Abstract
Strain-level epidemiology is a key approach to understanding the mechanisms underlying establishment of any host-microbe association. The squid-vibrio light organ symbiosis has proven to be an informative and tractable experimental model in which to discover these mechanisms because it involves only one bacterial species, Vibrio fischeri. In this horizontally transmitted symbiosis, the squid presents nutrients to the bacteria located in a bilobed light-emitting organ, while the symbionts provide bioluminescence to their host. To initiate this association, V. fischeri cells go through several distinct stages: from free-living in the bacterioplankton, to forming a multicellular aggregation near pores on the light organ’s surface, to migrating through the pores and into crypts deep in the light organ, where the symbiont population grows and luminesces. Because individual cells must successfully navigate these distinct regions, phenotypic differences between strains will have a strong impact on the composition of the population finally colonizing the squid. Here we review recent advances in our understanding of behavioral characteristics that differentially drive a strain’s success, including its effectiveness of aggregation, the rapidity with which it reaches the deep crypts, and its deployment of type VI secretion.
Graphical Abstract
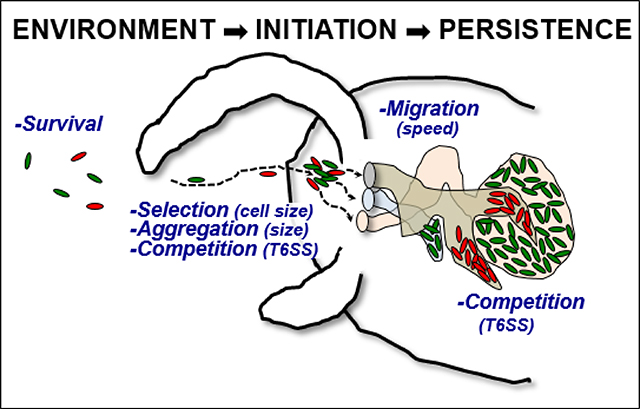
Summary figure: Vibrio fischeri cells (green ovals) are found at different stages: free in the environmental bacterioplankton, colonizing the squid, and persisting in the crypts of the light organ (pink, brown and light blue). The dashed arrows indicate the trajectory of the bacterial strains during colonization of the juvenile squid. Different activities (dark blue) influence the bacteria, depending on which stage they are in. In this figure, only the left side of the light organ is shown.
Introduction
The squid-vibrio symbiosis has been a powerful model for identifying and deciphering the mechanisms by which strain-level differences ultimately impact the horizontal transmission of a symbiont. The newly hatched bobtail squid, Euprymna scolopes, has an aposymbiotic light organ, which becomes colonized by harvesting a few V. fischeri cells present in the ambient seawater [1]. Different bacterial species, including V. fischeri, enter the mantle cavity as seawater is drawn across the gills and the ciliated surface of the nascent light organ. Ciliary activity delivers bacterium-sized particles to a location near the light organ’s pores, where they form aggregates (Figure 1) [1,2]. V. fischeri cells pass into the pores on the nascent light organ’s surface, and migrate through different microenvironments until a few cells reach and colonize the crypts (Figure 1) [3]. Strains isolated from wild-caught E. scolopes have shown distinct phenotypic traits [4,5], genomic composition [6] and competition behaviors [7,8]. Here, we present an overview of recent discoveries explaining the roles these differences play in determining colonization efficiency and effectiveness that drive symbiont population biology in the host.
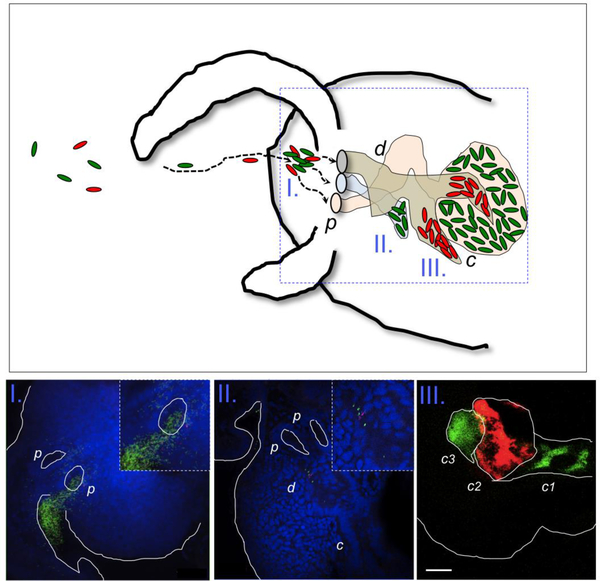
Fig. 1.
