Abstract
In breast cancer, the increased stiffness of the extracellular matrix (ECM) is a key driver of malignancy. Yet little is known about the epigenomic changes that underlie the tumorigenic impact of ECM mechanics. Here, we show in a 3D culture model of breast cancer that stiff ECM induces a tumorigenic phenotype through changes in chromatin state. We found that increased stiffness yielded cells with more wrinkled nuclei and with increased lamina-associated chromatin, that cells cultured in stiff matrices displayed more accessible chromatin sites, which exhibited footprints of Sp1 binding, and that this transcription factor acts along with the histone deacetylases 3 and 8 to regulate the induction of stiffness-mediated tumorigenicity. Just as cell culture on soft environments or in them rather than on tissue-culture plastic better recapitulates the acinar morphology observed in mammary epithelium in vivo, mammary epithelial cells cultured on soft microenvironments or in them also more closely replicate the in vivo chromatin state. Our results emphasize the importance of culture conditions for epigenomic studies, and reveal that chromatin state is a critical mediator of mechanotransduction.
Tumor ECM is substantially remodeled from normal tissue microenvironments, leading to changes in the composition and density of the ECM network. These modifications of the microenvironment result in changes in mechanical properties, such as increased matrix stiffness1. Differences in ECM stiffness have been broadly studied and are known to cause changes in gene expression by modulation of integrin binding and downstream signaling, cytoskeletal tension, conformational changes and activation of mechanosignaling complexes, and transcription factor activation and localization1–7. Stiff matrices even promote malignant phenotypes in non-malignant mammary epithelial cells4, 8. However, despite the broadly appreciated role of the epigenome in gene regulation and its recognized misregulation in cancer, little is known about how changes in chromatin state regulate the impact of ECM mechanics9.
Transcription factors bind regulatory DNA elements to control gene expression and cellular phenotypes10, and this binding is dictated by the chromatin state. Regulatory elements bound by transcription factors typically exhibit signatures of accessible chromatin11. Chromatin accessibility can be altered by several enzymatic modifications, such as acetylation and methylation of specific histone tail residues, that are also associated with activation or silencing of genes12. The nuclear lamina is the nexus of the cytoskeleton and chromatin and is known to be mechanoresponsive13, thus serving as a likely link between mechanical cues and chromatin remodeling. Indeed, HDACs have been shown to be responsive to mechanical properties and culture dimensionality through interactions with the nuclear lamina14, 15. Intriguingly, biological processes associated with changes in chromatin state, like stem cell differentiation and breast cancer progression, are also known to be mechanoresponsive16, 17. However, whether chromatin state mediates mechanotransduction in these processes is unclear. Further, direct evidence for mechanically-induced chromatin remodeling in general is limited, and it is unknown by what mechanisms these changes might occur.
Results
To address whether ECM mechanics can drive phenotypic shifts through changes to chromatin state, we utilized a well-studied, mechanosensitive breast cancer 3D culture model1, 4, 8 (Fig. 1a). Interpenetrating networks (IPNs) of reconstituted basement membrane (rBM) matrix and alginate, a material system in which matrix stiffness can be tuned independently of ligand density, matrix pore size, and matrix architecture, were used as matrices for 3D culture8. MCF10A breast epithelial cells were encapsulated in IPNs with elastic moduli similar to normal mammary tissue (~100 Pa, soft) or malignant tumor tissue (~2000 Pa, stiff) and cultured for fourteen days (Supplementary Figure 1). As in prior reports, the MCF10As formed organotypic acinar structures in the soft matrices, but exhibited a tumorigenic phenotype in stiff matrices, marked by an increase in invasive sites presenting higher levels of vimentin and phosphorylated focal adhesion kinase (pFAK), an associated decrease in roundness, matrix remodeling, filled acinar lumens, loss of normal β4 integrin containing hemidesmosomal adhesions to an intact basement membrane, and aberrant E-cadherin and N-cadherin localization (Fig. 1b–e, Supplementary Figures 2,3)4, 8. A prior report determined that enhanced stiffness is transduced through a ß4 integrin-phosphoinositide 3-kinase (PI3K) signaling pathway to drive the malignant phenotype8. A tumorigenic phenotype was also observed when hTERT-HME1 cells (HME1s), another cell line used to model normal mammary epithelium, were encapsulated in stiff matrices, while clusters in soft matrices were rounded and less invasive (Supplementary Figure 4).
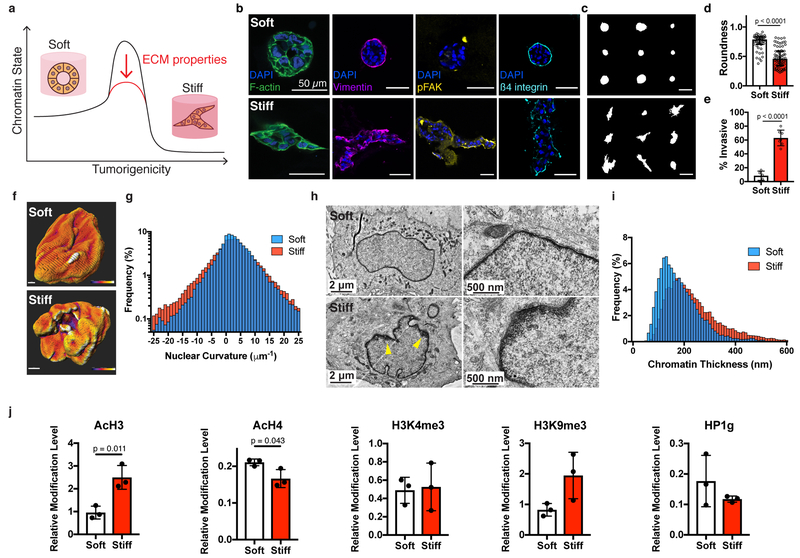
Fig. 1 |. Nuclear and chromatin alterations accompany phenotypic changes induced by ECM stiffness.
a, The hypothesis underlying this study is that ECM properties can stabilize normal phenotypes or make phenotypic transitions more permissive through chromatin alterations. b, Immunofluorescence staining for F-actin, vimentin, phosphorylated FAK (Y397), and β4 integrin in MCF10A acini after 14 days in soft (top) or stiff (bottom) matrices (representative images selected from 15, 3, 5, and 5 images per group respectively). c, Representative outlines of cell clusters from soft (top) and stiff (bottom) matrices. d, Quantification of roundness of cell clusters from at least three independent replicates (median ± 95% C.I.). Significance determined by Mann-Whitney test. e, Quantification of invasive clusters (n ≥ 5, mean ± S.D.) Significance determined by unpaired t-test. f, Color map of nuclear curvature for cells in soft (top) and stiff (bottom) matrices. Scale bar represents 2 μm and color bar ranges from −20 to 20 μm−1. g, Distribution of curvature values, showing more regions of extreme curvature for cells in stiff matrices. Distributions were significantly different by Kolmogorov-Smirnov test (p < 0.0001, n ≥ 11). h, TEM micrographs of nuclei from cells in soft or stiff matrices at 2,000X (left) and 10,000X (right) magnification (representative images from at least 16 images per group). i, Distribution of measured chromatin thickness at each pixel around nuclear boundary for at least six nuclei in each group. Distributions were significantly different by Kolmogorov-Smirnov test (p < 0.0001). j, Western blot quantification of histone modifications normalized to total H3 levels (mean ± S.D.). Three independent replicates were used and significance was tested using unpaired t-tests.
