hospital of Zhengzhou University between 2014 and 2017. None of the patients had previously undergone chemotherapy. All samples were preserved at −80°C until analyzed. This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and met international standards for patient confidentiality. Written informed consent was obtained from all patients.
Total RNA Extraction
Total RNA was extracted from plasma samples using TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The purity and concentration of RNA samples were determined with a Nanodrop 1000 spectrophotometry (Thermo Scientific, Wilmington, DE, USA).
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
cDNA was synthesized by reverse transcription using the PrimeScript RT Master Mix with random primers, and qRT-PCR was performed according to the manufacturer’s protocols (TaKaRa Bio, Shiga, Japan).
Cell Culture
HEK293T cells and the human MCL cell line, Z138, were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) and Iscove's modified Dulbecco's medium (IMDM) respectively (Invitrogen, California, USA) containing 10% fetal bovine serum (Clark Bioscience, Richmond, VA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, California, USA) at 37°C with 5% CO2.
Plasmid Construction and Stable Infection
Human circCDYL cDNA was synthesized and cloned into the GV493 vector (Genechem, Shanghai, China) to construct three short hairpin RNA plasmids. Plasmid constructions were confirmed by sequencing, then infected into HEK293T cells for lentiviral packaging. Z138 cells were then infected and selected for 7 days with 3 μg/mL puromycin.
Assay of Proliferation Using the Cell Counting Kit-8 (CCK-8) Assay
Z138 cells were cultured in 96-well plates coated with 20 μL of CCK-8 assay solution (Dojindo, Tokyo, Japan) per well, and incubated at 37°C with 5% CO2 for 2 hrs. Cell viability was assessed by measuring the absorbance at 450 nm with a Multiskan FC microplate reader (Thermo Scientific, Waltham, MA, USA).
CircRNA-micro (mi)RNA-mRNA/Long Non-Coding (lnc)RNA Network
The circRNA-miRNA-mRNA-lncRNA network was drawn through Cytoscape (http://www.cytoscape.org/) based on a predicting analysis of target miRNAs, lncRNAs and mRNAs using starBase v2.0 (http://starbase.sysu.edu.cn/),13 and MiRTarBase (http://mirtarbase.mbc.nctu.edu.tw/).14 We only selected mRNA targets with experimental evidence of the top five predicted targets of circCDYL.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
KEGG enrichment analyses were performed using the DAVID analysis tool (https://david.ncifcrf.gov/) to show related pathways among the target genes.15 We also conducted a gene ontology analysis to classify the biological functional roles of the target genes.
Statistical Analyses