(Upper) Different V. fischeri strains, present in the environment, (I.) form aggregates around the three pores (p) of the light organ, then detach and (II.) migrate through the duct (d), and finally (III.) arrive and colonize deep in the crypts (c) of the light organ. Green and red ovals represent different V. fischeri strains in the environment, and the dashed arrows are their trajectories. (Lower) Confocal images corresponding to the different stages of bacterial behavior along the colonization path. Bacteria are GFP or RFP labeled; nuclei of host tissue (blue) were stained with TOTO-3. Cells of V. fischeri are aggregated (in I.), migrating through the ducts (in II.), and colonizing the light organ crypts (in III.); bar = 50 mm. Note that each crypt is typically colonized by a single bacterium (green or red) that initiates the symbiont population therein.
Dominant and Sharing strains
Swiftness of colonization
When squid are experimentally co-inoculated with different symbiont strains, two behaviors were observed: (i) a dominant strain (“D” strain; corresponding to the previously described “A-type” strains [4]) would be found as the only one colonizing the squid, or (ii) a sharing strain (“S” strain) would share the light organ with another S strain [6]. Animals collected in the field are typically colonized by 6 to 8 strains [5]; surprisingly, in spite of this hierarchy of colonization dominance, some wild-caught squid harbor a mixture of both D and S strains. The co-occurrence of these two kinds of strains may be explained by a sequential encounter of different strains during the initial colonization of the juvenile host [7]. In the laboratory, a D strain needed a shorter exposure time than an S strain to colonize >50% of exposed hosts [7]. In addition, a D strain required less time than an S strain to migrate into the crypts, conferring a competitive priority effect (Figure 1). Such a priority effect has also been reported when two strains of Borrelia burgdorferi were co-inoculated into ticks [9], but has principally been described when different species compete for colonization of a host. For example, in the cnidarian-dinoflagellate symbiosis, a prior exposure to one species of alga gives it an advantages over the subsequent colonization by a more thermo-resistant species, even under elevated-temperature selection [10]. Similarly, among Bacteroides spp, a ‘commensal colonization factor’ is involved in priority effects of the bacteria during colonization of the mouse gut [11,12]. The presence and importance of competitive priority effects is understudied in many other models of simple or complex community symbioses, such as in the bee gut [13] and among and between the communities making up the human microbiome [14,15]. Thus, if the squid encounters an S strain sufficiently sooner than a D strain, it may become colonized by both [7]. The mechanism(s) underlying this timing advantage remains unknown, especially because it is not simply a case of swimming speed (D strains swim more slower in a soft-agar medium [5]), and the bacterial migration pathway passes through several tissue microenvironments [3] that are not yet possible to reproduce experimentally.
Population dynamics between the planktonic and symbiotic environments
The first parameter that influences the composition of the light organ population is the strain diversity in the environment [16]. Both D and S strains are typically found in adult squid [5] and, thus, will be in the ambient bacterioplankton to which the hatchling squid are exposed. However, if the D strains have such an advantage over the S strains during colonization [6], and the light organ is essentially the bacterium’s only growth environment [16], one would predict that D strains will eventually sweep the bacterioplankton population. One mitigating factor would be if D strains are less fit in the environment, and decrease in relative abundance over time; this hypothesis was experimentally supported when cells of D (but not S) strains become non-culturable after 48 h in natural ocean water [4]. Thus, an ecological trade-off may occur in which S strains survive better in the bacterioplankton, but D strains are more effective colonizers. As a result, even in an environment containing more S than D strains, a sequential encounter with the juvenile squid would create conditions for stochastic colonization by both types of strains [7]. Finally, it is ecologically significant that only a miniscule portion of the millions of symbionts that an adult releases each dawn into the bacterioplankton will have an opportunity to colonize a juvenile squid, emphasizing the importance of the symbiont’s population biology both in the host and in seawater [16,17].