We first examined the changes in nuclear morphology and chromatin architecture associated with the transition to a tumorigenic phenotype in the stiff matrices. An increase in nuclear wrinkling was observed for cells cultured in stiff matrices, and quantitative analysis revealed that these nuclei had a significantly higher proportion of extreme curvature than nuclei in cells cultured in soft matrices (Fig. 1f,g). As the nuclear lamina can bind chromatin in lamina-associated domains in a mechanoresponsive manner, chromatin organization was next analyzed using transmission electron microscopy (TEM)18, 19. Numerous heterochromatin bundles were observed at the nuclear periphery for cells in stiff matrices, in contrast to a thin chromatin boundary for cells in soft matrices (Fig. 1h). Heterochromatin thickness increased on average in stiff matrices, with substantially more heterochromatin bundles, indicated by occurrences in the right-tail of the distribution (Fig. 1i). Having found that stiffness can alter chromatin organization, several histone modifications associated with activation or repression were investigated. Interestingly, stiff matrices produced significantly higher levels of AcH3, but decreased levels of AcH4, and neither H3K4me3 (active), H3K9me3 (repressive) marks nor HP1γ (repressive) levels were significantly different (Fig. 1j). Together, these results indicate that chromatin state was broadly misregulated with increased stiffness, though not consistently toward a more open or closed architecture, motivating the use of a more specific assay to characterize chromatin organization.
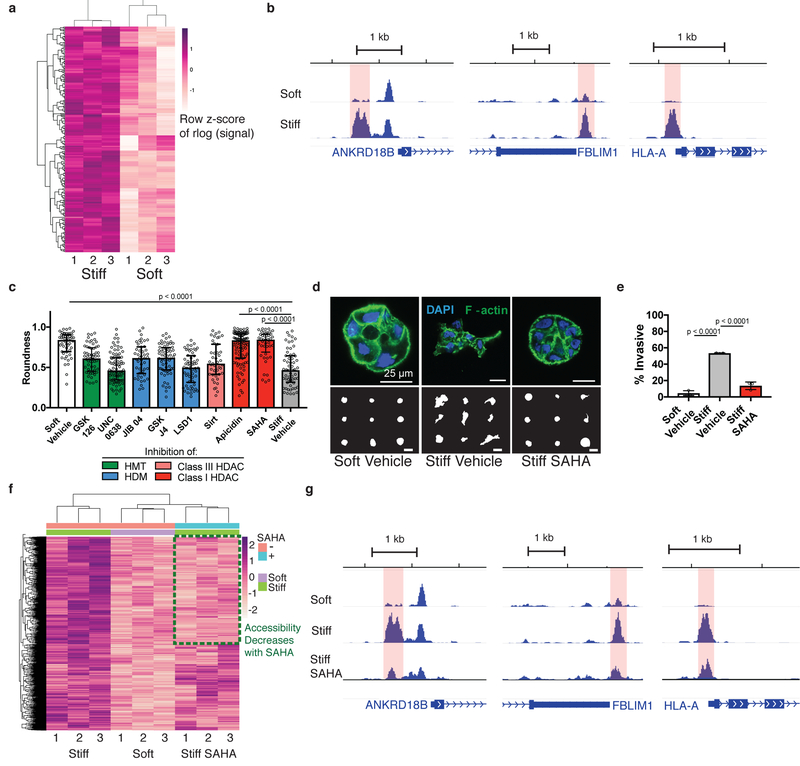
To gain a genome-wide, site-specific perspective of alterations in chromatin organization in response to increased stiffness, we performed the assay for transposase-accessible chromatin with sequencing (ATAC-seq)20. ATAC-seq utilizes a hyperactive Tn5 transposase that preferentially cleaves and inserts sequencing adapters in accessible chromatin. Sequencing and mapping the fragments enables genome-wide profiling of putative regulatory elements bound by transcription factors exhibiting signatures of chromatin accessibility. ATAC-seq can be carried out with low cell numbers (500–50,000), making it amenable to 3D cultures. Accessibility profiles from independent biological replicates for both soft and stiff matrices were highly correlated, enrichment at transcription start sites (TSS) was prevalent for both conditions, and signal within other genomic features was similar (Supplementary Table 1–3). Differential analysis of ATAC-seq peaks revealed 1658 significantly more accessible peaks for cells cultured in stiff matrices, with no regions found to be significantly more accessible in soft matrices (Fig. 2a,b).
Fig. 2 |. Chromatin accessibility changes are associated with normal and tumorigenic phenotypes.
a, Heatmap of 1658 regions with differential accessibility. Each row represents a differential region, each column is one of three biological replicates of soft or stiff conditions. b, Representative genome browser tracks of significantly differentially accessible regions (highlighted). c, Quantification of roundness for small molecule inhibitors of histone modifiers. Three independent replicates were used for each condition, significance was determined by Kruskal-Wallis test followed by Dunn’s multiple testing correction (median ± 95% C.I.). d, Confocal immunofluorescence images of clusters in control matrices and treated with SAHA (top, from at least 22 images per group) and representative outlines of clusters (bottom). e, Quantification of invasive clusters (n = 3, mean ± S.D.). Significance determined by one-way ANOVA followed by Dunnett’s multiple testing correction. f, Heatmap of 1658 differentially accessible regions between soft and stiff control matrices with signal for SAHA-treated cells in stiff matrices. Green box represents regions that are more accessible in stiff control and less accessible after SAHA treatment. g, Representative genome browser tracks of regions that are more accessible in stiff control matrices than soft control matrices and are less accessible upon SAHA-treatment (differential region highlighted).
We next sought to determine chromatin modifiers that may be associated with the changes in chromatin accessibility. Pharmacological inhibition of four major classes of histone modifying enzymes including histone methyltransferases, histone demethylases, class I histone deacetylases, and class III histone deacetylases was performed. Only inhibition of class I histone deacetylases caused cells cultured in stiff matrices to form rounded clusters similar to cells cultured in soft matrices (Fig. 2c,d). Clusters of cells cultured with either suberoylanilide hydroxamic acid (SAHA) or apicidin, two structurally distinct class I HDAC inhibitors, were significantly more rounded than controls in stiff matrices but similar to clusters cultured in soft matrices. A similar result was found in HME1s treated with SAHA (Supplementary Figure 4). HDAC inhibition by SAHA treatment also significantly reduced the fraction of invasive clusters for MCF10A cultured in stiff matrices (Fig. 2e). Others have shown class I HDACs to be differentially activated by matrix stiffness and actomyosin contractility14, 15. To clarify whether SAHA-treatment prevented the stiffness-induced tumorigenic phenotype by chromatin changes, ATAC-seq was performed on cells from this group as well. Differential peak calling followed by clustering identifies a group of peaks that are more accessible in stiff control matrices than soft control matrices, but that have decreased accessibility upon SAHA treatment, mirroring the observed phenotypic changes (Fig. 2f,g). Peaks from the stiff-SAHA group with z-values closer to soft matrices than stiff matrices were identified as a reverted subset, consisting of 660 regions from the total 1658 regions. This result is ostensibly contradictory to the conventional role for HDACs; deacetylation of histones to decrease chromatin accessibility. However, several reports have found that HDAC inhibition can induce both acetylation and deacetylation to alter chromatin accessibility in both directions21–26. These reports are in agreement with the broad effects we observe upon SAHA treatment in either soft or stiff matrices (Supplementary Figure 5), illustrating the complex roles HDACs play in epigenetic regulation.
