Abstract
Lipopolysaccharides (LPS) provide the outer membrane (OM) of Gram-negative bacteria with a strong protective barrier. The periplasm-spanning Lpt machinery is responsible for the transport of LPS molecules across the periplasm, culminating in insertion by the outer-membrane proteins LptD and LptE. In order to elucidate the mechanisms of LPS insertion by LptDE, we performed over 14 microseconds of equilibrium molecular dynamics simulations. Bilayer-dependent differences in the fluctuations and secondary structure of LptD’s extracellular loops are observed for a pure DMPE membrane vs. a model of the OM. Furthermore, LptD’s periplasmic N-terminal domain is highly dynamic, which may help to maintain the integrity of the periplasm-spanning complex amidst relative motion of the inner-membrane and outer-membrane anchored domains. In addition, our simulations demonstrate that binding of LPS substrate activates a switching between the associated and dissociated states of two lumenal loops at the interface between the β-barrel and the N-terminal domain as well as LptD’s lateral gate on the microsecond timescale, neither of which is observed for the apo state. Placement of a substrate LPS molecule also causes an increase in the average separation of the LptD lateral gate strands and a lowering of the energetic barrier to lateral gate opening.
Graphical abstract
Introduction
Gram-negative bacteria are characterized by their unique cell-envelope structure, which contains two membranes. The inner membrane is a symmetric phospholipid bilayer and the outer membrane consists of a phospholipid inner leaflet and an outer leaflet composed of lipopolysaccharide (LPS) molecules [1, 2]. The space between the membranes, called the periplasm, is host to a large number of proteins as well as the peptidoglycan cell wall [3]. LPS molecules possess a zwitterionic portion called lipid A consisting of four to eight acyl tails, a core oligosaccharide region of phosphate groups and sugars, and a long O-antigen sugar chain whose composition varies considerably between species [4–6]. These LPS molecules form an external barrier for Gram-negative bacteria, protecting them from environmental antagonists such as immunity factors and antibiotics [7, 8].
The assembly of LPS molecules begins at the cytosolic side of the inner membrane where biosynthesis of lipid A is first performed by the Lpx protein family and core oligosaccharide is added by Waa proteins [4, 9, 10]. The partially assembled LPS is flipped across the inner membrane by the ABC transporter MsbA, joined with an O-antigen chain, and finally transported across the periplasm by the LPS transport (Lpt) machinery. The Lpt machinery is a complex consisting of LptA–G [11], all of which are essential [12]. At the inner membrane, LptB2FG is an ABC transporter that hydrolyzes ATP to transfer LPS from the inner membrane to LptC [13, 14]. LptA, LptC, LptF, LptG, and the N-terminal domain of LptD each possess a β-jellyroll fold with a hydrophobic interior [15–18]. LptF, LptC, one or more repeats of LptA, and the N-terminal domain of LptD link together to span the periplasmic space, sequestering the aliphatic tails of lipid A within their hydrophobic groove [19, 20]. Finally, LptDE provides LPS with a pathway to the outer membrane by separating its N- and C-terminal β-strands [21, 22].
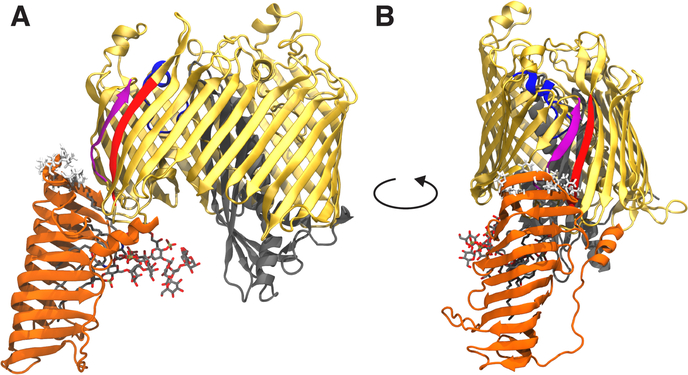
Crystal structures of LptD from Shigella flexneri (PDB ID: 4Q35) and Salmonella typhimurium (PDB ID: 4N4R), first released in 2014, exhibit a 26-stranded β-barrel domain and an N-terminal β-jellyroll domain [16, 21] (Fig. 1). At the proximal tip of the N-terminal domain are several hydrophobic residues in a region predicted to embed into the outer membrane (Fig. 1, shown in white sticks). This embedded region is the putative LPS insertion site, which, coincidentally, is also near the β-barrel seam between β1 and β26. Simulations of LptDE from S. typhimurium at a high negative pressure by Dong et al. [21] showed a separation between β1 and β26, which was posited to act as the gate through which LPS would pass. Through incorporation of the photocrosslinking amino acid pBPA, LPS was later shown to crosslink to several residues in the β1–β26 seam, giving further evidence that these strands separate in order to allow LPS passage [22]. However, many uncertainties surrounding the molecular details of this insertion process remain. For instance, it is currently unknown what initiates the insertion cascade.
Figure 1:
Front (A) and side (B) views of LptDE with an LPS molecule bound in the N-terminal β-jellyroll domain. The structure is taken from the Sf-LPS simulation at approximately 1900 ns into the simulation. The LptD β-barrel domain is shown in gold with the β1 strand in purple, β26 strand in red, extracellular loop L4 in blue and N-terminal domain in orange. LptE is shown in gray. The LPS molecule is shown in sticks with C, O, N, and P atoms in gray, red, blue, and tan, respectively. Hydrophobic residues predicted to embed into the outer membrane (based on Orientations of Proteins in Membranes database [23]) are shown in white sticks. The main dynamical features of LptDE observed in the equilibrium simulations were a large change in the angle of the N-terminal domain, a separation of the LptD β-barrel seam strands induced by the presence of the substrate, and a significant movement of L4 toward the periplasm.
