Abstract
Recent advances in whole genome and exome sequencing have dramatically increased the database of human gene variations. There are now enough sequenced human exomes and genomes to begin to identify gene variations that are notable because they are NOT observed in sequenced human genomes, apparently because they are subject to “purifying selection”, exemplifying genetic intolerance. Such “dysprocreative” gene variations are embryonic lethal or prevent reproduction through any one of a number of possible mechanisms. Here we review an emerging quantitative approach, “Missense Tolerance Ratio” (MTR) analysis, that is used to assess protein-encoding gene (cDNA) sequence intolerance to missense mutations based on analysis of the >100K currently available human genome and exome sequences. This approach is already useful for analyzing intolerance to mutations in cDNA segments with a resolution on the order of 90 residues. Moreover, as the number of sequenced genomes/exomes increases by orders of magnitude it may eventually be possible to assess mutational tolerance in a statistically robust manner at or near single site resolution. Here we focus on how cDNA intolerance analysis complements other bioinformatic methods to illuminate structure-folding-function relationships for the encoded proteins. A set of disease-linked membrane proteins is employed to provide examples.
Keywords: genome, exome, missense tolerance ratio, purifying selection, intolerance, missense mutation, variations, gene, protein, membrane, Alzheimer’s, neurodegeneration, γ-secretase, amyloid precursor protein, Notch, DAP12, TREM2, KCNQ1, KCNQ2, KCNQ3, KCNQ4
1. INTRODUCTION
The sequencing of the first human genome was plausibly described as “the single most important project in the biomedical sciences” of its era [1]. The achievements of the original project include creating the first human genome map, developing new approaches for sequencing DNA, advancing genomic training, and building tools for interpreting and comparing genetic data [2]. Two decades after the publication of the first human genome, the challenges and opportunities afforded in interpreting human genetic data remain impressive [3].
Next generation sequencing techniques have potentiated sequencing of an increasing number of whole human genomes and exomes at ever-decreasing cost. The largest currently available human dataset is the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org), which as of mid-2019 contains sequence information from over 125,000 exomes and over 15,000 whole genomes [4]. This and related databases provide public access to gene-specific average gene sequencing coverage, known gene variations, consequent protein mutations, and so forth. With the pool of known human gene variants growing rapidly, an important objective is to use these data to document and analyze observed gene variations, in part to help assess which variants are benign versus those that are disease-causing or predisposing [3, 5]. Various bioinformatic tools have been developed to predict the impact of individual mutations, such as SIFT [6], PolyPhen-2 [7], and CADD [8], etc. However, the prediction of the impact of gene variations on the encoded proteins and linked physiology remains a challenging problem [9]. We note that these methods all focus on observed gene variations.
Here we review bioinformatic approaches that fall into a distinct genre, “intolerance analysis”, which focuses on variations that would be expected to be seen in human gene databases based on biological, biochemical, and statistical considerations, but are not. These variations seem to have been filtered out of the analyzed human gene pool by natural selection. “Purifying selection” implies that such variants prevent procreation and inheritance of genetic information because they are embryonic lethal or interfere with human reproduction at any one of variety of possible levels, such as disruption of conception or defects in embryonic development. For the sake of simplicity in this paper, we will henceforth use the term “dysprocreative” as a catchall phrase for these possible mechanisms. Dysprocreative mutations are different from ordinary heritable disease mutations, in that the latter variants are passed on from generation to generation in a Mendelian manner. We note that dysprocreative mutations can occur in humans as germline (“de novo”) variations. Provided they are not embryonic lethal such mutations may be detected in living humans, but will be very rare because they cannot be passed on to the next generation. Otherwise-intolerant gene variations might also be detected when they occur as somatic mutations, for example in cancer tumor cells.
Importantly, for many genes the dysprocreative effect of these non-observed gene variations will be manifest even under heterozygous conditions in which the “toxic” variant is co-expressed with a wild type variant. With this in mind, we note that there are three general mechanisms by which such heterozygous variants can exert their dominant effect. First, a variant may induce loss of function of the encoded protein. This means there will be a 50% reduction in the function of that protein relative to WT/WT homozygotes under physiological conditions, which in some case will be dysprocreative. Secondly, a variant may induce a toxic “gain-of-function” effect such as mis-regulated native function or formation of toxic aggregates such as amyloids or prion-like structures, either one of which could be responsible for its dysprocreative effect. A third mechanism is the case where not only is the protein encoded by the variant allele functionally inactive, but it induces loss of function of the WT protein as well (most often through mechanisms that involve formation of WT/mutant oligomers).
We focus here on bioinformatic tools for assessing intolerance to gene variation in the human population that were pioneered by the David B. Goldstein lab at Duke and Columbia Universities, and further developed by Slavé Petrovski, now in the University of Melbourne. We begin this paper by reviewing the key concepts underlying these bioinformatic tools [10–13]. We then focus on assessment of the “missense tolerance ratio” (MTR) [13] and explore the results of MTR analysis when applied to a set of human membrane proteins, most known to be linked to human diseases. We write this review based on the working assumption that intolerant gene variations exert their dysprocreative effect via their impact on the encoded protein. However, we recognize that some of these mutations likely exert their effect by other mechanisms. These include variations that result in conversion of dinucleotides into or out of CpG sequences. Because CpG sequences are subject to DNA methylation (methylation of the cytosine), missense mutations that introduce or eliminate them can alter the DNA methylation patterns. While DNA methylation is best known when it occurs in gene regulatory sequences, CpG sites in protein-encoding exons are also subject to DNA methylation, which can profoundly impact RNA splicing and the protein sequence encoded by the final mature mRNA [14]. It seems likely that inappropriate RNA splicing may in some cases be dysprocreative. Indeed, missense mutations in exons that do not impact DNA methylation can also alter RNA splicing [15]. Yet other missense mutations could conceivably exert a dyprocreative outcome by altering mRNA stability [16] or even rendering the mutant mRNA toxic [17].
2. MEASUREMENT OF TOLERANCE TO GENE VARIATION
Scientists have long analyzed multiply aligned sequences of a given protein from different species to identify sites and segments that are highly conserved, an approach that was enabled by widespread genome sequencing of organisms from all domains of life. This has been a fantastically successful approach and does provide insight into protein sequence intolerance across species. Sites that are intolerant of mutation across species are often important for protein folding, structure, interactions, function, or some combination of these properties [18, 19]. On the other hand, just because a site in a protein varies from species to species does not mean that it is not important—in some cases residues are under “positive selective pressure” to adapt the protein to meet organism-specific conditions or physiological needs.
The availability of well over 200,000 human genome/exome sequences now enables the identification of human proteins for which entire alleles are observed to be deleted, that include gene variations that encode an early stop codon in the open reading frame (ORF), or that contain a frameshift mutation early in the ORF, in all three cases resulting in complete loss of function for the affected protein [4]. In most cases only a single allele of the protein will be subject to these dramatic mutations, resulting in a 50% reduction in the wild type (WT) expression level. In cases where both alleles are affected there will be complete loss of expression of the protein in question. In either circumstance, detection of lesions in adult humans that cause underexpression (haploinsufficiency) or complete elimination of expression of a protein reveals some degree of tolerance to a reduction or complete loss of that protein in humans.
The database of human gene variations can also be used to examine gene-to-gene patterns in missense mutations that alter single amino acid changes in the encoded protein. Genes or gene segments that harbor very few observed missense variants in the human population are presumed to be those that that are under high purifying selection, meaning that the consequence of each mutation-not-seen on the encoded protein is dysprocreation [10, 18, 20]. We emphasize that variations that are subject to purifying selection should not be thought of as generic “disease mutations”. A great many gene variations that are linked to human disease do not prevent human reproduction. For example, the cystic fibrosis transmembrane regulator (CFTR) is subject to over 1000 different missense mutations, any one of which can result in cystic fibrosis, but that do not appear to be subject to strongly purifying selection. Conversely, if an allele is resolutely intolerant of variation in the human population, it will not be associated with a known human disease because it is dysprocreative. Clearly, identifying intolerant sites in a protein is not the same thing as identifying disease-linked mutation sites. Nevertheless, as has been previously shown, genes that are relatively intolerant of variations are very often also subject to disease-causing mutations, such that identifying the partially intolerant regions of a gene sometimes facilitates interpretation of de novo mutations in that gene [12, 13, 21].