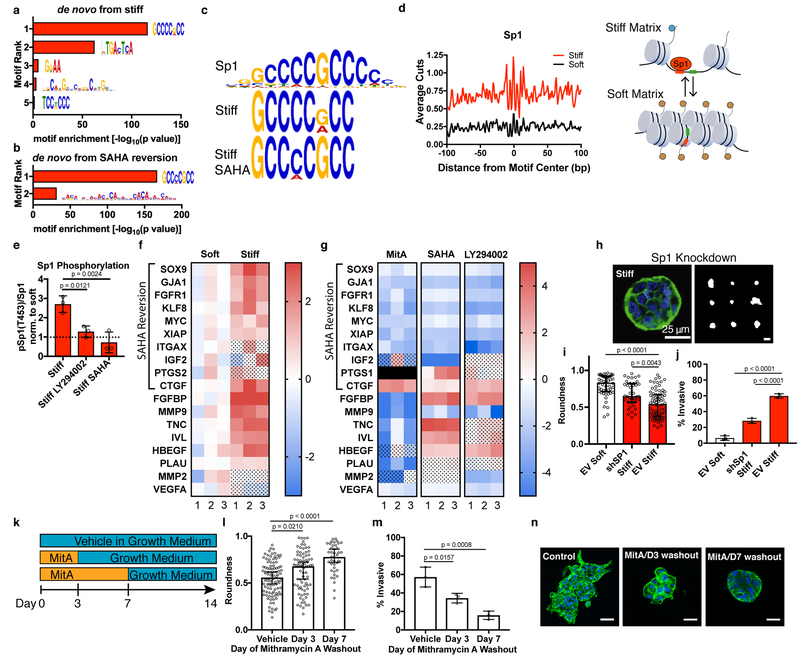
To identify candidate transcription factors that may bind in the differentially accessible chromatin sites, we performed de novo motif analysis on the regions that were more accessible in stiff matrices. The best match among known transcription factor motifs for the highest ranked de novo motif by MEME-ChIP analysis was Sp1 (Fig. 3a,c). The Homer motif-discovery tool identified Sp1 among the most enriched candidate motifs as well (Supplementary Figure 6). Additionally, motif analysis on the subset of peaks in Figure 2f that lose accessibility upon SAHA treatment (reversion) also implicated Sp1 (Fig. 3b,c, Supplementary Figure 7). Notably, the Sp1 motif is not enriched in the non-reverted set of peaks. ATAC-seq based footprinting analysis at the Sp1 motifs revealed a distinct signature for cells in stiff matrices, with higher Tn5 cut frequency (more accessibility) flanking the binding site and a deeper footprint at the motif center, where bound Sp1 protects the site from Tn5 cleavage (Fig. 3d). This footprint shape is consistent with those previously reported from ATAC-seq data27.
Fig. 3 |. Sp1 mediates the stiffness-induced tumorigenic phenotype.
a, MEME-ChIP de novo motif analysis for differentially accessible regions between soft and stiff matrices. b, MEME-ChIP de novo motif analysis for regions of decreased accessibility in SAHA-treated cells in stiff matrices c, Sp1 motif logo (top) best matches the top ranked MEME-ChIP de novo motifs from soft vs. stiff comparison (middle) and from regions of decreased accessibility in SAHA-treated cells in stiff matrices. d, Transcription factor footprint of 200 bp region centered on Sp1 motifs in differentially accessible regions. Cartoon illustrating an accessible chromatin region displaying the Sp1 motif in stiff ECM that is inaccessible to Sp1 in soft ECM. e, Quantification of Sp1 phosphorylation levels at Thr453 for cells in stiff matrices and treated with PI3K inhibitor (LY294002) or class I HDAC (SAHA). All values are normalized by the value for soft matrices of the same treatment condition (mean ± S.D.). Significance was determined by one-way ANOVA followed by Dunnett’s multiple comparison correction. f, Heatmap of gene expression for Sp1 target genes associated with malignant neoplasm of breast (n = 3, two-way ANOVA with multiple comparison correction, FDR = 0.05, color scale represents log2 fold change, dot pattern indicates no significance, black box represents no data). g, Heatmap of gene expression for Sp1 target genes associated with malignant neoplasm of breast for stiff matrices with indicated inhibitors compared to vehicle control (n = 3, two-way ANOVA with multiple comparison correction, FDR = 0.05, color scale represents log2 fold change, dot pattern indicates no significance). h, Confocal immunofluorescence of Sp1-knockdown cells in stiff matrices (left, from 38 total images) and representative outlines of shSp1 clusters. i, Quantification of roundness for shSp1 clusters in stiff matrices vs. empty vector controls in soft and stiff matrices (n = 3, median ± 95% C.I.). j, Quantification of invasive clusters (n = 3, mean ± S.D.). k, Schematic timeline of small molecule inhibitor of Sp1 washout experiment. l, Quantification of roundness of cell clusters in soft or stiff matrices treated with mithramycin A or vehicle control for the indicated period of time (n = 3, median ± 95% C.I.). m, Quantification of invasive clusters (n = 3, mean ± S.D.). n, Confocal immunofluorescence of cells in soft or stiff matrices treated for the duration indicated and imaged after 14 total days (representative images from 15 images per group). Significance was determined by Kruskal-Wallis test followed by Dunn’s multiple testing correction for roundness and by one-way ANOVA followed by Dunnett’s multiple testing correction for invasion.
We then analyzed Sp1 activation in the stiff matrices and its relevance to breast cancer. Sp1 is a potent transcription factor, known to regulate proliferation, apoptosis, differentiation, and malignant transformation28. Malignant neoplasm of breast is the most significant specific disease found for Sp1 target genes in a search of the TRRUST v2 database (Supplementary Table 4)29. Interestingly, this is the stage of breast cancer that our high stiffness model system most closely recapitulates in terms of cell-type, ligand-type, and stiffness. Sp1 interacts with a number of chromatin modifying proteins and can function as both an activator or repressor of transcription depending on which proteins it recruits to regulatory complexes. Importantly, Sp1 phosphorylation at Thr 453, a marker of Sp1 activation32, was assessed and was found to be significantly higher in stiff matrices relative to soft matrices (Fig. 3e, Supplementary Figure 7). Expression levels of Sp1 target genes associated with malignant neoplasm of breast were then examined, including ten genes within the subset of peaks reverted by SAHA treatment. Stiff matrices induced significantly increased expression in thirteen of the eighteen genes, consistent with the conclusion of enhanced Sp1 activity (Fig. 3f). As expected, Sp1 inhibition with mithramycin A, which diminishes the tumorigenic phenotype, significantly reduced expression of fourteen of the target genes compared to cells in stiff matrices treated with a vehicle control (Fig. 3g). Intriguingly, treatment with SAHA generally resulted in reduced expression for genes in regions with decreased chromatin accessibility (reverted subset), but SAHA increased expression for genes not found in the reverted subset (Fig. 3g). Thus, the gene expression profile largely mirrors the chromatin accessibility profile.
In the 3D culture model used here, it is known that increased stiffness promotes the tumorigenic phenotype through PI3K-mediated signaling, and PI3K has been shown by others to phosphorylate Sp1 to alter HDAC association, chromatin binding, and gene activation8, 30, 31. PI3K signaling was also the pathway most significantly associated with Sp1 target genes by TRRUST analysis (Supplementary Table 5). Sp1 phosphorylation levels were significantly reduced by inhibition of PI3K (with LY294002) or class I HDACs (with SAHA) (Fig. 3e, Supplementary Figure 7). The expression profile of Sp1 target genes upon PI3K inhibition was similar to SAHA treated groups (Fig. 3g). For genes within the SAHA-reverted subset, expression was generally reduced, while for regions not found in the reverted subset, expression was increased. That inhibition of either HDACs or PI3K results in similar effects on both Sp1 gene expression and Sp1 phosphorylation suggests that they may be regulating Sp1 through a common pathway, though the exact mechanism is still uncertain. Taken together, these data demonstrate increased activation of Sp1 in stiff matrices and indicate that Sp1 activation may be prominently involved in the establishment of the tumorigenic phenotype in response to stiff matrices through altered chromatin accessibility.