In order to better clarify the insertion process, we have performed over 14 μs of equilibrium simulations of two LptDE crystal structures from Shigella flexneri and Pseudomonas aeruginosa (PDB IDs 4Q35 and 5IVA, respectively). Simulations of outer-membrane (OM) proteins are becoming increasingly common, especially in asymmetric membranes containing LPS [24, 25]. Here, we simulated LptDE in both a symmetric phospholipid bilayer (DMPE) and an outer membrane (OM) model. We chose to use DMPE in addition to OM because it has been used in previous simulation studies as an outer membrane mimic [21, 26–31] and affords a greater degree of flexibility for the study of LptD β-barrel seam dynamics. Our simulations have revealed several conformational changes that may be vital to the function of LptDE and act as prerequisites for lateral gate opening and substrate insertion, some of which are dependent on the presence of substrate. Additionally, free-energy calculations display a significant reduction in the barrier to lateral-gate opening when an LPS molecule is placed in the N-terminal domain near it.
Methods
System construction
Two models of LptDE were generated for S. flexneri (SfLptDE) using its crystal structure (PDB ID: 4Q35), which includes the full N-terminal β-jellyroll domain. One system (Sf-LPS; see Table 1 for nomenclature) included a substrate LPS molecule bound in its N-terminal domain and another did not (Sf-apo). In order to generate the LPS-bound state, an LPS molecule was placed with its lipid A acyl tails near the hydrophobic interior of the N-terminal domain. Both systems were embedded in a DMPE bilayer. DMPE has been used as a more pliable outer membrane mimic in several previous simulation studies [21, 26–31]. These models were ionized with KCl to a concentration of 150 mM (96 K+ and 73 Cl− ions) and contained ~120,000 atoms in total, including ~26,000 water molecules. An additional 11 K+ were added to the Sf-LPS system as counterions for the substrate LPS molecule. The membrane included 145 DMPE molecules in the outer leaflet and 122 DMPE molecules in the inner leaflet. Fewer DMPE molecules were required in the inner leaflet due to the partial protrusion of the N-terminal domain.
Table 1:
Summary of simulations performed. Names starting with “Sf” pertain to the S. flexneri structure in PDB ID 4Q35 and “Pa” to the P. aeruginosa structure in PDB ID 5IVA. SfLptDE includes the β-barrel and N-terminal domains, while PaLptDE includes only the β-barrel domain. Sf systems were either simulated with (Sf-LPS) or without (Sf-apo) an LPS molecule bound in the N-terminal domain of LptD. Pa systems were simulated in either a symmetric membrane environment (DMPE) or an asymmetric outer membrane model (OM) with two replicas run for each. All simulations were performed at a temperature of 310 K.
| name | membrane | PDB | length (ns) |
|---|---|---|---|
| Sf-apo | DMPE | 4Q35 | 4040 |
| Sf-LPS | DMPE | 4Q35 | 4080 |
| Pa-DMPE-1 | DMPE | 5IVA | 1540 |
| Pa-DMPE-2 | DMPE | 5IVA | 1550 |
| Pa-OM-1 | OM | 5IVA | 1530 |
| Pa-OM-2 | OM | 5IVA | 1730 |
Two models of LptDE from P. aeruginosa (PaLptDE) were also generated by embedding its crystal structure (PDB ID: 5IVA) into either a symmetric bilayer (DMPE) or an Escherichia coli outer membrane (OM) model used in a previous study [32]. Two replicas of each of these systems were simulated. Pa-DMPE-1 and −2 contained ~129,000 atoms with ~29,000 water molecules and 150 mM KCl (102 K+ and 83 Cl− ions). The membrane included 137 DMPE molecules in the outer leaflet and 132 DMPE molecules in the inner leaflet. Pa-OM-1 and −2 contained ~129,000 atoms with ~25,000 water molecules and 150 mM NaCl (74 Na+ and 76 Cl− ions). The membrane included 44 LPS molecules (E. coli K-12 RaLPS chemotype) in the outer leaflet and 135 POPE molecules in the inner leaflet. To neutralize and stabilize the highly charged regions of LPS, 260 divalent cations were added (255 Mg2+ and 5 Ca2+). Preparation of the systems was carried out using VMD [33].
MD simulations
Nearly all simulations of LptDE systems (see Table 1) were carried out on the Anton-1 supercomputer hosted at the Pittsburgh Supercomputing Center [34]. Some simulations were extended using Amber on GPUs [35]. All simulations used the CHARMM36 force fields for proteins [36] and lipids [37]. A 2-fs time step was used on Anton and long-range electrostatic interactions were evaluated every three time steps. For Amber runs, a 4-fs time step was used with hydrogen mass repartitioning [38], which has been demonstrated to be valid for membrane and membrane-protein simulations [39]. A temperature of 310 K was maintained using a Nosé-Hoover thermostat [40, 41] on Anton and Langevin dynamics in Amber. A pressure of 1 atm was maintained using the semi-isotropic Martyna-Tuckerman-Klein barostat [42] on Anton and the Monte Carlo barostat in Amber. Lennard-Jones interactions were truncated at 11–13 Å on Anton and at 12 Å in Amber with a force-based switching function applied starting at 10 Å. Long-range electrostatics were evaluated using the k-Gaussian Split Ewald method [43] on Anton and the particle-mesh Ewald method [44] in Amber.
Free-energy calculations
Free-energy calculations of lateral gate opening for Sf-apo and Sf-LPS were performed using replica-exchange umbrella sampling (REUS) [45, 46]. To obtain the open states, steered molecular dynamics (SMD) was employed on the distance between pairs of closest Cα atoms in strand β1 (residues 228 to 237) and β26 (residues 750 to 759) at the LptD β-barrel seam. SMD was employed on the pairwise distance collective variables to drive them each to 25 Å over the course of 40 ns. Umbrella sampling windows were centered at center-of-mass distance values between 6.0 and 15.5 Å. The first 9 of 25 windows were placed at 6.0, 6.25, 6.375, 6.5, 6.625, 6.75, 7.0, 7.25, and 7.5 Å with force constants of 10.0, 20.0, 20.0, 40.0, 20.0 20.0 20.0, and 20.0 kcal/mol·Å2, respectively. The remaining 16 windows were spaced 0.5 Å apart from 8.0 Å to 15.5 Å each having a force constant of 5.0 kcal/mol·Å2. Higher force constants with smaller spacing between windows were used to ensure overlap of windows at low stand separation. Each window was simulated for 20.0 ns (500 ns per system). PMFs were calculated from the REUS sampling data using the Weighted Histogram Analysis Method (WHAM) [47].