Petrovski, Goldstein, and coworkers [10] introduced a metric called the Residual Variation Intolerance Score (RVIS) to identify genes that are relatively intolerant of variation at the whole-gene level. The RVIS scoring system assesses the level of genetic intolerance by regressing the observed total number of variants in a given gene against the number of common (minor allele frequency > 0.1% in human samples sequenced) nonsynonymous variants observed for a given human gene across available sequences. The studentized residual, i.e. the measure of deviation from the expected score based on evolutionary neutrality, is the RVIS score. A negative RVIS score indicates purifying selection and a positive score indicates that variations in that gene are seen more frequently than expected based on neutral evolutionary drift. Analysis of 16,956 genes revealed that genes with lower RVIS values (i.e. less tolerant to mutations and undergoing purifying selection) are more likely than others to be associated with Mendelian diseases [10]. This approach was later extended to include the proximal regulatory sequence of genes, termed as ncRVIS (noncoding RVIS), and used to evaluate how the intolerance of noncoding sequences can predict disease-causing genes through gene dosage fluctuation [22]. A very low (strongly negative) RVIS score does not imply that a protein is a particularly important protein relative to other proteins, even those with zero or positive RVIS scores. Rather, it simply means that the collection of human sequences for that protein exhibits a relatively high number of sites at which missense mutations are observed less often than expected based on neutral evolutionary drift, most probably because some variations are dysprocreative. A protein could have a small number of absolutely intolerance sites and still yield a near-0 to positive RVIS score. An example is the microtubule-associated protein tau, where the final quarter of the protein sequence is seen to be mutationally intolerant while the gene as whole is highly tolerant, resulting in an overall positive RVIS score [11]. Some proteins related to immunological diseases exhibit a positive average RVIS (meaning they appear to be under pressure to mutate), as might be expected for proteins whose function is to help the body to adapt to foreign antigens. On the other hand, proteins with variations that are related to developmental disorders tend to have a negative average RVIS [10]. This is reasonable because severe development disorders will often be dysprocreative. Akin to gene RVIS values, Samocha et al. developed a similar metric for measuring “gene constraint”, the missense Z score [23]. They also found that the constrained genes, i.e. genes that are depleted of missense variants, were enriched for known Mendelian diseases. While these scoring systems are useful in ranking genes based on overall sequence intolerance of missense variations, they do not identify the specific regions of the gene that are intolerant. They also do not identify proteins that are overall tolerant of mutations, but that may have one or a few absolutely intolerant segments.
To evaluate intolerance at the level of protein domains (in multi-domain proteins) and/or exons, the Goldstein group developed a “domain” and exon level of analysis [11]. Using the same RVIS approach, they focused on individual protein domains as defined by conserved domains database (CDD) and/or exons as the unit/s of “subRVIS” analysis. This new framework successfully identified a significant relationship between both domain and exon subRVIS scores and their respective pathogenic variants within the entire gene [11]. A similar approach was also developed by Havrilla, Quinlan, and coworkers to generate a map of constrained coding regions (CCR) within a given protein that are under purifying selection [24] and by Samocha, Daly and coworkers to develop a regional missense constraint metric [25]. Hayeck et al. [26] further improved the tool by introducing a Bayesian hierarchical model, which they called the localized intolerance model using Bayesian regression (LIMBR), that accounts for the effects of whole gene and sub-regions (domains) of gene, resulting in more stable estimates.
Genetic analysis at the whole-gene, domain, and exon levels proved successful in identifying protein units that are overall tolerant or intolerant of genetic variation. However, it is also possible that a purifying constraint may be acting only on a small subset of residues within a domain or exon. In other words, i.e. a short segment in a protein may be undergoing extreme purifying selection, but the whole protein or the whole domain or exon may exhibit a positive RVIS (or subRVIS) score. To increase the resolution of genetic intolerance analysis, Traynelis et al. [13] introduced the missense tolerance ratio (MTR) to identify intolerant segments in proteins, an approach that provides a much higher degree of sequence resolution than RVIS or subRVIS scores. Moreover, MTR analysis is independent of the boundaries created by domains and exons. This review focuses on MTR analysis rather than either RVIS or subRVIS because while the latter analyses provides insight into whether or not a protein is intolerant as a whole, or whether a domain or exon is intolerant to missense mutation (for subRVIS), MTR analysis can identify shorter segments that are missense intolerant and does not depend in foreknowledge of domain and exon boundaries.
The current default mode of MTR analysis involves a sliding window approach that calculates intolerance in a window spanning cDNA mutations 15 residues ahead and 15 residues behind the codon encoding each individual amino acid in a protein sequence [13, 21]. According to equation 1 below, for each 93 nucleotide frame, the MTR gives the ratio of the observed (obs) fraction of the number of missense DNA variations relative to the number of all observed (missense + synonymous) single base variants divided by the expected (exp) fraction of the number of missense mutations relative to the number of all possible variations in that segment [13, 21]. The expected values are simply the number of missense and synonymous variants for all possible single site base substitutions over the analyzed gene segment.
| (Equation 1) |
A key concept behind equation 1 is the assumption that synonymous mutations do not result in a change in the amino acid sequence of the protein and are therefore assumed to be evolutionarily neutral. While it is now clear that this assumption is not always correct [15, 27, 28], it is a reasonable working approximation. This equation also assumes that within a segment (usually 31 codons long) each possible single base change is equally likely within that segment, which is, again, a reasonable approximation. The MTR therefore provides the fraction of evolutionarily allowed missense mutations relative to the maximum possible fraction of mutations that are missense in the ideal case where there is no evolutionary pressure. An MTR value of 1 indicates overall selective neutrality for the segment, whereas an MTR value greater than 1 suggests positive selection and an MTR value less than 1 indicates purifying selection in the analyzed segment against missense mutations. When the purifying selection in the gene fragment encoding the protein segment is uniformly severe, then there will be no observed non-synonymous missense mutations in that segment, such that MTR → 0.
One factor that is not directly taken into account by equation 1 is the fact that, while single base mutation rates may be similar for all possible substitutions within short segments, different segments within the genome can exhibit very different intrinsic mutation rates for reasons that are not closely linked to natural selection [29–31]. The intrinsic mutation rate is expected to correlate for a given segment with the number of synonymous mutations seen in that segment in the current database of human sequences. In other words, MTR values from segments that have few observed synonymous mutations may not be statistically significant: MTR values that are lower than 1.0 might therefore represent statistically insignificant “false positives” for segments where the intrinsic mutation rate is low. Petrovski and co-workers therefore devised a method to calculate the false discovery rates (FDRs) for MTR values so as to flag cases where an MTR might deviate significantly from 1.0, but not be reliable as an indicator of true evolutionary intolerance. Details for calculation of the FDR are found in [21]. For the application of MTR analysis using the on-line Webserver prepared for this purpose, an FDR value of < 0.10 is used as the default cutoff to confirm the statistical significance of MTR values. In other words, FDR provides validation of truly intolerant segments in which <10% of the seemingly intolerant sites are false positives. One important implication of the FDR is that for any given protein segment, the FDR will go down as the number of available human genome/exome sequences included in that segment goes up.
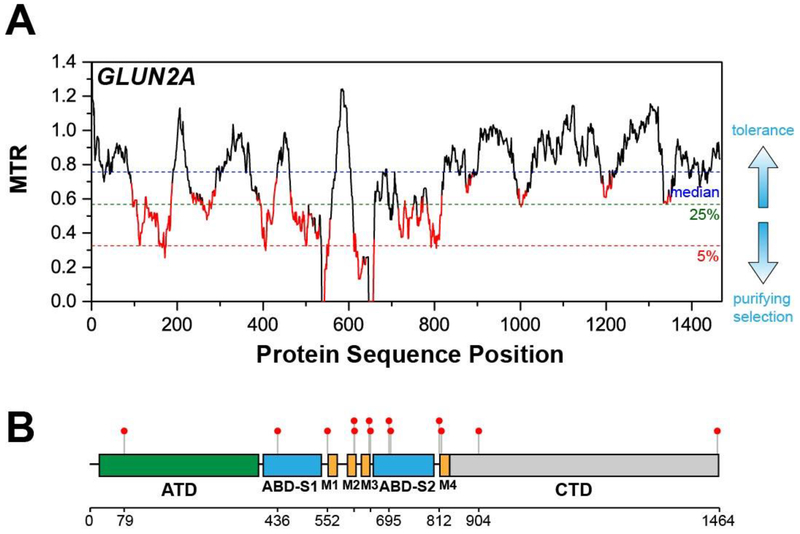
For this review, all MTR and FDR calculations were carried using the original version of the MTR-Viewer (http://mtr-viewer.mdhs.unimelb.edu.au/) [13]. We emphasize that a new version (V2) of this web server was very recently rolled out at: http://biosig.unimelb.edu.au/mtr-viewer/ [21]. Both servers are very user friendly and yield plots such as that provided in Figure 1A, which has been adapted from [12, 13]. This plot is for subunit 2A of the NMDA type glutamate ionotropic receptor (GluN2A) and is based on analysis of the cDNA segments of all possible 31 residue amino acid windows. A corresponding analysis for all possible 21 residue amino acid segments yields a similar plot (not shown), but is more noisy and with reduced statistical validity (based on higher FDRs).
