Abstract
Dysregulated leukocyte responses underlie the pathobiology of sepsis, which is a leading cause of death. However, measures of leukocyte function are not routinely available in clinical care. Here we report the development and testing of an inertial microfluidic system for the label-free isolation and downstream functional assessment of leukocytes from 50 μl of peripheral blood. We used the system to assess leukocyte phenotype and function in serial samples from 18 hospitalized patients with sepsis and 10 healthy subjects. The sepsis samples had significantly higher levels of CD16dim and CD16− neutrophils and CD16+ ‘intermediate’ monocytes, as well as significantly lower levels of neutrophil-elastase release, O2− production and phagolysosome formation. Repeated sampling of sepsis patients over 7 days showed that leukocyte activation (measured by isodielectric separation) and leukocyte phenotype and function were significantly more predictive of the clinical course than complete-blood-count parameters. We conclude that the serial assessment of leukocyte function in microlitre blood volumes is feasible and that it provides significantly more prognostic information than leukocyte counting.
Subject terms: Immunology, Diseases, Pathogenesis
The serial assessment of the functional parameters of leukocytes isolated via an inertial microfluidic system from 50 μl of peripheral blood from sepsis patients provides significantly more prognostic information than leukocyte counting.
Main
Dysregulated inflammation underlies the pathobiology of sepsis1, which is a leading cause of death from infection2. In healthy patients, neutrophils and monocytes are early responders that are important for combating a pathogen, limiting tissue damage and promoting the return to organ function and homeostasis3. These leukocytes must integrate diverse environmental signals to inform their protective functional responses4. Sepsis is characterized by immune cell dysfunction, which is linked to the pathophysiology of the disease with consequent organ injury and increased associated mortality1. In experimental sepsis in mice, regulating leukocyte function decreases organ injury and lessens the severity of the infection5. The assessment of leukocyte activation and function may enable new therapeutic strategies for the management of human sepsis and inflammatory diseases.
Sepsis patients are commonly monitored by clinical criteria and peripheral blood leukocyte counts; however, measures of leukocyte function are not routinely available for clinical care. The monitoring of leukocyte counts does not reliably reflect patients’ clinical and treatment responses. Sepsis-3 guidelines define sepsis as life-threatening organ dysfunction caused by a dysregulated host response to infection, where end-organ injury and disease severity are defined by the sequential organ failure assessment (SOFA) score6. These criteria carry an ~30% misdiagnosis rate7 and offer little insight into the underlying pathobiology. Multiple point-of-care microfluidic devices have been used to investigate circulating cytokines and other mediators of inflammation as potential biomarkers in sepsis, but no device that reliably predicts severity and outcomes has emerged8–14. Since leukocytes (white blood cells) are sensors for multiple environmental cues and pathogens4, their activation and function could prove useful as cellular biomarkers of illness. To this end, polymorphonuclear neutrophil (PMN) dysmotility in microfluidic chambers correlates with sepsis diagnosis15. Importantly, repeated measurement of the complete blood count (CBC) can contribute to anaemia, which is often present in hospitalized critically ill patients16. Ideally, new methods for the assessment of leukocyte function during the course of a patient’s illness should be sample sparing to decrease the risk of iatrogenic anaemia.
The sorting of leukocytes using spiral inertial microfluidics was introduced in 2012 (refs. 17,18). We modified our previous device and developed a closed-loop inertial microfluidic platform for the label-free isolation of adequate leukocyte numbers from microlitre blood volumes. A sample sparing approach would enable the acquisition of multiple biospecimens in one day, or during a hospital stay, for the repeated assessment of the host response without leading to iatrogenic anaemia. Here we demonstrate the power of preparative inertial microfluidics and cell-based assays of leukocyte activation and function in a proof-of-concept cohort of human patients with sepsis. Despite a relatively small sample size, this innovative integrative approach was more highly correlated to sepsis severity than the current clinical practice of measuring peripheral blood leukocyte counts and differentials.
Results
Inertial microfluidic separation of leukocytes from blood
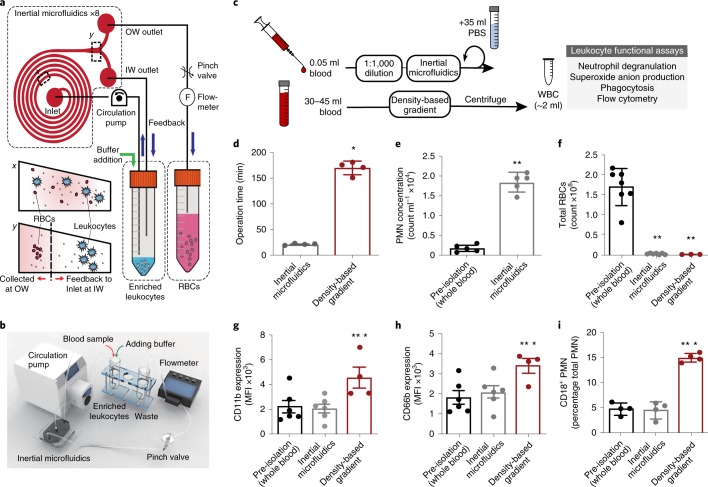
To enable the assessment of in vivo leukocyte activation and to minimize sample volume, we first developed an inertial microfluidics system (Fig. 1 and Supplementary Fig. 1) for the label-free isolation of leukocytes from microlitre (50 μl) quantities of human peripheral blood. Eight spiral microchannels with a trapezoidal cross-sectional design (Fig. 1a,b) were connected in parallel with a circulation pump (see Methods) to a sample tube containing diluted blood (1:1,000 volume/volume (v/v))19. As the diluted blood passed through the spiral microchannels, the system ordered and aligned the cells by size at each lateral equilibrium position secondary to a balance between the cells’ inertial lift and Dean drag forces20,21. This inertial microfluidics system was compared to an established method for leukocyte isolation by density-based gradient separation (see Methods, Fig. 1c). Relative to density-based gradient separation, the inertial microfluidics system had a shorter operation time (~22 min versus ~175 min; Fig. 1d). This system also increased PMN concentration by approximately eightfold relative to input sample (Fig. 1e). Both leukocyte isolation methods significantly decreased erythrocytes (red blood cells) to a similar extent (Fig. 1f).
Fig. 1. Closed-loop inertial microfluidic separation of leukocytes from whole blood.
a, A schematic of the closed-loop operation of the spiral microfluidics system; x-cross section of inertial microfluidics near inlet, and y-cross section near outlet. b, The experimental set-up for the closed-loop inertial microfluidics system. c, A comparison between a sample sparing approach using inertial microfluidics and a standard density-based gradient separation for leukocytes, and their functional assessment. d, The operation time for inertial microfluidics (n = 4) compared to density-based gradients (n = 4). e, The PMN concentration in peripheral whole blood (pre-isolation; n = 5) and after inertial microfluidics separation (n = 5). f, The erythrocyte count pre-isolation (n = 7) and after inertial microfluidics (n = 7) or density-based gradient separation (n = 3). g–i, Markers of PMN activation were assessed pre-isolation and after inertial microfluidics or density-based gradient separation, namely the expression of CD11b (MFI; n = 4–6; g), CD66b (MFI; n = 4–6; h) and CD18 (percentage total PMN; n = 4; i). The values are expressed as the mean ± s.e.m. *P < 0.05 for density-based gradient versus inertial microfluidics; **P < 0.05 for inertial microfluidics or density-based gradient versus pre-isolation by unpaired Student’s two-tailed t test. IW, inward; MFI, median fluorescence intensity; OW, outward; PBS, phosphate-buffered saline; RBC, red blood cell; WBC, white blood cell.
To assess the effects of inertial microfluidics on ex vivo cell activation, the PMN expressions of CD11b, CD66b and CD18 were determined. With inertial microfluidics separation, the mean expression of these cell adhesion molecules was similar to pre-isolation whole blood and significantly lower than the density-based gradient method (Fig. 1g–i). In pre-isolation whole blood and after inertial microfluidic separation, PMN subsets were determined by flow cytometry (see Methods, Supplementary Fig. 2). The relative percentages of the PMN subsets (CD16bright, CD16dim, CD16−) in pre-isolated whole blood significantly correlated with PMNs after isolation by inertial microfluidics (Supplementary Fig. 3a). The PMN functions in whole blood and after isolation were also similar when phagocytosis and phagolysosome formation were tested using Escherichia coli pHrodo bioparticles (Supplementary Fig. 3b). In addition, there was significant concordance for the detection of the PMN activation markers CD62L (Supplementary Fig. 3c) and CD69 (Supplementary Fig. 3d). Notably, no significant correlation was identified for CD11b (Supplementary Fig. 3e) and CD42b expression on PMNs (Supplementary Fig. 3f). Together, these findings indicate that the inertial microfluidic separation of leukocytes from whole blood is fast and results in less ex vivo activation than the density-based gradient method.