Results
N-terminal domain flexibility
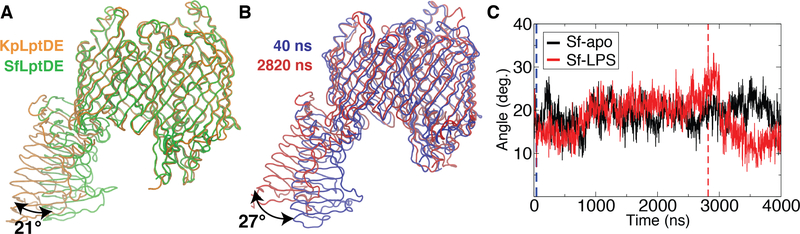
The current model of the Lpt complex assembly is that LptB2FG in the inner membrane is connected to LptDE at the outer membrane by a transperiplasmic bridge formed by the β-jellyroll domains of LptC, LptA, and the N-terminal domain of LptD [16, 19, 48]. To date, two crystal structures of LptDE that include the full N-terminal domain have been published: KpLptDE (PDB ID: 5IV9) [48] and SfLptDE (PDB ID: 4Q35) [16]. An alignment of the β-barrels of these structures reveals that the N-terminal domain is shifted by about 23 Å at the distal end, a rotation of 21° about its connection to the β-barrel (Fig. 2A). While attributed to an inherent flexibility of the domain [48], it remained possible that the shift was due to the different species used or to crystal packing effects. A high level of flexibility has been previously suggested to be an important feature to help maintain the integrity of the periplasm-spanning Lpt complex by withstanding relative motions of its inner and outer membrane components [48].
Figure 2:
(A) Alignment of SfLptDE (PDB ID: 4Q35) and KpLptDE (from Klebsiella pneumoniae; PDB ID: 5IV9) crystal structures, demonstrating the difference between their N-terminal domain positions. (B) Snapshots at 40 ns and 2820 ns illustrating a similar range of N-terminal-domain motion observed in the Sf-LPS simulation. (C) Angle between the membrane normal and the principal axis of the LptD N-terminal domain vs. time for SfLptDE with LPS bound (Sf-LPS, red) and Sf without LPS bound (Sf-apo, black). For reference, location of snapshots used in (B) are highlighted in blue and red dotted lines.
In order to address whether the difference seen in the N-terminal domain positions in crystal structures is reflected in the dynamics of this domain, we carried out over 8 μs of equilibrium simulation of the full-length SfLptDE crystal structure. The trajectories of SfLptDE demonstrated considerable dynamics of the N-terminal domain in both the Sf-apo and Sf-LPS simulations resulting in a maximal displacement of ~22 Å at the distal end (27°; Figs. 2B and C). This displacement is similar to the difference observed in the crystal structures (Fig. 2A), indicating that the N-terminal domain is inherently flexible about a hinge between it and the β-barrel domain.
Extracellular loop structure and dynamics
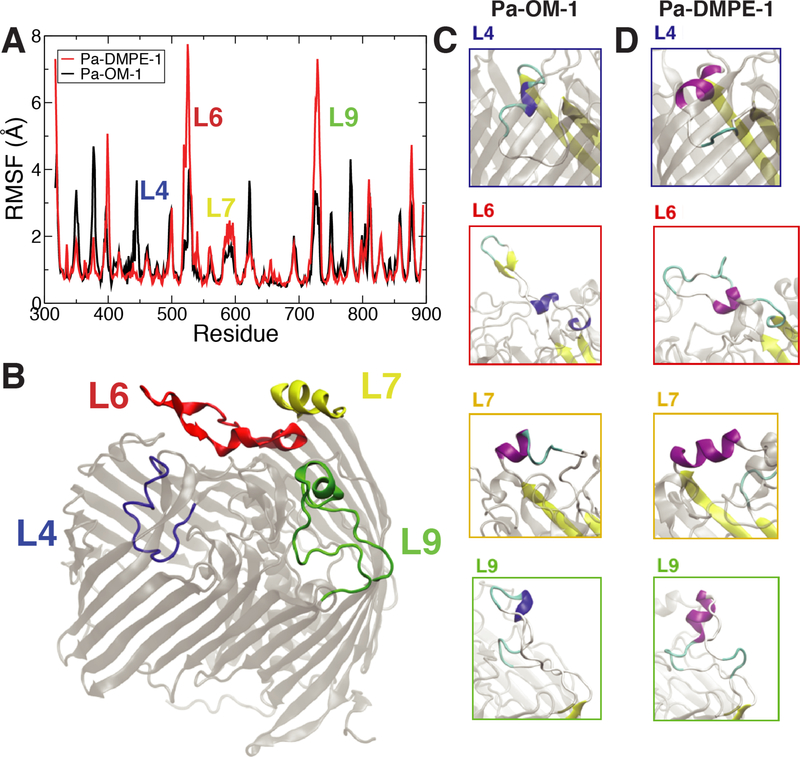
Of all the solved crystal structures, that of PaLptDE diverges the most from the others with less than 28% sequence identity compared to 65% to 91% between the others (YpLptDE from Yersinia pestis, KpLptDE, SfLptDE, and StLptDE from Salmonella typhimurium) [48]. The majority of unique structural features of PaLptDE lie in its extracellular loops. For example, a 23-residue insertion in L6 folds back into the barrel lumen, while an 11-residue insertion in L9 adopts a helical structure. L11 also adopts a unique β-turn motif. However, it remains uncertain how stable the conformations of the loops are in the OM environment. The OM creates a notably different electrostatic profile compared to a symmetric phospholipid membrane [25], which is likely to affect the protein’s structure, in addition to direct interactions between the protein and the large LPS molecules. Such differences in the behavior of proteins in symmetric membranes compared to LPS-containing membranes have been explored extensively in a number of studies [32, 49–52].