Figure 1. Segmental Intolerance Analysis of GluN2A.
(A) MTR plot of GluN2A (adapted from Ogden K. et al. (2017) PLoS Genet. 13, e1006536)). (B) Lollipop plot of the pathogenic variants of GluN2A superimposed on its domain organization. ATD – amino terminal domain, ABD – agonist binding domain, M1, M2, M3, and M4 – transmembrane domains, CTD – carboxy terminal domain (adapted from Traynelis J. et al. (2019) Genome Res. 27, 1715–1729)). As for all MTR plots presented in this paper, the sliding window for which MTR values was calculated is 31 amino acids long (93 nucleotides in the cDNA). The cut-off for the 5% most intolerant segments is indicated by the horizontal dashed red line. The MTR value for which the 5% cutoff occurs will vary from protein to protein. MTR values highlighted in red are those for which the false discovery rate (FDR) is less than 0.1, indicating the deviation of the MTR value from 1 is statistically robust. For the purposes of this review we describe MTR values as reflecting “high intolerance” if the MTR value is 0.5 or less and if the FDR is <0.1.
As illustrated in Fig. 1A, dashed horizontal lines mark the gene-specific median MTR value (blue), the cut-off for the lowest 25th percentile MTR values (green), and the cutoff for the lowest 5th percentile (red, most intolerant) MTR segments. All segments of a gene may be relatively tolerant of mutations, but there will always be a most intolerant (lowest MTR values) 5% percentile. FDR values for 31 residue segments are not directly plotted, but MTRs for which the FDR is < 0.10 are highlighted in red, highlighting their “statistically robust” status. For the purpose of this review we classify a segment as “highly intolerant” if its MTR falls below the exome-wide 5% percentile cutoff (corresponding to an MTR of ≤ ~0.5, see http://mtr-viewer.mdhs.unimelb.edu.au/) and if the associated FDR value is <0.10.
As illustrated in Figure 1B and now directly incorporated in the new on-line server [21], complementary insight to MTR analysis may be gleaned by using a sequence-aligned “lollipop plot” [32] to enable visual comparison of how MTR patterns correspond to the locations of known disease mutation sites in the protein being analyzed. Figure 1B illustrates the sites of clinically relevant epilepsy mutations in the GluN2A receptor (adapted from [13]). This plot enabled Ogden et al. [12] to successfully identify the previously unappreciated pre-M1 helix located between the ABD-S1 and M1 domains (Figure 1A) as a key player in receptor function, with at least one mutation in this domain being associated with a neurological disorder (Figure 1B). The MTR plot of GluN2A receptor shows that the pre-M1 helix is highly intolerant to missense mutation (MTR value of 0), suggesting that there are no non-syonymous missense mutations in this segment found in the >165,000 available human genomes/exomes. However, a de novo germline variant P552R has recently been reported for a patient presenting with epilepsy and developmental delay. This prompted Ogden and coworkers to biochemically test the functional importance of this helix [12], revealing both a critical role for this segment in channel gating and also that the P552R de novo variant results in aberrant receptor gain-of-function. Traynelis and coworkers also used MTR and lollipop plots in studies demonstrating that pathogenic variants are preferentially concentrated in the relatively intolerant segments of the protein for 6 out of 11 dominant epilepsy genes [13]. MTR has been used recently to interpret a number of clinically observed variants of genes associated with neurodevelopmental disorders [33–35] and to analyze the role of rare variants in the regulators of G protein signaling proteins in human disease [36].
3. SEGMENTAL INTOLERANCE VERSUS INTERSPECIES SEQUENCE CONSERVATON PATTERNS.
Does intra-human missense tolerance analysis yield the same information as phylogenetic sequence conservation analysis (interspecies)? These related methods of analysis employ different data sets. Evolutionary approaches based on evaluating sequence conservation between species do not provide a direct window into intraspecies intolerance of protein segments/sites to mutation. Moreover, a region in a protein may have evolved to satisfy a more specialized function specific to humans that is not conserved across species. Sites in such a segment may not exhibit high sequence conservations across species and yet may be seen to be intolerant to mutations in the human genome/exome database. Previous studies have demonstrated that there is no strong correlation between gene intolerance and conservation [10, 11, 23, 24], suggesting that some selective pressures are specific to the human lineage. Accordingly, sequence intolerance analysis provides a tool that complements and extends the insight provided by sequence homology.
In most work published to date, use of MTR plots has been based mainly on exploiting the rough correlation that protein segments relatively intolerant of variation are often seen to also be associated with disease mutations in that segment of the protein [12, 13, 21, 33–35]. Heritable mutations that cause or promote disease are less severe than gene variations-not-seen because the latter are dysprocreative. However, as explored in the next section, disease mutations are not always found in protein segments that are highly intolerant. Conversely, just because a segment is rich in disease mutations does not mean that it will be intolerant to mutation in MTR plots. These considerations indicated that while interspecies tolerance analysis and intra-species homology relationships may often provide complementary information, intolerance analysis is expected to sometimes provide insight that cannot be provided by interspecies sequence homology analysis, as is explored in the next section.
4. SEGMENTAL MISSENSE TOLERANCE ANALYSIS OF DISEASE-LINKED MEMBRANE PROTEINS
Here, we examine the results of segmental MTR analysis for different disease-linked membrane proteins to provide vignettes regarding what interesting information can be gleaned from MTR plots. For this purpose, disease-linked membrane proteins are chosen both because of the membrane focus of the journal as well as because of the personal interests of the authors. While transmembrane proteins are subject to some distinctive evolutionary pressures [37] and also have distinctive cellular trafficking pathways and folding quality control systems[38], what we learn about MTR analysis based on application to these proteins is broadly extendable to all proteins. We remind the reader that we are taking the somewhat arbitrary tack in this paper of defining as “highly intolerant” any segment that has an MTR of 0.5 or less and that is color-coded red in the plots, indicating the segmental FDR is less than 0.1.
The MTR plots of this section (Figures 2–7) were generated using the online MTR gene viewer (http://mtr-viewer.mdhs.unimelb.edu.au/) based on sliding window (31 codon) analyses of the >135,000 human exomes and whole genomes current available at https://gnomad.broadinstitute.org/. Lollipop plots [32] were constructed using the scripts available at https://github.com/pbnjay/lollipops. The pathogenic mutation lists used for the lollipop plots were obtained from ClinVar [39] (https://www.ncbi.nlm.nih.gov/clinvar/, accessed May 6, 2019), HGMD [40] (HGMD2019.1, accessed March 20, 2019), and www.alzforum.org/mutations (accessed May 6, 2019; for Alzheimer’s related proteins). For ClinVar, we selected the missense mutations that are classified as “Pathogenic”, “Likely Pathogenic”, or “Likely Pathogenic; Pathogenic”, while for HGMD, we selected the missense mutations that were classified as disease-causing mutations or “DM”. We generally pooled the pathogenic or disease-causing missense mutations from the three sources, but we removed the mutations classified as “DM” in HGMD that have conflicting annotations in ClinVar or are classified as “not pathogenic” in www.alzforum.org/mutations.
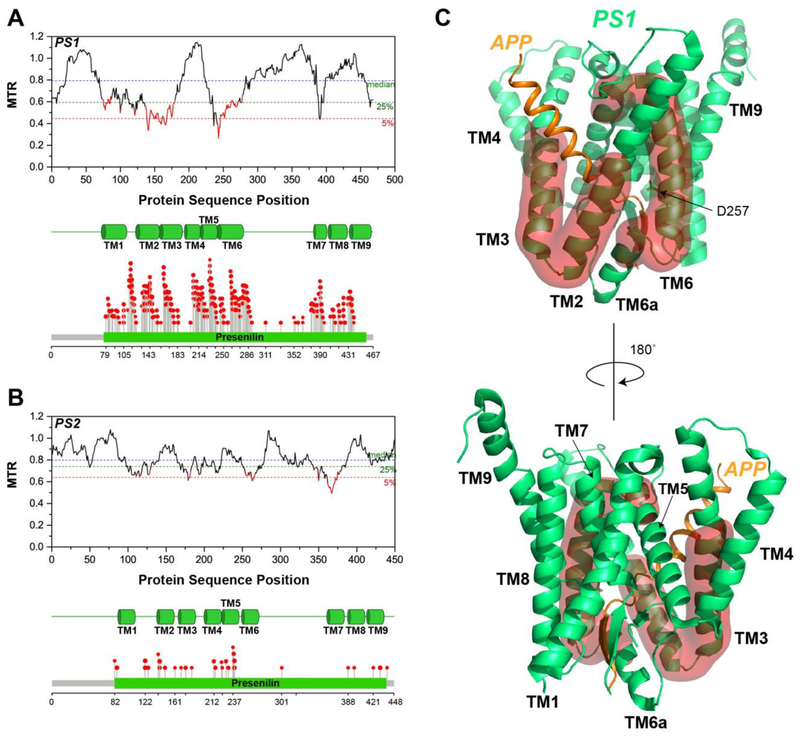
Figure 2. Segmental Intolerance Analysis of Presenilin.