To directly assess the role of Sp1 in phenotypic determination, we performed genetic and pharmacological inhibition of Sp1. Knockdown of Sp1 using shRNA caused cells to form rounded clusters despite being cultured in stiff matrices (Fig. 3h,i). Compared to empty vector (EV) control cells cultured in stiff matrices, Sp1 knockdown cells were significantly more rounded and similar to EV control cells in soft matrices. Knockdown of Sp1 also resulted in a significant reduction in the fraction of invasive clusters for cells cultured in stiff matrices (Fig. 3j). Sp1 knockdown cells were still able to proliferate in soft and stiff matrices, as cell clusters contained dozens of cells, but the tumorigenic characteristics typically associated with stiff matrices were not observed. In addition, pharmacological inhibition of Sp1 with mithramycin A in HME1s abrogated the invasive phenotype in stiff matrices (Supplementary Figure 4). Further, inhibition of Sp1 was performed for breast cancer cells lines MCF7 and MDA-MB-231 in soft and stiff 3D matrices. Sp1 inhibition altered the morphology of these cancer cell lines in a stiffness-dependent manner (Supplementary Figure 8). Both MCF7 and MDA-MB-231 adopted invasive, proliferative phenotypes in stiff matrices, but Sp1 inhibition resulted in significantly more rounded and smaller clusters, indicative of diminished invasion and proliferation respectively. Interestingly, no difference in cluster size was observed for either cell line cultured in soft matrices with or without mithramycin A, suggesting that for these breast cancer cell lines, Sp1-driven proliferation and invasion is stiffness-dependent. These studies confirm the role of Sp1 in mediating the impact of stiffness on the tumorigenic phenotype.
Next, we examined the timeline for Sp1 regulation of tumorigenicity. Our previous experiments revealed a role for Sp1 in cultures lasting 14 days. However, it was not clear whether stiff matrices initiated Sp1-induced effects in the early stages of induction of the tumorigenic phenotype, or whether the effects of Sp1 occurred only after the phenotype was well established. To determine when Sp1 was active within our system, mithramycin A was applied for the first 3 or 7 days of culture, and then washed out with growth medium, while culture was continued for another 11 or 7 days, respectively (Fig. 3k). Inhibition of Sp1 for just 3 days resulted in a significant increase in cluster roundness and a significant decrease in invasiveness (Fig. 3 l–n). These results indicate that Sp1 must be involved in the earliest stages of tumorigenic conversion.
As Sp1 is known to recruit chromatin modifiers33, 34, we next investigated what chromatin modifiers might cooperate with Sp1 to mediate the phenotypic transition. Assessment of known protein-protein interactions with Sp1 revealed a strong association with class I HDACs (Fig. 4a). Motivated by the combination of this result with the prior results showing that class I HDAC inhibition was effective at preventing the tumorigenic phenotype from arising in stiff matrices, reduced Sp1 phosphorylation and altered Sp1 target gene expression profiles, shRNA knockdowns were performed on the four known members of class I HDACs (HDAC 1, 2, 3, and 8). Interestingly, knockdowns of HDAC3 and HDAC8 were highly effective at preventing the tumorigenic phenotype from arising in stiff matrices, while knockdowns of HDAC1 and HDAC2 had no effect (Fig. 4b–d). To determine whether HDACs 3 and 8 are involved early in the stiffness-induced tumorigenic conversion, or are further downstream, small molecule inhibitor wash out experiments were performed (Fig. 4e). Inhibition of class I HDACs for only 3 or 7 days resulted in significantly more rounded, less invasive clusters (Fig. 4f–h), supporting their involvement in early mechanosignaling pathways. Together, these data implicate Sp1 and HDACs in driving the tumorigenic phenotype in response to stiffness by altering chromatin state, potentially through interactions or overlapping signaling pathways (Fig. 4i), though we have not established a direct interaction here.
Fig. 4 |. HDACs 3 and 8 regulate the stiffness-induced tumorigenic phenotype.
a, String-DB protein-protein interaction network of top ten interactors with Sp1. b, Confocal immunofluorescence of shHDAC1, shHDAC2, shHDAC3, and shHDAC8 cells in stiff matrices (top, from at least 12 total images per group) and representative outlines of clusters (bottom). Scale bars represent 100 μm. c, Quantification of roundness for HDAC knockdowns in stiff matrices compared to empty vector controls in soft or stiff matrices and SAHA-treated cells in stiff matrices (n = 3, median ± 95% C.I.). d, Quantification of invasive clusters (n = 3, mean ± S.D.). e, Schematic timeline of small molecule inhibitor of Sp1 washout experiment. f, Confocal immunofluorescence of cells in soft or stiff matrices treated for the duration indicated and imaged after 14 total days (representative images from 15 images per group). g, Quantification of roundness of cell clusters in soft or stiff matrices treated with SAHA or vehicle control for the indicated period of time (n = 3, median ± 95% C.I.). h, Quantification of invasive clusters (n = 3, mean ± S.D.). i, Schematic illustrating sequential events from ECM mechanical properties to phenotype via mechanically-induced chromatin remodeling. Significance was determined by Kruskal-Wallis test followed by Dunn’s multiple testing correction for roundness and by one-way ANOVA followed by Dunnett’s multiple testing correction for invasion.