Aggregation behavior during colonization
Bacterial specificity
On either side of the nascent light organ of a newly hatched E. scolopes, there are two surface appendages that are covered by ciliated fields [3]; the activity of these fields moves seawater in the mantle cavity, winnowing bacteria-sized particles into an accumulation zone near the light organ’s pores [1]. At bacterial concentrations typical of seawater, V. fischeri and certain other Gram-negative species attach to the short cilia found in this zone [18] forming aggregates of a few cells (Figure 1). If these cells are V. fischeri, they specifically induce host responses and chemotax toward the light organ’s pores [19]. In contrast, all tested Gram-positive bacteria did not form such aggregates [2,20]. If V. fischeri cells are added at a concentration above that found in seawater, the bacteria begin to attach to each other, forming aggregates of hundreds to thousands of cells [2]. Some Gram-negative species (e.g., Vibrio campbellii strain KNH1) form larger aggregates than symbiotic V fischeri strains [20]; however, when co-occurring with other species, V. fischeri cells interfere in an unknown way with aggregation by the other species [2]. Thus, while there is no direct correlation between the size of an aggregate and the colonization capability or efficiency of a bacterium, this step, together with flagellar motility and chemotaxis, is a necessary step in the selection of the correct bacterial species [21–23]. Although the symbionts appear to be passive participants in their accumulation at the pores [1], they must subsequently detach from the aggregates and proceed to and through the pores to reach the crypts [18,24].
Aggregation behavior has been observed for a number of different strains of symbiotic V. fischeri, revealing a range of sizes and speeds of aggregation [20]. While there is no direct correlation between the number of cells in an aggregate and the ability to compete for colonization, strains producing an aggregate above a certain size seem to have an advantage. In fact, while the more rapid detachment and subsequent migration of cells from the aggregates into the crypts [7] appears to explain at least some of the dominant behavior, the expression of other as yet unknown adaptability traits specific to D strains may also play a role.
Regulation of aggregate formation
In V. fischeri aggregation is dependent on the expression of the symbiosis polysaccharide (syp) locus, which encodes capsule-synthesis genes, and is under a complex regulation that includes a number of factors that function upstream of the proximal regulatory protein, SypG (Table 1) [26,27]. Studies of this regulatory pathway have been confined to the V. fischeri symbiont strain ES114 and its mutant derivatives ([21]). The first regulator of symbiont aggregation (and biofilm formation in culture) to be identified was RscS [28,29]. In various V. fischeri strains, rscS, is either present, absent or frameshifted [30]. Specifically, RscS is not required for host colonization by several symbiont strains, since they colonize the squid even though they don’t encode RscS, or their rscS gene contains an inactivating frame-shift; nevertheless, they remain dependent on the downstream syp locus. For instance, the D strain MB13B2 forms large aggregates even with a frame-shifted rscS, but loses that phenotype if the structural gene sypQ is mutated [20]. This finding indicated that RscS isn’t the only factor controlling Syp-dependent biofilm formation [21]. In fact, a second key regulator is HahK, which, like RscS, activates the SypF sensor kinase upstream of SypG. In addition to these positive regulators, the negative regulator BinK, which reduces SypG function and antagonizes RscS action [31], appears to be present and functional in all V. fischeri strains examined [30]. Even at this stage in our understanding of this tightly controlled pathway, the complexity of the system indicates V. fischeri aggregate/biofilm production is sensitive to both recognized, and as yet unknown, signals from the abiotic and biotic environment. Niche colonization by many symbionts involves specific aggregation behavior as described, for example, in the reproducible spatial distribution of distinct gut microbes in the zebrafish [32]. Aggregation-driven colonization resistance by the normal microbiota has also been reported to protect against vaginal infection by inhibiting Trichomonas vaginalis adhesion to host cell [33]. Interestingly, Fusobacterium nucleatum protects itself from clearance by adhering to a specific previously attached species as part of its integration into the oral microbial community [34].
Table 1.