To determine the effects of the OM on PaLptDE’s extracellular loops, we performed over 6 μs of equilibrium simulations of PaLptDE in symmetric (DMPE) and asymmetric (OM) membrane models. In these simulations we observed multiple conformational differences (Fig. 3). The root mean-square fluctuations (RMSF), a measure of each residue’s fluctuation about an average position, were higher overall in the DMPE membrane compared to the OM, with the largest differences observed in L6 and L9. Secondary structure differences were also found in extracellular loops L4, L6, L7, and L9 (Figs. 3B and C). These differences were persistent, with loops forming more secondary structure in DMPE than in OM, akin to what was observed for BtuB in the same membranes [32]. For example, a 310-helix formed in L4 in OM while a longer α-helix formed in the same region in DMPE; similarly, L7 forms a longer helix in DMPE than in the OM. L6, which initially points into the barrel, is pulled out through interactions with LPS in the OM model. The helical insert in L9 also partially unravels in the OM. The β-turn in L11 remains in both the OM and DMPE environments. Some of these differences are less pronounced in the second runs of each system, shown in Figs. S2 and S3. While the functional relevance of most of the observed structural changes is not readily apparent, they do indicate the importance of considering the native environment when interpreting results from both simulations and experiment.
Figure 3:
PaLptDE structure and dynamics for the first runs in DMPE and in OM. (A) Root-mean-square fluctuation (RMSF). Higher RMSF is seen for the DMPE system compared to the OM system for a number of extracellular loops, especially L6 and L9. (B) Location of key loops with respect to the overall structure. (C, D) Prominent secondary structure differences in L4 (blue), L6 (red), L7 (yellow) and L9 (green). Timeline of secondary structure is shown in Fig. S1. RMSF and timeline for the second runs are in Fig. S2 and S3, respectively.
Because of its high charge, LPS condenses a large number of divalent ions on its surface [8]. We found in both Pa-OM simulations that 15–16 divalent ions on average were within 4 Å of the proteins, most of them near the extracellular loops. These ions bridge interactions between LPS and the loops, stabilizing them. We also observe three clusters of negatively charged or polar residues binding an ion stably over both Pa-OM simulations. The first is formed by residues D335 in L1 with D871 and D885 in L13, although only the last residue is conserved [48]. The second cluster is formed by D616, T629, and E631 betweeen β14 and 15 on the interior of the barrel near the periplasmic side; all three are highly conserved. Finally, the third cluster is formed by E719, D733, and D735, all on L9, although only the latter two are weakly conserved.
Due to its position over the barrel lumen, a movement of L4 has been suggested to be necessary for passage of the LPS oligosaccharide region [53]. Two residues in LptE (R91 and K136) shown to be important for binding of LPS are near LptD’s L4 [54], and several well-known OM assembly defect alleles, including LptD4213, contain deletions in L4, indicating that this loop is important for proper delivery of LPS [55, 56]. In the Pa-OM-1 simulation, we observe L4 to move deeper into the barrel. This movement is precipitated by the unwinding of a small helix, which is still partially retained in Pa-OM-2 by the end of the 1.5-μs simulation and fully retained in both Pa-DMPE-1 and Pa-DMPE-2 (Figs. 3 and S1–S3). In Sf-apo, on the other hand, L4 moves toward the extracellular space (Sf-apo). The difference between the maximum and minimum positions is ~15 Å (Fig. 4; see also Fig. S4). This movement may enhance interactions between the oligosaccharide and O-antigen regions of LPS and L4 at an early stage of insertion.
Figure 4:
Distance from center of membrane vs. time of L4 for (A) Pa systems (residues 436 to 448) and (B) Sf systems (residues 341 to 353). An increase in distance indicates movement towards the extracellular space (such as Sf-apo) and a decrease indicates movement toward the periplasm (such as Pa-OM-1). The center of the membrane is defined here using the reference point predicted by the OPM (Orientations of Proteins in Membrane) server [23].
Conversely, we also examined the effects of LptD on the OM. Specifically, we compared the order parameters of the LPS acyl tails within 12 Å of LptD’s lateral seam with those from all LPS. We find a nearly consistent decrease in order for those LPS tails near the seam (Fig. S5), similar to what was previously observed for the lateral gate of BamA [29]. This disorder of LPS tails near the gate may make it easier for another LPS molecule to insert into the membrane from LptD.
Position of the lumenal loops
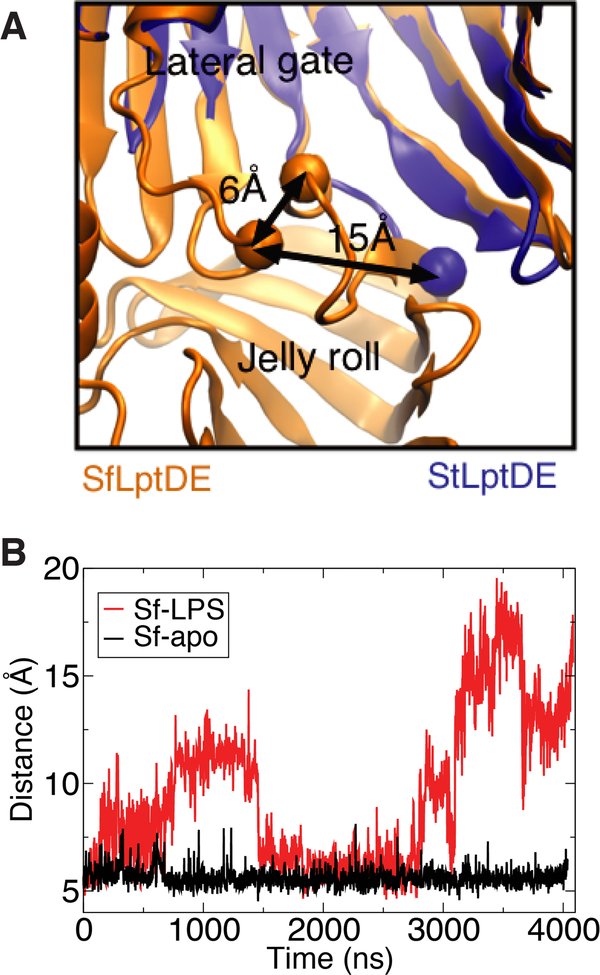
Two so-called “lumenal loops” associate at the interface between the LptD N-terminal domain and lateral seam. Gu et al. previously noted that these loops demonstrate either an open or closed position in crystal structures [57]. They are in a closed position in the S. flexneri (PDB ID: 4Q35) [16] and the K. pneumoniae (PDB ID: 5IV9) [48] structures, which are the only two with the N-terminal domain resolved. For the structures without the N-terminal domain, none of them possess sufficient resolution in both lumenal loops to fully characterize the state, but for the resolved loop, each state is represented. For example, in the truncated K. pneumoniae structure (PDB ID: 5IV8), loop 1 is disordered and loop 2 is in the open state, while in the P. aeruginosa structure (PDB ID: 5IVA), loop 1 is disordered and loop 2 is in the closed state (see Fig. S6 for remaining structures) [48]. Deletion of either of these lumenal loops was shown to be lethal in E. coli as was locking them together with a double cysteine mutation (R225C/S762C) [57].