PMN function in sepsis and healthy patients
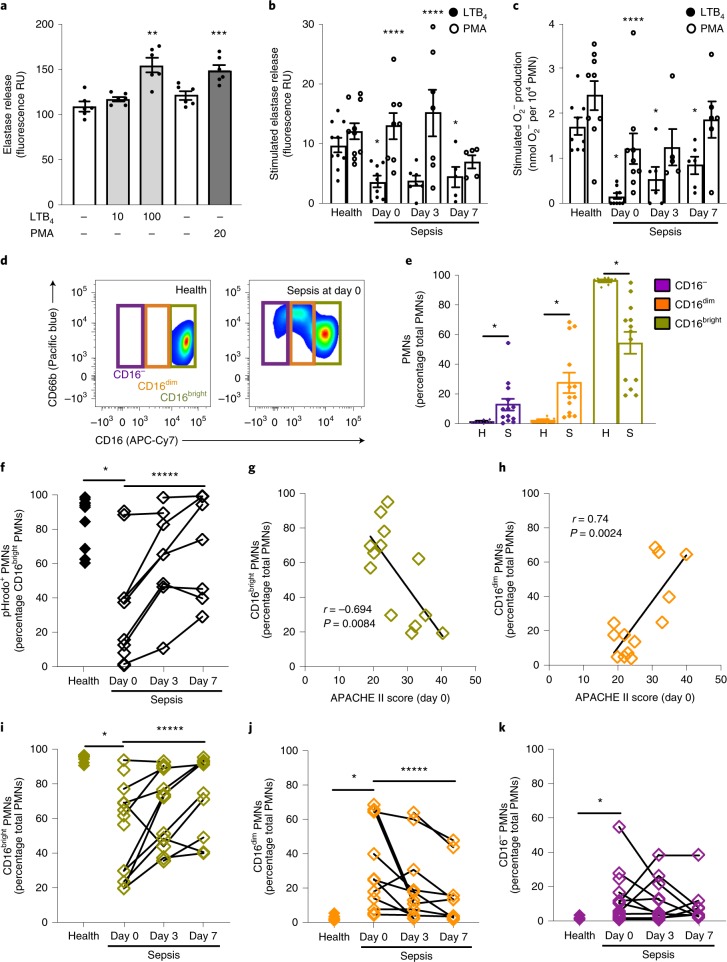
To determine PMN functional responses after inertial microfluidic isolation, we assessed degranulation, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase assembly and phagolysosome formation. Cell dose dependency for degranulation was assessed by phorbol myristate acetate (PMA)-stimulated elastase release (see Methods). Significant PMA-stimulated elastase release was detectable at PMN numbers as low as 5,000 cells from healthy donors (Supplementary Fig. 4a). A receptor-mediated stimulus, leukotriene B4 (LTB4; 10 nM, 100 nM), increased elastase release from these cells in a dose-dependent manner (Fig. 2a).
Fig. 2. Neutrophil subsets and function in sepsis and healthy patients.
PMNs were isolated from 50 μl freshly obtained peripheral blood using the inertial microfluidics system. a, The elastase released from healthy subject (n = 6) PMNs (5,000 per well) exposed to LTB4 (light grey), phorbol-12-myristate-13-acetate (PMA; dark grey) or vehicle (ethanol 0.02% v/v for LTB4, dimethylsulfoxide 0.002% v/v for PMA; white). b,c, The increase in the elastase release (b; 5,000 PMNs per well) and O2− production (c; 10,000 PMNs per well) of cells from healthy subjects (n = 9–10) or patients with sepsis (n = 21 sequential samples of 9 sepsis patients from days 0, 3 and 7 of hospitalization) when exposed to LTB4 (100 nM) or PMA (20 nM for elastase release; 100 nM for superoxide anion production) relative to vehicle. The results are shown as stimulus − vehicle. In a–c, the values are expressed as the mean ± s.e.m. d, A representative flow cytometry zebra plot of PMN (CD45+FSC+SSC+) subsets as determined by CD16 and CD66b surface expression. e, The frequency of CD16−, CD16dim and CD16bright PMNs from healthy subjects (n = 10) or sepsis patients (n = 13) (day 0). The values are expressed as the mean ± s.e.m. f, The frequency of CD16bright PMNs expressing pHrodo-labelled E. coli bioparticles (n = 10 healthy donors, n = 25 sequential samples of 10 sepsis patients). g,h, The relationship between the APACHE II score and PMN subsets CD16bright (g) and CD16dim (h) in sepsis patients (n = 13) at presentation (day 0). The Pearson coefficient of correlation (r value) and significance are noted and regression lines are shown. i–k, The frequencies of CD16bright (i), CD16dim (j) and CD16− (k) PMN subsets from individual healthy subjects (n = 10) and 13 sepsis patients on days 0, 3 and 7 (n = 32). **P < 0.05 for LTB4 versus vehicle (ethanol), ***P < 0.05 for PMA versus vehicle (dimethylsulfoxide), *P < 0.05 for healthy subjects versus sepsis patients by unpaired Student’s two-tailed t test; ****P < 0.05 for LTB4 versus PMA by paired two-tailed t test; *****P < 0.05 by one-way analysis of variance (ANOVA) for n = 6 sepsis patients at days 0, 3 and 7 of hospitalization. H, health; S, sepsis.
To determine PMN responses in sepsis, we obtained peripheral blood from 18 sepsis patients within 72 h of admission to Brigham and Women’s Hospital medical intensive care unit (defined as protocol day 0) and on hospital days 3 and 7 thereafter, as well as from 10 healthy control subjects. The subject characteristics are shown in Table 1. The LTB4-stimulated elastase release from PMNs isolated from patients with sepsis (3.67 ± 0.91 fluorescence RU (relative units), mean ± s.e.m.) was significantly lower than that from cells from healthy subjects (9.77 ± 1.21 fluorescence RU, mean ± s.e.m.) (Fig. 2b) at all time points. By contrast, PMA-stimulated elastase release was similar for PMNs from patients with sepsis and healthy subjects (Fig. 2b). The mean LTB4-stimulated PMN elastase release in sepsis patients increased from day 0 to day 7 (Fig. 2b). Similar to PMN elastase release, LTB4 exposure resulted in significantly lower values for superoxide anion production in PMNs from sepsis patients (day 0) (0.15 ± 0.07 nmol O2− per 104 PMNs, mean ± s.e.m.) relative to healthy donors (1.58 ± 0.21 nmol O2− per 104 PMNs, mean ± s.e.m.) (see Methods, Fig. 2c). The mean LTB4-stimulated superoxide anion production also increased from day 0 to day 7 in sepsis patients (Fig. 2c) and PMA-stimulated PMNs from healthy donors produced significantly higher levels of O2− production relative to sepsis patients at day 0 (Fig. 2c).
Table 1.
Subject characteristics
| Characteristic | Healthy patients (n = 10) | Sepsis patients (n = 18) |
|---|---|---|
| Sex (%) | Male: 60; female: 40 | Male: 88.9; female: 11.1 |
| Race, ethnicity (%) |
Asian (30) Black (0) Hispanic (0) White (70) |
Asian (0) Black (5.6) Hispanic (16.7) White (77.8) |
| Age (mean (range)) | 31.2 (23–40) | 59.7 (19–92) |
| Pneumonia (%) | – | 27.8 |
| ARDS (%) | – | 27.8 |
| Malignancy (%) | – | 61.1 |
| APACHE II (mean (range)) | – | 25.4 (11–40) |
| SOFA score (mean (range)) | – | 7.89 (3–17) |
| Vasopressors (%) | – | 22.20 |
| Mechanical ventilation (%) | – | 38.9 |
| Mortality (%) | – | 22.2 |
| White blood cells (×1,000 per mm3) (mean (range)) | 6.9 (5–10) | 14.27 (0.08–40.6) |
PMN subsets in sepsis and healthy patients
Next, PMN subsets were identified by flow cytometry (CD45+FSC+SSC+CD66b+ and CD16 status; see Methods, Supplementary Figs. 2 and 4b). In sepsis patients at day 0, the percentages of CD16dim and CD16− cells were increased and the frequency of CD16bright was decreased (Fig. 2d,e). In contrast to sepsis patients, the vast majority of PMNs identified in healthy patients were CD16bright (Fig. 2d,e). We next determined relative PMN activity for the phagocytosis of E. coli pHrodo bioparticles (see Methods). In sepsis patients, the mean percentage of CD16bright PMNs that were pHrodo+ was significantly lower at day 0 (Supplementary Figs. 2f and 4c). Notably, the uptake of pHrodo by CD16bright PMNs increased significantly from day 0 to day 7 in sepsis patients (Fig. 2f). CD16bright PMNs isolated from sepsis patients (day 0) displayed decreased cell surface CD62L relative to cells from healthy subjects (Supplementary Fig. 4d), but no significant differences were seen for the surface expression of CD69 (Supplementary Fig. 4e).
To investigate whether the CD16 PMN subsets were related to disease severity, we next determined their correlation with the Acute Physiology and Chronic Health Evaluation II (APACHE II) score. The percentage of CD16bright PMNs was inversely correlated to the APACHE II score (Pearson’s correlation coefficient r = −0.694, P = 0.0084; Fig. 2g). Conversely, the percentage of CD16dim PMNs was positively correlated to the APACHE II score (r = 0.74, P = 0.0024; Fig. 2h). Over the week of monitoring, the percentages of these PMN subsets in sepsis patients evolved towards the values in healthy patients (Fig. 2i–k).