Regulation of the V. fischeri syp locus
| Regulatory factor | Type* | Effector / activator(s) | Effect on syptranscription | Other effects† | Homologs present in: | Reference | |
|---|---|---|---|---|---|---|---|
| SypG | VFA1026 | RR | SypF; BinK | + | Nd†† | other Vibrio spp | 25 |
| SypE | VFA1024 | RR | SypF | − | post- transcr. | V. fischeri | 21 |
| SypA | VFA1020 | STAS | SypE | − | post- transcr. | other Vibrio spp | 25 |
| SypF | VFA1025 | HSK | RscS; HahK | + | nd | other Vibrio spp | 25 |
| RscS | VFA0237 | HSK | nd | + | nd | certain V. fischeri | 30 |
| HahK | VFA0072 | SK | HnoX | + | nd | other Proteobacteria | 36 |
| BinK | VFA0360 | SK | nd | − | nd | other Vibrio spp | 31 |
| HnoX | VFA0071 | S | nitric oxide | − | HahK (−) | other Proteobacteria | 24 |
| --- | --- | Ca2+ | + | Bcs (+) | --- | 36 |
HSK (hybrid sensor kinase); RR (response regulator); S (sensor); SK (sensor kinase); STAS (anti-sigma factor antagonist and sulfate transporter domain)
Downstream targets other than the syp locus; Bcs = bacterial cellulose synthesis
nd (none determined)
In addition to these internal regulatory proteins, external nutrient and salt conditions play a modulating role in biofilm formation [35]. In particular, two specific extracellular factors have been shown to control the extent of biofilm formation: calcium is an activator of syp gene expression [36], while nitrite oxide (NO) is an inhibitor [24]. Interestingly, the presence of BinK is sufficient to prevent calcium’s ability to induce biofilm in wild-type cells [36]. In addition, calcium increased the expression of a newly discovered bacterial cellulose-based biofilm through the bcs locus, which was also dependent on the regulator SypF. Nitric oxide synthase is secreted by the host and is present in the mucus, where it produces NO and affects bacterial production of NO-detoxifying activity in the aggregates [37,38]. In addition, host-derived NO serves as a signal to inhibit the formation of Syp polysaccharide, allowing the bacteria to dissociate and migrate from the aggregate and into the crypts of the light organ [24,27]. The degree to which symbiotic V. fischeri cells aggregate and/or dissociate varies from strain to strain, a difference that is likely to contribute significantly to their relative success at squid colonization.
Type VI secretion system (T6SS) activity
The T6SS is a cell-contact mechanism that can be deployed by one bacterium to kill another [39]; such antagonistic behavior provides a fitness advantage during strain-strain competition for nutrients or ecological niches, especially during symbiosis [40]. T6SS also has a role both in shaping the normal host microbiota and in pathogenesis in complex communities [41]: for example, V. cholerae uses a T6SS to attack the pre-existing gut microbiota to facilitate its colonization [42]. The impact of T6SSs in shaping bacterial communities raises the question of its involvement in the colonization of the squid by its symbiont. Two T6SSs have been identified in V. fischeri [8]. Homologs of one system, T6SS1, are present in all strains studied but their specific function has not been described yet [8]. The other one, referred to as T6SS2, is required for killing of conspecific strains when tested in a culture-based co-incubation assay [8]. Homologs of genes encoding this system have been demonstrated in half of the 32 V. fischeri strains examined and, intriguingly, only some of the strains isolated from a given light organ would harbor it [8]. Significantly, the carriage of this T6SS2 and the dominance phenotype are not correlated; in fact, this system is encoded by certain sharing or dominant strains, as well as some strains of V. fischeri that are incapable of colonizing the light organ. At present, six groups of compatible strains have been identified, where incompatible strains must be spatially separated from each other or else one will eliminate the other [8]. These antagonistic interactions can effect colonization of the squid: in the rare event where two incompatible V. fischeri strains reached the same light organ crypt (Figure 1), the T6SS was involved in killing the strain that didn’t encode it [8]. A question of future interest is whether the T6SS2 is involved in strain selection as early as the aggregation step of the squid colonization. Except for when they are attached to particles [43], planktonic V. fischeri cells will rarely encounter each other in the environment; however, because the host concentrates them there ([1,2]), an aggregate provides a time and location at which they would likely come into contact.