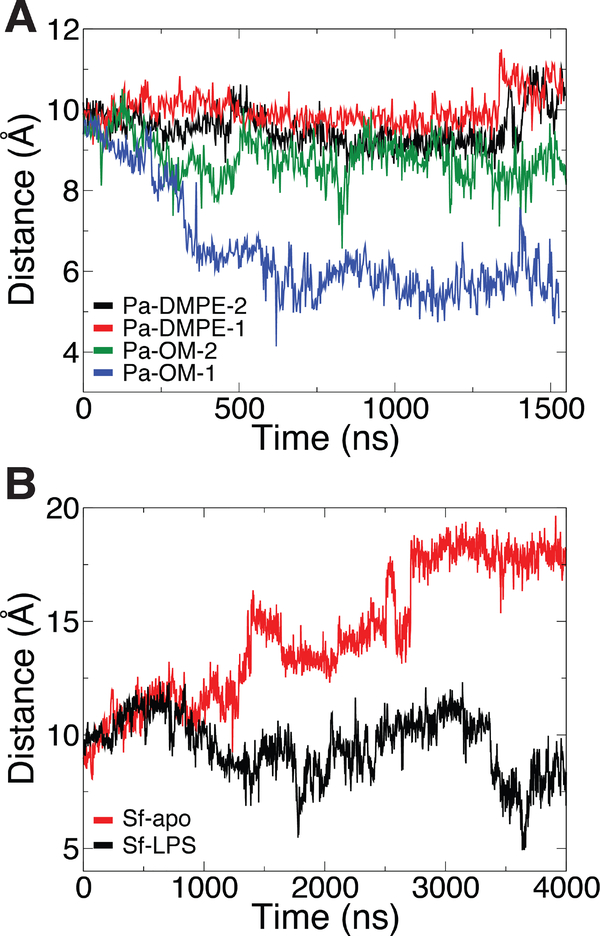
In order to quantify the dynamics of the lumenal loops, we measured the distance between two residues on the loops, R225 and S762, in our simulations of SfLptDE with and without an LPS molecule present in the N-terminal domain (Fig. 5). When no LPS is present in the N-terminal domain, the loops remain firmly in the closed position (distance of ~6 Å). However, when the LPS substrate is present, the loops switch between closed and open states, with the latter, at a loop-loop distance of 10–20 Å, persisting for over 500 ns.
Figure 5:
(A) Alignment of SfLptDE (closed) and StLptDE (open) crystal structures demonstrating the difference in the position of the lumenal loops. (B) Distance between lumenal loops (measured by the Cα atoms of R225 and S762) demonstrating a gate near the barrel seam. Discrete switching occurs between a closed state (6–8 Å) and an open state (10–20 Å).
Gating at the lateral β-barrel seam
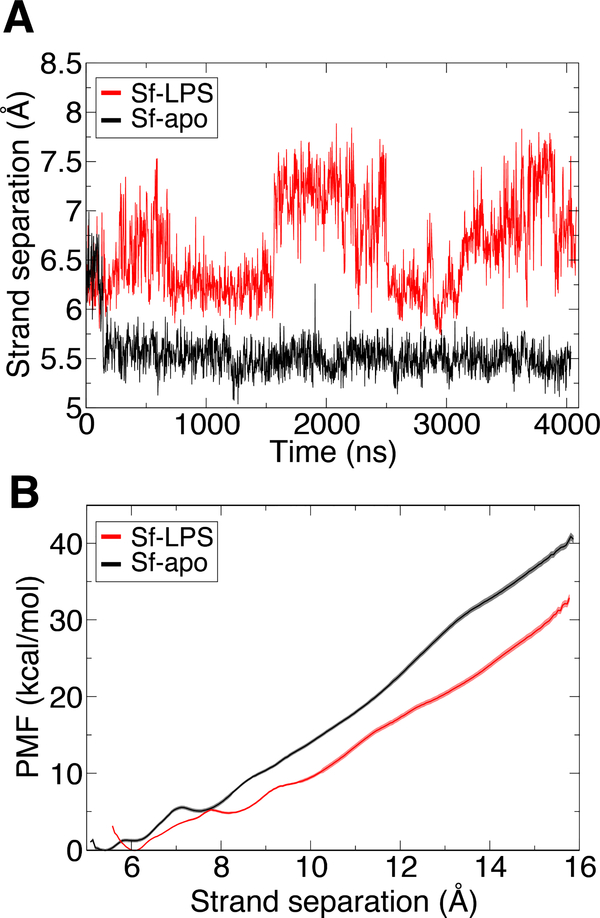
LPS follows a periplasmic bridge along the hydrophobic groove of LptC/A/D [57, 58]. Upon reaching LptD’s β-barrel, the LPS is believed to insert into the OM by breaching the seam formed by the N- and C-terminal β-strands of LptD (β1 and β26, respectively). Both crosslinking experiments and simulations indicate that these strands can separate, forming a lateral gate [21, 22]. Separation of the strands is made easier by the existence of conserved, essential prolines in β1 and β2 [48].
