Abstract
HIV-1 Vif promotes degradation of the antiviral APOBEC3 (A3) proteins through the host ubiquitin-proteasome pathway to enable viral immune evasion. Disrupting Vif-A3 interactions to reinstate the A3-catalyzed suppression of HIV-1 replication is a potential approach for antiviral therapeutics. However, the molecular mechanisms by which Vif recognizes A3 proteins remain elusive. Here we report a cryo-EM structure of the Vif-targeted C-terminal domain of human A3F in complex with HIV-1 Vif and its cellular cofactor CBFβ, at 3.9 Å resolution. The structure shows that Vif and CBFβ form a platform to recruit A3F, revealing a direct A3F-recruiting role of CBFβ beyond Vif stabilization, and captures multiple independent A3F-Vif interfaces. Together with our biochemical and cellular studies, our structural findings establish the molecular determinants that are critical for Vif-mediated neutralization of A3F and provide a comprehensive framework of how HIV-1 Vif hijacks the host protein degradation machinery to counteract viral restriction by A3F.
INTRODUCTION
The apolipoprotein B mRNA editing catalytic polypeptide-like 3 (APOBEC3, A3) family of proteins are host intrinsic immunity factors that potently restrict a wide variety of viruses, including human immunodeficiency virus type 1 (HIV-1). As deaminases, A3 enzymes convert cytosine to uracil in the minus strand of viral single-stranded DNA (ssDNA) during reverse transcription, causing lethal hypermutation in the viral genome 1–3. To evade this host defense, HIV-1 virion infectivity factor (Vif), facilitated by the cellular cofactor core-binding factor beta (CBFβ) 4,5, hijacks a host Cullin-RING E3 ubiquitin ligase complex to target A3s for proteasomal degradation 6–10. Humans express seven A3 proteins that are homologous in sequence and structure, each containing one or two zinc-containing deaminase-like domains 11. The di-domain A3s normally have only their C-terminal domain (CTD) catalytically active while their N-terminal domain (NTD) is responsible for encapsidation 12. Of these, A3G and A3F are the most efficient restrictors of HIV-1 13. They interact with HIV-1 Vif through distinct domains, NTD for A3G and CTD for A3F 14–17. Although the two domains are structurally conserved, they are proposed to interact with Vif via two separate interfaces 18,19. Similarly, Vif is predicted to rely on different, partially overlapping motifs from three clustered regions to recognize A3G and A3F 18,20.
The Vif-hijacked E3 ubiquitin ligase is composed of the scaffold protein Cullin5 (Cul5), the E2-binding Rbx2, and adaptor proteins Elongin B (EloB) and Elongin C (EloC) 9, while the specific cofactor CBFβ plays a critical role in stabilizing Vif and its assembly with the ligase 21,22. Although the recent crystal structure of the Vif-containing E3 ligase provided the molecular details of this Vif hijacking event 23, the fundamental question regarding how Vif recruits A3s to the E3 ligase remains unelucidated due to the absence of a Vif-A3 complex structure.
Here we present the cryo-EM structure of A3F CTD in complex with Vif and CBFβ, which reveals the structural basis of how Vif and CBFβ recruit A3F to the E3 ligase complex. This structure, together with our biochemical and virological mutagenesis observations, provides new insights into the molecular mechanism of Vif-mediated degradation of A3s.
RESULTS
Vif and CBFβ form a platform for interaction with A3F
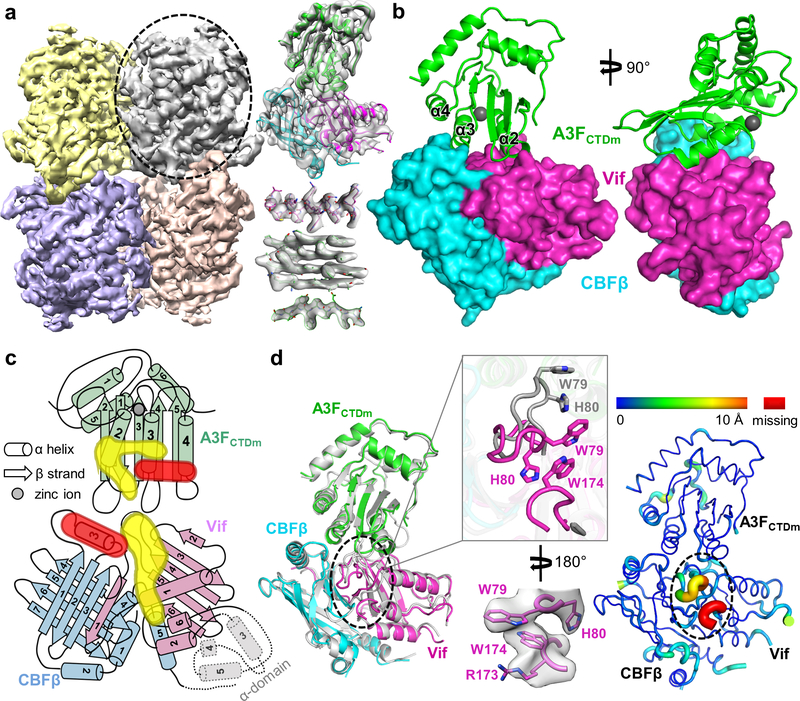
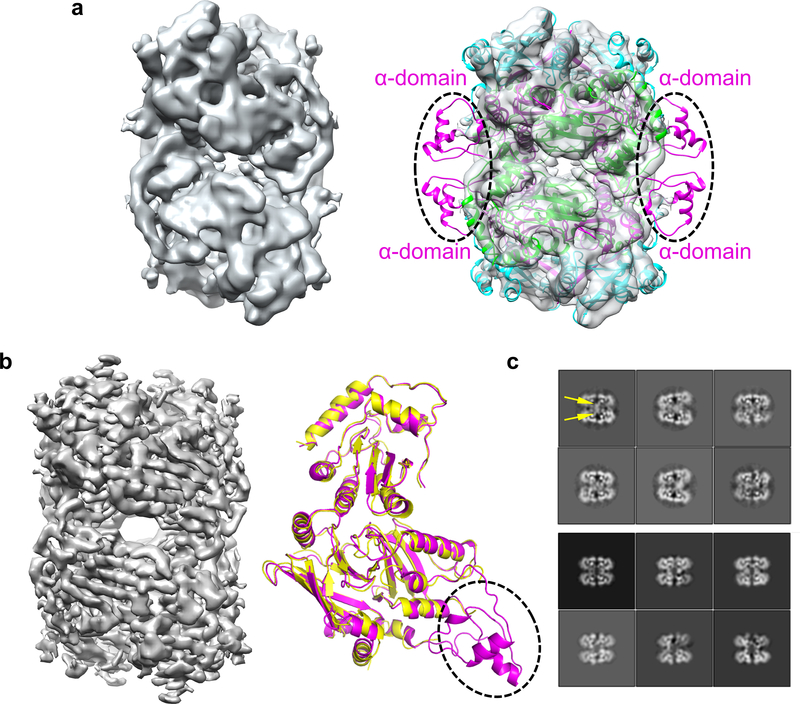
We overcame various technical challenges to obtain cryo-EM reconstructions of Vif–A3F complexes (Fig.1). We first assembled a 7-component Vif–CBFβ–Cul5–EloB–EloC–Rbx2–A3FCTD complex by fusing a previously well characterized A3F CTD with solubility-enhancing mutations 24 (referred to as A3FCTDm hereafter for simplicity, see Methods) and CBFβ, which produced unreliable 3D reconstructions due to problems involving preferred particle orientations and flexible A3F binding. We then investigated the ternary Vif–CBFβ–A3FCTDm complex containing the same fusion. Interestingly, it stabilized a weak tetrameric form of the unfused ternary complex (Extended Data Fig. 1a). This tetramer complex is likely capturing multiple Vif–A3F interfaces, as a consequence of in vitro complex formation. The three-dimensional (3D) cryo-EM reconstruction of this complex at 5 Å resolution showed flexible regions including the Vif α-domain 23 protruding away from its molecular core and the corresponding interacting CBFβ C-terminal regions (Fig. 1c and Extended Data Fig. 2a), whose removal improved the 3D reconstruction to 3.9 Å resolution without affecting the complex architecture (Extended Data Fig. 2b, right) and allowed reliable model building (Table 1, Fig. 1a and Extended Data Fig. 2b, left).
Fig. 1: Vif and CBFβ form a platform for interaction with A3F.
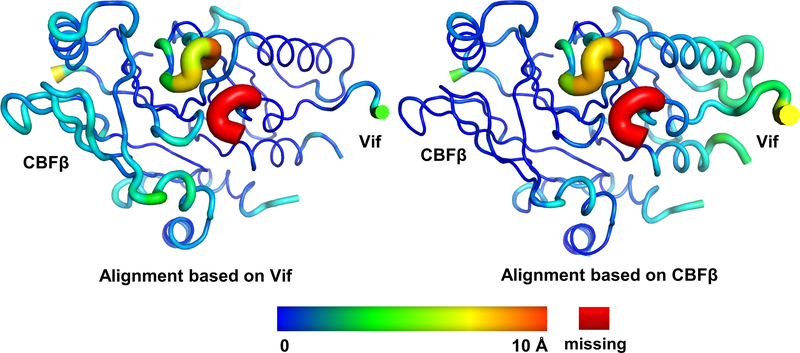
a, Left, 3D cryo-EM reconstruction of the Vif–CBFβ–A3FCTDm complex in the tetrameric state at 3.9 Å resolution, with each ternary complex protomer colored differently. Right, the cryo-EM map of one Vif–CBFβ–A3FCTDm ternary complex (oval on left) is shown on top, with examples of high-quality α helix and β sheet/strand regions at the bottom. b, Overall structure of one Vif–CBFβ–A3FCTDm ternary complex in two orthogonal views. Vif (magenta) and CBFβ (cyan) form a wedge-like platform for A3FCTDm (green) to dock primarily via one end of its α-helical surface, distal from the zinc-containing catalytic site (red sphere). c, Cartoon illustration of the A3FCTDm and Vif/CBFβ secondary structures, with the CBFβ-A3FCTDm interface highlighted in red and the Vif-A3FCTDm interface in yellow. The α helices are represented by cylinders, β strands by arrows, and the zinc ion by a circle. The deleted small α-domain (grey) of Vif and the corresponding interacting CBFβ C-terminus are contoured by dash lines. The detailed annotations of the secondary structure elements are illustrated in Extended Data Fig. 10. d, Majority of Vif–CBFβ and A3FCTDm maintain their conformations upon ternary complex formation. Left, superposition of the ternary structure with individual A3FCTD (PDB 3WUS, grey) and Vif–CBFβ portion from the Vif–E3 ligase structure (PDB 4N9F, grey). The major local conformational changes are circled, with details (top) and the observed cryo-EM map (bottom) shown in the middle inset. Right, the rainbow putty representation of the superposition. The color spectrum and the coil thickness represent the deviation of the aligned Cα atoms in the structures, which varies from 0 Å (blue) to ~10 Å (orange). The Vif C-terminal region residues 173–176 missing in the Vif-E3 ligase structure 23 are colored in red.