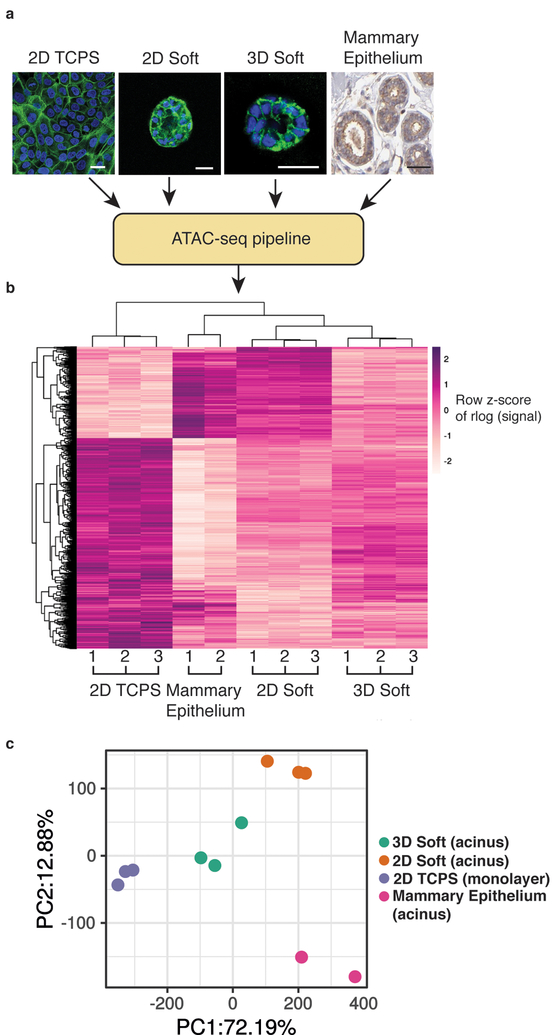
Finally, we examined the implications of our results for the use of three-dimensional cell culture versus traditional culture on tissue culture plastic for recapitulation of the in vivo chromatin landscape. It has been well established that mammary epithelial cells cultured on or in soft microenvironments yield phenotypes that mimic healthy mammary tissue35, while stiff matrices induce phenotypes similar to invasive tumors1, 8. However, chromatin state profiling experiments, to date, have largely been carried out in samples cultured on 2D tissue culture polystyrene (TCPS) or in vivo specimens. To evaluate whether soft microenvironments reproduce in vivo chromatin state more faithfully than culture on TCPS, ATAC-seq data from MCF10A cultured on 2D TCPS, on soft 2D polyacrylamide substrates (150 Pa) coated with rBM matrix, or in 3D soft matrices, as well as non-malignant human mammary epithelial tissue (ENCODE) were compared. Morphologically, acini generated from soft matrices in vitro resemble the hollow glandular structure found in mammary epithelium in vivo (Human Protein Atlas36), and both are distinct from the monolayer that forms on TCPS (Fig. 5a). Peaks with significantly different accessibility were identified between 2D TCPS and mammary epithelium samples (1314 peaks, Figure 5b). Accessible regions from soft matrices, either 2D or 3D, cluster with mammary epithelium. Interestingly, while the majority of differential regions are less accessible in mammary epithelium or soft microenvironments, a substantial subgroup (~30%) have increased accessibility compared to 2D TCPS culture. Principal component analysis also shows a clear separation of 2D TCPS samples from the soft matrices, which are much closer along the PC1 axis that accounts for 72% of the variance (Fig. 5c). Thus, just as culture of mammary epithelial cells on soft matrices better replicate the morphology and phenotype of in vivo mammary epithelium, the chromatin landscape is also remodeled in response to soft matrices in a manner that much more accurately represents in vivo tissue.
Fig 5 |. Soft hydrogel culture produces more physiologically representative chromatin accessibility profiles than standard tissue culture.
a, Representative morphologies of MCF10A cells cultured on tissue culture polystyrene (TCPS, left), on soft 2D matrices, in soft 3D matrices, and from human mammary tissue (image credit: Human Protein Atlas). Scale bars represent 25 μm. b, Heatmap of significantly differentially accessible regions between the different culture conditions and mammary epithelium, demonstrating the similarity in accessibility with cultures from soft matrices. Each row represents a differential region, each column is one biological replicates of the indicated condition. c, Principal components analysis reveals that accessible regions in soft matrices cluster closer to mammary epithelium than 2D TCPS along the first PC that accounts for 72% of variance. Three biological replicates were used for 2D TCPS, 2D soft and 3D soft, and two were used for in vivo mammary epithelium.
Discussion
This work demonstrates that alterations in ECM stiffness can drive changes to the nucleus and chromatin state, which in turn regulate phenotypic changes. Enhanced matrix stiffness alters both lamina-associated chromatin and accessible chromatin regions to promote the tumorigenic phenotype in mammary epithelium through Sp1-HDAC3/8 mediated pathways. Our findings highlight a major, previously undescribed, role of chromatin in mediating mechanotransduction and identify changes in chromatin organization in response to matrix stiffness genome wide at a level of detail previously unexplored. Several recent studies have shown that mechanical cues can alter nuclear mechanics, nuclear lamina components, and global chromatin condensation13, 37. Extracellular mechanical signals can also be transmitted to the nucleus through the cytoskeleton, allowing for forces on the cell surface to directly remodel chromatin to alter gene expression18. Prior studies on 2D substrates have demonstrated that class I HDACs are differentially active in response to actomyosin contractility15, 38. Additionally, HDAC3 has been shown to be bound and activated by emerin on the nuclear lamina39 and emerin regulates heterochromatin in response to mechanical strain40. These studies complement our findings that enhanced stiffness in 3D matrices promotes chromatin remodeling through the class I HDACs HDAC3 and HDAC8. Enhanced stiffness induces increased accessibility in chromatin regions throughout the genome, and these accessible sites frequently present Sp1-binding motifs. Increased activation of Sp1, increased expression of key Sp1-target genes, and abrogation of tumorigenicity by knockdown or inhibition of Sp1, point to increased Sp1 activity as a key event in regulation of the stiffness-induced tumorigenic phenotype in this 3D culture model of mammary epithelium. The known increase in stiffness during breast cancer progression combined with the known association of Sp1 target gene expression with malignant neoplasm of the breast suggest the relevance of this stiffness-Sp1-HDAC3/8 pathway to the earliest stages of breast cancer progression. More broadly, our work reveals a pathway by which mechanical signals can be transduced to the nucleus to alter chromatin through a complex of regulatory proteins to drive different phenotypic outcomes.
In addition, our work highlights the role of culture conditions in epigenomic studies, finding that soft matrices match not only the phenotypic state, but also the chromatin state of mammary epithelium more closely than conventional 2D culture on rigid TCPS. These experiments build upon foundational studies showing the effects on mammary epithelial cells from culture in recombinant basement membrane (rBM) protein matrices compared to conventional 2D TCPS. A previous study found that MEC culture on 2D TCPS causes loss of tissue specific function through chromatin remodeling41. Additionally, culture in the presence of rBM causes a reduction in histone acetylation, an increase in chromatin condensation, and a global decrease in gene expression42; all indicative of less accessible chromatin. Here we employed ATAC-seq to reveal site-specific changes in chromatin accessibility and found broad agreement with prior work demonstrating chromatin changes based on bulk chromatin assays. However, we were able to distinguish a subgroup of approximately 30% of the significantly differentially accessible regions between 2D TCPS and mammary epithelium that have increased accessibility in in vivo samples. These chromatin changes are mirrored by soft microenvironments. The ability to differentiate chromatin accessibility in a site-specific, genome-wide manner highlights the power of next generation sequencing techniques when applied to studies of mechanotransduction, as conventional bulk assays would mask the specific effects of mechanical cues by averaging signal across the epigenome. This work underscores the need for chromatin profiling experiments in biomimetic culture systems when they become available, particularly in models that are known to be mechanically responsive, such as cancer progression and stem cell differentiation43, 44.
Methods
Hydrogel formation
Hydrogel matrices were composed of interpenetrating networks (IPNs) of alginate (5 mg/ml final, Pronova LF20/40) and rBM matrix (Matrigel, 4.4 mg/ml final, Corning)8, 45. Calcium sulfate was used to crosslink alginate matrices (1 mM Ca2+, and 21 mM Ca2+ final concentrations). To form IPNs, alginate and rBM solutions were first mixed on ice, then added to a cooled Luer lock syringe. A calcium sulfate slurry was diluted in DMEM/F12 basal medium and added to a second Luer lock syringe. The two syringes were connected with a coupler and mixed by passing the solutions back and forth six times. The IPNs were then deposited into a well plate pre-coated with rBM and incubated at 37°C for 30 minutes to gel.