Results are reported as means ± standard deviation for triplicate measurements. Significant differences between the two groups were estimated by Student’s t or Mann–Whitney tests, depending on the normality test. The receiver operating characteristic (ROC) and the respective area under the curve were used to estimate the diagnosis potential. Kaplan–Meier curves were used to estimate overall survival. All tests were conducted using SPSS v.21.0 software (IBM, Chicago, IL, USA) or Prism version 6.0 (GraphPad, La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Results
CircCDYL Was Upregulated in MCL Patients
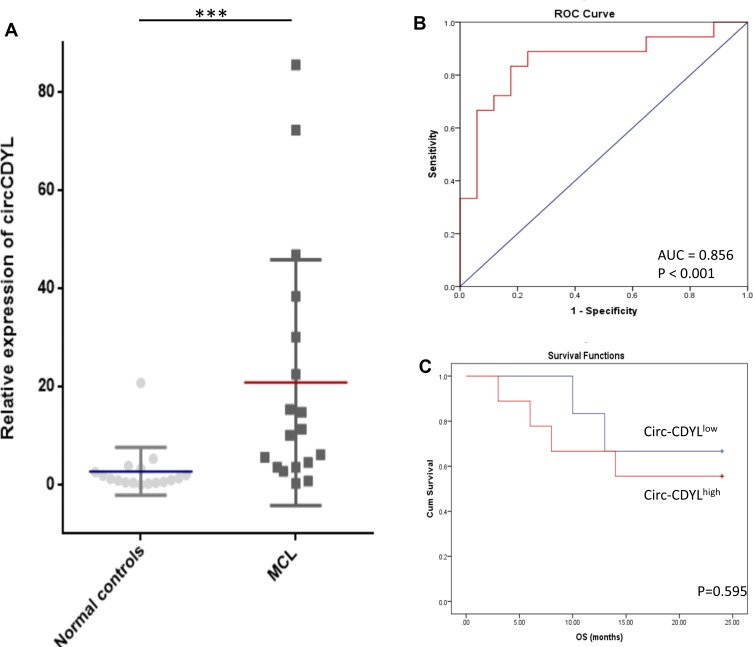
We first examined the expression levels of circCDYL in plasma from MCL patients and healthy donors using qRT-PCR. CircCDYL expression was significantly upregulated in MCL patients compared to healthy donors (P < 0.001) (Figure 1A). To estimate whether circCDYL could serve as a biomarker for diagnosis or prognosis in MCL patients, we performed ROC and Kaplan–Meier curve analyses. The ROC result indicated that the expression of circCDYL in plasma could significantly distinguish MCL patients from healthy controls (area under the curve (AUC) = 0.856; P < 0.001; 95% confidence interval, 0.724–0.989), with 66.67% sensitivity and 94.12% specificity (Figure 1B). In addition, examining 18 MCL patients with exact overall survival results from a Kaplan–Meier curve analysis showed that circCDYLlow patients may have a better prognosis, though the result was not statistically significant (Figure 1C) (P = 0.595).
Figure 1.
Circ-CDYL was upregulated in MCL patients. (A) qRT-PCR analysis showed that Circ-CDYL was upregulated in MCL patients (n = 18) compared with normal controls (n = 17). (B) ROC curve showed that Circ-CDYL expression could serve as a diagnostic biomarker in MCL. (C) Kaplan–Meier OS curves for MCL patients stratified according to circ-CDYL expression. ***p < 0.001.
CircCDYL Knockdown Inhibited MCL Cell Proliferation
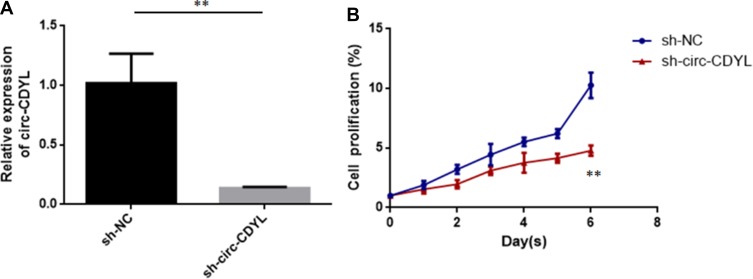
To assess the biological function of circCDYL in MCL, we established a loss-of-function model in the MCL cell line, Z138, by transfecting a short hairpin circCDYL lentivirus and conducting qRT-PCR to determine the expression of circCDYL (Figure 2A). After stable inhibition of circCDYL, we conducted the CCK-8 assay to detect MCL cell proliferation. The results showed that decreasing the expression of circCDYL significantly suppressed the proliferation of Z138 cells (Figure 2B).
Figure 2.
Circ-CDYL knockdown inhibited MCL cell proliferation. (A) Relative expression of circ-CDYL in MCL cells (Z138) transfected with sh-circ-CDYL or sh-NC (negative control). (B) Cell proliferation was quantified in MCL cells using the CCK8 kit. **p < 0.01.