TABLE 1.
Cryo-EM data collection, refinement and validation statistics
| Vif176Δ114–157–A3FCTDm-40-CBFβ151 (EMD-9380, PDB 6NIL) | Vif176–A3FCTDm-40-CBFβ187 (EMD-9381) | ||
|---|---|---|---|
| Data collection and processing | 1 | 2/3 | 1 |
| Magnification | 130,000 | 130,000 | |
| Voltage (kV) | 300 | 300 | |
| Electron exposure (e–/Å2) | ~56 | 61/56 | 59 |
| Defocus range (μm) | −1.2 to −2.9 | −1.7 to −3.7/ −1.5 to −3.2 | −0.9 to −2.9 |
| Pixel size (Å) | 1.05 | 1.05 | |
| Stage tilting (°) | 0 | −30/−30 | −30 |
| Symmetry imposed | D2 | D2 | |
| Initial particle images (no.) | 1243243 (untilt) / 1197365 (tilt) | 1140691 | |
| Final particle images (no.) | 337256 | 165629 | |
| Map resolution (Å) | 3.9 | 5 | |
| FSC threshold | 0.143 | 0.143 | |
| Map resolution range (Å) | 3.7–4.6 | - | |
| Refinement | |||
| Initial model used (PDB code) | 4N9F, 3WUS | - | |
| Model resolution (Å) | 3.9 | - | |
| FSC threshold | 0.5 | ||
| Model resolution range (Å) | 3.9–113.4 | - | |
| Map sharpening B factor (Å2) | −223 | −359 | |
| Model composition | |||
| Nonhydrogen atoms | 15216 | - | |
| Protein residues | 1800 | - | |
| Ligands | Zn, 4 | - | |
| B factors (Å2) | |||
| Protein | 113 | - | |
| Ligand | 343 | - | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.005 | - | |
| Bond angles (°) | 1.0 | - | |
| Validation | |||
| MolProbity score | 1.67 | - | |
| Clashscore | 3 | - | |
| Poor rotamers (%) | 0.99 | - | |
| Ramachandran plot | |||
| Favored (%) | 90.4 | - | |
| Allowed (%) | 9.6 | - | |
| Disallowed (%) | 0 | - | |
Strikingly, Vif and CBFβ together provide a rigid platform by forming two sides of a shallow wedge-like structure, and A3FCTDm lodges into the wedge mainly through its α-helical surface (α2, α3, and α4) (Fig. 1b). The catalytic site of A3F is located on the other end of the α-helices away from the interface (Fig. 1b). The interactions between Vif–CBFβ and A3FCTDm bury a surface area of 1004 Å2. CBFβ, which was previously postulated to partially block A3F-binding motifs and need to be displaced for Vif-A3F interaction 18,21, in fact does not change the conformation of how it associates with Vif at all upon A3F binding. Instead it directly interacts with A3F, with its overall conformation and interaction with Vif largely unperturbed. Therefore, CBFβ not only serves as a critical cofactor stabilizing Vif and its assembly with the Cul5-E3 ligase 21,22, but also directly participates in A3F recruitment.
Both Vif–CBFβ and A3FCTDm maintain their overall architectures upon ternary complex formation (Fig. 1d). A3FCTDm undergoes little conformational changes upon binding to Vif-CBFβ (RMSD ~0.8 Å). Similarly, the majority of Vif and CBFβ remain structurally rigid (overall RMSD ~1.3 Å), with only a slight relative orientation shift between the two upon A3FCTDm binding (Extended Data Fig. 3). The largest local conformational change occurs in the Vif β4-β5 loop at the edge of the Vif/CBFβ interface (Fig. 1d, inset), which is mostly disordered in the reported Vif-E3 ligase structure 23. This loop flips towards the Vif core to contact A3F, in a conformation further stabilized by the C-terminal region of Vif. Residues in this loop have been shown to be critical for A3F degradation 25–28. This interaction also highlights the contribution of the Vif C-terminal region in recruiting A3F and explains its importance in neutralizing A3F 27,29. For A3FCTDm, the major local conformational change occurs in loops located on the opposite side of the interaction interface (Fig. 1d), which is a result of a packing interaction from a neighboring Vif molecule in the tetramer complex (Extended Data Fig. 4a, left). These observations demonstrate that Vif and CBFβ can associate into a preformed structure poised to recognize A3F with minor structural adjustments.
Biological importance of the CBFβ-A3F interface
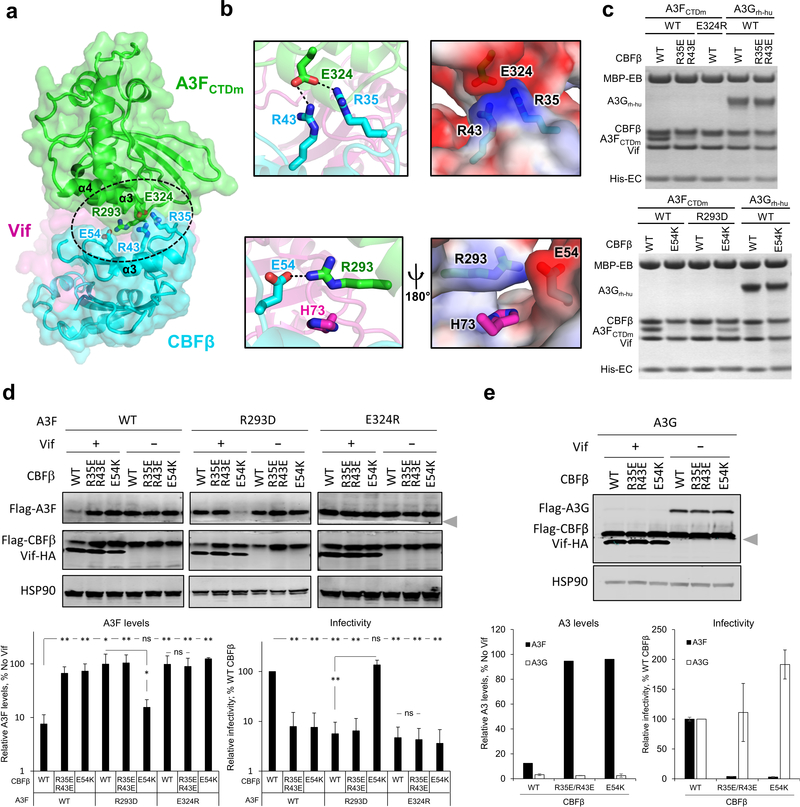
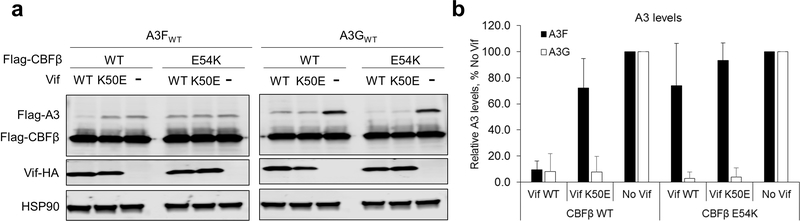
The newly identified interaction between CBFβ and A3F plays a pivotal role in Vif antagonization of A3F. This interface is primarily stabilized by electrostatic interactions between CBFβ residues F32-E54 and the C-terminal regions of A3FCTDm α3 and α4 helices (Fig. 2a). These A3F residues were previously considered to interact with Vif 16,27,30–32. Particularly, CBFβ residues R35 and R43 create a strong positively charged surface abutting a large negatively charged surface at the C-termini of the α3 and α4 helices of A3FCTDm, formed by E324 and multiple main chain carbonyls (Fig. 2b, top). Similarly, the negatively charged CBFβ E54 interacts with the positively charged A3F R293, whose conformation is further stabilized by Vif H73 (Fig. 2b, lower). Mutating any of these residues effectively disrupted the binding between the Vif–CBFβ–EloB–EloC complex and A3FCTDm in vitro (Fig. 2c), rendered A3F resistant to Vif-mediated degradation in cells, and inhibited viral infectivity even in the presence of HIV-1 Vif (Fig. 2d). Remarkably, the charge swapped CBFβ E54K/A3FCTDm R293D double mutation rescued the complex formation in vitro (Fig. 2c), restored the Vif-mediated degradation of A3FR293D in vivo, and reestablished the viral infectivity against the otherwise Vif-resistant A3FR293D mutant (Fig. 2d), confirming that CBFβ E54 and A3F R293 are physically interacting. These data firmly validate the importance of the observed CBFβ-A3F interface in Vif-mediated degradation of A3F and enhancement of viral infectivity.
Fig. 2: The CBFβ-A3F interface is important for Vif–CBFβ–A3F complex formation, Vif-mediated A3F degradation, and viral infectivity.
a, Overview of the CBFβ-A3FCTDm interface (oval) with the critical interacting residues highlighted as sticks. The zinc atom is shown as red sphere. b, Detailed illustrations of the interacting residues and the electrostatic complementation at the interface. Blue, positively charged; red, negatively charged. c, Mutational analysis of the interactions by an in vitro binding assay using MBP-tagged Vif–CBFβ–EloB–EloC variants to pulldown A3FCTDm variants or A3Grh-hu. WT, wild type. The loading controls are shown in Supplementary Fig. 1a & c. d, Analysis of the critical interacting residues at the CBFβ-A3F interface in cell-based Vif-mediated A3F degradation and infectivity assays (mean ± s.d.; n=4 biologically independent experiments). The blot was cut as indicated (gray arrow), with one part probed for Flag-A3F and HSP90, and the other for Flag-CBFβ and Vif-HA. e, The A3F-interacting CBFβ residues are not critical for Vif-mediated A3G degradation (mean ± s.d.; n=3 biologically independent experiments for A3G). The blot was analyzed as in d; the Flag-CBFβ band was partially cut and could be detected in both parts by the same anti-Flag antibody. Statistical significance of A3 degradation and infectivity was assessed by two-tailed t-test assuming equal variance; *, P < 0.05; **, P < 0.005; ns, p-value not significant. Uncropped images and data behind graphs are available as Source Data.