Isodielectric separation (IDS) of human PMNs in sepsis and healthy patients
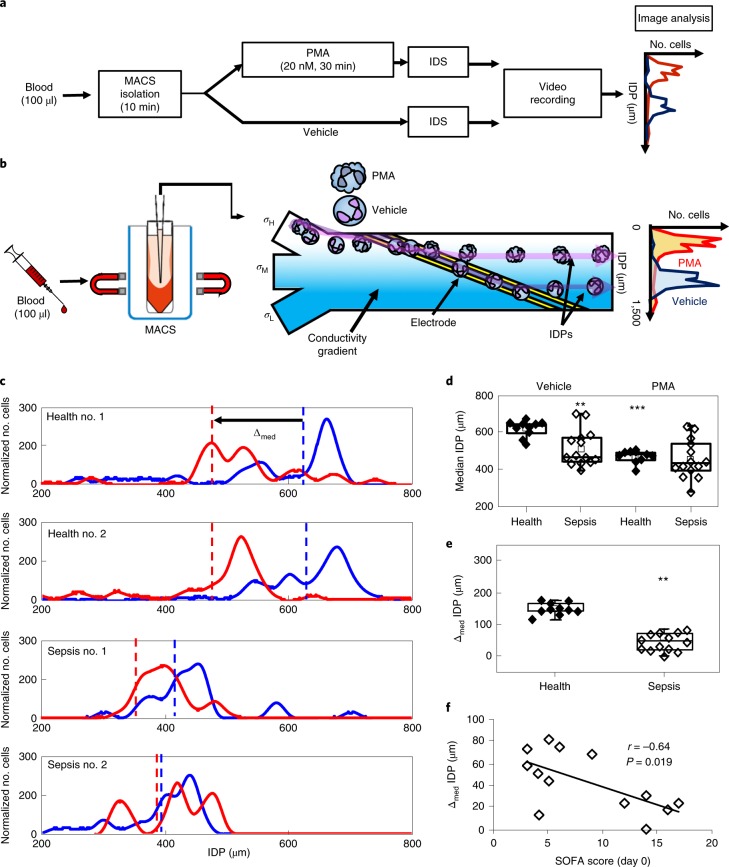
IDS can determine an isodielectric position (IDP) for PMNs that distinguishes between resting and activated cells22. To determine whether IDS can identify differences between PMNs from sepsis and healthy donors, we next measured the IDP for freshly isolated PMNs exposed to PMA (20 nM) or vehicle (ethanol 0.02%) (Fig. 3a; see Methods). For these assays, the PMNs were isolated from 100 μl peripheral blood by negative selection (rather than the inertial microfluidics system) before introduction into the IDS chamber. PMN IDP was inversely correlated to electrical voltage (Supplementary Fig. 5a). When exposed to an electrical voltage of 7 MHz, PMNs incubated with PMA had a lower IDP (activated PMNs) and flow in the lower conductivity channel (σL) (Fig. 3a,b). By contrast, PMNs incubated with vehicle have a higher IDP (non-activated PMNs) and flow in the higher conductivity channel (σH) (Fig. 3a,b). When exposed to vehicle, PMNs from patients with sepsis demonstrated a significantly lower median IDP relative to PMNs from healthy donors (Fig. 3c,d). With PMA stimulation, the median IDP of PMNs from healthy donors significantly decreased to a level that approximated that of PMNs from sepsis patients (Fig. 3c,d, Supplementary Fig. 5b). The difference between the median IDP of PMNs exposed to PMA or vehicle can be expressed as Δmed IDP (Δmed = median IDPveh − median IDPPMA) (Fig. 3c). The Δmed IDP of PMNs from sepsis patients (day 0) was significantly lower than the Δmed IDP of PMNs in healthy patients (Fig. 3e). The Δmed IDP of PMNs from sepsis patients significantly increased from day 0 to day 7 (Supplementary Fig. 6a), but the median IDP from sepsis patients had no significant changes from day 0 to day 7 (Supplementary Fig. 6b). To investigate whether PMN IDP was related to disease severity, we next determined whether the IDP correlated with clinical severity scores obtained on admission. The Δmed IDP was inversely correlated with the SOFA score (r = −0.64, P = 0.019; Fig. 3f). There was a similar but not significant trend for Δmed IDP and APACHE II score (r = −0.38, P = 0.29; Supplementary Fig. 6c) and median IDP was not correlated with the SOFA score (r = 0.08, P = 0.77; Supplementary Fig. 6d) and APACHE II score at day 0 (r = 0.25, P = 0.40; Supplementary Fig. 6e).
Fig. 3. IDS of activated from non-activated human PMNs.
PMNs were isolated from 100 μl freshly obtained peripheral blood using magnetic activated cell sorting (MACS). a, The experimental workflow for PMN activation by PMA before IDS and the determination of the cell’s IDP as an electrical signature. b, A schematic of IDS and IDP determination. c, After exposure to PMA (20 nM, 30 min, 25 °C RT) or vehicle, the IDPs of PMNs from healthy donors or sepsis patients were distributed at 7 MHz electrical voltage. The shift in median IDP values was determined for PMA-activated cells (red) relative to vehicle (blue). The dashed lines represent the median values of each IDP distribution. Representative IDP distributions are shown for PMNs from representative healthy subjects (top two distributions) and sepsis patients (bottom two distributions). d,e, Box and whisker plots (median, 25th and 75th percentiles) of the median IDP in vehicle- or PMA-exposed PMNs (d) and Δmed IDP (e) from healthy subjects (n = 10) or patients presenting with sepsis (n = 13). f, The relationship between the SOFA score and Δmed (μm) was determined in sepsis patients at presentation (day 0). The Pearson correlation r value is noted and a regression line is shown. **P < 0.05 in healthy versus sepsis patients by unpaired two-tailed Student’s t test; ***P < 0.05 in vehicle verus PMA by paired two-tailed Student’s t test.
Monocyte subsets in sepsis and healthy patients
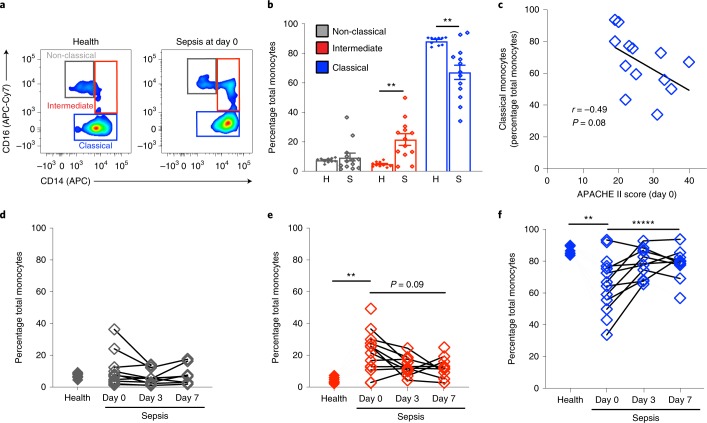
Peripheral blood monocytes play essential roles in host responses during sepsis1 and were isolated from whole blood using inertial microfluidics (Fig. 4a). To assess the effects of inertial microfluidics on ex vivo cell activation, monocytes in pre-isolation whole blood and after isolation were determined by flow cytometry (see Methods, Supplementary Figs. 7 and 8a). The relative percentages of monocyte subsets (classical, intermediate and non-classical) in the isolated cells was concordant with those in whole blood and had a significant correlation by linear regression (Supplementary Fig. 7a). There was also a significant correlation between isolated cells and whole blood for the monocyte activation markers CD62L (Supplementary Fig. 7b), CD69 (Supplementary Fig. 7c) and CD11b (Supplementary Fig. 7d). Monocyte function in whole blood and after isolation was similar when phagolysosome formation was tested using E. coli pHrodo bioparticles (Supplementary Fig. 7e). Similar to PMNs, no significant correlation for monocyte CD42b expression was present (Supplementary Fig. 7f). Classical monocytes isolated from sepsis patients (day 0) displayed no differences in cell surface CD62L relative to cells from healthy subjects (Supplementary Fig. 8b).
Fig. 4. Monocyte subsets in sepsis and healthy patients.
Monocytes were isolated from 50 μl peripheral blood using the inertial microfluidic system. a, A representative flow cytometry zebra plot of monocyte (CD45+FSC+SSC−) subsets as determined by CD16 and CD14 surface expression. b, The frequency of non-classical (CD16highCD14low), intermediate (CD16highCD14high) and classical (CD16lowCD14high) monocytes in healthy subjects (n = 10) or sepsis patients (n = 13) (day 0). The values are expressed as the mean ± s.e.m. c, The relationship between the APACHE II score and classical monocytes (CD16lowCD14high) in sepsis patients (n = 13) (day 0). Pearson correlation r value and significance are noted and a regression line is shown. d–f, The frequencies of non-classical (CD16highCD14low; d), intermediate (CD16highCD14high; e) and classical (CD16lowCD14high; f) monocyte subsets in individual healthy subjects (n = 10) and 13 sepsis patients on days 0, 3 and 7 (n = 32). **P < 0.05 in healthy versus sepsis patients by unpaired two-tailed Student’s t test; *****P < 0.05 by one-way ANOVA for n = 6 sepsis patients at day 0 to day 7 of hospitalization.
Next, monocyte subsets were assessed in sepsis and healthy patients by flow cytometry (CD45+FSC+SSC− and CD14 and CD16 status; see Methods, Fig. 4a). Relative to healthy patients, patients with sepsis had significantly increased percentages of intermediate monocytes and significantly decreased percentages of classical monocytes; no significant differences were identified for non-classical monocytes (Fig. 4a,b). To investigate whether monocyte subsets were related to disease severity, we then determined whether their percentages correlated with APACHE II score. There was a trend for an inverse correlation between classical monocyte percentage and APACHE II score (r = −0.49, P = 0.08; Fig. 4c). Notably, classical monocyte percentages changed significantly over time towards the values for healthy patients (Fig. 4d–f).