miRNA Prediction and Bioinformatics Analyses
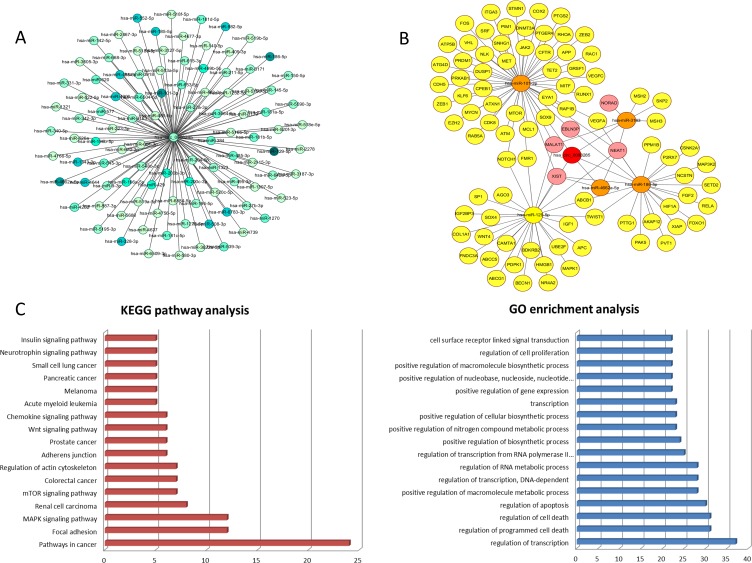
Bioinformatics analyses showed that circCDYL may harbor 116 miRNA response elements, primarily hsa-miR-129-5p, hsa-miR-3163, hsa-miR-4662a-5p, hsa-miR-101-3p, and hsa-miR-186-5p (Figure 3A). We also assessed the circCDYL-miRNA–mRNA-lncRNA network by Cytoscape to examine the molecular mechanism. This network included the top five miRNAs (hsa-miR-129-5p, hsa-miR-3163, hsa-miR-4662a-5p, hsa-miR-101-3p, and hsa-miR-186-5p) and five lncRNAs (MALAT1, NEAT1, EBLN3P, NORAD, and XIST) (Figure 3B). Among these, the associations were complex because several miRNAs had target lncRNAs in common, including lncRNAs MALAT1, NEAT1, EBLN3P, and XIST. Similarly, miRNAs shared some mRNAs in common, such as NOTCH1, FMR1, ABCB1, TWIST1, and VEGFA.
Figure 3.
Bioinformatics analyses of Circ-CDYL and miRNAs, lncRNAs and mRNAs. (A) Relative miRNA response elements. Darker color represents stronger relevance. (B) CircRNA-miRNA-mRNA/lncRNA co-expression network. The red, orange, pink, and yellow nodes represent circRNA, miRNA, lncRNA, and mRNA, respectively. (C) KEGG pathway analysis and GO enrichment analysis of related mRNAs. MAPK: mitogen- activated protein kinase
We conducted gene ontology and KEGG pathway enrichment analyses for these miRNAs and their potential target mRNAs and determined the top relationships (Figure 3C). The strongest relationships were regulation of transcription, programmed cell death, cell death, and apoptosis. The strongest associations involved pathways in cancers, followed by focal adhesion and the mitogen-activated protein kinase signaling pathway.
Discussion
MCL is a rare non-Hodgkin lymphoma characterized by the translocation, t (11;14), and cyclin D1 overexpression.16 However, the mechanism of MCL remains unclear and there is an urgent need to identify novel biomarkers for early diagnosis. Sry-related high-mobility-group box 11 has been identified as a diagnostic biomarker of MCL,17 and a recent study showed that lncRNA GATA6-AS may have potential in the early diagnosis of this disease.18
CircRNAs are new members of the non-coding RNA family and potential molecular markers for cancers.19,20 The stable presence of circRNAs in human peripheral blood makes them potentially easily accessible disease biomarkers.21,22
CDYL serves as a histone methyllysine reader and transcriptional corepressor.23 A recent study reported that circCDYL was derived from exon 4 of CDYL through back-splicing.24 A study focused on bladder cancer reported that circCDYL was downregulated in both tumor tissue and cell lines.24 In bladder cancer, circCDYL inhibited cell growth through the downregulation of Myc.24 In contrast, one study demonstrated that circCDYL was highly expressed in the early stages of hepatocellular carcinoma, and inhibition of circCDYL in HCCLM3 cells inhibited their proliferation, tumor spheroid growth, and ameliorated chemotherapy resistance.25 Overexpression of circCDYL in SMMC-7721 cells promoted malignant proliferation, self-renewal, and chemoresistance.25 Moreover, it functioned as a sponge of miR-892a and miR-328-3p and inhibited hepatoma-derived growth factor and hypoxia-inducible factor asparagine hydroxylase, respectively.25
In our study, circCDYL was highly expressed in the plasma of MCL patients compared to healthy donors. The ROC results suggested that circCDYL could serve as a potential biomarker for the diagnosis or auxiliary diagnosis of MCL patients. In addition, a combination of biomarkers might increase diagnostic accuracy. For example, Lin et al found a panel of three circRNAs (circ-CCDC66, circ-ABCC1, and circ-STIL) was more accurate than traditional diagnostic biomarkers, carcinoembryonic antigen, and carbohydrate antigen 19-9, in colorectal cancer.26 Similarly, Yu et al identified a plasma circRNA panel containing three circRNAs (hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897) that showed higher accuracy than alpha‐fetoprotein in the diagnosis of hepatitis B virus-related hepatocellular carcinoma.27
However, we conducted a survival analysis of MCL patients and found no significant difference between the high and low circCDYL expression groups, possibly due to an insufficient follow-up time and limited sample pool. We also found that inhibition of circCDYL suppressed cell proliferation in vitro, consistent with its functions in regulating transcription, programmed cell death, cell death, and apoptosis.