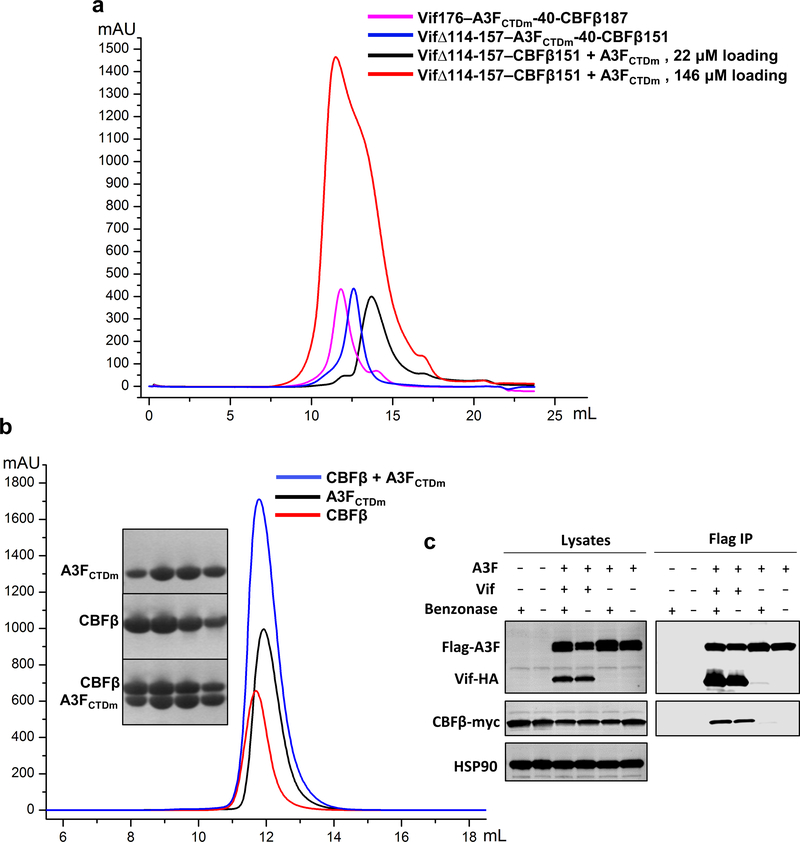
The observed CBFβ interface appears to uniquely interact with A3F, but not A3G. The same CBFβ mutations (R35E/R43E or E54K), which had critical effects on A3F, did not affect the Vif interaction with a solubility-enhanced A3G chimera construct (referred to as A3Grh-hu, see Methods) in vitro (Fig. 2c), Vif-mediated human A3G degradation in vivo, or Vif-enhanced infectivity in the presence of A3G (Fig. 2e). The fact that these CBFβ mutants are functional against A3G also demonstrates that the mutations do not impair protein folding or the functional interactions between Vif and CBFβ, substantiating the direct role of CBFβ in A3F degradation. CBFβ has been found to be required for the expression of the A3 gene repertoire 33. It is intriguing to speculate about the intrinsic cellular function of the A3F-CBFβ interaction at the protein level; however, their direct association in the absence of Vif was not detected in vitro or in vivo (Extended Data Fig. 1b,c). In contrast, a substantially enhanced interaction between A3F and CBFβ was found in cells in the presence of Vif (Extended Data Fig. 1c), demonstrating the importance of Vif for mediating the A3F-CBFβ interaction.
Biological importance of the Vif-A3F interface
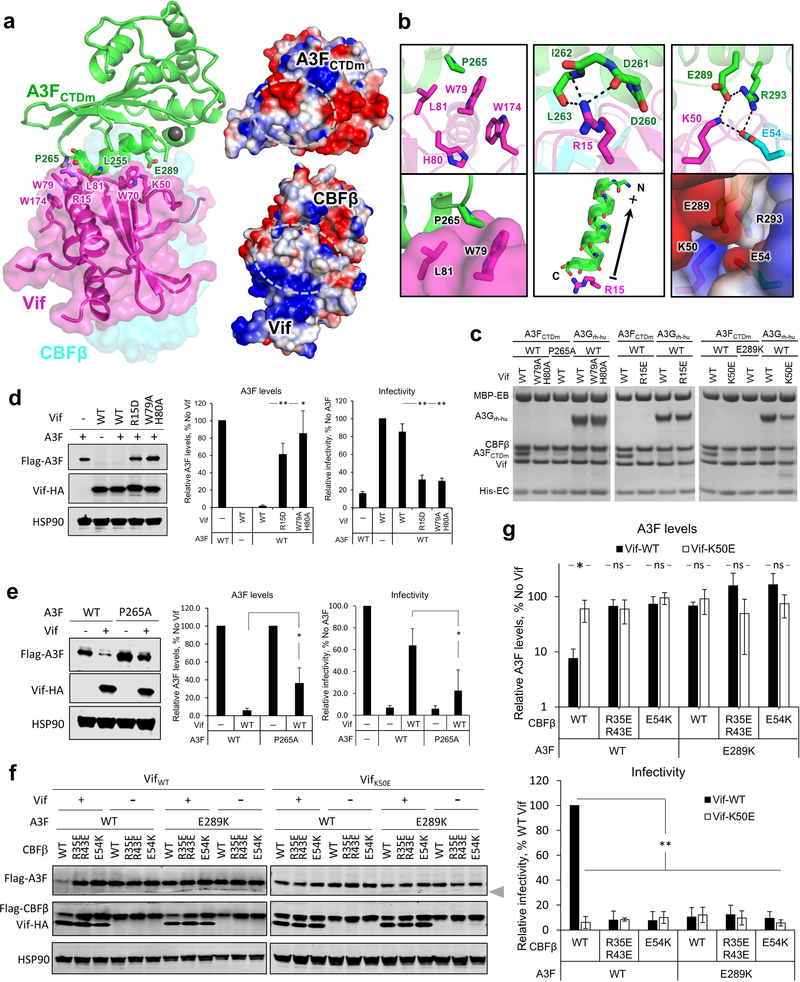
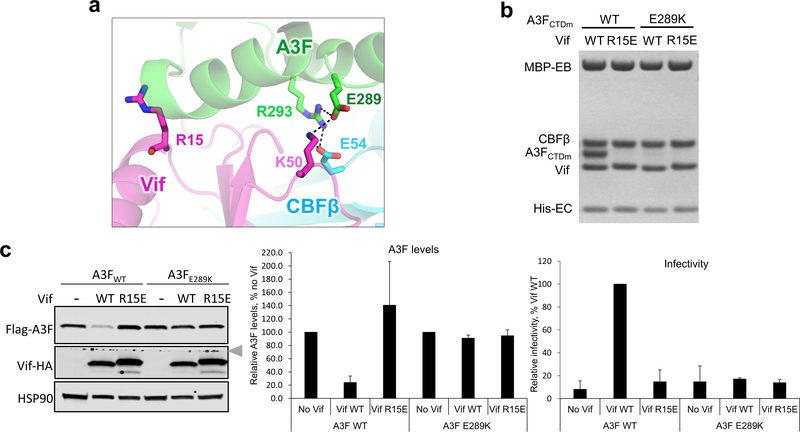
The observed interface between HIV-1 Vif and A3F is also essential for viral evasion of A3F restriction. The Vif-A3FCTDm interface is mainly formed between two helices (α2 and α3) of A3FCTDm and multiple loop regions of Vif on the opposite side of its Cul5/EloC binding interface 23 (Fig. 3a, left and 4b). Both electrostatic and hydrophobic interactions are involved in stabilizing the interface (Fig. 3a, right). Specifically, a major stacking interaction occurs between Vif W79 and A3F P265 (Fig. 3b, left), which causes the aforementioned large conformational change of the Vif β4-β5 loop (Fig. 1d). Vif H80, which stacks with W79 in the absence of A3F (Fig. 1d), flips away to release W79 for A3F interaction (Fig. 3b, left top). W79, whose conformation is further stabilized by W174 at the Vif C-terminus, forms a small hydrophobic cleft with L81 to anchor A3F P265 (Fig. 3b, left lower). A modest stacking interaction also occurs between Vif W70 and A3F L255 (Fig. 3a). Besides hydrophobic interactions, two important electrostatic interactions occur at the interface. First, Vif R15 forms a strong interaction with the negatively charged main-chain carbonyls of A3F residues 260–263 located at the C-terminus of helix α2, whose α-helix dipole effect results in a net negative charge further contributing to the interaction with R15 (Fig. 3b, middle). Second, Vif K50, together with CBFβ E54 at the Vif/CBFβ interface, engages in electrostatic interactions with A3F E289 and R293, respectively, forming a ternary interface of the three proteins (Fig. 3b, right).
Fig. 3: The Vif-A3F interface is critical for Vif–CBFβ–A3F complex formation, Vif-mediated A3F degradation, and viral infectivity.
a, Left, overview of the Vif-A3FCTDm interface with the interacting residues highlighted as sticks. The zinc atom is shown as red sphere. Right, the overall electrostatic potential surfaces for Vif/CBFβ and A3FCTDm with the Vif-A3FCTDm interaction interfaces circled. Blue, positively charged; red, negatively charged; white, hydrophobic. b, Detailed illustrations of the critical interacting residues at the Vif-A3FCTDm interface. The electrostatic interactions are indicated with dashed lines. The cumulative dipole effect of A3FCTDm α2 helix from C-terminus to N-terminus is indicated by an arrow (lower middle). c, Mutational analysis of the interactions by an in vitro binding assay using MBP-tagged Vif–CBFβ–EloB–EloC variants to pulldown A3FCTDm variants or A3Grh-hu. WT, wild type. The loading controls are shown in Supplementary Fig. 1b & c. d, e, Mutagenesis analysis of critical interacting residues at the Vif-A3F interface in cell-based Vif-mediated A3F degradation and infectivity assays (mean ± s.d.; n=3 biologically independent experiments). Vif mutants are shown in d, A3F mutant is shown in e. f, g, Analysis of A3FE289K, VifK50E and CBFβE54K mutants on Vif-mediated A3F degradation and infectivity (mean ± s.d.; n=4 biologically independent experiments). The blot was cut as indicated (gray arrow), where one part was probed for Flag-A3F and HSP90, and the other for Flag-CBFβ and Vif-HA. Statistical significance of A3 degradation and infectivity was assessed by two-tailed t-test assuming equal variance; *, P < 0.05; **, P < 0.005; ns, p-value not significant. Uncropped images and data behind graphs are available as Source Data.