Polyacrylamide gels were prepared as the 2D soft substrates for ATAC-seq comparative analyses. Briefly, coverslips were cleaned with ethanol, immersed in 0.5% (3-Aminopropyl)trimethoxysilane (in dH2O) at room temperature for 30 min, and washed with dH2O. Then, coverslips were treated with 0.5% glutaraldehyde in dH2O at room temperature for 30 min. A solution was prepared containing 3% acrylamide, 2% N,N′-methylenebis-acrylamide, 1/100 volume of 10% ammonium persulfate (APS), and 1/1000 volume of 4 N,N,N’,N’-Tetramethylethylenediamine (TEMED) to produce 150 Pa polyacrylamide gels. After gentle mixing, the solution was deposited on a Sigma-cote treated glass plate, covered with the activated coverslips, and allowed to polymerized between the glass plate and coverslips. Once polymerization was completed, gels were gently detached from the plate. The surface of gels was activated by adding a solution containing 1mg/ml sulphosuccinimidyl 6-(4’-azido-2’-nitrophenylamino)hexanoate (sulfo-SANPAH) dissolved in 50 mM HEPES pH 8.5. The gels were then exposed to UV light (wavelength 365 nm, 4 mW/cm2), washed with the HEPES solution, and incubated in 100 ug/ml of rBM in HEPES solution overnight at 4°C. The gels were washed with PBS before use.
Matrix Deformation Calculation
To calculate matrix deformation, MCF10As were embedded in alginate-rBM IPNs containing 21 mM calcium and 7.5% FluoSpheres Carboxylate-Modified Microspheres, 0.2 μm, dark red fluorescent (660/680) (Thermo Fisher Scientific). Seven days post encapsulation, the hydrogels were imaged every 30 minutes for 24 hours to track microbead displacements. The acquired images were corrected for drift using an image registration plug-in from ImageJ46. Then, matrix deformation was calculated by tracking the microbeads using a standard particle image velocimetry algorithm (PIVlab; open source code for MATLAB) with three passes (128 × 128, 64 × 64 and 32 × 32 pixel-size interrogation window with 50% overlap)47. Microbead displacement for each time frame was calculated relative to the bead position from initial time point, so the matrix deformation maps display cumulative displacements. Finally, brightfield images were overlaid over the matrix deformation maps.
Encapsulation and Cell Culture
MCF10A mammary epithelial cells were obtained from ATCC and cultured according to established protocols48. DMEM/F12 basal medium (ThermoFisher Scientific) was supplemented with 5% horse serum (ThermoFisher Scientific), 1% penicillin/streptomycin (ThermoFisher), 20 ng/ml epidermal growth factor (Peprotech), 0.5 mg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), and 10 μg/ml insulin (Sigma). HME1 cells were obtained from ATCC and cultured in the medium described above without cholera toxin. MCF7 and MDA-MB-231 cell lines were obtained from ATCC and cultured in DMEM basal medium (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (GE Healthcare), and 1% penicillin/streptomycin. Cells were encapsulated at 50,000 cells/ml final concentration in hydrogel matrices and cultured for 14 days.
Immunofluorescence, Confocal Imaging and Analysis
Cells in hydrogel matrices were fixed in 4% paraformaldehyde for 45 minutes, then washed twice in DPBS with Ca2+/Mg2+ for 15 minutes. The matrices were dehydrated in 30% a sucrose solution in DPBS overnight, then incubated in a 1:1 mixture of 30% sucrose and OCT solution (Fisher Scientific). The matrices were then frozen on dry ice and 40 μm cryosections were adhered to slides.
Slides were blocked in a solution of 10% goat serum (ThermoFisher Scientific), 1% bovine serum albumin (Sigma), 0.1% Triton X-100 (Sigma), and 0.3 M glycine (Sigma) for 1 hour at RT. Primary antibodies were diluted in the blocking solution (1:100) and incubated at 4°C overnight. Alexa Fluor 488-phalloidin (1:100 dilution, ThermoFisher Scientific) and DAPI (1 μg/ml) were diluted in the blocking solution and incubated for 1 hour. Fluorescently conjugated secondary antibodies were incubated in blocking solution for 1 hour at RT. The slides were then washed 3 times for 5 minutes in DPBS, and coverslips were applied with Prolong Gold antifade reagent (ThermoFisher Scientific). Slides were imaged on a Leica SP8 laser scanning confocal microscope with a 63X objective. Antibodies used were anti-phosphoFAK (Y397, Invitrogen #700255), anti-E-cadherin (BD Biosciences, #610181), anti-N-cadherin (BD Biosciences, #610920), anti-vimentin (Abcam, ab92547), anti-β4 integrin (ThermoFisher, 439–9B), Alexa Fluor 488 goat anti-mouse IgG2a (#A21131), Alex Fluor 488 goat anti-mouse IgG2b (#A21141), Alexa Fluor 647 goat anti-mouse IgG1 (#A21240), and Alexa Fluor 647 goat anti-rabbit (#A21244).
Image Analysis
A semi-automated image processing pipeline was created as an ImageJ macro to determine cluster roundness. The phalloidin signal intensity was used to segment cell clusters from background, and the Particle Analysis feature was used to outline clusters. The roundness metric available in ImageJ was used without modification. Invasive clusters were identified manually, based on the presence of sharp protrusions emanating from the clusters. The area of MCF7 and MDA-MB-231 clusters was determined using the metric available in ImageJ Particle Analysis feature.
Nuclear curvature was analyzed using Bitplane Imaris software with the Lumen Curvature extension. Z-stacks of DAPI-stained nuclei were imported into Imaris and a 3D volume was rendered. The curvature extension determines the mean curvature for every vertex on the surface. Curvature magnitudes were plotted as color-coded spheres on the surface of the 3D nucleus rendering.
TEM Preparation, Imaging, Quantification
Hydrogel matrices for transmission electron microscopy were fixed in 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4. The samples were embedded in resin, sectioned, mounted on TEM grids and stained with uranyl acetate. The samples were imaged at 2,000X and 10,000X with a JEOL JEM1400 transmission electron microscope.
To quantify lamina-associated chromatin thickness, raw image intensity histograms were normalized and equalized, then segmented. Seg3D software was used to generate connected components, representing the entirety of the lamina-associated chromatin. These masks were dilated and eroded, and then boundaries were identified. A MATLAB script was used to measure chromatin thickness for each inner boundary pixel, defined as the shortest distance from the inner boundary to the outer boundary.
Western Blotting
Cells were extracted from matrices by incubating gels in ice cold 50 mM EDTA in PBS for 10 minutes with pipette mixing. The suspensions were centrifuged at 500 g at 4°C for 10 mins to pellet cells. The pellets were incubated in 1 ml of 0.25% trypsin/2.21 mM EDTA at 37°C for 5 minutes to digest any remaining rBM, then centrifuged at 500 g for 10 mins again. Pellets were lyzed in RIPA lysis buffer with protease and phosphatase inhibitors (ThermoFisher Scientific) and the protein concentration was determined using the BCA assay. Samples were diluted in Laemmli Sample Buffer (Bio-Rad) to 2.5 μg/μl and 25 μg was loaded in 4–20%, 15 well gels. The gels were run for 35 minutes and the protein was then transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked in 5% non-fat milk for 1 hour then incubated with primary antibody overnight. A fluorescent secondary antibody against the primary antibody was then incubated for 1 hour. The blots were imaged using a Licor Odyssey imaging system. The primary antibodies used were anti-Sp1 (Millipore, 07–645), anti-Sp1 (phospho T453, Abcam, ab37707), anti-H3K4me3 (Abcam, ab1012), anti-H3K9me3 (Abcam, ab8898), anti-AcH3 (Millipore, 06–599), anti-AcH4 (Millipore, 06–866), anti-HP1γ (Santa Cruz, sc-398562), anti-histone 3 (Cell Signaling Technologies, 4499), and anti-GAPDH (Abcam, ab181602). The secondary antibodies used were IRDye 680CW donkey anti-mouse (925–68072) or anti-rabbit (925–68073), and IRDye 800CW donkey anti-mouse (925–32212) or anti-rabbit (925–32213).