CircCDYL is associated with various pathways in cancers and thus affects their progression. CircRNAs can compete with endogenous RNAs, and the miRNA sponge effect is currently an area of intense research interest. For example, CDR1-as can bind to miR-7.11 We conducted bioinformatics analyses and identified a co-expression network of circCDYL-miRNA–mRNA-lncRNA, highlighted by five miRNAs (hsa-miR-129-5p, hsa-miR-3163, hsa-miR-4662a-5p, hsa-miR-101-3p, and hsa-miR-186-5p), three lncRNAs (MALAT1, NEAT1, and XIST), and five mRNAs (NOTCH1, FMR1, ABCB1, TWIST1, and VEGFA).
Previous research showed that lncRNA MALAT1 was overexpressed in MCL tissues and may be a prognostic factor.26 Functionally, MALAT1 inhibited p21 and p27, mediated by an enhancer of zeste homologue 2 (EZH2)-associated mechanism in MCL.28 p21 and p27 are both cyclin-dependent kinase suppressors that function during cell cycle progression. Low expression of p21 or p27 is correlated with a poor prognosis in MCL patients.29,30 EZH2, a component of the polycomb group complex, is involved in hematopoiesis, balancing cell self-renewal and differentiation.31 In addition, inhibiting EZH1/EZH2 in MCL up-regulated the expression of the cell cycle regulators, CDKN1C and TP53INP1, and showed an anticancer effect.32 Consistently, miR-101 can inhibit cell proliferation and induce apoptosis by targeting EZH2 in MCL.33 Accordingly, circCDYL might serve as a sponge for miR-101, targeting EZH2 and further regulating p21 and p27.
NOTCH1 mutations were present in 12% of MCL clinical samples and 20% of the cell lines and were associated with poor survival.34 Inhibition of NOTCH1 suppressed the proliferation of the MCL cell line, Jeko-1, induces apoptosis, and regulated the Akt/mTOR signaling pathway.35 Thus, circCDYL may regulate the expression of NOTCH1 in MCL, though the detailed mechanism still needs further study.
Conclusion
Overall, the results of the current study reveal the clinical significance and relevant functions of circCDYL in MCL. These findings indicate that circCDYL might serve as a potential diagnostic biomarker in clinical practice.