Principal component analysis for clinical and leukocyte parameters in sepsis patients
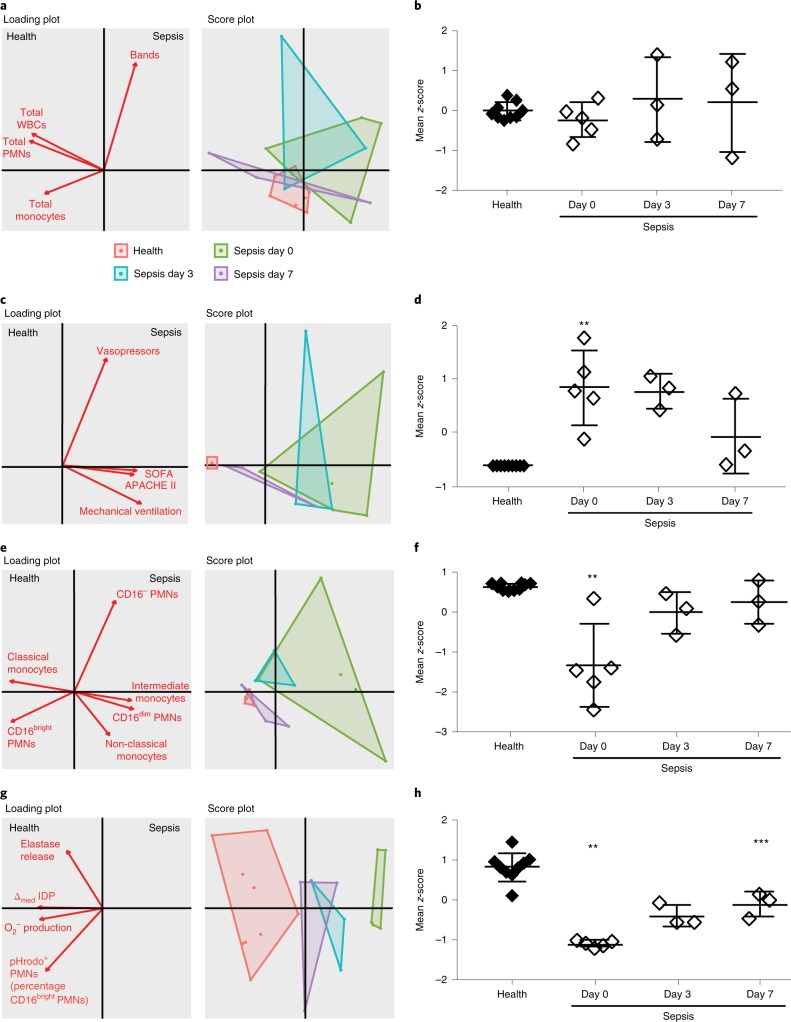
Next, the relationships between clinical CBC parameters (total white blood cells, PMNs, band cells and monocytes), clinical severity indicators, leukocyte phenotype subsets and leukocyte function in sepsis and healthy patients were determined using multivariate principal component analysis (see Methods). Clinical CBC parameters (Fig. 5a) did not distinguish sepsis from healthy patients at presentation or at subsequent time points during hospitalization as noted by the overlap in the sample distribution on the score plot (Fig. 5a). The standard scores (that is z-scores) for CBC measures were aggregated and indicated no statistically significant difference between healthy and sepsis patients (day 0 to day 7) (Fig. 5b). As expected, the aggregate scores of clinical severity indicators, including the need for vasopressors and mechanical ventilation, as well as the APACHE II and SOFA scores did distinguish sepsis patients at presentation from healthy donors but did not significantly change until day 7 (Fig. 5c,d). Most notably, the aggregate scores of leukocyte phenotype markers (Fig. 5e,f, Supplementary Fig. 9a) and PMN functional responses (Fig. 5g,h) distinguished sepsis from healthy patients and further distinguished sepsis patients by day of illness. The aggregate scores of CD16bright PMNs and classical monocytes and PMN functional responses demonstrated a significant decrease at day 0 relative to healthy donor scores and a return to values that approximate health at day 7 (Fig. 5e–h). PMN functional responses that incorporated median IDP also distinguished between sepsis and healthy patients at presentation and by day of illness (day 0 to day 7) (Supplementary Fig. 9b-c).
Fig. 5. Principal component analyses for routine CBC measures, clinical severity and leukocyte phenotype and function in sepsis and healthy patients.
a–h, Two-dimensional score plots and loading plots from multivariate principal component analyses and mean Z score were determined for routine clinical CBC parameters (a,b), clinical severity indicators (c,d), leukocyte phenotype markers (e,f; CD16bright PMNs and classical monocytes; see Supplementary Fig. 7 for CD16dim PMNs, CD16− PMNs, intermediate monocytes and non-classical monocytes) and neutrophil functional responses (g,h). The mean Z scores were derived from individual variable Z scores in a given subject (n = 9 healthy subjects and n = 11 sequential samples from 5 sepsis patients). **P < 0.05 in sepsis patients (day 0) versus healthy patients, ***P < 0.05 in sepsis patients day 7 versus day 0 by unpaired Student’s two-tailed t-test. Horizontal lines and error bars in b,d,f,h indicate mean ± s.e.m. Values for O2− production and elastase release were obtained from PMNs exposed to LTB4.
Correlation between PMN responses and clinical severity
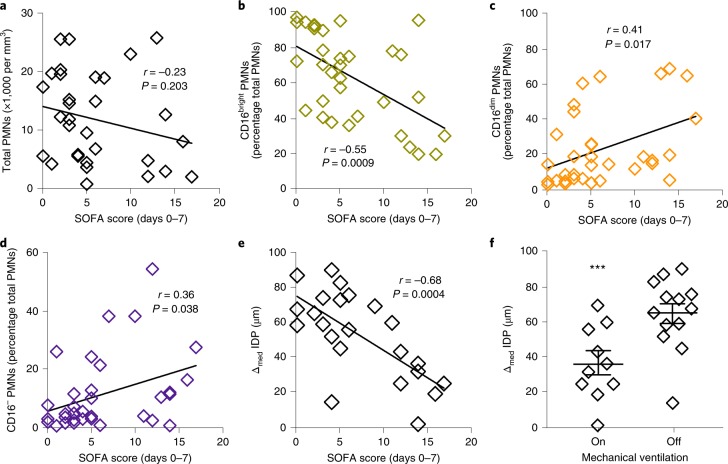
To investigate whether individual measures of leukocyte phenotype or function were related to the severity of sepsis during hospitalization, we determined their correlations to indicators of sepsis severity (SOFA score and the need for mechanical ventilation). While the total PMN count was not significantly correlated to the SOFA score (Fig. 6a), the percentage of CD16bright PMNs was inversely correlated and the percentages of CD16dim and CD16− PMNs were positively correlated with the SOFA score (Fig. 6b–d). In addition, the Δmed IDP for PMNs from sepsis patients demonstrated a significant inverse correlation with SOFA score (Fig. 6e). The median IDP of PMNs from sepsis patients had no significant correlation with SOFA score (Supplementary Fig. 10a). Moreover, there was no significant correlation identified between severity indicators and other PMN functional responses (Supplementary Fig. 10b–e) or monocyte subsets (Supplementary Fig. 11). The Δmed IDP of PMNs in sepsis patients was also significantly lower in patients requiring mechanical ventilation (Fig. 6f). In this limited sample size, neither current clinical severity indicators (SOFA and APACHE II scores) nor leukocyte phenotype and functional responses were associated with mortality (Supplementary Fig. 12a–k).
Fig. 6. Correlation between PMN responses and measures of clinical severity during sepsis.
a–e, The relationships between SOFA score (days 0–7) and total PMNs (n = 32 separate biospecimens from 14 sepsis patients; a), CD16bright PMNs (n = 33 from 13 sepsis patients; b), CD16dim PMNs (n = 33 from 13 sepsis patients; c), CD16− PMNs (n = 33 from 13 sepsis patients; d) and Δmed IDP (n = 23 from 13 sepsis patients; e) from sepsis patients at days 0, 3 and 7. The Pearson correlation r value and significance are noted and regression lines are shown. f, The relationship of Δmed IDP with mechanical ventilation in sepsis patients at days 0, 3 and 7 (n = 23 from 13 sepsis patients). The values are expressed as the mean ± s.e.m. ***P < 0.05 for on versus off mechanical ventilation by unpaired Student’s two-tailed t test.