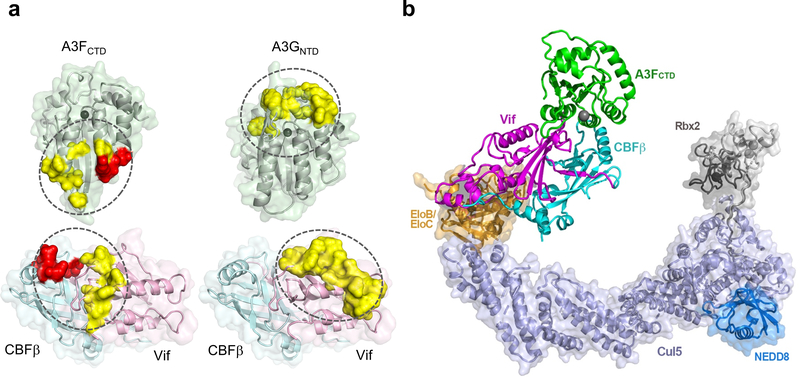
Fig. 4: Models of A3-Vif/CBFβ interactions and the Vif–CBFβ–Cul5 E3–A3FCTD complex.
a, Comparison of the Vif/CBFβ-A3 interfaces (ovals) for A3FCTD (left) and A3GNTD (homology model built from rhesus A3GNTD (PDB 5K81), right). Critical residues observed at the CBFβ-A3F interface are highlighted as red surfaces, critical residues at the observed Vif-A3F interface and the predicted Vif-A3G interface 39–45 are highlighted in yellow. The zinc ion at the ssDNA binding or catalytic site is shown as a dark gray sphere. b, A comprehensive picture of the fully assembled Vif–CBFβ–Cul5 E3–A3FCTD complex structure by overlaying the Vif–CBFβ portion from the current ternary complex and that of the Vif–E3 ligase complex (PDB 4N9F) structures, along with the separately determined Cul5CTD–Rbx2 (PDB 3DPL) and Cul5CTD–NEDD8–Rbx1 (PDB 3DQV) structures. The N-terminal of A3FCTD labelled as “N”, is exposed in a geometry that can accommodate A3FNTD.
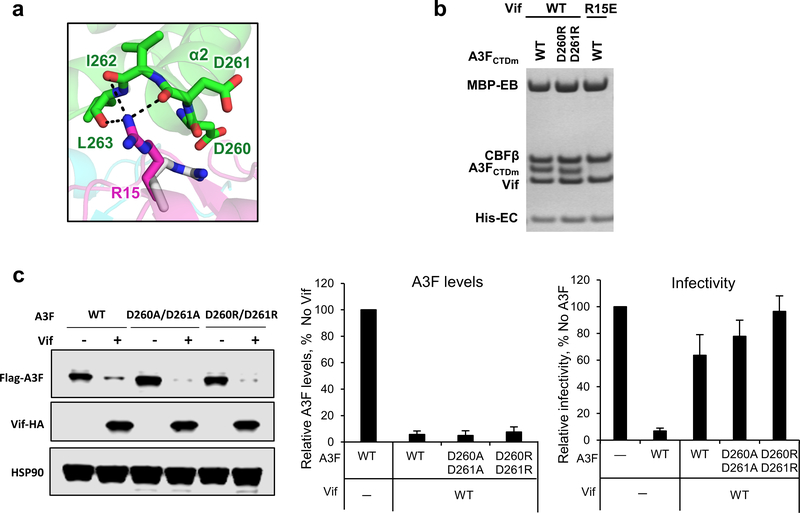
We also validated the biological relevance of the observed Vif-A3F interface. As expected, the W79A/H80A double mutation of Vif completely abolished its A3F binding capacity in vitro (Fig. 3c), while greatly inhibiting the A3F degradation and restricting viral infectivity in vivo (Fig. 3d). The A3F P265A mutant was partially resistant to Vif recognition and degradation (Fig. 3c,e). Demonstrating the importance of the electrostatic interactions, A3F E289K lost the ability to bind Vif, conferred resistance against Vif-mediated degradation, and retained viral restriction in the presence of Vif (Fig. 3c,f). Furthermore, the Vif R15D/E and K50E mutants were also severely defective in binding and neutralizing A3F (Fig. 3c,d,f,g). Consistent with the observed main chain and helix dipole interactions (Fig 3b, middle), the A3F D260R/D261R mutation had no effect in vitro or in vivo (Extended Data Fig. 5). In contrast to the effects on A3F, both Vif W79A/H80A and R15E mutants retained the ability to bind A3Grh-hu in vitro (Fig. 3c). The Vif K50E mutant had impaired binding efficiency to A3Grh-hu in vitro (Fig. 3c), which, however, was still sufficient to induce the degradation of human A3G in vivo (Extended Data Fig. 6).
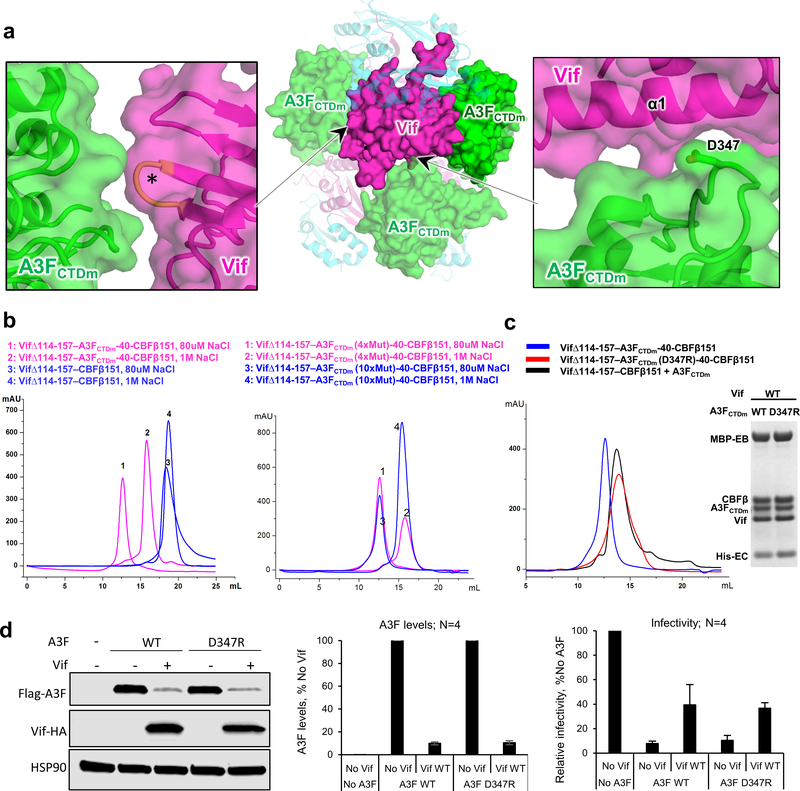
The major Vif-A3FCTDm interface described above is located within one ternary complex, which further docks onto neighboring molecules in the observed tetramer through two other much smaller inter-ternary complex interfaces (Extended Data Fig. 4a), which have been implicated in A3F binding by several mutagenesis studies 20,25,27,28,34,35. One such interface involves the Vif α1 helix close to the major interface (Extended Data Fig. 4a, right), while the other interface involves Vif residues in the 55VxIPLx4–5L64 motif (Extended Data Fig. 4a, left) located on the opposite side of the major A3F interface on Vif (Extended Data Fig. 4a, center), indicating a single A3F molecule would not be able to cover all interacting surfaces on Vif simultaneously. The structure shows that the tetramer interfaces are mediated by A3FCTDm, consistent with our observation that the Vif–CBFβ subcomplex alone does not form tetramers (Extended Data Fig. 4b, left). We also confirmed that the tetramer was not induced by the solubility-enhancing mutations in A3FCTDm, as reverting those located near the tetramer interfaces back to wild type residues did not affect the tetramer formation (Extended Data Fig. 4b, right). However, when we mutated A3F D347 located at one of the tetramer interface (Extended Data Fig. 4a, right) to Arg, the D347R substitution disrupted the tetramer formation but did not affect the formation of the Vif–CBFβ–A3FCTDm-D347R ternary complex in vitro (Extended Data Fig. 4c). Consistently, our functional data showed that the D347R mutant of the full-length A3F behaved the same as WT A3F in terms of its antiviral activity and sensitivity to Vif-mediated degradation, which resulted in rescue of viral infectivity (Extended Data Fig. 4d). These results demonstrate that the major interface observed in the Vif–CBFβ–A3FCTDm ternary complex should represent the primary interaction mode of A3F recruitment to the Vif-containing E3 ligase, and argue against the possible biological significance of the tetramer. However, we cannot rule out the possibility that some of the observed tetramer interface interactions may be involved during the Vif antagonization of A3F.
DISCUSSION
Our structure establishes an unambiguous, comprehensive interaction model, enabling the reevaluation and interpretation of the extensive but sometimes confusing mutagenesis observations 16,20,25–32,36,37. It allows for the delineation between residues directly participating in the interactions, and those contributing indirectly by maintaining the conformation required for the interactions. In fact, many of the residues previously speculated to be at the A3F-Vif interface are either buried in the molecular core or serve indirect structural roles at the Vif-CBFβ interface. Moreover, a previous interaction model predicted an electrostatic interaction between Vif R15 and A3F E289 based on genetic and computational analysis 37. However, these two residues are located far away from each other in our structure (Extended Data Fig. 7a), and VifR15E mutant failed to restore the degradation and viral infectivity against A3FE289K mutant (Extended Data Fig. 7c), contradictory to the prior charge-swapping mutagenesis result 37. In contrast, our structural, biochemical, and cellular data unambiguously elucidate the correct interactions at the interfaces between Vif–CBFβ and A3F (Fig 2 and 3). These advances demonstrate the power of the structure in synthesizing and clarifying the existing data to further our understanding of the molecular basis of Vif-mediated degradation of A3F.