Conclusion
Strain variation is a key factor in understanding the epidemiology of host colonization. Here, we described three behaviors – dominance, aggregation, and antagonism – that are employed to different degrees by strains of V. fischeri that are successful light organ symbionts. Other behaviors, such as bioluminescence and chemotaxis, while not described here, also play critical roles in the squid-vibrio association. Each of these mechanisms can be shown to contribute to a strain’s fitness in laboratory assays, but because the symbionts are horizontally transmitted, and encounter different sets of conditions and selective pressures in both their symbiotic and their planktonic niches, evolutionary success is not dependent on only one mechanism. In addition, while this review focuses on strain-dependent behaviors during the initiation of symbiosis, there are distinct and equally stringent behaviors required for bacterial persistence in the association [44,45]. Understanding the dynamics of these interacting mechanisms, and the trade-offs different strains make, is fundamental to understanding the population biology of symbiosis.
Acknowledgements
We thank the Ruby and McFall-Ngai laboratory members, especially Silvia Moriano Gutierrez and Eric Koch, for their helpful discussions and support. We also are grateful to Karen Visick and Alice Tischler for their advices on syp regulation. Funding was provided by NIH grants from NIGMS (GM099507), NIAID (AI050661) and Office of the Director (OD011024).
Footnotes
Conflict of interest
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, Ruby EG, Dabiri JO, Kanso E, McFall-Ngai M: Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci 2017, 114:9510–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ: Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 2000, 97:10231–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai MJ: The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 2014, 68:177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wollenberg MS, Ruby EG: Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 2012, 6:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollenberg MS, Ruby EG: Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 2009, 75:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, McFall-Ngai M, Polz MF, Ruby EG: A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J 2016, 10:2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bongrand C, Ruby EG: Achieving a multi-strain symbiosis: strain behavior and infection dynamics. ISME J 2018, 13:698–706.** This study demonstrates how reaching the light-organ crypts quickly provides a basis for the dominant colonization phenotype expressed by a clade of Vibrio fischeri symbiotic strains, and indicates how multi-strain symbioses can result from a sequential infection of the crypts.
- 8.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN: Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci 2018, 115:8528–8537.** In this break-through report, the presence of two Type Vi Secretion Systems (T6SSs) are reported in V. fischeri strains, one of which is ubiquitous and the other is carried by strains that can kill other non-carrying V. fischeri, creating different groups of compatible symbionts.
- 9.Devevey G, Dang T, Graves CJ, Murray S, Brisson D: First arrived takes all: Inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains Ecological and evolutionary microbiology. BMC Microbiol 2015, 15:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabay Y, Parkinson JE, Wilkinson SP, Weis VM, Davy SK: Inter-partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian-dinoflagellate symbiosis. ISME J 2019, doi: 10.1038/s41396-019-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK: Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013, 501:426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. : Gut microbiota utilize immunoglobulin a for mucosal colonization. Science 2018, 360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson KE, Rodrigues PAP, Mott BM, Maes P, Corby-Harris V: Ecological succession in the honey bee gut: Shift in Lactobacillus strain dominance during early adult development. Microb Ecol 2016, 71:1008–1019. [DOI] [PubMed] [Google Scholar]
- 14.Sprockett D, Fukami T, Relman DA: Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol 2018, 15:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA: Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere 2018, 3:e00441–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KH, Ruby EG: Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 1994, 60:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankey SM, Foxall RL, Ster IM, Perry LA, Schuster BM, Donner RA, Coyle M, Cooper VS, Whistler CA: Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria. eLife 2017, 6:e24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altura MA, Heath-Heckman EAC, Gillette A, Kremer N, Krachler AM, Brennan C, Ruby EG, Orth K, McFall-Ngai MJ: The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol 2013, 15:2937–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer N, Philipp EER, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, et al. : Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 2013, 14:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehler S, Gaedeke R, Thompson C, Bongrand C, Visick K, Ruby E, McFall-Ngai M: The model squid-vibrio symbiosis provides a window into the impact of strain-and species-level differences during the initial stages of symbiont engagement. Environ Microbiol 2018, doi: 10.1111/1462-2920.14392.** This paper presents the observation that the range of V. fischeri symbiont strains are characterized by a span of aggregate sizes with a thousand-fold difference in cell number, and that the hyperaggregation process requires carriage of the syp-capsule locus.