Discussion
Dysregulated immune responses to infection and tissue injury underlie sepsis pathobiology1; yet these immune responses are not directly measured in routine clinical management. Multiple groups have developed point-of-care microfluidic devices to investigate potential biomarkers in sepsis diagnosis. C-reactive protein, procalcitonin, interleukin-6, neopterin and many others have been studied extensively but provided limited utility in sepsis severity assessment and outcomes8–14. In a large cohort of patients, using a point-of-care microfluidic device, CD64 surface receptor expression on neutrophils was found to be predictive of patient diagnosis but was of limited use in sepsis severity assessment9. A recent study demonstrated that the measurement of patterns of neutrophil spontaneous motility in microfluidic chambers enabled the accurate diagnosis of sepsis, which suggests that the measurement of leukocyte function (rather than number) may represent a potential approach for the development of sepsis severity biomarkers15. Here we modified and demonstrated a closed-loop microfluidics platform to isolate leukocytes using microlitre quantities of peripheral blood volumes to provide an innovative biomedical engineering approach to both the label-free sample preparation of leukocytes and the study of leukocyte activation and function in a clinical context. Our approach provides evidence that leukocyte responses: are measurable in small aliquots of peripheral blood; are distinct in sepsis and healthy patients; are dynamic over time; reflect sepsis clinical severity; are more predictive of the clinical course than leukocyte count; and are differential by repeated CBC (the current clinical practice). This sample sparing approach would enable the repeated assessment of leukocyte activation and function without substantially increasing the risk for iatrogenic anaemia, which is a common problem in intensive care units16. The features described here enable its potential clinical integration.
Recently, microfluidic devices have been developed to enable clinical sample preparation. For example, a surface acoustic wave assisted microfluidics platform was proposed as a method to separate blood components, for example, circulating tumour cells23,24, leukocytes25,26, mesenchymal stromal cells27, bacteria28 and microvesicles29. The different electrical properties between cells can also enable the fractionation of fluid components with electrode-embedded microfluidics, for example stem cells from enzyme-digested adipose tissue30, the profiling of yeast genotypes31 and neutrophils from white blood cells22. These platforms have the advantage of obtaining a high purity separation from a small volume of sample without the use of a membrane filter. External stimuli can cause leukocyte activation, which may lead to inaccuracies in the assessment of the host response. Inertial microfluidics was introduced as a passive, label-free separation method. The fluid flow in an intermediate Reynold’s number regime (~1 < Re < ~100) through a curved microchannel was used to order and align particles by their size at each lateral equilibrium position due to the balance between the inertial lift force and Dean drag force21,26,32–34. This technique has been applied to various clinical samples, for example the separation of circulating tumour cells35–39, leukocytes17,18, mesenchymal stem cells40, viruses41,42, bacteria42 and antigen-specific lymphocytes from human blood43,44. To overcome the limitations of the syringe-based operation of inertial microfluidics, our group introduced the closed-loop operation of spiral microfluidics for clinical sample preparation (neutrophil extraction from patient airway secretions)17 and virus isolation from patient blood cell contents41. Here we modified the closed-loop microfluidics platform to demonstrate the isolation of leukocytes using microlitres of peripheral blood. This approach was designed to fulfil two aspects of sample preparation: the extraction of functionally intact and relatively pure PMNs from aliquots of human blood; and the isolation of appropriate numbers of leukocytes from minimal blood volumes from humans to facilitate clinical integration.
Bacteraemia during sepsis was previously shown to activate the complement system that reduces PMN superoxide anion production, phagocytosis45 and degranulation46. In this study, we found that patients with sepsis had relatively decreased LTB4-induced PMN superoxide anion production and elastase release on admission. By contrast, PMN responses to PMA were similar in sepsis and healthy patients. These findings suggest that, in sepsis patients, PMN responses to receptor-mediated soluble stimuli were uncoupled with a preserved capacity for NADPH oxidase assembly and degranulation in the presence of receptor independent stimuli. The detection of this functional distinction emphasizes the potential value of a cellular biomarker. There was a time-dependent increase in LTB4-initiated responses in sepsis from day 0 to day 7 that reflected decreasing clinical severity.
According to previous studies, the composition of circulating PMNs can change in sepsis patients47. Leukocyte differential counting commonly identifies PMN band cells in sepsis patients that can be distinguished from mature PMNs by morphology and flow cytometry criteria47. Here we identified PMN subsets using flow cytometry with a focus on CD16 surface expression. PMNs from healthy subjects were predominantly CD16bright. Patients with sepsis had increased percentages of CD16dim and CD16− PMNs and a decreased percentage of CD16bright PMNs relative to healthy patients. Notably, the percentage of CD16bright and CD16dim PMNs significantly changed over time. The dynamic changes in PMN subsets correlated to clinical severity scores at presentation (APACHE II score) and over time (SOFA score).
Cell activation can be determined by monitoring the IDP after IDS22. Previous quantifiable methods for the electrical characterization of neutrophil activation, such as patch clamping48 and electrorotation49, have a low throughput that makes it difficult to continuously characterize thousands of cells to provide clinically meaningful results. Other high-throughput electrical characterization techniques, such as microfluidic impedance cytometry50, have not been shown to be sensitive to PMN activation and, furthermore, are sensitive to size variations that may confound the analysis. The rapid quantification of activated PMNs using IDS can provide a timely indicator of host responses to sepsis progression, thus allowing faster stratification of the severity of illness and improved administration of targeted treatment. In the course of the research presented here, we found that the electrical properties correlated better with the SOFA score and mechanical ventilation than the current measures of leukocyte enumeration. Differences in the electrical properties of human PMNs are due to electrical changes in both membrane capacitance and cytoplasm conductivity during the degranulation process and a substantial increase in reactive oxygen species generation. The IDS platform enables the characterization of PMN activation in sepsis patients by monitoring the cells’ natural (high-conductivity) media. In preclinical animal models, electrical cell profiles correlate with established assessments of cell activation (flow cytometry immunophenotyping)22. Here, IDS distinguished ex vivo human PMNs activated by PMA from non-activated PMNs. We used the Δmed IDP, as it provides the quantitative IDP change that reflects the changing electrical properties according to PMN activation, with the addition of a positive control to account for variabilities in the device and sample. In healthy patients, a decreased median IDP with PMA relative to the vehicle was expressed as a Δmed IDP. The Δmed IDP in sepsis patients was significantly smaller because the median IDP for vehicle-exposed PMNs from patients with sepsis was already reduced compared to PMNs from healthy subjects, consistent with the notion that PMNs from sepsis patients were activated in vivo. The Δmed IDP for PMNs from sepsis patients significantly increased from day 0 to day 7, approaching the Δmed IDP for PMNs in healthy patients at hospital day 7. In addition, the SOFA score was inversely correlated with Δmed IDP. Notably, the differences in flow rate through the designed inertial microfluidic and IDS devices precluded their integration, so PMNs were isolated by negative selection before IDS. However, there are a variety of ways to connect a high-flow-rate device to a low-flow-rate device. One approach, for example, is to store isolated PMNs from the spiral device in an injection loop, which can be loaded at a high flow rate, and then inject that sample into the IDS device at a lower flow rate for electrical profiling. In this fashion, the injection loop serves to bridge the flow rate mismatch between the two devices. Such integration will be one focus of future work.
Monocytes in peripheral human blood can be classified into classical CD16lowCD14high, intermediate CD16highCD14high and non-classical CD16highCD14low immune phenotypes51. In acute inflammation, the levels of circulating classical monocytes can rapidly decrease with increased intermediate monocytes and non-classical monocytes52,53. The intermediate monocyte population is of interest as, relative to other monocyte subsets, these cells display proinflammatory properties54 and are associated with risk for acute inflammatory diseases, such as acute respiratory distress syndrome (ARDS)52. Consistent with these reported changes, we identified an expansion of intermediate monocytes and concomitant reduction in the percentages of classical monocytes in patients admitted with sepsis (day 0). The APACHE II score was inversely correlated with the classical monocyte percentage. The percentages of classical and intermediate monocytes changed over time, returning towards the values seen in healthy patients. Thus, bedside determinations of monocyte subsets in patients with sepsis may reflect disease severity and convey prognostic information.
In clinical practice, clinical assessment of the immune system during disease is based almost entirely on clinical criteria and the peripheral blood leukocyte count and differential6. Using a multivariate principal component analysis, the score plot of routine CBC measures and clinical severity indicators determined a notable overlap between healthy subjects and sepsis patients (day 0 to day 7) and thus they were not powerful identifiers of ‘sick’ versus ‘not sick’ clinical status. By comparison, the score plot and aggregation of the mean z scores of leukocyte phenotype markers and PMN functional responses differentiated healthy and sepsis patients significantly at day 0. Measures of PMN function changed significantly from day 0 to day 7 and correlated with the SOFA score. The Δmed IDP was significantly lower for the most critically ill patients who required mechanical ventilation. Together, these findings suggest the measurement of leukocyte function (rather than number) as a potential biomarker of sepsis severity.
While individual inflammatory mediators can be significantly correlated with sepsis55 and its severity, they have not proved clinically useful secondary to the heterogenous nature of sepsis-associated infections and host responses. Single cell transcriptomics approaches show promise56 but are limited by expense and availability. Our findings in this study support cell-based measures as a potential clinical tool for assessing sepsis severity. Here the measures of cell activation and function outperformed routine CBC enumeration for correlation with sepsis severity and distinguishing sepsis patients from healthy patients. In addition, only microlitre quantities of blood were required to ascertain these functional measures. Here the percentages of PMN subsets and the Δmed IDP of PMNs best correlated with sepsis severity. Several factors may explain the relatively low r values for some of these correlations, including: variance in the clinical prediction tools (SOFA and APACHE II scores); variance in the clinical disorders and comorbidities leading to sepsis in the volunteer subjects; and the limited sample size in this proof-of-concept clinical cohort. While the limitations of the SOFA score are acknowledged, it is the best available measure at present57. In addition to the prediction tool, we identified a correlation between leukocyte function (Δmed IDP) and the clinical need for mechanical ventilation as a surrogate for disease severity.