Our Vif–CBFβ–A3FCTDm structure not only provides the structural basis of A3F targeting by HIV-1 Vif, but also enables a deeper understanding of Vif interactions with other A3s. Notably, many of the Vif residues proposed to participate in both A3F and A3G interactions based on mutagenesis studies 18 are not found at the Vif-A3F interfaces. As mentioned above, they play indirect structural roles in the Vif core or its interaction with CBFβ, indicating that Vif recruits A3F and A3G through distinct interfaces with considerably less overlapping regions than expected (Fig. 4a). In addition, our structure shed light on a similar Vif interface speculated for A3C, which together with A3DCTD, falls into the same A3 zinc domain group as A3FCTD 16,30,38. Nonetheless, differences exist within the group such as the Vif C-terminal residues E171 and R173, previously found to be less critical for the degradation of A3C 27, are important for A3F interaction. Our work also substantiates the prediction that A3F uses a surface opposite from that of A3G to interact with Vif 39–45, while A3F and A3C contact Vif using similar regions 16,27,30. These comparisons show that Vif is a versatile A3 binder, targeting a variety of A3s with regions of various degrees of overlap.
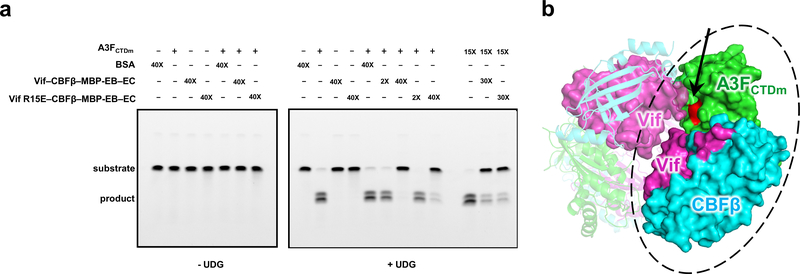
Our Vif–CBFβ–A3FCTDm structure also provide insights into the potential degradation-independent mechanisms of Vif-mediated inhibitions on A3s. The ternary structure shows that A3FCTDm contacts Vif/CBFβ through an interface away from its catalytic site (Figs. 1b and 4a), indicating that this interaction should not block the A3F deamination activity. However, our ssDNA deamination assay showed that the catalytic activity of A3FCTDm at low concentration (5 μM) was not affected by two-fold excess of the Vif–CBFβ–EloB–EloC subcomplex, but was inhibited by a large excess (40-fold) of the subcomplex (Extended Data Fig. 8a). This inhibition was not revoked by the Vif R15E mutation (Extended Data Fig. 8a) that disrupted the Vif-A3FCTDm major interface and the Vif-A3F interaction both in vitro and in vivo (Fig. 3c, Extended Data Fig. 7c). In contrast, at high A3FCTDm concentration (75 μM), two-fold excess of either Vif WT or R15E variants was sufficient to inhibit the A3FCTDm deamination activity (Extended Data Fig. 8a), potentially due to the formation of a higher oligomer state at the high concentration (Extended Data Fig. 1a). These results indicate that interactions other than those at the major interface were involved in the inhibition of the A3F deamination activity. Interestingly, one of the tetramer interfaces observed in our structure involving the Vif 55VxIPLx4–5L64 motif does have the potential to block the A3F catalytic site for DNA substrate access (Extended Data Fig. 8b). Even though tetramer formation may not necessarily occur in cells, this observed interface points to the propensity for such an interaction at very high (local) protein concentration leading to inhibition. Several enzymatic studies have also shown that Vif can directly attenuate the deaminase activities of A3G without inducing its proteasomal degradation 46,47, primarily by disrupting the A3G interactions with viral ssDNA, whose binding site is postulated to be adjacent to the major Vif-binding interface in A3GNTD (Fig. 4a, top right). These findings emphasize that Vif antagonizes A3s through multifarious mechanisms regulated by their specific interactions. Further elucidation of the degradation-independent inhibition mechanisms must await future studies.
The work described herein establishes a detailed biochemical and structural framework of how A3F is targeted by HIV-1 Vif and its cellular cofactor CBFβ. This, together with the previous Vif-E3 ligase structure 23, allows a complete mechanistic understanding of Vif-mediated recruitment of A3F to the ubiquitin-proteasome degradation pathway (Fig. 4b). The E2 ubiquitin-conjugating enzyme bound to the flexible Rbx2 component in the NEDD8 activated E3 ligase 48 potentially allows ubiquitination of multiple positions on A3F, as well as the bound Vif molecule 49. This advance provides critical insights into the molecular interactions enabling the viral evasion of a major host defense, which is also conserved between other known lentiviruses and their hosts 50. Furthermore, it has been found that HIV-1 can evade A3G, but not A3F restriction in the absence of Vif 51,52, highlighting that the Vif-A3F interaction is essential for HIV-1 survival in cells. Therefore the structural details obtained in this work can effectively facilitate the development of novel anti-HIV therapeutics by specifically targeting the interfaces between Vif–CBFβ and A3F.
ONLINE METHODS
Plasmid construction
HIV-1 Vif residues 1–176 (Vif176) from pNL4–3 and Vif176 with the α-domain (residues 114–157) replaced with a six amino-acid (a.a.) linker (EASEGS) (Vif176Δ114–157) were cloned into the pETDuet vector. 6×His tagged human CBFβ residues 1–187 (CBFβ187) and C-terminal truncated CBFβ residues 1–151 (CBFβ151) were cloned into pCDFDuet vector. MBP-tagged EloB residues 1–118 (EloB118) and 6×His tagged EloC residues 17–112 (EloC17–112) were cloned into the pACYCDuet vector. 6×His tagged human A3F residues 185–373 with ten solubility mutations (Y196D, H247G, C248R, F302K, W310K, Y314A, Q315A, K355D, K358D, and F363D) 24, which has been thoroughly validated to retain wild type-like deamination activities, Vif responsiveness, and structure, termed as A3FCTDm in this study (the other A3FCTDm variants were constructed on this background) was fused to CBFβ151 or CBFβ187 through a 40 a.a. linker (GVDGSDEASELACPTPKEDGLAQQQTQLNLRSQATGSGSG) to stabilize Vif binding. Each of the A3FCTDm-CBFβ fusion protein variants or the 6×His tagged A3FCTDm alone was cloned into the pCDFDuet vector. An A3G chimera was created in which a modified rhesus A3GNTD 53 was fused to human A3GCTD, with a K128D mutation to enable binding to HIV-1 Vif. This chimera (referred to as A3Grh-hu in this study) showed a solution property superior to that of wild-type human A3G and was used in the in vitro binding assays. The 6×His tagged A3Grh-hu was cloned into the pETDuet vector. All Duet vectors were from Novagen, Inc. All mutants were constructed following the protocol of QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies).
For the cell-based assays, the following previously described plasmids or their derivative mutants were used, including, pFlag-CBFβ, pFlag-A3F, pFlag-A3G, pVif-HA, pHDV-eGFP, and pHCMV-G, which expresses vesicular stomatitis virus glycoprotein (VSV-G) 16,20,54–58. Vif, CBFβ, and A3F mutants were generated by site-directed mutagenesis using a QuickChange Lightening Multi Site-directed mutagenesis kit (Agilent Technologies) and verified by sequencing.
Protein expression and purification
The Escherichia coli BL21 (DE3) cells (New England BioLabs) were used for protein expressions. Vif176 and 6×His tagged A3FCTDm-40-CBFβ187 constructs were co-expressed to obtain the Vif–CBFβ–A3FCTDm complex. Vif176Δ114–157 and 6×His tagged A3FCTDm-40-CBFβ151 constructs were co-expressed to obtain the truncated Vif–CBFβ–A3FCTDm complex. In addition, the truncated Vif176Δ114–157 and 6×His tagged CBFβ151 constructs were co-expressed, and 6×His tagged A3FCTDm was separately expressed to reconstitute the unfused Vif–CBFβ–A3FCTDm complex. Vif176 variants, 6×His tagged CBFβ187 variants, MBP-tagged EloB118/EloC were co-expressed to form various MBP-tagged Vif–CBFβ–EloB–EloC variants, while the 6×His tagged A3FCTDm variants, 6×His tagged CBFβ187, 6×His tagged A3Grh-hu were separately expressed for in vitro binding assays. The protein expression was induced by 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C for 16 hours in Terrific Broth.
Cells were harvested and lysed by a microfluidizer. The lysate was clarified by centrifugation and then applied to a Ni-NTA (Qiagen) column. The fused Vif–CBFβ–A3FCTDm complexes or the Vif–CBFβ subcomplexes were then purified by two rounds of size exclusion chromatography (SEC) (HiLoad Superdex 200, GE healthcare), first in a buffer containing 30 mM Tris, 1 M NaCl, 0.2 mM TCEP, pH 8.0 to remove bound nucleic acids, and followed by a second SEC run at a low salt of 80 mM NaCl. The purification of all other proteins were performed following the Ni-NTA step by anion exchange (HiTrap Q HP, GE healthcare) chromatography in a buffer of 30 mM Tris, 0.2 mM TCEP, pH 8.0 with a gradient NaCl concentration from 20 mM to 1M, and subsequently SEC (HiLoad Superdex 200 or 75, GE healthcare) in a buffer of 30 mM Tris, 300 mM NaCl, 0.2 mM TCEP, pH 8.0 (A3Grh-hu in 1M NaCl). Purity of the proteins was analyzed by SDS-PAGE after each step.
Binding assays of Vif–CBFβ–A3FCTDm complexes in vitro
SEC binding assay: Various amounts of the truncated Vif–CBFβ subcomplex were incubated with A3FCTDm at 1:1 molar ratio in the buffer of 30 mM Tris, 100 mM NaCl, 0.2 mM TCEP, pH 8.0 at 4 °C for 2 hours; the ternary complex formation was then analyzed by SEC (Superdex 200). Similarly, high amount of CBFβ and A3FCTDm were incubated at 1:1 molar ratio in the same buffer at 4 °C for 6 hours. The retention volumes of individual CBFβ, A3FCTDm, and their mixture at equal concentrations were analyzed by SEC (Superdex 75), and the peak fractions of all three species were analyzed by SDS-PAGE. The Vif–CBFβ–Cul5 E3 ligase complex was formed by incubating the Vif–CBFβ–EloB–EloC complex and Cul5–Rbx2 complex with 1:1 molar ratio at 4°C for 2 hours and further purified by SEC (Superdex 200).