Acknowledgment
I would like to show gratitude to all members in the Oncology Department of the First Affiliated Hospital of Zhengzhou University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Herrmann A, Hoster E, Zwingers T, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27(4):511–518. doi: 10.1200/JCO.2008.16.8435 [DOI] [PubMed] [Google Scholar]
- 2.Li W, Xue W, Wang X, et al. MiR-199a mediated the dissemination of human mantle cell lymphoma by interacting with the CCR7/CCL21 pair. Anticancer Drugs. 2018;29(9):861–870. doi: 10.1097/CAD.0000000000000656 [DOI] [PubMed] [Google Scholar]
- 3.Vose JM. Mantle cell lymphoma: 2017 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2017;92(8):806–813. doi: 10.1002/ajh.24797 [DOI] [PubMed] [Google Scholar]
- 4.Maddocks K. Update on mantle cell lymphoma. Blood. 2018;132(16):1647–1656. doi: 10.1182/blood-2018-03-791392 [DOI] [PubMed] [Google Scholar]
- 5.Mei M, Zhang M. Non-coding RNAs in natural killer/T-cell lymphoma. Front Oncol. 2019;9:515. doi: 10.3389/fonc.2019.00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- 7.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA (New York, NY). 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 10.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 12.Dahl M, Daugaard I, Andersen MS, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest. 2018;98(12):1657–1669. doi: 10.1038/s41374-018-0108-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–247. doi: 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 16.Vogt N, Dai B, Erdmann T, Berdel WE, Lenz G. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma. 2017;58(7):1530–1537. doi: 10.1080/10428194.2016.1248965 [DOI] [PubMed] [Google Scholar]
- 17.Xu W, Li JY. SOX11 expression in mantle cell lymphoma. Leuk Lymphoma. 2010;51(11):1962–1967. doi: 10.3109/10428194.2010.514968 [DOI] [PubMed] [Google Scholar]
- 18.Fan Z, Wang X, Li P, Mei C, Zhang M, Zhao C. Overexpression of lncRNA GATA6-AS inhibits cancer cell proliferation in mantle cell lymphoma by downregulating GLUT1. Oncol Lett. 2019;18(3):2443–2447. doi: 10.3892/ol.2019.10540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10(2):187–197. doi: 10.2217/epi-2017-0109 [DOI] [PubMed] [Google Scholar]
- 22.Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Lai S, Ma W, et al. CDYL suppresses epileptogenesis in mice through repression of axonal Nav1.6 sodium channel expression. Nat Commun. 2017;8(1):355. doi: 10.1038/s41467-017-00368-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Zhang H, Tao D, et al. CircCDYL inhibits the expression of C-MYC to suppress cell growth and migration in bladder cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):1349–1356. doi: 10.1080/21691401.2019.1596941 [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology (Baltimore, Md). 2019. doi: 10.1002/hep.30795 [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Cai D, Li W, et al. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem. 2019. doi: 10.1016/j.clinbiochem.2019.10.012 [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Ding WB, Wang MC, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int J Cancer. 2019. doi: 10.1002/ijc.32647 [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14(1):346. doi: 10.1186/s12967-016-1100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letestu R, Ugo V, Valensi F, et al. Prognostic impact of p27KIP1 expression in cyclin D1 positive lymphoproliferative disorders. Leukemia. 2004;18(5):953–961. doi: 10.1038/sj.leu.2403337 [DOI] [PubMed] [Google Scholar]
- 30.Pinyol M, Hernandez L, Cazorla M, et al. Deletions and loss of expression of p16INK4a and p21Waf1 genes are associated with aggressive variants of mantle cell lymphomas. Blood. 1997;89(1):272–280. doi: 10.1182/blood.V89.1.272 [DOI] [PubMed] [Google Scholar]
- 31.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28(1):44–49. doi: 10.1038/leu.2013.288 [DOI] [PubMed] [Google Scholar]
- 32.Li W, Bi C, Han Y, et al. Targeting EZH1/2 induces cell cycle arrest and inhibits cell proliferation through reactivation of p57CDKN1C and TP53INP1 in mantle cell lymphoma. Cancer Biol Med. 2019;16(3):530–541. doi: 10.20892/j.issn.2095-3941.2018.0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YL, Zou ZK, Su HY, Huang YQ. [Expression of MiR101 and EZH2 in patients with mantle cell lymphoma and its clinical significance]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2019;27(3):820–826. doi: 10.19746/j.cnki.issn.1009-2137.2019.03.029 [DOI] [PubMed] [Google Scholar]
- 34.Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. doi: 10.1182/blood-2011-11-391474 [DOI] [PubMed] [Google Scholar]
- 35.Huang YQ, Huang XL, Ma XD. [Effect of silencing NOTCH1 gene by shRNA interference on AKT/mTOR pathway in mantle cell lymphoma]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22(6):1616–1620. doi: 10.7534/j.issn.1009-2137.2014.06.021 [DOI] [PubMed] [Google Scholar]