While this system is amenable to the microbiological determination of bacteria and their responses to antibiotics17,18, here we did not focus on the diagnosis of sepsis. Rather, this project focused on determining the host response to critical illness and we have translated its use to sepsis as the most common clinical condition of leukocyte activation in the intensive care unit. In sepsis there is only rarely a failure to obtain source control of the infection. More commonly, an overly exuberant host immune response leading to multiorgan system injury causes excess morbidity and mortality in sepsis. We lack clinical diagnostics for the early and rapid assessment of the nature and intensity of the host response, so this project was designed to monitor circulating leukocyte activation and function as relevant indicators of the host response to address this important unmet clinical need and, in the longer term, to enable new therapeutic strategies that target the host response. In addition, this study has identified several leukocyte parameters that correlate with sepsis severity and could serve as leukocyte biomarkers for early stratification, on initial assessment, and for disease progression, with repeated measurement. The correlation of these parameters with early stratification and clinical outcomes will need validation and further study in a larger clinical cohort that incorporates patients at more than a single clinical centre.
We have demonstrated that the assessment of specific leukocyte subsets and functional responses using a sample sparing closed-loop inertial microfluidic system and IDS is feasible and that it enabled the predictive assessment of host responses to sepsis and its severity. Measures of leukocyte activation and function outperformed the current measures of leukocyte enumeration (CBC). The bedside assessment of the host immune response to sepsis required only microlitre quantities of blood, which would reduce the risk for iatrogenic anaemia in hospitalized patients. Further research is needed to determine the ultimate clinical utility of the diagnostic testing of host leukocyte function as a window into the immunopathogenesis of disease and its resolution.
Methods
Design of the inertial microfluidic chip
A spiral microchannel with a trapezoidal cross-section was made of polydimethylsiloxane (PDMS) with soft lithography, as previously described18,20,21. The mould was designed with three dimensional computer-aided design and micromachined on aluminium (Whits Technologies). Four spiral channels were parallelized with individual inlets and common outlets (Fig. 1a). In this study, double-layered microfluidics (eight spiral microchannels in total) were used to enhance the throughput and match the range of the flow sensor. Further enhancement of throughput was achieved by stacking more channels within the tolerable dead volume limits of the assay. The first (thick) and second (thin) layers of the spiral channel were constructed by casting degassed PDMS (10:1 mixture of base and curing agent, Sylgard 184, Dow Corning Inc.) onto the aluminium mould and curing on the hotplate for 10 min at 150 °C before peeling the cured PDMS off the mould. Two layers were plasma bonded with the channel side facing up. A thin film of PDMS (BISCO HT-6240 40 Duro silicone sheet, 0.25 mm thick, Rogers Corp.) was also plasma bonded on the glass slide. Both complexes were placed into the oven and annealed for 6 h. Holes for the fluidic access of microchannels were punched with 4 mm biopsy punches (Integra Miltex). The punched two-layered PDMS was then plasma bonded to a PDMS glass slide and placed in a 60 °C oven for over 6 h to strengthen the bonding. The microfluidic chips were then sealed in an autoclavable bag and sterilized before use.
Preparation of the closed-loop inertial microfluidic separation system
Inertial microfluidic cell separation was performed using a closed-loop system that included a circulation pump, a microfluidic chip (see above), a pinch valve and a flowmeter. The circulation pump (peristaltic, diaphragm and piezoelectric pump, Cole–Parmer) was connected to the sample and to the microfluidic chip via silicone tubing (Cole–Parmer). To evenly load a sample into each channel, a 0.16 cm barbed five-way connector was used and an equal length of tubing was directly plugged into the inlet holes of the microfluidic chip from the connector. Both the inner-wall and outer-wall outlet tubing were also of equal lengths. The inner-wall outlet was connected to the sample for recirculation and the outer-wall outlet was connected to a pinch valve and flowmeter to adjust the fluidic resistance, before ending in the waste tube (Fig. 1a,b and Supplementary Fig. 1). After preparation, PBS−/− was introduced at a flow rate of 4 ml min−1 to prime the system before sample loading.
Sample collection and processing for inertial microfluidic separation system
This study was reviewed and approved by the Partners Healthcare Institutional Review Board (IRB no. 2002P000272 for healthy donors; IRB no. 2008P000495 for sepsis patients in the Registry of Critical Illness intensive care unit population that was previously described in detail58; and IRB no. 2017P000367 for patients admitted to the hospital floor services who had a positive blood culture result within 24 h and were adjudicated to have sepsis based on the Sepsis-3 criteria as determined by an adjudication committee of critical care physicians, as for the Registry of Critical Illness population58). After obtaining informed consent, peripheral blood (500 µl) was freshly obtained from 10 healthy volunteers and 18 patients with sepsis within 72 h of admission to the Brigham and Women’s Hospital medical intensive care unit (day 0) and on days 3 and 7 thereafter of their hospitalization; it was collected in EDTA-containing vacutainer tubes. Sepsis was defined according to the Sepsis-3 guidelines6 and SOFA scores were calculated as previously described59. Healthy volunteers reported general health and no prescription or over-the-counter medications within the 2 weeks before enrolment. A fraction of the freshly obtained peripheral blood (50 μl) was added to the sample tube containing 50 ml buffer solution (PBS without Mg2+ and Ca2+, Gibco) and connected to the circulation pump (see above). The pump flow was set to 12 ml min−1 and the flow resistance was adjusted to a 4.0 ml min−1 outer-wall outlet flow rate using the pinch valve. When the sample volume reached 7.5 ml, an additional 35 ml of PBS−/− was added to the sample tube for further leukocyte enrichment. When the sample volume reached 1 ml, the flow resistance was increased and the circulation pump was stopped. Next, air was introduced to recover the residual fluid in the tubing and channels. For comparison with microfluidic cell sorting, granulocytes were isolated from healthy subjects by density gradient centrifugation over Histopaque 1077 (Sigma–Aldrich) and Histopaque 1119 (Sigma–Aldrich) as in ref. 60.
Flow cytometry analysis
For flow cytometry analysis, 50 μl peripheral venous blood was used to obtain 100,000 leukocytes via the inertial microfluidic system (Fig. 1). Leukocytes were incubated for 20 min at RT with the following antibodies to human proteins, with clones noted in parentheses: anti-CD45 PerCP (HI30), anti-CD66b Pacific blue (G10F5), anti-CD16 APC-Cy7 (3G8), anti-CD69 FITC (FN50), pHrodo (PE), anti-CD62L Brilliant Violet 510 (DREG-56), anti-CD42b Alexa Fluor 700 (HIP1) (all from BioLegend) and anti-CD14 APC (61D3) and anti-CD11b PE-Cy7 (ICRF44) (from Thermo Fisher). After staining, the cells were lysed and fixed with 2 ml 1:4 dilution of Lyse/Fix Buffer 5× (BD Phosflow) with distilled water for 15 min at RT. Data were acquired from the BD LSR Fortessa flow cytometer and analysed using FlowJo version 10.1 (Tree Star). PMN (CD45+SSCHiFSCHi) subsets were determined by CD16 and CD66b surface expression. Monocyte (CD45+SSCLowFSCHigh) subsets were determined by CD14 and CD16 surface expression. The molecules of leukocyte activation were assessed, namely the expression of CD62L, CD11b, CD69 and CD42b.
PMN O2− production
PMNs were isolated from 50 μl peripheral blood using the inertial microfluidic system (Fig. 1). PMNs (10,000 per well) were transferred to a 96-well plate and were exposed to cytochalasin B at 10 μM for 5 min at 37 °C. After incubation, the cells were stimulated with LTB4 at 100 nM or vehicle (ethanol 0.02% v/v) and exposed to cytochrome C reduction from equine heart (Sigma c2506; 0.0007% v/v) for 10 min at 37 °C. In addition, PMNs were incubated for 10 min at 37 °C after being stimulated with either PMA at 100 nM or vehicle (ethanol 0.02% v/v) and exposed to cytochrome C reduction from equine heart (0.0007% v/v). After incubation, the plate was immediately put in an ice water bath to stop the reaction. The cells were pelleted at 600g for 8 min at 0 oC and 90 μl supernatant was transferred to another 96-well plate. The plate was read on the absorbance plate reader (uQuant Biotek Instrument) using the 500–600 nM spectrum.
PMN elastase degranulation
PMNs were isolated from 50 μl peripheral blood using the inertial microfluidic system (Fig. 1). PMNs (5,000 per well) were transferred to a 96-well plate and exposed to LTB4 at 100 nM, PMA at 20 nM or vehicle (ethanol 0.02% v/v for LTB4; DMSO 0.002% v/v for PMA) for 3 h at 37 °C. After incubation, the cells were pelleted at 1,200 r.p.m. for 10 min and 2.5 μl supernatant was transferred to a fluorescence plate containing non-fluorescent elastase substrate (Z-Ala-Ala-Ala-Ala)2Rh110. After incubating the plate for 90 min, the elastase released cleaved Z-Ala-Ala-Ala-Ala 2Rh110 to a highly fluorescent compound, R110, that can be measured using the Flx-700 with an excitation wavelength of 485 nm and emission wavelength of 525 nm. More information on this commercially available assay can be found at Cayman Chemicals61.