Using the simulations of SfLptDE with and without an LPS molecule present in the N-terminal domain, we measured the separation between β1 and β26 over time. When no LPS molecule is present in the N-terminal domain, the lateral gate remains closed over 3 μs (distance of ~5.5 Å; Fig. 6A); the gate also remains closed in all PaLptD simulations (Fig. S7). However, when an LPS molecule is present, the lateral gate in SfLptD becomes more dynamic, fluctuating from 6 to 7.5 Å separation, suggesting that the presence of a substrate can trigger gate opening. We note that on the 4-μs time scale, the LPS molecule moves only ~4 Å toward the gate (Fig. S8).
Figure 6:
(A) Separation of β1 and β26 strands from SfLptD vs. time. β1 was defined as the Cα atoms of residues 228 to 237 and β26 as those of residues 750 to 759. An increase in strand separation is observed in LptD with LPS bound (red) vs. that without LPS bound (black). (B) Potential of mean force (PMF) vs. strand separation for LPS-bound and apo systems. A significant reduction in energy is found for the bound state (red) with respect to the unbound state (black). Statistical error is shown for each PMF as a light red and grey shaded region for Sf-LPS and Sf-apo, respectively.
To better quantify the impact of an LPS molecule in LptD’s N-terminal domain, we carried out free-energy calculations for gate opening. The cost for opening both with and without an LPS molecule present was relatively large: at least 30 kcal/mol for an increase of 10 Å (Figs. 6B and S9). Notably, the cost was reduced by 10 kcal/mol when the LPS molecule was present, despite it not having any interactions with β1 and β26 directly (Fig. 1). This cost may be further reduced as the LPS molecule comes into contact with the gate strands. Comparison of the PMFs with the equilibrium simulation data in Fig. 6A suggests that the energetic cost of opening to small (~7.5 Å) separations is overestimated. We have also calculated the free energy as a function of separation from the equilibrium simulations (Fig. S10). Although only covering a range of 5–8 Å separation, the key result still holds, namely that the presence of the LPS substrate enhances gating.
Conclusions
The results presented here reveal some of the structural and dynamical features of LptDE that may be required for LPS insertion. Our simulations demonstrate that the N-terminal domain, which delivers LPS to LptD’s β-barrel, possesses a flexible hinge, likely to facilitate cohesion of the transperiplasmic bridge between the inner and outer membranes [20]. Upon presentation of the LPS substrate, there is a separation of the lumenal loops, a necessary step in the insertion process. At the same time, extracellular loop 4 is dynamic within the β-barrel, possibly to guide the oligosaccharide region of LPS through. In simulations of P. aeruginosa LptDE, for which the N-terminal domain was not fully resolved, a number of extracellular loops are stabilized in the presence of an asymmetric outer-membrane model containing LPS in the outer leaflet, although certain secondary structures are sometimes lost (Figs. 3 and S1–S3). Loss of these structures, which were observed in the crystal structure and maintained in simulations in a symmetric DMPE membrane, demonstrate the importance of simulating in a native-like environment [25].
Our free-energy calculations of lateral gate opening indicate that presence of a bound LPS substrate in the N-terminal domain significantly lowers the barrier for lateral gate opening in LptD (by at least 10 kcal/mol), despite not being in direct contact with the gate. However, the cost to open the gate (~30 kcal/mol) still remains higher than could plausibly be achieved, at least by a single inserting LPS molecule. In contrast, BamA and TamA, both of which aid insertion of outer-membrane proteins, require only ~10 kcal/mol to open to the same distance [59, 60]. However, our equilibrium simulations suggest that the free-energy cost with the bound LPS may yet be overestimated (Fig. S10). It remains to be seen if deeper insertion of an LPS molecule into the LptD β-barrel would further reduce the energetic cost.
Although our work focused only on the end of the Lpt pathway, namely insertion of LPS by LptDE, structures of the LptB2FGC inner-membrane complex have recently been determined [17, 18]. LptB2FGC transfers LPS to one or more copies of LptA, which bridge the gap between the inner-membrane complex and LptDE in the OM [20]. With high-resolution structures of all components now determined, modeling and simulation of the full transperiplasmic complex would allow us to test some of the ideas presented here, e.g., that the flexibility of the N-terminal domain of LptD is important for stability of the connection. Furthermore, the energetics of other steps, such as the hand-off of an LPS substrate from each component to the next in the bridge, could be calculated. It is possible that formation of the bridge and/or the presence of multiple LPS substrates could reduce the energetic barrier to lateral gate opening of LptD.
Given its essential role in building the Gram-negative bacterial cell envelope, LptDE presents an attractive target for the development of novel antibiotics [61]. So far, promising lead molecules are either peptides [62] or peptidomimetics [63, 64]. Analysis indicates that these molecules form β-hairpin-like structures and bind to the periplasmic N-terminal domain, possibly interrupting the transperiplasmic bridge [61]. Precise localization of where they bind and how they prevent bridge formation remain to be determined. How this bridge interacts with the cell wall, which lies between the two membranes, is also unknown [3].
Supplementary Material
Highlights.
The Gram-negative bacterial outer membrane contains lipopolysaccharides, which must be inserted by the outer-membrane protein LptD.
Simulations demonstrate that LptD’s N-terminal domain in the periplasm is dynamic, possibly aiding connection to the rest of the Lpt complex.
Equilibrium simulations and free-energy calculations reveal that the lateral gate in LptD becomes easier to open when a substrate LPS molecule is present in the N-terminal domain.