(A) MTR plot for human presenilin 1 (PS1). The transmembrane (TM) domains of PS1 and the lollipop plot showing the locations of its familial Alzheimer’s disease missense variants are at the bottom of this panel. (B) MTR plot for presenilin 2 (PS2), along with the transmembrane domains and the lollipop plot for its familial Alzheimer’s disease mutations. (C) Mapping of the most intolerant segments of PS1 onto its cryo-EM structure (PDB: 6IYC) [32], as highlighted using a red surface representation.
4.1. THE γ-SECRETASE COMPLEX
Gamma-secretase is a heterotetrameric intramembrane protease, whose function and dysfunction are involved in the development of Alzheimer’s disease (AD). It catalyzes the proteolytic cleavage of single span membrane proteins, such as the amyloid precursor protein (APP) and the Notch receptor. Successive cleavage of the transmembrane C-terminal “C99” domain of APP by γ-secretase leads to production of β-amyloid (Aβ) peptides of varying lengths [41]. Aggregation of Aβ peptides, such as Aβ42, result in the formation of toxic oligomers and amyloid plaques, which are generally thought to contribute to the etiology of AD [42]. The human γ-secretase complex is comprised of four subunits: presenilin (PS), presenilin enhancer-2 (PEN-2), anterior pharynx-defective 1 (APH-1), and nicastrin [43, 44]. PS, an aspartyl protease containing nine transmembrane helices, serves as the catalytic subunit of γ-secretase. PEN-2 binds to PS and is essential for the maturation and proteolytic activity of PS. APH-1 helps to stabilize the complex, while the heavily glycosylated nicastrin is believed to be involved in substrate recognition.
We used MTR plots to examine segmental intolerance to variation in the genes coding for each of the subunits of γ-secretase (Figures 2A and 2B). For PS1, the two highly intolerant regions in the protein are the segment spanning the second half of TM2 to the first half of TM3, and TM6 (Figure 2A). These regions appear to undergo structural rearrangement upon substrate binding although flexible and disordered in the substrate-free enzyme, TM2 and TM3 become ordered upon substrate binding, while TM6 unravels into a rigid loop followed by a short helix (TM6a) (Figure 2C) [45, 46]. The TM helices housing the catalytic aspartate residues, D257 (TM6) and D385 (TM7), are intolerant to missense mutation. Interestingly, both the loop between TM6 and TM7 that forms antiparallel β strands with the substrate and the PAL motif (residues 433-435) thought to be essential for substrate recognition [47, 48] are generally tolerant to missense mutation.
While there is much overlap between the locations of pathogenic mutations in PS1 and the highly intolerant regions, many pathogenic mutations are seen in segments that exhibit high segmental tolerance—see residues 200-225, for example (Figure 2A). While only additional data can provide a clear explanation, it could be that even the most severe mutations at these sites result in disease only later in life, providing an abundance of time for reproduction before the age of disease onset. In contrast, variations in the intolerant segments are dysprocreative, an outcome that appears unrelated to Alzheimer’s disease. Nonetheless, these same segments do include some Alzheimer disease mutations. It is not yet clear whether these genetically dominant Alzheimer’s mutations act by causing γ-secretase loss of function, by causing aberrant gain of function (such as increasing the Aβ42 to Aβ40 production ratio) or both (see review in [42]).
PS2, the other isoform of PS, exhibits the same general pattern of MTR as PS1 (Figure 2B). However, PS2 does not undergo the same extent of purifying selection as PS1—its lowest MTR values are not as low as those seen for PS1. This may be explained by the notion that PS1 is thought to be the major “all-purpose” isoform of PS, with PS2 playing a more niche role in cell physiology [42]. Indeed, among the reported Alzheimer’s mutations (www.alzforum.org/mutations. accessed May 6, 2019), roughly 270 of the known mutations are situated in PS1, while only 48 are in PS2. It is intriguing that the first part of TM7 of PS2 is highly intolerant and yet is not associated with any known disease mutations (Figure 2B).
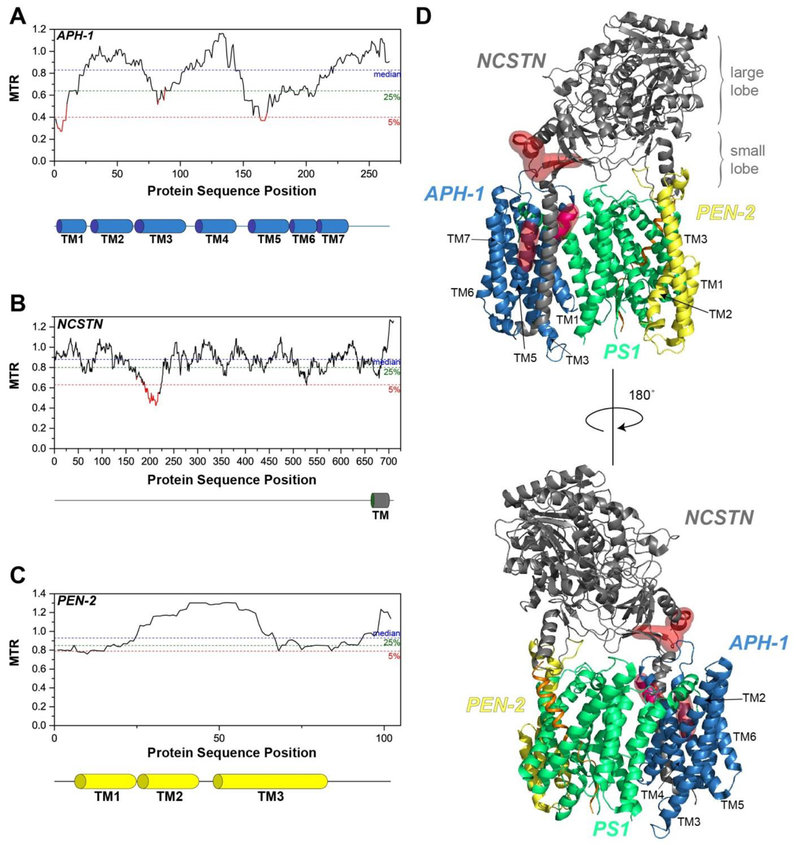
Among the seven transmembrane helices of APH-1, the highly intolerant regions are found in TM1 and TM5 (Figure 3A). In the cryo-EM structure of the human γ-secretase complex, TM1 and TM5 are packed with the TM domain of nicastrin (Figure 3D) [32]. Neither APH-1 nor nicastrin are subject to disease mutations, yet they contain highly intolerant segments.
Figure 3. Segmental Intolerance Analysis of APH-1, Nicastrin and PEN-2.
MTR plots for (A) APH-1, (B) Nicastrin (NCSTN), and (C) PEN-2. The transmembrane (TM) domains of APH-1, NCSTN, and PEN-2 are shown below their respective MTR plots. (D) The intolerant regions of nicastrin (NCSTN) and APH-1 are mapped onto the structure (PDB: 6IYC) using red surfaces.
Nicastrin has a large extracellular domain (ECD) and a single transmembrane domain. The highly intolerant region of nicastrin spans residues 197-220, situated in the small lobe of the ECD of nicastrin (Figures 3B and 3D). Most of these sites are at the interface with the other subunits of the γ-secretase complex (Figure 3D). The precise roles of these residues of nicastrin in the catalytic mechanism or stability of γ-secretase are not yet well-characterized; hence, it will be worth investigating how missense mutations in this region could impact the function and/or folding of nicastrin. It is notable that the highly conserved Trp164 near the lid, as well as the hydrophilic residues in the substrate-binding pocket including the DYIGS motif (residues 336-340), do not seem to be undergoing purifying selection, suggesting that the high intolerance seen for residues 197-220 may be unrelated to the native function of γ-secretase.
The gene coding for PEN-2, PSENEN, is generally tolerant to mutations. Indeed, the region containing residues 30-61, which covers TM2 and the first third of TM3, appears to be undergoing positive selection (MTR > 1.0) (Figure 3C). This is an example of a protein that is tolerant of missense-encoded mutations.