PMN phagolysosome acidification assay
PMN phagocytic capacity was determined using the pHrodo red E. coli bioparticles conjugate. pHrodo was resuspended with 2 ml PBS (without Ca2+ and Mg2+) and sonicated for 5 min. After incubation, 2 ml of FBS was added to the pHrodo and incubated for 30 min at 37 °C. After incubation, the particles were pelleted at 1,600g for 5 min and then resuspended with 2 ml PBS (with Ca2+ and Mg2+) and sonicated for 10 min before use. PMNs (100,000 per FACS tube) from 50 μl peripheral venous blood were isolated using the inertial microfluidic system (see Fig. 1). PMNs were incubated for 20 min at 37 oC after exposing cells with 25 μl pHrodo and a total of 4 μl antihuman antibodies to human proteins: anti-CD45 (PerCP), anti-CD66b (Pacific blue), anti-CD16 (APC-Cy7), anti-CD14 (APC), anti-CD69 (FITC), anti-CD11b (PE-Cy7), anti-CD62L (Brilliant Violet 510) and anti-CD42b (Alexa Fluor 700). After staining, the cells were lysed and fixed with 2 ml 1:4 dilution of Lyse/Fix Buffer 5× (BD Phosflow) with dH2O for 15 min at RT. Data were acquired from the BD LSR Fortessa flow cytometer and analysed using FlowJo version 10.1 (Tree Star).
Device fabrication and pretreatment
The double-sided IDS device consisted of a microfluidic channel covered by a double-sided tape and cut by two electrode-patterned glass slides. We e-beam evaporated 10 nm of Ti and 200 nm of Au and performed a lift-off to pattern the electrodes on the glass. The gap and width of the interdigitated electrodes were 15 and 50 μm, respectively. We cut a 2-mm-wide microchannel using a laser cutter in 25-μm-thick double-sided tape (silicon transfer film adhesive, Adhesive Research) and aligned it to the glass substrate. It was then pressed to bond the channel to one of the substrates. The 1.5-mm-wide inlets were punched using diamond bits on the top glass slide to allow fluidic access to the microchannel. The top glass slide was then aligned and pressed onto the top side of the tape on a hotplate at 90 °C for 60 min with a 1 kg weight on top to ensure a proper seal. Finally, we attached the wires to electrode contact pads on both glass slides using a two-part, conductive silver epoxy (MG Chemicals).
The surface treatment of the device was performed by applying a preflow treatment of 1% (weight/volume) Pluronic F-127 solution (Sigma–Aldrich) to prevent the attachment of bioparticles to the device surface. After 15 min of Pluronic treatment, we washed the device by loading fresh PBS into the inlets and flushing for 5 min. After IDS operation, we washed the device with Tryple Select 10× (Life Technologies, A12177–01) and incubated it for 10 min at 37 °C in 5% CO2 to remove the attached non-specific cells. Following the incubation step, we washed the device 2–3 times with deionized water.
Sample processing for IDS
PMNs were isolated from 100 μl whole blood by magnetic bead-based negative selection using the MACSxpress Neutrophil Extraction Kit (Miltenyi Biotec) and a strong magnet (MACSxpress Separator, Miltenyi Biotec). Harvested PMNs were resuspended in PBS (with Ca2+ and Mg2+) by centrifugation at 1,000 r.p.m. for 5 min. PMNs were stimulated with PMA at 20 nM (Cayman Chemical Co. Inc.) or vehicle (ethanol v/v 0.02%) and incubated for 30 min at 37 °C in 5% CO2. After incubation, PMNs were resuspended in PBS buffer by a single centrifugation at 1,000 r.p.m. for 5 min.
IDS experimental set-up
MACS-separated PMNs suspended in high-conductivity media were allowed to flow into the microfluidic chip. The other two inlets containing intermediate and low conductivity media provided a parallel laminar coflow that generated a transverse conductivity gradient. The slanted planar electrodes that were patterned at the top and bottom guided the PMNs in the direction of decreasing medium conductivity until the DEP force became sufficiently small. The PMNs escaped the dielectrophoretic barrier at their IDP, at which point they were at a dielectric equilibrium with the surrounding medium. The PMNs continued downstream unobstructed for collection and imaging. A camera near the end of the microchannel continuously took images of the deflected PMNs. We detected these cells and computed the resulting IDP distribution by image processing.
A function generator (Agilent 33220A) and an amplifier applied a 15 V peak-to-peak AC field at various frequencies from 1 to 10 MHz. PMNs were observed using a camera (Image QE, LaVision) with fully automated microscopy (Axio Imager Mu1m, Zeiss). We analysed the images to detect the cells in each frame and determine the centroid of each cell using a MATLAB script. Then we created the cell position distributions that correspond to the resulting IDP distribution. To drive the flow process, external syringe pumps controlled the flow in all three inlets. BD Luer-Lok 1 ml syringes were attached to 150 μm diameter Tygon tubes and inserted into the device inlets. We used cell suspensions (1 × 106 cells per ml) in PBS buffer with a conductivity of 1.2 S m−1 (PBS and 1% BSA). We mixed the PBS buffer with an isotonic sucrose solution to make an intermediate conductivity buffer (σ = 0.8 S m−1) and a low conductivity buffer (σ = 0.4 S m−1). To avoid fouling and the injection of air bubbles, we used a custom-made fluidic switch that allowed device priming and cleaning between samples.
Image analysis for IDS
All of the image processing and analysis was performed in MATLAB R2018b. We obtained background images through a time-domain median filter. We calculated the difference between the raw images and background and transformed the images into binary images with a threshold of 0.05. We identified the cell objects in the binary images and extracted their morphometric properties. The extracted cells’ centroid locations in each frame were then applied to the customized multiobject tracking algorithm, thus creating the cell position distributions that correspond to the resulting IDP distribution.
Statistics
All of the data were expressed as the mean ± s.e.m, unless otherwise indicated. For statistical analysis, comparisons of patients with sepsis and healthy subjects were conducted by unpaired two-tailed Student’s t test, whereas comparisons of matched samples were conducted using a paired two-tailed Student’s t test. For non-normal distributions, a Mann–Whitney U test or Wilcoxon matched-pairs signed rank test was used. Multiple group comparisons were conducted using one-way ANOVA. The correlations were evaluated using Pearson’s correlation coefficient r. A value of P < 0.05 was considered statistically significant. The statistical analyses were performed using Prism 6.0 for Mac (GraphPad). For multivariate statistical analysis, principal component analysis was performed for samples with complete datasets using R software and the mean z score was calculated using SPSS.
Study approval
Written informed consent from subjects or their surrogates was obtained after Institutional Review Board approval (IRB no. 2002P000272 for healthy subjects, IRB no. 2008P000495 for intensive care unit subjects with sepsis58 and IRB no. 2017P000367 for floor subjects with sepsis59), before inclusion in the study at the Brigham and Women’s Hospital.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary figures.
Acknowledgements
The authors acknowledge the contributions of G. Zhu, L. A. Cosimi and additional members of the Brigham and Women’s Registry of Critical Illness (including L. Fredenburgh, P. Dieffenbach, S. Ash and J. Englert). The work was supported by grant nos. U24-AI118656 (B.D.L., J.V.), K08-HL130540 (R.E.A.) and K12-HD047349 (M.G.D).
Author contributions
B.J., H.R. and D.-H.L. contributed equally to this study. J.H., J.V. and B.D.L. coconceived the study. B.J., H.R., D.-H.L., R.E.A., B.D.E., M.G.D., J.L., R.M.B., N.K., J.H., J.V. and B.D.L. designed the experiments and interpreted the results. B.J., D.-H.L., R.E.A., B.D.E, J.L., J.V. and B.D.L. performed and analysed the functional biological experiments. H.R. and J.H. designed and performed experiments using the inertial microfluidic system. A.H., M.P.-V., M.E.B. and R.M.B. provided blood samples and clinical data from patients with sepsis. The manuscript was written by B.J., H.R., D.-H.L., R.E.A., B.D.E., M.G.D., J.L., N.K., R.M.B., J.H., J.V. and B.D.L.
Data availability
The data supporting the results in this study are available in the Article and Supplementary Information. The raw patient data are available from the authors, subject to approval from the Institutional Review Board of Partner’s Healthcare.
Code availability
The custom code used in this study is available at https://github.com/bdlevylab/BDLevy-Lab.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bakr Jundi, Hyunryul Ryu, Do-Hyun Lee
These authors jointly supervised this work: Joel Voldman, Bruce D. Levy.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41551-019-0473-5.
References
- 1.Delano MJ, Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J. Clin. Invest. 2016;126:23–31. doi: 10.1172/JCI82224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus DC, van der Poll T. Severe sepsis and septic shock. N. Engl. J. Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 3.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majno, G. & Joris, I. Cells, Tissues, and Disease 2nd edn (Oxford University Press, 2004).