Pull-down assay: 0.15 mg of MBP-tagged Vif–CBFβ–EloB–EloC variants were first incubated with A3FCTDm variants or A3Grh-hu at 1:2 molar ratio in 100 μl binding buffer containing 30 mM Tris, 100 mM NaCl, 0.2 mM TCEP, pH 8.0 at 4 °C for 2 hours, subsequently mixed with 50 μl of amylose resin (New England BioLabs) in a Pierce spin column (Thermo Fisher) and incubated for one additional hour. After removing the supernatant by centrifugation, the resin was washed with 300 μl of binding buffer for three times. 80 μl of the elution buffer (binding buffer plus 0.2 mM maltose) was then added to the resin and incubated at 4 °C for 10 minutes before centrifugation. The loading and elution fractions were analyzed by SDS-PAGE.
A3 degradation assays, virus production, and determination of infectivity
TZM-bl and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Corning Cellgro) containing 10% fetal bovine serum (Hyclone), and 1% penicillin-streptomycin (GIBCO). To produce virus, 293T cells were seeded at 4 × 105 cells/well in six-well plates and co-transfected using Lipofectamine2000 (Invitrogen) with the following plasmids: pHDV-eGFP (2 μg), phCMV-G (200 ng), WT or mutant pFlag-A3F (250 ng) or pFlag-A3G (500 ng), and WT or mutant pVif-HA (2.5 μg). After 48 hours, the virus-containing supernatants were filtered through 0.45-μm filter and kept at −80 °C until use. Producer cell lysates were also harvested for immunoblotting analyses to detect the steady-state levels of A3F and A3G in the absence or presence of Vif. Flag-A3F and Flag-A3G were detected using a rabbit anti-Flag polyclonal antibody or a mouse monoclonal anti-Flag antibody (Sigma; both at a 1:5000 dilution). CBFβ was detected using a rabbit anti-CBFβ polyclonal antibody (Abcam), a mouse monoclonal anti-Flag antibody, or a rabbit anti-myc polyclonal antibody (Sigma; all of them used at a 1:5000 dilution), Vif was detected using a mouse anti-HA monoclonal antibody (Sigma) or a rabbit monoclonal anti-HA antibody (Cell Signaling); both at 1;5000 dilution, and HSP90 was detected using mouse anti-HSP90 antibody (Santa Cruz Biotechnology; at 1:20000 dilution). Rabbit and mouse primary antibodies were detected using an IRDye® 800CW-labeled goat anti-rabbit secondary antibody at 1:5000 dilution (Licor) or an IRDye® 680-labeled goat anti-mouse secondary antibody at 1:5000 dilution (Licor). Protein bands were visualized and quantified using Odyssey® Infrared Imaging System and Image Studio™ Lite version (Licor).
To determine infectivity, virus stocks were quantified using a p24 CA enzyme-linked immunosorbent assay (XpressBio) and TZM-bl luciferase reporter assay was performed as previously described 59.
CBFβ knockdown and back complementation with CBFβ mutants
To knockdown endogenous CBFβ and complement with exogenous CBFβ variants, 293T cells were seeded at 2.5 × 105 cells per well in six-well plates a day before transfection. Next day, cells were transfected using Lipofectamine RNAiMAX (Invitrogen) with Stealth CBFβ siRNA targeting 3-UTR region (Invitrogen; NM_022845.2_stealth_865) as previously described 59. Next day, using the Lipofectamine2000 transfection method (Invitrogen), the following plasmids were co-transfected: pHDV-eGFP (2 μg), phCMV-G (200 ng), Flag-CBFβ variants (500 ng), and Vif (2.5 μg) in combination with the CBFβ siRNA and pFlag-A3F (250 ng) or pFlag-A3G (500 ng). Producer cell lysates and supernatants were collected 48 hours later for western blotting and infectivity assays, respectively.
Co-immunoprecipitation assays
Flag Co-IP assays were carried out as previously described 59. Briefly, 293T cells were seeded at 4 × 106 cells per 10-cm dish and transfected the next day using standard methods. The following DNA amounts were used to transfect 293T cells: 4 μg Flag-A3F, 4 μg CBFβ-myc, and 2 μg Vif-HA expression plasmids. Next day, the cells were treated with MLN-4924 (BioVision; Cat # 2566) at 2 μM final concentration for 24 hours to prevent Vif-mediated proteasomal degradation of A3F. 48 hours post-transfection, total cell lysates were harvested in 1 mL of RIPA lysis buffer (Sigma) supplemented with proteinase cocktail inhibitors EDTA-free tablets (Roche) with or without benzonase® nuclease (250U/ml final concentration; Sigma) addition and Flag Co-IP was performed at 4°C for overnight. To detect eluted complexes as well as the input cell lysates, western blotting was performed. CBFβ was detected using a rabbit anti-myc polyclonal antibody at a 1:5000 dilution, Vif was detected using a rabbit anti-HA monoclonal antibody at a 1:5000 dilution, and A3F detected using a mouse anti-Flag antibody at a 1:5000 dilution. HSP90 was used as loading control and detected with mouse anti-HSP90 antibody at a 1:20000 dilution. Protein bands were visualized using an Odyssey® Infrared Imaging System (Licor) as described above.
UDG-based deamination assay
Various amounts (5 μM or 75 μM) of 6×His tagged A3FCTDm was incubated with MBP-tagged Vif–CBFβ–EloB–EloC at 1:2 or 1:40 molar ratio in 10 μl buffer of 30 mM Tris pH 7.10, 50 mM NaCl, 1mM MgCl2, 5% glycerol, 0.5 mM EDTA and 0.2 mM TCEP at 4°C for 2 hours, followed by the addition of 5 μM of 6-FAM labeled ssDNA oligo substrate containing a TC hotspot (5’-TAAGAAAGAATTCAGAAGAGGAA-3’). The reaction mixtures were then incubated at room temperature for overnight (for 5 μM A3FCTDm) or 3 hrs (for 75 μM A3FCTDm) and terminated at 95°C for 5 minutes. 5 units of uracil DNA glycosylase (UDG, New England BioLabs) were added into the reactions for incubation at 37°C for 1 hour, and then treated with 0.25 M NaOH at 37°C for 30 minutes. 10 μL of 1 M Tris pH 8.0 was then added to neutralize the pH of the samples and equal volume of formamide was added to the samples as loading buffer. The samples were analyzed on a 15% TBE-Urea gel (Invitrogen), which was imaged on a BioRad ChemiDoc.
Cryo-EM sample preparation, data collection and processing
Purified protein complex (4 μl) was applied to a C-Flat 2/1 3C copper grid (Electron Microscopy Sciences) pretreated by glow-discharging at 8 mA for 20 seconds. The grid was blotted at 10 °C with 100% humidity and plunge-frozen in liquid ethane using FEI Vitrobot Mark IV (Thermo Fisher). The grids were stored in liquid nitrogen before data collection.
Images were acquired on a FEI Titan Krios electron microscope (Thermo Fisher) equipped with Gatan K2 Summit direct detector in super-resolution mode, at a calibrated magnification of 130,000× with the physical pixel size corresponding to 1.05Å. Detailed data collection statistics for the Vif–CBFβ–A3FCTDm complexes have been indicated in Table 1. Automated data collection was performed using SerialEM 60.
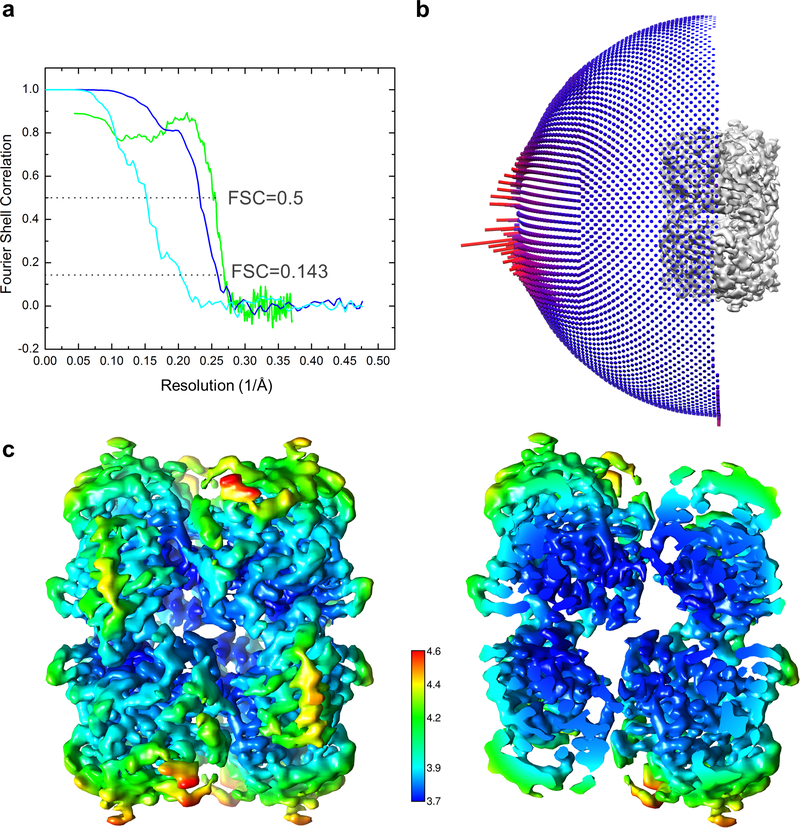
A total of 3486 movie series without tilting the microscope stage and a total of 3359 movie series with the stage tilted by −30° were collected for the truncated Vif–CBFβ–A3FCTDm complex, and a total of 2768 movie series with the stage tilted by −30° were collected for the Vif–CBFβ–A3FCTDm complex. Tilting the stage helped alleviate modest preferred orientation problem of the samples. The same data processing procedures were carried out for each complex described as below. Motion correction of each micrograph was determined using MotionCor2 61 and contrast transfer function (CTF) estimation was calculated using Gctf 62. Around 5000 particles were manually picked from selected micrographs to generate initial 2D class averages by RELION 63 as the templates for automatic particle picking of the entire dataset by Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/). All particles were included directly for 3D classification in RELION to include those at rare views that may not have sufficient numbers to generate good 2D classes. For the Vif–CBFβ–A3FCTDm complex, the 3D classification without imposing symmetry produced one good 3D class out of fifteen, containing 165629 particles. Conformational flexibility was detected in 3D classes (Extended Data Fig. 2c). The good 3D class was used for gold standard refinement in RELION with D2 symmetry. The final resolution of the reconstruction was 5.0 Å, based on the Fourier shell correlation (FSC) cutoff at 0.143 between the two half maps 64, after a soft mask was applied to mask out solvent region (Extended Data Fig. 9a). The final map was corrected for K2 detector modulation and sharpened by a negative B-factor within RELION 65. Particles of the truncated Vif–CBFβ–A3FCTDm complex was processed in a similar way. The truncated complex showed less conformational flexibility and obeyed the D2 symmetry better (Extended Data Fig. 2c). A good 3D class of 337256 particles was identified after combining 1243243 titled and 1197365 untilted particles. The particles showed some orientation preferences (Extended Data Fig. 9b), which resulted in a final reconstruction of 3.9 Å resolution (Extended Data Fig. 9a). Local resolution variation was estimated by using RELION (Extended Data Fig. 9c).