4.2. AMYLOID PRECURSOR PROTEIN (APP)
The β-amyloid precursor protein (APP) is a transmembrane protein best known as the source of the β-amyloid (Aβ) polypeptides that form amyloid plaques in the brains of patients with Alzheimer’s disease (AD) [41]. APP consists of a long ectodomain, a transmembrane domain, and a short cytoplasmic tail. It has three major isoforms, composed of 770, 751, and 695 residues, which are the consequence of differential splicing of APP mRNA [49–51]. APP can be cleaved by either α-secretase or β-secretase to generate the C-terminal transmembrane fragment, C83 or C99, respectively [52, 53], which will be subsequently cleaved by γ-secretase to produce the p3 and Aβ polypeptides, respectively, together with APP-intracellular domain (AICD) [54]. The aberrant aggregation of neurotoxic Aβ peptides is a hallmark feature of AD. While APP has been extensively studied in the context of its role in amyloid production, it may play roles in cell proliferation, differentiation, neurite outgrowth, and synaptogenesis [55].
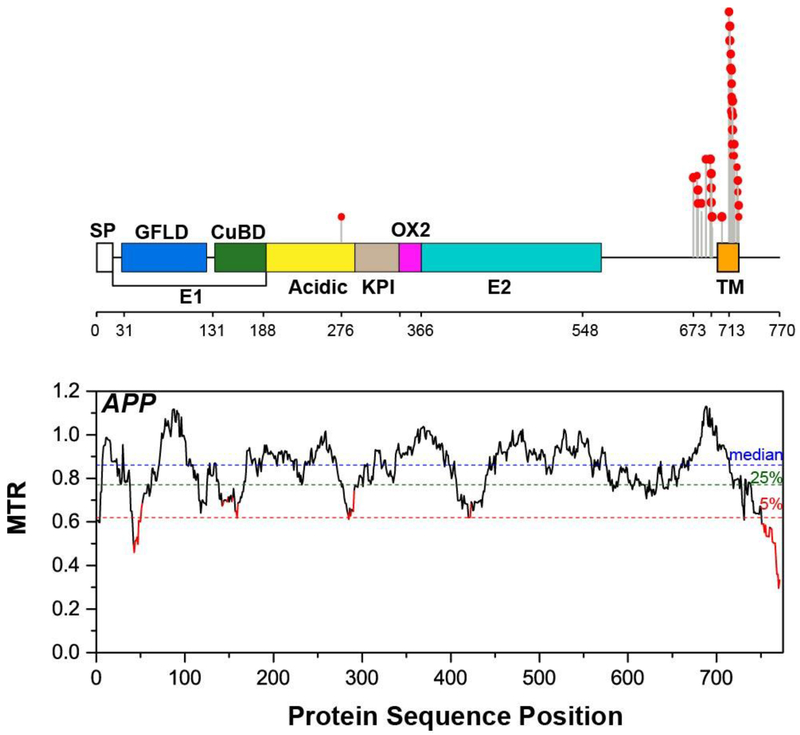
APP exhibits a couple of highly intolerant segments (Figure 4). The presence of highly intolerant segments means that many variations in APP are dysprocreative even under conditions heterozygous WT/variant co-expression. This is surprising in light of the fact that APP (-,-) knockout mice are reasonably healthy and are able to breed [56]. This suggests that some mutations in APP are highly toxic to the host cell, even when co-expressed with WT. Given that Alzheimer’s is thought to be associated with the toxicity of a fragment of APP late in life, it is perhaps not surprising that some APP mutations are not tolerated by evolution. However, it seems likely that the mechanism of toxicity associated with Alzheimer’s disease may be different from the mechanism that results in dysprocreation.
Figure 4. Segmental Intolerance Analysis of the Amyloid Precursor Protein (APP).
MTR plot for human APP. A lollipop plot showing locations of the familial Alzheimer’s disease mutations superimposed on the domain organization of APP are shown on top of the MTR plot. SP – signal peptide, GFLD – growth factor-like domain, CuBD – copper-binding domain, KPI - Kunitz-type protease inhibitor, TM – transmembrane domain.
The most intolerant domain of APP is the cytoplasmic tail of APP. This domain is not subject to Alzheimer’s disease mutations, providing another example where intolerance seems to be decoupled from disease mutations. This portion of APP has been shown to interact with a plethora of adaptor proteins [57, 58]. It contains the highly conserved YENPTY sorting motif (residues 757-762) that is believed to be involved in APP trafficking and internalization via clathrin-mediated endocytosis [59–61]. Being a part of the AICD protein released from C99 by γ-secretase, it has been reported to bind Fe65 and then possibly translocate to the nucleus to activate gene transcription, similar to Notch [62, 63]. The multifarious roles of the cytoplasmic tail of APP could help to explain its extreme intolerance to missense variations. It would be expected that the non-tolerated variations must NOT be loss-of-function in nature (because we know that APP knockout mice are reasonably healthy), but instead somehow alters the interactions of this cytosolic tail with other molecules (or itself) in a way that ultimately results in dysprocreation.
4.3. NOTCH Receptor
The Notch receptor is a single-pass transmembrane protein, whose signaling pathway plays a vital role in cell-to-cell communication, rendering it a master regulator of cell development and differentiation [64–66]. It consists of a large extracellular N-terminal domain (NECD), a transmembrane domain, and an intracellular domain (NICD). Upon ligand binding, Notch signaling is initiated by two proteolytic cleavage events: it is first cleaved by an ADAM metalloprotease [67, 68] followed by γ-secretase cleavage, ultimately releasing the NICD that then translocates to the nucleus to activate target gene expression [69–72]. Dysregulation of the Notch receptor is implicated in various cancers [73–75].
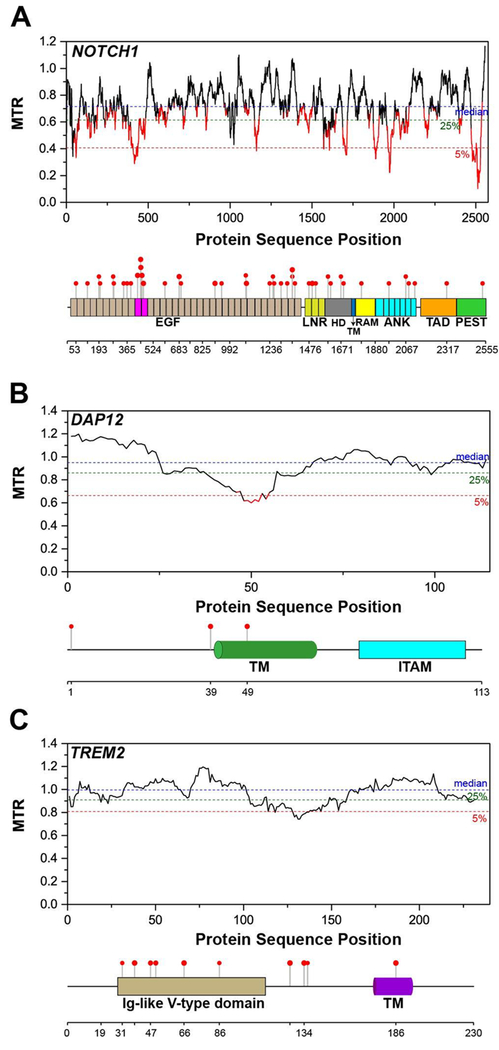
The Notch1 MTR plot shows that this protein has an unusually high number of severely intolerant regions (Figure 5A), some of which are located in the NICD. This accentuates the pivotal role that NICD plays in the overall signaling pathway. Once released after γ-secretase cleavage, NICD translocates to the nucleus and interacts with the DNA-binding transcription factor CSL and a coactivator, Mastermind, to form a complex that activates gene transcription [76–78]. The NECD also contains some intolerant regions, most noteworthy of which is the region spanning residues 381-479, comprised mainly of EGF domains 11 and 12 (highlighted in magenta in Figure 5A). EGF domains 11 and 12 are known to be centrally involved in ligand binding [79]. A modest number of reported pathogenic mutations are spread throughout Notch1, including in EGF domains 11 and 12 (Figure 5A).
Figure 5. Segmental Intolerance Analysis of Notch1, DAP12, and TREM2.
MTR plots for (A) the NOTCH1 receptor, (B) DAP12 (TYROBP), and (C) TREM2. Lollipop plots showing the positions of associated pathogenic missense variants for each protein are mapped on top of the domain organization below each MTR plot. EGF - epidermal growth factor-like domain, LNR – Lin-12/Notch Repeat, HD – heterodimerization domain, TM – transmembrane domain, RAM - RBPJ-associated module, ANK – ankyrin repeats, TAD – transactivation domain, PEST – proline, glutamic acid, serine, and threonine domain, ITAM – immunoreceptor tyrosine-based activation motif.
4.4. DAP12
The DNAX-activating protein of 12 kDa (DAP12, also known as TYROPB) is a homodimer expressed by natural killer (NK) and myeloid cells [80]. DAP12 consists of a single transmembrane domain with a short extracellular region and a cytoplasmic tail containing “immunoreceptor tyrosine-based activation” (ITAM) motifs, which when phosphorylated mediate downstream signaling resulting in activation of NK cells [81, 82]. As a homodimer it must assemble with ligand-binding co-receptors, termed the “DAP12-associated receptors”, in order to transduce the signal from the receptor to its downstream effectors [83]. One such co-receptor is the triggering receptor expressed on myeloid cells 2 (TREM2), for which mutations are now appreciated to be risk factors for several neurodegenerative disorders [84], including late-onset Alzheimer’s disease (see below).