Acknowledgments
This work was supported by the National Institutes of Health (R01-GM123169). Computational resources were provided through the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant OCI-1053575. Anton computer time was provided by the Pittsburgh Supercomputing Center (PSC) through NIH Grant R01-GM116961. The Anton machine at PSC was generously made available by D.E. Shaw Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ruiz N, Kahne D, Silhavy TJ, Advances in understanding bacterial outer-membrane biogenesis, Nat. Rev. Microbiol 4 (2006) 57–66. [DOI] [PubMed] [Google Scholar]
- [2].Bos MP, Robert V, Tommassen J, Biogenesis of the gram-negative bacterial outer membrane, Annu. Rev. Microbiol 61 (2007) 191–214. [DOI] [PubMed] [Google Scholar]
- [3].Hwang H, Paracini N, Parks JM, Lakey JH, Gumbart JC, Distribution of mechanical stress in the Escherichia coli cell envelope, Biochim. Biophys. Acta Biomembr 1860 (2018) 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Whitfield C, Trent MS, Biosynthesis and export of bacterial lipopolysaccharides, Annu. Rev. Biochem 83 (2014) 99–128. [DOI] [PubMed] [Google Scholar]
- [5].Erridge C, Bennett-Guerrero E, Poxton IR, Structure and function of lipopolysaccharides, Microbes Infect. 4 (2002) 837–851. [DOI] [PubMed] [Google Scholar]
- [6].Raetz CR, Whitfield C, Lipopolysaccharide endotoxins, Annu. Rev. Biochem 71 (2002) 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hancock RE, Alterations in outer membrane permeability, Annu. Rev. Microbiol 38 (1984) 237–264. [DOI] [PubMed] [Google Scholar]
- [8].Nikaido H, Molecular basis of bacterial outer membrane permeability revisited, Microbiol. Mol. Biol. Rev 67 (2003) 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ruiz N, Kahne D, Silhavy TJ, Transport of lipopolysaccharide across the cell envelope: the long road of discovery, Nat. Rev. Microbiol 7 (2009) 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang G, Meredith TC, Kahne D, On the essentiality of lipopolysaccharide to Gram-negative bacteria, Curr. Opin. Microbiol 16 (2013) 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Dehò G, Bolognesi M, Kahne D, Polissi A, The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains, J. Bacteriol 195 (2013) 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chng SS, Gronenberg LS, Kahne D, Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex, Biochemistry 49 (2010) 4565–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Narita S, Tokuda H, Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides, FEBS Lett. 583 (2009) 2160–2164. [DOI] [PubMed] [Google Scholar]
- [14].Sperandeo P, Villa R, Martorana AM, Samalikova M, Grandori R, Dehò G, Polissi A, New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex, J. Bacteriol 193 (2011) 1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tran AX, Dong C, Whitfield C, Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export path-way in Escherichia coli, J. Biol. Chem 285 (2010) 33529–33539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y, Structural basis for lipopolysaccharide insertion in the bacterial outer membrane, Nature 511 (2014) 108–111. [DOI] [PubMed] [Google Scholar]
- [17].Li Y, Orlando BJ, Liao M, Structural basis of lipopolysaccharide extraction by the LptB2FGC complex, Nature 567 (2019) 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D, Structural basis of unidirectional export of lipopolysaccharide to the cell surface, Nature 567 (2019) 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D, Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge, Science 359 (2018) 798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bishop RE, Ratcheting up lipopolysaccharide transport, Nature 567 (2019) 471–472. [DOI] [PubMed] [Google Scholar]
- [21].Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C, Structural basis for outer membrane lipopolysaccharide insertion, Nature 511 (2014) 52–56. [DOI] [PubMed] [Google Scholar]
- [22].Li X, Gu Y, Dong H, Wang W, Dong C, Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane, Sci. Rep 5 (2015) 11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL, OPM database and PPM web server: resources for positioning of proteins in membranes, Nucleic Acids Res. 40 (2012) D370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cournia Z, Allen TW, Andricioaei I, Antonny B, Baum D, Brannigan G, Buchete NV, Deckman JT, Delemotte L, Del Val C, Friedman R, Gkeka P, Hege HC, Henin J, Kasimova MA, Kolocouris A, Klein ML, Khalid S, Lemieux MJ, Lindow N, Roy M, Selent J, Tarek M, Tofoleanu F,Vanni S, Urban S, Wales DJ, Smith JC, Bondar AN, Membrane Protein Structure, Function, and Dynamics: a Perspective from Experiments and Theory, J. Membr. Biol 248 (2015) 611–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pavlova A, Hwang H, Lundquist K, Balusek C, Gumbart JC, Living on the edge: Simulations of bacterial outer-membrane proteins, Biochim. Biophys. Acta 1858 (2016) 1753–1759. [DOI] [PubMed] [Google Scholar]
- [26].Carpenter T, Khalid S, Sansom MS, A multidomain outer membrane protein from Pasteurella multocida: modelling and simulation studies of PmOmpA, Biochim. Biophys. Acta 1768 (2007) 2831–2840. [DOI] [PubMed] [Google Scholar]
- [27].Gumbart J, Wiener MC, Tajkhorshid E, Coupling of calcium and substrate binding through loop alignment in the outer-membrane transporter BtuB, J. Mol. Biol 393 (2009) 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barnard TJ, Gumbart J, Peterson JH, Noinaj N, Easley NC, Dautin N, Kuszak AJ, Tajkhorshid E, Bernstein HD, Buchanan SK, Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter, J. Mol. Biol 415 (2012) 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK, Structural insight into the biogenesis of β-barrel membrane proteins, Nature 501 (2013) 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK, Lateral opening and exit pore formation are required for BamA function, Structure 22 (2014) 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leong SW, Lim TS, Tye GJ, Ismail A, Aziah I, Choong YS, Assembly and stability of Salmonella enterica ser. Typhi TolC protein in POPE and DMPE, J. Biol. Phys 40 (2014) 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Balusek C, Gumbart JC, Role of the Native Outer-Membrane Environment on the Transporter BtuB, Biophys. J 111 (2016) 1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics, J. Mol. Graph 14 (1996) 33–38. [DOI] [PubMed] [Google Scholar]