Model building and refinement
The structures of the Vif–CBFβ portion (extracted from PDB 4N9F) and A3FCTD (PDB 3WUS) were docked into the cryo-EM maps of the truncated and full Vif–CBFβ–A3FCTDm complex using Chimera 66 and refined with phenix.real_space_refine module in PHENIX with secondary structure restraints and Ramachandran restraints 67. The model of the truncated Vif–CBFβ–A3FCTDm complex was further adjusted in COOT 68 manually with intervening cycles of refinement in PHENIX and Refmac5 69. The final model with good geometry and fit to the map was validated using the comprehensive cryo-EM validation tool implemented in PHENIX 70 (Table 1). All structural figures were generated using PyMol 71 and Chimera 66.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The model of the truncated Vif–CBFβ–A3FCTDm complex has been deposited in the wwPDB with accession code PDB 6NIL. The cryo-EM maps of the truncated and full-length Vif–CBFβ–A3FCTDm complexes have been deposited in EMDB with accession codes EMD-9380 and EMD-9381, respectively. Source data for Fig. 2c,d,e, Fig. 3c,d,e,f,g, Extended Data Fig. 1b,c, Extended Data Fig. 4c,d, Extended Data Fig. 5b,c, Extended Data Fig. 6a,b, and Extended Data Fig. 7b,c are available with the paper online. Other data are available from corresponding authors upon reasonable request.
Extended Data
Extended Data Fig. 1. Biochemical and cellular characterizations of various Vif–CBFβ–A3FCTDm assemblies and the A3FCTDm-CBFβ interaction.
a, The Vif–CBFβ–A3FCTDm fusion complex with or without the Vif α-domain and corresponding interacting CBFβ C-terminus stays as tetramer in low salt solution. The unfused Vif–CBFβ–A3FCTDm without these regions switches from monomer to tetramer at high protein concentration (146 μΜ loading concentration). b, No obvious shift for the elution peak was observed upon incubation of CBFβ and A3FCTDm compared to the CBFβ alone or A3FCTDm alone. The SDS-PAGE analysis of the peak fractions of CBFβ alone, A3FCTDm alone and CBFβ/A3FCTDm mixture is indicated. c, Co-immunoprecipitation (Co-IP) analysis of the interaction between A3F and CBFβ in the presence or absence of Vif in cells. Flag-A3F and CBFβ-myc were co-transfected with or without Vif-HA and co-immunoprecipitated using anti-Flag antibody. In the absence of Vif, no binary A3F and CBFβ binding was observed. A representative blot from two independent experiments was shown.
Extended Data Fig. 2. Cryo-EM study of the Vif–CBFβ–A3FCTDm complex with or without the Vif α-domain and the corresponding interacting region of the CBFβ C-terminus.
a, The 5 Å cryo-EM reconstruction of Vif–CBFβ–A3FCTDm with (right) or without (left) the docked-in model (ribbon). The density corresponding to the flexible Vif α-domain and the corresponding interacting CBFβ C-terminus (circled) is not visible. b, Left, the 3.9 Å cryo-EM reconstruction of the truncated Vif–CBFβ–A3FCTDm. Right, overlay of the cryo-EM models of Vif–CBFβ–A3FCTDm ternary complexes with (magenta) and without (yellow) the Vif α-domain and the corresponding interacting CBFβ C-terminus shows that the removal of these regions does not affect the architecture of the ternary complex. c, Central slices of the top 3D classes of the Vif–CBFβ–A3FCTDm complex with (upper) or without (lower) the Vif α-domain and the corresponding interacting CBFβ C-terminus indicate that removing these flexible regions reduces the tetramer flexibility. The location of the Vif α-domain and the corresponding interacting CBFβ C-terminus is marked by yellow arrows in the first class average.
Extended Data Fig. 3. The relative conformational changes between Vif and CBFβ upon A3FCTDm binding, shown as rainbow putty representations of superpositions.
The color spectrum and the coil thickness represent the deviation of the aligned Cα atoms in the structures, which varies from 0 Å (blue) to ~10 Å (orange). The Vif C-terminal residues 173–176 missing in the Vif–E3 ligase structure are colored in red. The Vif–CBFβ structure without A3FCTDm binding used for superposition is extracted from the Vif–E3 ligase structure (PDB 4N9F).
Extended Data Fig. 4. Effects of Vif–CBFβ–A3FCTDm tetramer on A3F-Vif interaction, Vif-mediated A3F degradation, and viral infectivity.
a, Inter-ternary complex interfaces between Vif and A3FCTDm detected in the tetrameric complex. Center, Overview of the two inter-ternary complex interfaces (pointed to by the arrows) involving one Vif molecule. Vif is shown in magenta, CBFβ in cyan, and A3FCTDm in green. One ternary complex containing the major Vif-A3F interface is marked by an oval. The detailed illustrations of the two interfaces are shown on the sides, with one involving the Vif α1 helix (right), and the other involves Vif residues (orange, marked by *) in the 55VxIPLx4–5L64 motif (left). b, SEC binding assays show that the tetramer formation depends on the presence of A3FCTDm (left) but not on the solubility-enhancing mutations of A3FCTDm (right). In contrast to the Vif–CBFβ–A3FCTDm complex which switched from monomer to tetramer when reducing salt concentration, Vif-CBFβ alone stayed as monomer. Six A3FCTDm residues (196, 247, 248, 310, 314, 315) located near the observed tetramer interfaces were reverted back to WT amino acids to verify that this A3FCTDm variant (with 4 remaining point mutations away from the interface and without any disturbance to A3F structure) retained the ability to form tetramers. c, SEC binding (left) and MBP pull-down (right) assays show that the A3FCTDm D347R mutation disrupts the tetramer formation but not the individual Vif–CBFβ–A3FCTDm ternary complex. The loading controls are shown in Supplementary Fig. 1b & c. d, The effect of D347R mutation on A3F sensitivity to Vif-mediated degradation indicated by western blot (left), quantified A3F levels relative to No Vif (mean ± s.d.; n=4 biologically independent experiments; middle), and relative infectivity (mean ± s.d.; n=4 biologically independent experiments; right). A3FD347R retained WT A3F-like sensitivity to Vif-mediated degradation, which resulted in rescue of viral infectivity.
Extended Data Fig. 5. Vif R15 is located at the C-terminus of A3FCTDm α2 helix interacting with the backbone carbonyls of the helix.
a, Upon A3F binding, Vif R15 flips away from the position (gray) pointing into the molecule core to electrostatically interact with the backbone carbonyls of A3FCTDm α2 helix rather than the side chain carboxylates of D260/D261. b, Mutational analysis of the interactions by in vitro binding assay using MBP-tagged Vif–CBFβ–EloB–EloC variants to pulldown A3FCTDm variants. The D260R/D261R double mutation did not affect the Vif interaction. The loading controls are shown in Supplementary Fig. 1b & c. c, The effect of D260A/D261A or D260R/D261R double mutants on A3F sensitivity to Vif-mediated degradation indicated by western blot (left), quantified A3F levels relative to No Vif (mean ± s.d.; n=3 biologically independent experiments; middle), and relative infectivity (mean ± s.d.; n=3 biologically independent experiments; right). Both alanine and arginine mutants did not confer resistance, indicating that the side chains of the residues are not involved in the Vif interaction.
Extended Data Fig. 6. Analysis of the effect of Vif K50E mutation on A3G degradation in cells.
Western blot (a) and quantified A3 levels relative to No Vif (b) show that the Vif K50E mutant could not induce A3F degradation in the presence of either CBFβ WT or CBFβ E54K but could induce A3G degradation (mean ± s.d.; n=3 biologically independent experiments).
Extended Data Fig. 7. HIV1-Vif R15 does not interact with A3F E289.
a, Vif R15 is located far away from A3F E289, which interacts with Vif K50 in the Vif–CBFβ–A3FCTDm structure. b, Mutational analysis of the interactions by in vitro binding assay using MBP-tagged Vif–CBFβ–EloB–EloC variants to pulldown A3FCTDm variants. The loading controls are shown in Supplementary Fig. 1b & c. The charge-swapped Vif R15E/A3F E289K double mutation did not rescue the Vif-A3F interaction in vitro. c, The effect of Vif R15E or A3F E289K mutation on A3F sensitivity to Vif-mediated degradation indicated by western blot (left), quantified A3F levels relative to No Vif (mean ± s.d.; n=3 biologically independent experiments; middle), and relative viral infectivity (mean ± s.d.; n=3 biologically independent experiments; right). In contrast to the prior report 37, the charge-swapped Vif R15E/A3F E289K double mutation did not restore the Vif-mediated A3F degradation or viral infectivity in cells. The blot was cut as indicated (gray arrow), where one half was used to detect Flag-A3F and HSP90, and the other half was used to detect Vif-HA.
Extended Data Fig. 8. Effect of Vif–CBFβ–A3FCTDm complex on A3FCTDm deaminase activity.
a, UDG-based deamination assay of A3FCTDm (+: 5 μM; 15×: 75 μM) in the presence or absence of different molar excesses (2×: 10 μM; 30×: 150 μM; 40×: 200 μM) of Vif–CBFβ–EloB–EloC variants. The inhibition of A3FCTDm deamination activity by a large excess of Vif–CBFβ–EloB–EloC variants was not caused by nonspecific binding interactions as the same molar amount excess (40×) of BSA did not trigger the inhibition.
b, One of the Vif–CBFβ–A3FCTDm tetramer interface involving the 55VxIPLx4–5L64 motif (Extended Data Fig. 4a, left) blocks the catalytic site of A3FCTDm (red and marked by an arrow). Vif is shown in magenta, CBFβ in cyan, and A3FCTDm in green. One Vif–CBFβ–A3FCTDm ternary complex with the major interface is circled with an oval.