DAP12 is overall tolerant to missense mutation but contains a one moderately intolerant region, centered around residues 48-54 (Figure 5B). This segment is part of the transmembrane domain of DAP12, which contains two residues, Asp50 and Thr54, which are known to be critical for the assembly of the DAP12 dimer with a DAP12-associated receptor. These residues, together with a basic residue located in the single TM segment of its co-receptors such as TREM2, form an electrostatic and H-bonding network that is essential for the stable assembly of the DAP12-receptor complex [85]. This also suggests that disruption of the complex of DAP12 with one or more of its co-receptors is dysprocreative.
4.5. TREM2
The triggering receptor expressed on myeloid cells 2 (TREM2) is an immune receptor that is expressed on the surface of immune cells, such as microglia, macrophages, and dendritic cells, in the brain. Mutations in TREM2 are associated with various neurodegenerative diseases, such as Nasu-Hakola disease, frontotemporal dementia, and Alzheimer’s disease [86]. It is a single pass transmembrane protein containing an extracellular Ig-like V-type domain that binds a number of different ligands [87–91], a short stalk, a transmembrane domain, and a short cytoplasmic tail. The transmembrane domain of TREM2 contains a lysine residue that associates with the homodimer of DAP12 to form the signaling complex [92, 93]. Upon ligand binding to TREM2, the tyrosine residues of the ITAM motifs of DAP12 will be phosphorylated, activating downstream signaling [81].
The MTR plot of TREM2 does not show any significant region in the protein that is intolerant to missense mutation (Figure 5C). However, there are known missense mutations in the ectodomain of TREM2 that are linked to neurodegenerative diseases (see lollipop plot in Figure 5C). TREM2 is an example of a protein where MTR analysis does not reveal the critical region of the protein where missense mutations are pathogenic. This is likely because the neurodegenerative diseases associated with TREM2 mutations are late age-onset disorders and are not dysprocreative.
4.6. KV7/KCNQ POTASSIUM CHANNELS
KV7 channel subtypes 1-5 are a family of voltage-gated potassium ion channels that are encoded by the KCNQ1-KCNQ5 genes. These channels have multiple physiological functions and are found throughout the body, including in neurons, cardiac myocytes, epithelia, smooth muscle cells, and the cochlea. For example, one of the important functions of the KV7.1/KCNQ1 channel is to partner with the KCNE1 channel-modulatory protein to form a complex that functions to generate the IKs current that is an essential component of the cardiac action potential. The KCNQ gene family is linked to a number of hereditary disorders that arise from dominant missense mutations, including arrhythmias such as long QT syndrome (KCNQ1), deafness (KCNQ1), and both mild and severe forms of epilepsy (KCNQ2 and KCNQ3) [94–100]. KV7 channels are comprised of six transmembrane helices (S1-S6) with a long C-terminal cytoplasmic domain. The first four transmembrane segments (S1-S4) of each subunit form a transmembrane voltage-sensing domain (VSD) while the last two (S5-S6) assemble into the pore-forming domain [101, 102]. The KV7 channels function as homotetramers and are also known to partner with any one of several single transmembrane span accessory proteins KNCE1-KCNE5, which act to profoundly modulate channel function [103–110].
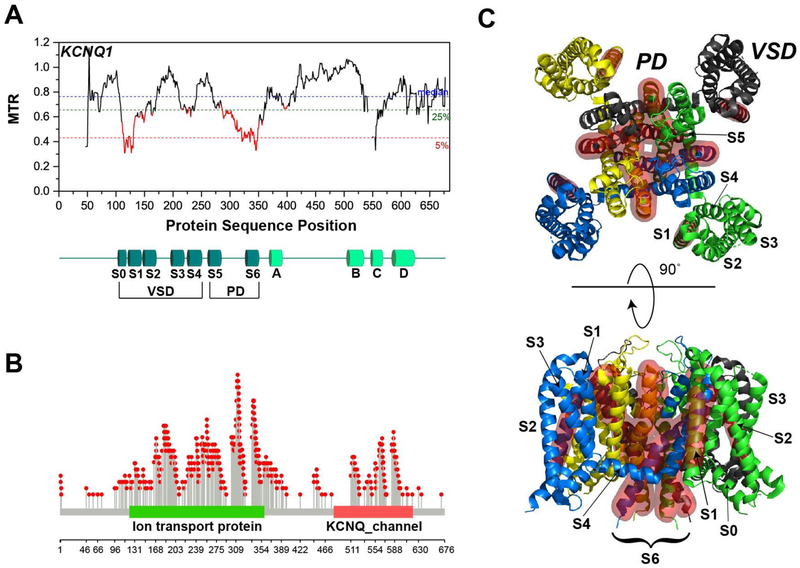
The pore forming domains (S5-S6) of all five KV7 channels exhibit the lowest MTR scores across the entire KCNQ family (Figures 6–7), with a few segments being seen to be absolutely intolerant (MTR = 0). Given that the not-tolerated mutations usually occur under genetically-dominant conditions, this poses the question of whether a 50% loss in channel function for each of these channels is sufficient to confer dysprocreation. This seems unlikely as it is known that many disease mutations in KCNQ1 cause complete loss of function of the affected allele; such variations are sufficient to cause disease but are not dysprocreative. One possibility is that channel encoded by the allele containing the non-tolerated variation is not only bereft of function, but still forms oligomers with the WT allele in a way that induces loss of the function of the WT allele product. Another possibility is that the non-tolerated mutations induce some sort of toxic gain of function, which could range from promoting unregulated channel conductance to causing misfolding to form toxic aggregates. Only future experiments can resolve which of these possibilities pertain or whether there are other possible mechanisms (see Section 5) that are the sources of the purifying genetic selection.
Figure 6. Segmental Intolerance Analysis for KV7.1/KCNQ1.
(A) MTR plot for human KCNQ1. The domain organization is shown below. VSD – voltage-sensing domain, PD – pore domain. S0-S6 – transmembrane helices, A-D – intracellular helices. Notice that there are gaps in the MTR plot (from residues 1-46, 542-548, and 552-553). MTR is not calculated in these windows because there currently are less than 3 observed variants in these windows, which indicates inadequate sequence coverage to calculate an MTR. (B) Lollipop plot showing the locations of long QT syndrome mutations in KCNQ1. (C) The intolerant regions of KCNQ1 mapped onto its cryo-EM structure (PDB: 5VMS) [88] using a red surface representation.
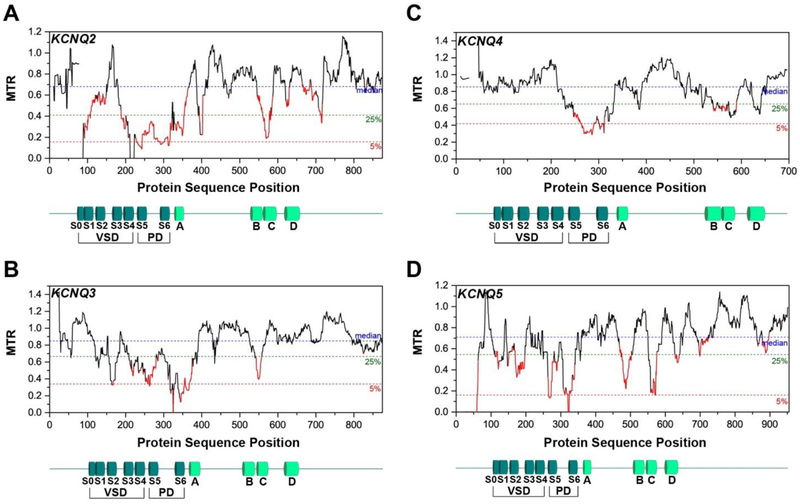
Figure 7. Segmental Intolerance Analysis for Kv7 Family Members 2-5 (KCNQ2-KCNQ5).
VSD – voltage-sensing domain, PD – pore domain, S0-S6 – transmembrane helices, A-D – intracellular helices. Gaps in MTR plots mean that MTR is not calculated in those windows because there are less than 3 observed variants the segments.