Quantitative polymerase chain reaction
Cells were extracted from matrices as described above. TRIzol (ThermoFisher Scientific) reagent was added and the cells were lysed by passing through a 30 G syringe. RNA was isolated by phenol-chloroform extraction and RNA extraction columns (Green Bio). One microgram of RNA was reverse transcribed into cDNA using the High-Capacity Reverse Transcription Kit (Applied Biosystems). Fast SYBR green master mix was used, with primers listed in Supplementary Table 6. Reactions were performed on an Applied Biosystems 7500 instrument.
ATAC-seq library preparation
MCF10A cells were extracted from the alginate matrices via chelation and digestion as described above or trypsinized from 2D TCPS, then processed according to published protocols49. DNA concentration and library quality were assessed with a Qubit fluorometer and a Bioanalyzer before sequencing. The libraries were sequenced on Illumina HiSeq 2500 or 4000 instruments in the Stanford Genome Sequencing Service Center using paired-end 101 bp reads. At least 3 biological replicates were sequenced for each experimental condition. The conditions include soft, stiff, soft with SAHA treatment, stiff with SAHA treatment, soft 2D and 2D TCPS.
TAC-seq analysis pipeline
The ATAC-seq data were QC-ed and processed using a publicly available pipeline that serves as the official specification of the ENCODE consortium (https://github.com/kundajelab/atac_dnase_pipelines version 1.0). Briefly, sequencing adapters were trimmed from the reads. The reads were mapped in paired-end mode to the hg19 reference genome using Bowtie250, and multimapping, duplicate and mitochondrial reads were discarded. For each experimental condition, peaks were called for each replicate, for a pooled sample (by pooling reads from replicates) and for a pair of virtual pseudo-replicates (by randomly splitting the reads from the pooled sample into two pseudo-replicates). Peaks were called using Macs2 with a relaxed p-value threshold of 0.0151. The peaks were ranked by p-value and only the top 300,000 peaks were retained. Irreproducible discovery analysis (IDR) was executed to determine high-confidence peak sets for all ATAC-seq samples. The IDR optimal set for all ATAC-seq samples was used for analysis. A consensus peak set for soft vs stiff analysis was generated by running bedtools merge on the IDR optimal peak set from the soft, stiff, stiff + SAHA, and soft + SAHA samples. The pipeline also produces several QC metrics for each experimental replicate (Supplementary Table 1).
A count matrix for the soft/stiff/SAHA samples was generated by computing the filtered read coverage for each replicate on the merged IDR peak set. This was done by running the bedtools coverage command on the filtered tagAlign files generated by the ENCODE analysis pipeline with duplicates, blacklisted regions, and mitochondrial reads removed (*nodup.tn5.no_chrM.25M.R1.tagAlign.gz).
Differential Accessibility Analysis for Soft, Stiff, and SAHA-treated Samples
The soft/stiff/SAHA count matrix served as the input for differential accessibility analysis via the DESeq2 analysis52. For DESeq2 analysis, a design matrix of Read Count ~ Stiffness+Stiffness:SAHA was used. Surrogate variable analysis was first performed with this design on the count matrix to determine batch effects. This was done with the svaseq function from the R sva library and one significant surrogate variable was identified and added to the model design. Size factor correction was performed using a set of housekeeping genes from https://www.tau.ac.il/~elieis/HKG/ whose promoters intersect with the set of IDR optimal peaks. After the custom size factor correction, DESeq2 analysis was performed and differential regions were identified using an FDR threshold of 0.05.
Normalized counts from the DESeq2 matrix were corrected for batch effects using the limma removeBatchEffect function. An rlogTransform of the normalized, filtered counts was then computed. The rlog data was used as the input for principal component analysis (PCA) and heatmap visualization. The row z-scores across soft, stiff, and SAHA treated samples were used for heatmap visualization.
It was determined that the DESeq2 model of soft and stiff samples was underpowered to detect the significantly differential peaks between the two conditions. Correcting for surrogate variables further reduced the power of the model. However, the rlog transformed, normalized reads from the DESeq2 object indicated a clear soft versus stiff difference on the PCA. Consequently, the R loadings function was used to compute the contribution of each IDR peak to PC1 in a PCA of soft/stiff samples. The distribution of peak loadings for PC1 was computed, and those peaks with a loading value greater than two standard deviations above the mean were considered differential. This set of peaks underwent further analysis with MEME-ChIP53 and HOMER54.
Row-z scores from the stiff/soft/SAHA heatmap were analyzed to determine the subset of differential peaks whose accessibility reverted to soft-like levels after treatment with SAHA. Motif analysis on this subset of peaks was performed using MEME-ChIP53 and HOMER54. For MEME-ChIP analysis, a FASTA file of the differentially accessible regions was used to search against a background of all other shared IDR optimal set regions. Similarly, the HOMER command findMotifsGenome.pl was run on a BED file of differentially accessible regions with the same background described above. Motifs discovered de novo were compared to known motifs to find the best matches.
Aggregate ATAC-seq footprint profiles at transcription factor motifs
The HOMER command scanMotifsGenome.pl was used to scan for Sp1 motif occurrences in differentially accessible regions (foreground) or consensus regions (background). The resulting motif coordinates were padded to 200 bases and intersected with the bed file of differential peaks between soft and stiff samples. The tagAlign files for soft and stiff samples from the ENCODE pipeline were split by strand, and the bedtools coverage command was used to compute the average number of 5’-end cuts and 3’-end cuts at each position in +/− 100 bp window centered on the motif.
Pharmacological Inhibition
For all small molecule inhibition studies, the drug was added on the day of encapsulation and replaced with each subsequent media change. The inhibitors used were GSK 126 (100 nM, Fisher Scientific), UNC 0638 (250 nM, Sigma), JIB-04 (1 μM, Sigma), GSK-J4 (10 μM, Abcam), GSK-LSD1 (100 nM, Sigma), Sirtinol (50 μM, Sigma), apicidin (1 μM, Sigma), mithramycin A (50 nM, Sigma), LY294002 (20 μM, Sigma) and SAHA (1 μM, Sigma). All drugs were dissolved in DMSO and diluted in basal medium before adding to the culture medium. DMSO alone was added to the culture medium as a vehicle control.
RNA interference
MISSION lentiviral transduction particles were purchased from Sigma and used according to the manufacturer’s instructions. Clone ID’s for the lentiviral particles were TRCN0000020445 (Sp1), TRCN0000004814 (HDAC1), TRCN0000004819 (HDAC2), TRCN0000194993 (HDAC3), TRCN0000004851 (HDAC8) and pLKO.1-puro Control Transduction Particles were used as empty vector controls. MCF10A cells were plated on 24 well plates and cultured until 80% confluency was reached. Hexadimethrine bromide (8 μg/ml, Sigma) and 2 × 107 transducing units (TU) per ml were added to the cells and incubated overnight. The transduction medium was removed and replaced with growth medium for 24 hours. Then puromycin-containing medium (1 μg/ml) was used to select for transduced cells. Puromycin-containing medium was used for expansion, 2D validation studies, and 3D encapsulations.
Protein-protein interaction screening
String-DB was used to search for proteins interacting with Sp155. The search was limited to experimental evidence only, gathered from protein-protein interaction databases, with a minimum interaction score of 0.4.
TRRUST Pathway Analysis
The Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) version 2 web platform was used to assess Sp1 target genes and pathways associated with them. A subset of Sp1 target genes that overlapped with genes associated with malignant neoplasm of breast were selected for gene expression analysis. The top 15 enriched diseases or pathways were included in Supplementary Tables 4 and 5.