- 21.Thompson CM, Marsden AE, Tischler AH, Koo J, Visick KL: Vibrio fischeri biofilm formation prevented by a trio of regulators. Appl Environ Microbiol 2018, 84:e01257–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG: Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. MicrobiologyOpen 2013, 2:576–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CA, DeLoney-Marino CR, Mandel MJ: Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl Environ Microbiol 2013, 79:1889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson CM, Tischler AH, Tarnowski DA, Mandel MJ, Visick KL: Nitric oxide inhibits biofilm formation by Vibrio fischeri via the nitric oxide sensor HnoX. Mol Microbiol 2019, 111:187–203.** Here, the authors demonstrate that the syp locus is regulated by the presence of host-derived NO, which is sensed by the V. fischeri regulatory protein HnoX, providing a long-awaited mechanism by which colonizing symbionts detach from the aggregate and migrate into host tissue.
- 25.Thompson CM, Visick KL: Assessing the function of STAS domain protein SypA in Vibrio fischeri using a comparative analysis. Front Microbiol 2015, 6:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visick KL: An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri: MicroReview. Mol Microbiol 2009, 74:782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stabb EV: Should they stay or should they go? Nitric oxide and the clash of regulators governing Vibrio fischeri biofilm formation. Mol Microbiol 2019, 111:1–5. [DOI] [PubMed] [Google Scholar]
- 28.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL: The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 2006, 62:1586–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG: A single regulatory gene is sufficient to alter bacterial host range. Nature 2009, 458:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotman ER, Bultman KM, Brooks JF, Gyllborg MC, Burgos HL, Wollenberg MS, Mandel MJ: Natural strain variation reveals diverse biofilm regulation in squid-colonizing Vibrio fischeri. J Bacteriol 2019, 201:e00033–19.** This comparative study of the evolution of the syp-locus’ regulatory histidine kinase, RscS, carried by certain V. fischeri strains, reveals that it is either present, frame-shifted or absent among different symbiosis-competent strains, which either use RscS or replace it with another mechanism by which to regulate the syp locus.
- 31.Brooks JF, Mandel MJ: The histidine kinase BinK Is a negative regulator of biofilm formation and squid colonization. J Bacteriol 2016, 198:2596–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlomann BH, Wiles TJ, Wall ES, Guillemin K, Parthasarathy R: Bacterial cohesion predicts spatial distribution in the larval zebrafish intestine. Biophys J 2018, doi: 10.1016/j.bpj.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phukan N, Brooks AES, Simoes-Barbosa A: A cell surface aggregation promoting factor from Lactobacillus gasseri contributes to inhibition of Trichomonas vaginalis adhesion to human vaginal ectocervical cells. Infect Immun 2018, 86:e00907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Hu W, Kaplan CW, Guo L, Shi W, Lux R: Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb Ecol 2012, 63:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsden AE, Grudzinski K, Ondrey JM, DeLoney-Marino CR, Visick KL: Impact of salt and nutrient content on biofilm formation by Vibrio fischeri. PLoS One 2017, 12:e0169521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischler AH, Lie L, Thompson CM, Visick KL: Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol 2018, 200:e00016–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG: Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 2010, 78:903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ: NO means “yes” in the squid-vibrio symbiosis: Nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 2004, 6:1139–1151. [DOI] [PubMed] [Google Scholar]
- 39.Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH: Rules of engagement: The type VI secretion system in Vibrio cholerae. Trends Microbiol 2017, 25:267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coyne MJ, Comstock LE: Type VI secretion systems and the gut microbiota. Microbiol Spectr 2019, 7:doi: 10.1128/microbiolspec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Yang X, Shen X: Confirmed and potential roles of bacterial T6SSs in the intestinal ecosystem. Front Microbiol 2019, 10:1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao W, Caro F, Robins W, Mekalanos JJ: Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 2018, 359:210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF: Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 2008, 320:1081–1085. [DOI] [PubMed] [Google Scholar]
- 44.Schwartzman JA, Koch E, Heath-Heckman EAC, Zhou L, Kremer N, McFall-Ngai MJ, Ruby EG: The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci 2015, 112:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG: Features governing symbiont persistence in the squid-vibrio association. Mol Ecol 2014, 23:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]