- [34].Shaw DE, Dror RO, Salmon JK, Grossman J, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, Chow E, Eastwood MP, Ierardi DJ, Klepeis JL, Kuskin JS, Larson RH, Lindorff-Larsen K, Maragakis P, Moraes MA, Piana S, Shan Y, Towles B, Millisecond-scale molecular dynamics simulations on Anton, in SC’09: Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis, ACM, New York, NY, USA, 39:1–39:11. [Google Scholar]
- [35].Case DA, Betz RM, Botello-Smith W, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, York DM, Kollman PA, Amber 2016, University of California, San Francisco. [Google Scholar]
- [36].Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, MacKerell AD Jr., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles, J. Chem. Theory Comput 8 (2012) 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr., Pastor RW, Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types, J. Phys. Chem. B 114 (2010) 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hopkins CW, Le Grand S, Walker RC, Roitberg AE, Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning, J. Chem. Theory Comput 11 (2015) 1864–1874. [DOI] [PubMed] [Google Scholar]
- [39].Balusek C, Hwang H, Lau CH, Lundquist K, Hazel A, Pavlova A, Lynch D, Reggio P, Wang Y, Gumbart JC, Accelerating membrane simulations with Hydrogen Mass Repartitioning, J. Chem. Theory Comput (2019), in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hoover WG, Canonical dynamics: Equilibrium phase-space distributions, Phys. Rev. A 31 (1985) 1695–1697. [DOI] [PubMed] [Google Scholar]
- [41].Martyna GJ, Klein ML, Tuckerman M, Nosé-Hoover chains - the canonical ensemble via continuous dynamics, J. Chem. Phys 97 (1992) 2635–2643. [Google Scholar]
- [42].Martyna GJ, Tobias DJ, Klein ML, Constant pressure molecular dynamics algorithms, J. Chem. Phys 101 (1994) 4177–4189. [Google Scholar]
- [43].Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE, Gaussian split Ewald: A fast Ewald mesh method for molecular simulation, J. Chem. Phys 122 (2005) 54101. [DOI] [PubMed] [Google Scholar]
- [44].Darden TA, York DM, Pedersen LG, Particle mesh Ewald: An N log N method for Ewald sums in large systems, J. Comput. Phys 98 (1993) 10089–10092. [Google Scholar]
- [45].Sugita Y, Okamoto Y, Replica-exchange molecular dynamics method for protein folding, Chem. Phys. Lett 314 (1999) 141–151. [Google Scholar]
- [46].Sugita Y, Kitao A, Okamoto Y, Multidimensional Replica-Exchange Method for Free-Energy Calculations, J. Chem. Phys 113 (2000) 6042–6051. [Google Scholar]
- [47].Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA, The weighted histogram analysis method for free-energy calculation on biomolecules., J. Comput. Chem 13 (1992) 1011–1021. [Google Scholar]
- [48].Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, Wojtowicz D, Barnard TJ, Gumbart JC, Buchanan SK, Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens, Structure 24 (2016) 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Straatsma TP, Soares TA, Characterization of the outer membrane protein OprF of Pseudomonas aeruginosa in a lipopolysaccharide membrane by computer simulation, Proteins 74 (2009) 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Holdbrook DA, Piggot TJ, Sansom MS, Khalid S, Stability and membrane interactions of an autotransport protein: MD simulations of the Hia translocator domain in a complex membrane environment, Biochim. Biophys. Acta 1828 (2013) 715–723. [DOI] [PubMed] [Google Scholar]
- [51].Piggot TJ, Holdbrook DA, Khalid S, Conformational dynamics and membrane interactions of the E. coli outer membrane protein FecA: a molecular dynamics simulation study, Biochim. Biophys. Acta 1828 (2013) 284–293. [DOI] [PubMed] [Google Scholar]
- [52].Wu EL, Fleming PJ, Yeom MS, Widmalm G, Klauda JB, Fleming KG, Im W, coli outer membrane E and interactions with OmpLA, Biophys. J 106 (2014) 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bishop RE, Emerging roles for anionic non-bilayer phospholipids in fortifying the outer membrane permeability barrier, J. Bacteriol 196 (2014) 3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Malojčić G, Andres D, Grabowicz M, George AH, Ruiz N, Silhavy TJ, Kahne D, LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface, Proc. Natl. Acad. Sci. U.S.A 111 (2014) 9467–9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sampson BA, Misra R, Benson SA, Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability, Genetics 122 (1989) 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Braun M, Silhavy TJ, Imp/OstA is required for cell envelope biogenesis in Escherichia coli, Mol. Microbiol 45 (2002) 1289–1302. [DOI] [PubMed] [Google Scholar]
- [57].Gu Y, Stansfeld PJ, Zeng Y, Dong H, Wang W, Dong C, Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD, Structure 23 (2015) 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Okuda S, Freinkman E, Kahne D, Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli, Science 338 (2012) 1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bamert RS, Lundquist K, Hwang H, Webb CT, Shiota T, Stubenrauch CJ, Belousoff MJ, Goode RJA, Schittenhelm RB, Zimmerman R, Jung M, Gumbart JC, Lithgow T, Structural basis for substrate selection by the translocation and assembly module of the β-barrel assembly machinery, Mol. Microbiol 106 (2017) 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lundquist K, Bakelar J, Noinaj N, Gumbart JC, C-terminal kink formation is required for lateral gating in BamA, Proc. Natl. Acad. Sci. U.S.A 115 (2018) E7942–E7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Robinson JA, Folded Synthetic Peptides and Other Molecules Targeting Outer Membrane Protein Complexes in Gram-Negative Bacteria, Front. Chem 7 (2019) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vetterli SU, Zerbe K, Muller M, Urfer M, Mondal M, Wang SY, Moehle K, Zerbe O, Vitale A, Pessi G, Eberl L, Wollscheid B, Robinson JA, Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli, Sci. Adv 4 (2018) eaau2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA, Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa, Science 327 (2010) 1010–1013. [DOI] [PubMed] [Google Scholar]
- [64].Martin-Loeches I, Dale GE, Torres A, Murepavadin: a new antibiotic class in the pipeline, Expert. Rev. Anti. Infect. Ther 16 (2018) 259–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.