Extended Data Fig. 9. Parameters of the cryo-EM reconstructions of Vif–CBFβ–A3FCTDm complexes and the final model.
a, Fourier shell correlation (FSC) curves of the half maps from gold standard refinements of the Vif–CBFβ–A3FCTDm complex with (cyan) or without (blue) the Vif α-domain and the corresponding interacting CBFβ C-terminus. The FSC curve of the map and final model of the truncated Vif–CBFβ–A3FCTDm complex is in green. Resolution of the maps are determined by the cutoff values at FSC = 0.143. b, The Euler angle distribution of the classified particles of the truncated Vif–CBFβ–A3FCTDm complex used for the final 3D reconstruction. c, Color coded local resolution estimation of the D2 symmetrized map of the truncated Vif–CBFβ–A3FCTDm complex.
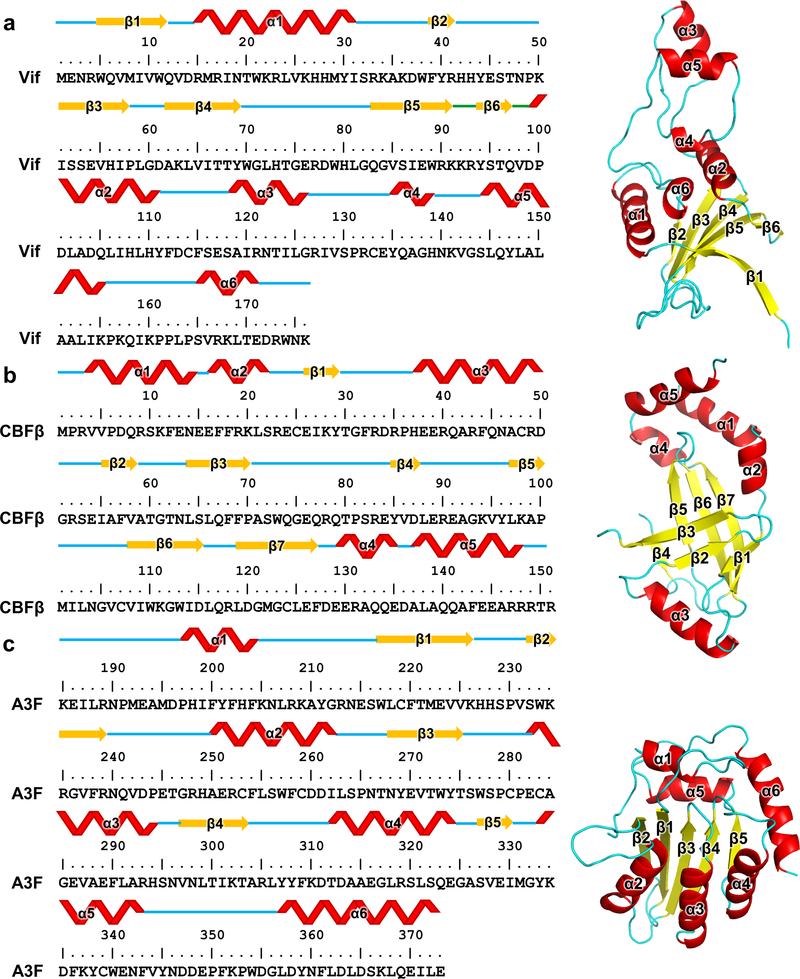
Extended Data Fig. 10. The detailed illustrations of the secondary structure elements of Vif176 (a), CBFβ151 (b) and A3FCTD (c).
The secondary structures are annotated on primary amino acid sequences (left) and tertiary structures (right). The tertiary structures for illustration are: Vif, extracted from PDB 4N9F; CBFβ151, extracted from our cryo-EM structure; A3FCTD, PDB 3WUS.
Supplementary Material
Acknowledgements
We thank S. Wu, M. Llaguno and X. Liu at Yale Cryo-EM facilities and C. Wang and J. Liu for assistance with data collection. We thank K. Knecht, O. Buzovetsky, S. C. Devarkar and other Xiong lab members for discussions. This work was supported by National Institutes of Health grant AI116313 (Y.X.). This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by Intramural AIDS Targeted Antiviral Program grant and Innovation Fund, Office of AIDS Research, NIH to V.K.P.
Footnotes
Competing interests
All authors declare no competing financial interests.
References
- 1.Lecossier D, Bouchonnet F, Clavel F & Hance AJ Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jager S, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Du J, Evans SL, Yu Y & Yu XF T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Marin M, Rose KM, Kozak SL & Kabat D HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9, 1398–1403 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Sheehy AM, Gaddis NC & Malim MH The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9, 1404–1407 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Stopak K, de Noronha C, Yonemoto W & Greene WC HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Molecular Cell 12, 591–601 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Conticello SG, Harris RS & Neuberger MS The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol 13, 2009–2013 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Conticello SG, Thomas CJ, Petersen-Mahrt SK & Neuberger MS Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22, 367–377 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Baig TT, Love RP & Chelico L Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol 5, 450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaipan C, Smith JL, Hu WS & Pathak VK APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol 87, 444–453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology 333, 374–386 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Russell RA, Smith J, Barr R, Bhattacharyya D & Pathak VK Distinct Domains within APOBEC3G and APOBEC3F Interact with Separate Regions of Human Immunodeficiency Virus Type 1 Vif. Journal of Virology 83, 1992–2003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JL & Pathak VK Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol 84, 12599–12608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hache G, Liddament MT & Harris RS The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem 280, 10920–10924 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Aydin H, Taylor MW & Lee JE Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure 22, 668–684 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Kitamura S, Ode H & Iwatani Y Structural Features of Antiviral APOBEC3 Proteins are Linked to Their Functional Activities. Front Microbiol 2, 258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell RA & Pathak VK Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol 81, 8201–8210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fribourgh JL, et al. Core binding factor beta plays a critical role by facilitating the assembly of the Vif-cullin 5 E3 ubiquitin ligase. J Virol 88, 3309–3319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DY, et al. CBFbeta stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol Cell 49, 632–644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Bohn MF, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure 21, 1042–1050 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Z, Zhang W, Chen G, Xu R & Yu XF Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol 381, 1000–1011 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Pery E, Rajendran KS, Brazier AJ & Gabuzda D Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol 83, 2374–2381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima M, et al. Structural Insights into HIV-1 Vif-APOBEC3F Interaction. Journal of Virology 90, 1034–1047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita T, Kamada K, Hatcho K, Adachi A & Nomaguchi M Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect 10, 1142–1149 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Dang Y, Davis RW, York IA & Zheng YH Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol 84, 5741–5750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura S, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol 19, 1005–1010 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Albin JS, et al. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem 285, 40785–40792 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu KK, Sultana A, Azimi FC & Lee JE Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun 4, 2593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson BD & Harris RS Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-beta/RUNX axis. Sci Adv 1, e1500296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang Y, Wang X, Zhou T, York IA & Zheng YH Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol 83, 8544–8552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, He Z, Wang T, Xu R & Yu XF A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol 83, 8674–8682 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DY The assembly of Vif ubiquitin E3 ligase for APOBEC3 degradation. Arch Pharm Res 38, 435–445 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Richards C, et al. The Binding Interface between Human APOBEC3F and HIV-1 Vif Elucidated by Genetic and Computational Approaches. Cell Reports 13, 1781–1788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83, 494–497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakashima M, et al. Mapping Region of Human Restriction Factor APOBEC3H Critical for Interaction with HIV-1 Vif. J Mol Biol 429, 1262–1276 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Huthoff H & Malim MH Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol 81, 3807–3815 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogerd HP, Doehle BP, Wiegand HL & Cullen BR A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A 101, 3770–3774 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangeat B, Turelli P, Liao S & Trono D A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem 279, 14481–14483 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Schrofelbauer B, Chen D & Landau NR A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc Natl Acad Sci U S A 101, 3927–3932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A 101, 5652–5657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouno T, et al. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat Struct Mol Biol 22, 485–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Britan-Rosich E, Nowarski R & Kotler M Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. J Mol Biol 410, 1065–1076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Love RP & Chelico L HIV-1 viral infectivity factor (Vif) alters processive single-stranded DNA scanning of the retroviral restriction factor APOBEC3G. J Biol Chem 288, 6083–6094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehle A, Goncalves J, Santa-Marta M, McPike M & Gabuzda D Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev 18, 2861–2866 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris RS & Anderson BD Evolutionary Paradigms from Ancient and Ongoing Conflicts between the Lentiviral Vif Protein and Mammalian APOBEC3 Enzymes. PLoS Pathog 12, e1005958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hache G, Shindo K, Albin JS & Harris RS Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol 18, 819–824 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albin JS, Hache G, Hultquist JF, Brown WL & Harris RS Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol 84, 10209–10219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 53.Xiao X, Li SX, Yang H & Chen XS Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun 7, 12193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell RA, Smith J, Barr R, Bhattacharyya D & Pathak VK Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol 83, 1992–2003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith JL, Izumi T, Borbet TC, Hagedorn AN & Pathak VK HIV-1 and HIV-2 Vif Interact with Human APOBEC3 Proteins Using Completely Different Determinants. J Virol (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen KL, et al. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319, 163–175 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Yee JK, Friedmann T & Burns JC Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods in cell biology 43 Pt A, 99–112 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Unutmaz D, KewalRamani VN, Marmon S & Littman DR Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J Exp Med 189, 1735–1746 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desimmie BA, Smith JL, Matsuo H, Hu WS & Pathak VK Identification of a tripartite interaction between the N-terminus of HIV-1 Vif and CBFbeta that is critical for Vif function. Retrovirology 14, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheres SH & Chen S Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Afonine PV, et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeLano W The PyMOL Molecular Graphics System DeLano Scientific, Palo Alto, CA, USA: (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The model of the truncated Vif–CBFβ–A3FCTDm complex has been deposited in the wwPDB with accession code PDB 6NIL. The cryo-EM maps of the truncated and full-length Vif–CBFβ–A3FCTDm complexes have been deposited in EMDB with accession codes EMD-9380 and EMD-9381, respectively. Source data for Fig. 2c,d,e, Fig. 3c,d,e,f,g, Extended Data Fig. 1b,c, Extended Data Fig. 4c,d, Extended Data Fig. 5b,c, Extended Data Fig. 6a,b, and Extended Data Fig. 7b,c are available with the paper online. Other data are available from corresponding authors upon reasonable request.