- 5.Spite M, et al. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 2009;461:1287–1291. doi: 10.1038/nature08541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer M, et al. The Third International Consensus definitions for sepsis and septic shock (Sepsis-3) J. Am. Med. Assoc. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee C, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. J. Am. Med. Assoc. 2017;318:1241–1249. doi: 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchegger, P. & Preininger, C. Point-of-care chip for diagnosis of sepsis: joining biomarker quantification and bacterial class identification. Biomed. Tech.10.1515/bmt-2013-4146 (2013). [DOI] [PubMed]
- 9.Hassan U, et al. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat. Commun. 2017;8:15949. doi: 10.1038/ncomms15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemmler M, Sauer U, Schleicher E, Preininger E, Brandenburg A. Biochip point-of-care device for sepsis diagnostics. Sens. Actuat. B. 2014;192:205–215. doi: 10.1016/j.snb.2013.10.003. [DOI] [Google Scholar]
- 11.Reddy B, et al. Point-of-care sensors for the management of sepsis. Nat. Biomed. Eng. 2018;2:640–648. doi: 10.1038/s41551-018-0288-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, et al. Detection of sepsis in patient blood samples using CD64 expression in a microfluidic cell separation device. Analyst. 2017;143:241–249. doi: 10.1039/C7AN01471F. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Multiparameter affinity microchip for early sepsis diagnosis based on CD64 and CD69 expression and cell capture. Anal. Chem. 2018;90:7204–7211. doi: 10.1021/acs.analchem.7b05305. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, et al. Detection of culture-negative sepsis in clinical blood samples using a microfluidic assay for combined CD64 and CD69 cell capture. Anal. Chim. Acta. 2019;1062:110–117. doi: 10.1016/j.aca.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 15.Ellett F, et al. Diagnosis of sepsis from a drop of blood by measurement of spontaneous neutrophil motility in a microfluidic assay. Nat. Biomed. Eng. 2018;2:207–214. doi: 10.1038/s41551-018-0208-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent JL, et al. Anemia and blood transfusion in critically ill patients. J. Am. Med. Assoc. 2002;288:1499–1507. doi: 10.1001/jama.288.12.1499. [DOI] [PubMed] [Google Scholar]
- 17.Ryu, H.et al. Label-free neutrophil enrichment from patient-derived airway secretion using closed-loop inertial microfluidics. J. Vis. Exp. 10.3791/57673 (2018). [DOI] [PMC free article] [PubMed]
- 18.Wu L, Guan G, Hou HW, Bhagat AA, Han J. Separation of leukocytes from blood using spiral channel with trapezoid cross-section. Anal. Chem. 2012;84:9324–9331. doi: 10.1021/ac302085y. [DOI] [PubMed] [Google Scholar]
- 19.Ryu H, et al. Patient-derived airway secretion dissociation technique to isolate and concentrate immune cells using closed-loop inertial microfluidics. Anal. Chem. 2017;89:5549–5556. doi: 10.1021/acs.analchem.7b00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amini H, Lee W, Di Carlo D. Inertial microfluidic physics. Lab Chip. 2014;14:2739–2761. doi: 10.1039/c4lc00128a. [DOI] [PubMed] [Google Scholar]
- 21.Di Carlo D. Inertial microfluidics. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 22.Prieto JL, et al. Monitoring sepsis using electrical cell profiling. Lab Chip. 2016;16:4333–4340. doi: 10.1039/C6LC00940A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez I, et al. A label free disposable device for rapid isolation of rare tumor cells from blood by ultrasounds. Micromachines. 2018;9:129. doi: 10.3390/mi9030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, et al. Acoustic separation of circulating tumor cells. Proc. Natl Acad. Sci. USA. 2015;112:4970–4975. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mach AJ, Di Carlo D. Continuous scalable blood filtration device using inertial microfluidics. Biotechnol. Bioeng. 2010;107:302–311. doi: 10.1002/bit.22833. [DOI] [PubMed] [Google Scholar]
- 26.Nivedita N, Papautsky I. Continuous separation of blood cells in spiral microfluidic devices. Biomicrofluidics. 2013;7:54101. doi: 10.1063/1.4819275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith AJ, et al. Rapid cell separation with minimal manipulation for autologous cell therapies. Sci. Rep. 2017;7:41872. doi: 10.1038/srep41872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ai Y, Sanders CK, Marrone BL. Separation of Escherichia coli bacteria from peripheral blood mononuclear cells using standing surface acoustic waves. Anal. Chem. 2013;85:9126–9134. doi: 10.1021/ac4017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl Acad. Sci. USA. 2017;114:10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PR. Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation. Lab Chip. 2008;8:1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vahey MD, Quiros Pesudo L, Svensson JP, Samson LD, Voldman J. Microfluidic genome-wide profiling of intrinsic electrical properties in Saccharomyces cerevisiae. Lab Chip. 2013;13:2754–2763. doi: 10.1039/c3lc50162k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl Acad. Sci. USA. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nivedita N, Ligrani P, Papautsky I. Dean flow dynamics in low-aspect ratio spiral microchannels. Sci. Rep. 2017;7:44072. doi: 10.1038/srep44072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Papautsky I. Size-based microfluidic multimodal microparticle sorter. Lab Chip. 2015;15:1350–1359. doi: 10.1039/C4LC00803K. [DOI] [PubMed] [Google Scholar]
- 35.Hou HW, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khoo BL, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS ONE. 2014;9:e99409. doi: 10.1371/journal.pone.0099409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warkiani ME, et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14:128–137. doi: 10.1039/C3LC50617G. [DOI] [PubMed] [Google Scholar]
- 38.Warkiani ME, et al. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst. 2014;139:3245–3255. doi: 10.1039/C4AN00355A. [DOI] [PubMed] [Google Scholar]
- 39.Warkiani ME, et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2016;11:134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 40.Yin L, et al. Microfluidic label-free selection of mesenchymal stem cell subpopulation during culture expansion extends the chondrogenic potential in vitro. Lab Chip. 2018;18:878–889. doi: 10.1039/C7LC01005B. [DOI] [PubMed] [Google Scholar]
- 41.Choi K, et al. Negative selection by spiral inertial microfluidics improves viral recovery and sequencing from blood. Anal. Chem. 2018;90:4657–4662. doi: 10.1021/acs.analchem.7b05200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou HW, Bhattacharyya RP, Hung DT, Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015;15:2297–2307. doi: 10.1039/C5LC00311C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdulla A, Liu W, Gholamipour-Shirazi A, Sun J, Ding X. High-throughput isolation of circulating tumor cells using cascaded inertial focusing microfluidic channel. Anal. Chem. 2018;90:4397–4405. doi: 10.1021/acs.analchem.7b04210. [DOI] [PubMed] [Google Scholar]
- 44.Nivedita N, Garg N, Lee AP, Papautsky I. A high throughput microfluidic platform for size-selective enrichment of cell populations in tissue and blood samples. Analyst. 2017;142:2558–2569. doi: 10.1039/C7AN00290D. [DOI] [PubMed] [Google Scholar]
- 45.Shen XF, Cao K, Jiang JP, Guan WX, Du JF. Neutrophil dysregulation during sepsis: an overview and update. J. Cell. Mol. Med. 2017;21:1687–1697. doi: 10.1111/jcmm.13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomkin JS, Cotta LA, Brodt JK, Hurst JM. Regulation of neutrophil superoxide production in sepsis. Arch. Surg. 1985;120:93–98. doi: 10.1001/archsurg.1985.01390250081013. [DOI] [PubMed] [Google Scholar]
- 47.Pillay J, et al. A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J. Clin. Invest. 2012;122:327–336. doi: 10.1172/JCI57990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lollike K, Lindau M. Membrane capacitance techniques to monitor granule exocytosis in neutrophils. J. Immunol. Methods. 1999;232:111–120. doi: 10.1016/S0022-1759(99)00169-6. [DOI] [PubMed] [Google Scholar]
- 49.Griffith AW, Cooper JM. Single-cell measurements of human neutrophil activation using electrorotation. Anal. Chem. 1998;70:2607–2612. doi: 10.1021/ac980070c. [DOI] [PubMed] [Google Scholar]
- 50.Holmes D, et al. Leukocyte analysis and differentiation using high speed microfluidic single cell impedance cytometry. Lab Chip. 2009;9:2881–2889. doi: 10.1039/b910053a. [DOI] [PubMed] [Google Scholar]
- 51.Ziegler-Heitbrock L. Monocyte subsets in man and other species. Cell. Immunol. 2014;289:135–139. doi: 10.1016/j.cellimm.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Abdulnour RE, et al. Early intravascular events are associated with development of acute respiratory distress syndrome. A substudy of the LIPS-A clinical trial. Am. J. Respir. Crit. Care Med. 2018;197:1575–1585. doi: 10.1164/rccm.201712-2530OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zawada AM, et al. SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood. 2011;118:e50–e61. doi: 10.1182/blood-2011-01-326827. [DOI] [PubMed] [Google Scholar]
- 55.Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit. Care. 2010;14:R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seymour CW. Assessment of clinical criteria for sepsis. JAMA. 2016;315:762. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolinay T, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am. J. Respir. Crit. Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fredenburgh LE, et al. A phase I trial of low-dose inhaled carbon monoxide in sepsis-induced ARDS. JCI Insight. 2018;3:e124039. doi: 10.1172/jci.insight.124039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barnig C, et al. Lipoxin A4 regulates natural killer cell and type 2 innate lymphoid cell activation in asthma. Sci. Transl. Med. 2013;5:174ra126. doi: 10.1126/scitranslmed.3004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neutrophil Elastase Activity Assay Kit. Cayman Chemicalhttps://www.caymanchem.com/product/600610/neutrophil-elastase-activity-assay-kit (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.
Data Availability Statement
The data supporting the results in this study are available in the Article and Supplementary Information. The raw patient data are available from the authors, subject to approval from the Institutional Review Board of Partner’s Healthcare.
The custom code used in this study is available at https://github.com/bdlevylab/BDLevy-Lab.