With regard to the voltage sensor domain (which includes the surface S0 helix and the transmembrane S1-S4 helices), there is not a consistent pattern across the KCNQs (Figures 6 and 7). KCNQ1 has a low MTR score spanning the S0 and S1 helices, consistent with the recent discovery that the S0 segment of KCNQ1 is a central stabilizing element of the VSD fold [111]. This fact, along with the observation that the helices most directly involved in the KCNQ1 voltage-sensing mechanism (S4 and to a lesser degree S2, [112–118]) exhibit only moderate intolerance, may be consistent with the notion that intolerant sites in KCNQ1 are sometimes associated with misfolding. KCNQ2, KCNQ3, and KCNQ5 all have complex patterns of intolerance in their VSD. On the other hand, S0-S3 of KCNQ4 exhibit MTR scores near 1.0 (no intolerance), with only S4 displaying moderate intolerance. We can only speculate that while all 5 isoforms of KV7 are homologous and function as voltage-gated potassium channels, each of their voltage sensing domains has been uniquely adapted to different host cell types, membrane compositions, physiological niches, transmembrane potentials, and other forms of regulation (PIP2 binding, phosphorylation, calmodulin binding, and so forth). The differing intolerance patterns seen for the VSD of these five isoforms are mysterious, yet compelling in suggesting that there are fundamental differences in the properties and functional roles of these VSDs that remain to be discovered.
The intolerance patterns in the cytosolic C-terminal domains of the five channels exhibit considerable variability, but also some common features (Figures 6–7). Calmodulin (CaM) is known to bind to helices A and B and is thought to be essential to proper protein trafficking and function, yet helices A and B do not achieve consistently low MTR scores across the KCNQ family. [102, 108, 119–123]. A common feature of the intolerance patterns of the C termini is the consistently low scores in the C helix, which forms a tetrameric coiled-coil helix that helps to stabilize the channel tetramer, as well as potentially playing a role in trafficking [102, 121, 124]. This suggest that the C helix may actually play a leading role in both channel oligomerization and trafficking.
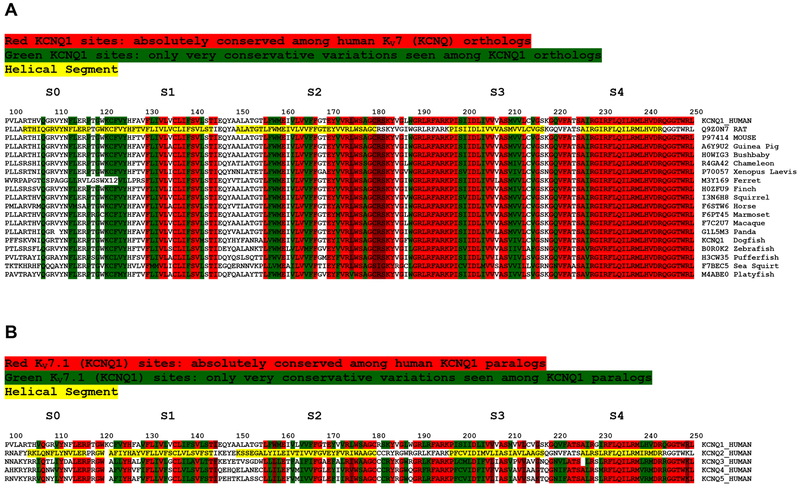
Studying the KV7/KCNQ channel family offers the unique opportunity to compare the MTR with the sequence homology pattern among both orthologs and paralogs. Both the set of human KV7 paralogs and the species to species orthologs of Kv7.1/KCNQ1 indicate generally high levels of conservation in segments exhibiting low (intolerant) MTR scores (Figure 8). However, MTR plots provide additional information. For example, while the S0 segment in KCNQ1 is highly intolerant compared to S2-S4 (Figure 8A), the sequences of S2 and S4 are much more highly conserved than S0. This exemplifies that the factors determining sequence conservation in the KCNQ1 VSD only partially overlap with the factors that determine the intolerance to mutation of segments within the VSD. The nature of the factors that are unique to determining intolerance is a matter that seems to call for discovery.
Figure 8. Sequence Alignments for (A) KCNQ1 VSD Orthologs and (B) Human KCNQ1 Paralogs.
Sequences highlighted in yellow denote the helical segments of the KCNQ1 VSD.
5. CONCLUSIONS
The missense tolerance ratio represents a new and intriguing approach to analyzing genetic information based on the growing body of human genome variation data. It offers insight into human genes and their encoded proteins that complements the information that can be gleaned from multiple sequence alignment and sequence conservation patterns. MTR values can also help identify specific regions in the protein that are most crucial for protein structure, function, folding and interactions. Intolerance analysis may also help to confirm and illuminate disease-linked gene variations. Perhaps most intriguingly, MTR analysis may point to important roles for protein segments that have evaded detection using homology-based analysis or other methods.
One note of caution to analyzing exon segmental intolerance in protein-centric terms is that some non-tolerated variants could conceivably exert their dysprocreative effect by impacting DNA methylation, RNA splicing, or mRNA stability [14–16]. Indeed some missense variants could even render the encoded mRNA toxic [17]. It is not yet clear how often one of these phenomena occur as mechanisms underlying genetic intolerance. However, these mechanisms should be kept in mind to avoid overanalysis of MTR values in protein-only terms. Indeed, it is intriguing to wonder if there are as-yet-undiscovered mechanisms by which gene variations might induce dysprocreative effects. If so, then MTR analysis may point the way to new fundamental discoveries in biology.
Highlights.
Genetic intolerance analysis provides insight into non-synonymous gene variations that are deemed important because they have been filtered out of human genome/exome sequences because they prevent human reproduction or early development.
A form of intolerance analysis is reviewed that calculates the “missense tolerance ratio” (MTR) that identifies intolerant sequence segments in gene exons, with a current resolution of roughly 90 nucleotides (30 amino acids).
MTR analysis can provide insight into the roles of protein segments in folding, structure, function, interactions, and potential mutant protein toxicity that is sometimes complementary or even orthogonal to insight provided by phylogenetic multiple sequence alignment and homology analysis.
ACKNOWLEDGEMENTS
The authors would like to thank Drs. Greg Sliwoski, John A. Capra, Emily Hodges, James Patton, and Tony Forster for helpful discussion. We also thank Dr. Slavé Petrovski for kindly answering e-mail questions about intolerance analysis. This work was supported by US NIH Grants RF1 AG056147, R01 HL122010, and R01 NS058815.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L, New goals for the U.S. Human Genome Project: 1998-2003, Science 282(5389) (1998) 682–9. [DOI] [PubMed] [Google Scholar]
- [2].Collins F, Galas D, A new five-year plan for the U.S. Human Genome Project, Science 262(5130) (1993)43–6. [DOI] [PubMed] [Google Scholar]
- [3].Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X, The sequence of the human genome, Science 291(5507) (2001) 1304–51. [DOI] [PubMed] [Google Scholar]
- [4].Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C, Analysis of protein-coding genetic variation in 60,706 humans, Nature 536(7616) (2016) 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kroncke BM, Vanoye CG, Meiler J, George AL Jr., Sanders CR, Personalized biochemistry and biophysics, Biochemistry 54(16) (2015) 2551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ng PC, Henikoff S, Predicting deleterious amino acid substitutions, Genome Res 11(5) (2001) 863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Adzhubei I, Jordan DM, Sunyaev SR, Predicting functional effect of human missense mutations using PolyPhen-2, Curr Protoc Hum Genet Chapter 7 (2013) Unit 7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J, A general framework for estimating the relative pathogenicity of human genetic variants, Nat Genet 46(3) (2014) 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gilissen C, Hoischen A, Brunner HG, Veltman JA, Disease gene identification strategies for exome sequencing, Eur J Hum Genet 20(5) (2012) 490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB, Genic intolerance to functional variation and the interpretation of personal genomes, PLoS Genet 9(8) (2013) e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB, The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes, Genome Biol 17 (2016) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, Su Z, Pecha J, Bhattacharya S, Petrovski S, Cohen AE, Aizenman E, Traynelis SF, Yuan H, Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology, PLoS Genet 13(1) (2017) e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, Balding DJ, Petrovski S, Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation, Genome Res 27(10) (2017) 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marina RJ, Sturgill D, Bailly MA, Thenoz M, Varma G, Prigge MF, Nanan KK, Shukla S, Haque N, Oberdoerffer S, TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing, EMBO J 35(3) (2016) 335–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cartegni L, Chew SL, Krainer AR, Listening to silence and understanding nonsense: exonic mutations that affect splicing, Nat Rev Genet 3(4) (2002) 285–98. [DOI] [PubMed] [Google Scholar]
- [16].Chang YF, Imam JS, Wilkinson MF, The nonsense-mediated decay RNA surveillance pathway, Annu Rev Biochem 76 (2007) 51–74. [DOI] [PubMed] [Google Scholar]
- [17].Mittal P, Brindle J, Stephen J, Plotkin JB, Kudla G, Codon usage influences fitness through RNA toxicity, Proc Natl Acad Sci U S A 115(34) (2018) 8639–8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG, Natural selection on protein-coding genes in the human genome, Nature 437(7062) (2005) 1153–7. [DOI] [PubMed] [Google Scholar]