Comparison to Mammary Epithelium
Publicly available ATAC-seq data from human mammary epithelium was obtained from the ENCODE database for two patients (ENCSR846ZBX, ENCSR65UYP). FASTQ files were processed through the same pipeline described above. A consensus peak set for the culture-specific analysis was generated by running bedtools merge on the optimal IDR peak sets for the mammary epithelium samples, the 2D TCPS samples, the 2D soft samples, and the 3D soft samples.
The DESeq2 analysis pipeline, as described above, was applied to the merged IDR optimal peak set for mammary epithelium, 2D TCPS, 2D soft, and 3D soft samples. Surrogate variable analysis was performed to identify sources of variation not captured by the DESeq2 model:
Read Count ~ Sample. Size factor correction was performed using the set of housekeeping genes, as described above. Differential peaks were determined with an FDR threshold of 0.01 and a log2 fold change of 1.
IDR peaks that overlapped between mammary epithelium and the three cultures (and were not found to be differential by the DESeq2 analysis above) underwent comparative analysis. Comparisons were performed using principal components analysis. The histology image was obtained from the Human Protein Atlas: https://www.proteinatlas.org/ENSG00000075624-ACTB/tissue/breast#img.
Gene and Genome Ontology
The HOMER54 command annotatePeaks.pl with the gene ontology flag was run on regions differentially accessible between 3D soft and 3D stiff matrices, and on the regions shared by two of mammary epithelium and one of 2D TCPS, 2D soft, or 3D soft. The genome ontology program was run on the naïve overlap peak set for 3D soft and 3D stiff matrices to evaluate differences in accessibility of genomic features.
Statistical Analysis
Statistical comparisons were performed with GraphPad Prism 7.0 software using the tests described in the figure captions. p-values less than 0.05 were considered statistically significant. Significant differences in gene expression were determined by setting the false discovery rate to 5% and used the two-stage step-up method of Benjamini, Krieger, and Yekutieli. Default significance thresholds were used for motif analyses, gene and genome ontologies, and protein-protein interactions.
Supplementary Material
Acknowledgements
The authors acknowledge helpful discussions with the Chaudhuri lab, Connie Hsueh for assistance with nuclear curvature analysis, Marianne Black for assistance with chromatin thickness analysis, the Bollyky lab for use of the Licor imager, the Stanford Genome Sequencing Service Center for sequencing (NIH S10OD020141), and the Stanford Cell Science Imaging Facility for assistance with confocal image analysis and TEM preparation and imaging (NIH 1S10RR026780–01). This work was supported by an NIH F32 fellowship to R.S. (F32CA210431), and an American Cancer Society grant (RSG-16-028-01) and NIH grant (R37-CA214136) to O.C.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reporting summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the main data supporting the results in this study are available within the paper and its Supplementary Information. ATAC-seq data generated in this study are available through the Gene Expression Omnibus under accession code GSE131968. Additional datasets are available from the corresponding author upon reasonable request.
Code availability
Custom analysis pipelines are available at https://github.com/kundajelab.
References
- 1.Levental KR et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Provenzano PP, Inman DR, Eliceiri KW & Keely PJ Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 28, 4326–4343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choquet D, Felsenfeld DP & Sheetz MP Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 88, 39–48 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Paszek MJ et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005). [DOI] [PubMed] [Google Scholar]
- 5.del Rio A et al. Stretching single talin rod molecules activates vinculin binding. Science 323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elosegui-Artola A et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Aragona M et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri O et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater 13, 970–978 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Uhler C & Shivashankar GV Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat Rev Mol Cell Biol 18, 717–727 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Maniatis T, Goodbourn S & Fischer JA Regulation of inducible and tissue-specific gene expression. Science 236, 1237–1245 (1987). [DOI] [PubMed] [Google Scholar]
- 11.Pique-Regi R et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21, 447–455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides T Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Swift J et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 341, 1240104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain N, Iyer KV, Kumar A & Shivashankar GV Cell geometric constraints induce modular gene-expression patterns via redistibution of HDAC3 regulated by actomyosin contractility. Proceedings of the National Academy of Sciences of the United States of America 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Tang CB & Kilian KA Matrix Mechanics Influence Fibroblast–Myofibroblast Transition by Directing the Localization of Histone Deacetylase 4. Cellular and Molecular Bioengineering 10, 405–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flavahan WA, Gaskell E & Bernstein BE Epigenetic plasticity and the hallmarks of cancer. Science 357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald OG, Wu H, Timp W, Doi A & Feinberg AP Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 18, 867–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajik A et al. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 15, 1287–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo M et al. MLL5 Orchestrates a Cancer Self-Renewal State by Repressing the Histone Variant H3.3 and Globally Reorganizing Chromatin. Cancer Cell 28, 715–729 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi JY et al. HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics 10, 418–430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rafehi H et al. Systems approach to the pharmacological actions of HDAC inhibitors reveals EP300 activities and convergent mechanisms of regulation in diabetes. Epigenetics 12, 991–1003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fork C et al. Epigenetic control of microsomal prostaglandin E synthase-1 by HDAC-mediated recruitment of p300. J Lipid Res 58, 386–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafehi H et al. Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res 24, 1271–1284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafehi H & El-Osta A HDAC Inhibition in Vascular Endothelial Cells Regulates the Expression of ncRNAs. Noncoding RNA 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen F et al. Inhibition of histone deacetylase reduces transcription of NADPH oxidases and ROS production and ameliorates pulmonary arterial hypertension. Free Radic Biol Med 99, 167–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu K et al. Chromatin Accessibility Landscape of Cutaneous T Cell Lymphoma and Dynamic Response to HDAC Inhibitors. Cancer Cell 32, 27–41 e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaczynski J, Cook T & Urrutia R Sp1- and Kruppel-like transcription factors. Genome Biology 4, 1–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han H et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46, D380–D386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stowers RS et al. Extracellular Matrix Stiffening Induces a Malignant Phenotypic Transition in Breast Epithelial Cells. Cellular and Molecular Bioengineering 10, 114–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liao M & Dufau ML Phosphatidylinositol 3-kinase/protein kinase Czeta-induced phosphorylation of Sp1 and p107 repressor release have a critical role in histone deacetylase inhibitor-mediated derepression of transcription of the luteinizing hormone receptor gene. Mol Cell Biol 26, 6748–6761 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu MC, Chang HC & Hung WC HER-2/neu represses the metastasis suppressor RECK via ERK and Sp transcription factors to promote cell invasion. J Biol Chem 281, 4718–4725 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Doetzlhofer A et al. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 19, 5504–5511 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 17, 333–347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen OW, Ronnov-Jessen L, Howlett AR & Bissell MJ Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A 89, 9064–9068 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Heo SJ et al. Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Sci Rep 5, 16895 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain N, Iyer KV, Kumar A & Shivashankar GV Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demmerle J, Koch AJ & Holaska JM The nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J Biol Chem 287, 22080–22088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le HQ et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 18, 864–875 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Xu R et al. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J Cell Biol 184, 57–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Beyec J et al. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp Cell Res 313, 3066–3075 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraga MF et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37, 391–400 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Bibikova M et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res 16, 1075–1083 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wisdom K & Chaudhuri O 3D Cell Culture in Interpenetrating Networks of Alginate and rBM Matrix. Methods Mol Biol 1612, 29–37 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Thielicke W & Stamhuis EJ Towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw 2 (2014). [Google Scholar]
- 48.Debnath J, Muthuswamy SK & Brugge JS Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machanick P & Bailey TL MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.