- [19].Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S, Identifying a high fraction of the human genome to be under selective constraint using GERP++, PLoS Comput Biol 6(12) (2010) e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB, A genome-wide comparison of the functional properties of rare and common genetic variants in humans, Am J Hum Genet 88(4) (2011) 458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Silk M, Petrovski S, Ascher DB, MTR-Viewer: identifying regions within genes under purifying selection, Nucleic Acids Res 47(W1) (2019) W121–W126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Petrovski S, Gussow AB, Wang Q, Halvorsen M, Han Y, Weir WH, Allen AS, Goldstein DB, The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity, PLoS Genet 11(9) (2015) e1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnstrom K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH Jr., Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale ΒΜ, Daly MJ, A framework for the interpretation of de novo mutation in human disease, Nat Genet 46(9) (2014) 944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Havrilla JM, Pedersen BS, Layer RM, Quinlan AR, A map of constrained coding regions in the human genome, Nat Genet 51(1) (2019) 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Samocha KE, Kosmicki JA, Karczewski KJ, O’Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, Neale BM, Daly MJ, Regional missense constraint improves variant deleteriousness prediction, bioRxiv (2017) 148353. [Google Scholar]
- [26].Hayeck TJ, Stong N, Wolock CJ, Copeland B, Kamalakaran S, Goldstein DB, Allen AS, Improved Pathogenic Variant Localization via a Hierarchical Model of Sub-regional Intolerance, Am J Hum Genet 104(2) (2019) 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lazrak A, Fu L, Bali V, Bartoszewski R, Rab A, Havasi V, Keiles S, Kappes J, Kumar R, Lefkowitz E, Sorscher EJ, Matalon S, Collawn JF, Bebok Z, The silent codon change I507-ATC->ATT contributes to the severity of the DeltaF508 CFTR channel dysfunction, FASEB J 27(11) (2013) 4630–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bali V, Bebok Z, Decoding mechanisms by which silent codon changes influence protein biogenesis and function, Int J Biochem Cell Biol 64 (2015) 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tian D, Wang Q, Zhang P, Araki H, Yang S, Kreitman M, Nagylaki T, Hudson R, Bergelson J, Chen JQ, Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes, Nature 455(7209)(2008)105–8. [DOI] [PubMed] [Google Scholar]
- [30].Harpak A, Bhaskar A, Pritchard JK, Mutation Rate Variation is a Primary Determinant of the Distribution of Allele Frequencies in Humans, PLoS Genet 12(12) (2016) el006489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nachman MW, Crowell SL, Estimate of the mutation rate per nucleotide in humans, Genetics 156(1) (2000) 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jay JJ, Brouwer C, Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine, PLoS One 11(8) (2016) e0160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kelly M, Park M, Mihalek I, Rochtus A, Gramm M, Perez-Palma E, Axeen ET, Hung CY, Olson H, Swanson L, Anselm I, Briere LC, High FA, Sweetser DA, Undiagnosed Diseases N, Kayani S, Snyder M, Calvert S, Scheffer IE, Yang E, Waugh JL, Lai D, Bodamer O, Poduri A, Spectrum of neurodevelopmental disease associated with the GNAOl guanosine triphosphate-binding region, Epilepsia (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hemati P, Revah-Politi A, Bassan H, Petrovski S, Bilancia CG, Ramsey K, Griffin NG, Bier L, Cho MT, Rosello M, Lynch SA, Colombo S, Weber A, Haug M, Heinzen EL, Sands TT, Narayanan V, Primiano M, Aggarwal VS, Millan F, Sattler-Holtrop SG, Caro-Llopis A, Pillar N, Baker J, Freedman R, Kroes HY, Sacharow S, Stong N, Lapunzina P, Schneider MC, Mendelsohn NJ, Singleton A, Loik Ramey V, Wou K, Kuzminsky A, Monfort S, Weiss M, Doyle S, Iglesias A, Martinez F, McKenzie F, Orellana C, van Gassen KLI, Palomares M, Bazak L, Lee A, Bircher A, Basel-Vanagaite L, Hafstrom M, Houge G, Group CRR, study DDD, Goldstein DB, Anyane-Yeboa K, Refining the phenotype associated with GNB1 mutations: Clinical data on 18 newly identified patients and review of the literature, Am J Med Genet A 176(11) (2018) 2259–2275. [DOI] [PubMed] [Google Scholar]
- [35].Szczaluba K, Chmielewska JJ, Sokolowska O, Rydzanicz M, Szymanska K, Feleszko W, Wlodarski P, Biernacka A, Murcia Pienkowski V, Walczak A, Bargel E, Krolewczyk K, Nowacka A, Stawinski P, Nowis D, Dziembowska M, Ploski R, Neurodevelopmental phenotype caused by a de novo PTPN4 single nucleotide variant disrupting protein localization in neuronal dendritic spines, Clin Genet 94(6) (2018) 581–585. [DOI] [PubMed] [Google Scholar]
- [36].Squires KE, Montanez-Miranda C, Pandya RR, Torres MP, Hepler JR, Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease, Pharmacol Rev 70(3) (2018) 446–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Oberai A, Joh NH, Pettit FK, Bowie JU, Structural imperatives impose diverse evolutionary constraints on helical membrane proteins, Proc Natl Acad Sci U S A 106(42) (2009) 17747–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Marinko JT, Huang H, Penn WD, Capra JA, Schlebach JP, Sanders CR, Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis, Chem Rev 119(9) (2019) 5537–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, Maglott DR, ClinVar: improving access to variant interpretations and supporting evidence, Nucleic Acids Res 46(D1) (2018) D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN, The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution, Curr Protoc Bioinformatics Chapter 1 (2012) Unit 1 13. [DOI] [PubMed] [Google Scholar]
- [41].Golde TE, Estus S, Younkin LH, Selkoe DJ, Younkin SG, Processing of the amyloid protein precursor to potentially amyloidogenic derivatives, Science 255(5045) (1992) 728–30. [DOI] [PubMed] [Google Scholar]
- [42].Castro MA, Hadziselimovic A, Sanders CR, The vexing complexity of the amyloidogenic pathway, Protein Sci (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ, Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2, Proc Natl Acad Sci U S A 100(11) (2003) 6382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fraering PC, Ye W, Strub JM, Dolios G, LaVoie MJ, Ostaszewski BL, van Dorsselaer A, Wang R, Selkoe DJ, Wolfe MS, Purification and characterization of the human gamma-secretase complex, Biochemistry 43(30) (2004) 9774–89. [DOI] [PubMed] [Google Scholar]
- [45].Yang G, Zhou R, Zhou Q, Guo X, Yan C, Ke M, Lei J, Shi Y, Structural basis of Notch recognition by human gamma-secretase, Nature 565(7738) (2019) 192–197. [DOI] [PubMed] [Google Scholar]
- [46].Zhou R, Yang G, Guo X, Zhou Q, Lei J, Shi Y, Recognition of the amyloid precursor protein by human gamma-secretase, Science 363(6428) (2019). [DOI] [PubMed] [Google Scholar]
- [47].Wang J, Brunkan AL, Hecimovic S, Walker E, Goate A, Conserved “PAL” sequence in presenilins is essential for gamma-secretase activity, but not required for formation or stabilization of gamma-secretase complexes, Neurobiol Dis 15(3) (2004) 654–66. [DOI] [PubMed] [Google Scholar]
- [48].Sato C, Takagi S, Tomita T, Iwatsubo T, The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the gamma-secretase, J Neurosci 28(24) (2008) 6264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B, The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor, Nature 325(6106) (1987) 733–6. [DOI] [PubMed] [Google Scholar]
- [50].Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL, Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer’s disease, Nature 331(6156) (1988) 528–30. [DOI] [PubMed] [Google Scholar]
- [51].Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K, Identification, biogenesis, and localization of precursors of Alzheimer’s disease A4 amyloid protein, Cell 57(1)(1989)115–26. [DOI] [PubMed] [Google Scholar]
- [52].Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ, Cleavage of amyloid beta peptide during constitutive processing of its precursor, Science 248(4959) (1990) 1122–4. [DOI] [PubMed] [Google Scholar]
- [53].Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M, Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE, Science 286(5440) (1999) 735–41. [DOI] [PubMed] [Google Scholar]
- [54].Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, Ihara Y, gamma-Secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment, J Neurosci 29(41) (2009) 13042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dawkins E, Small DH, Insights into the physiological function of the beta-amyloid precursor protein: beyond Alzheimer’s disease, J Neurochem 129(5) (2014) 756–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zheng H, Koo EH, Biology and pathophysiology of the amyloid precursor protein, Mol Neurodegener 6(1) (2011) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kerr ML, Small DH, Cytoplasmic domain of the beta-amyloid protein precursor of Alzheimer’s disease: function, regulation of proteolysis, and implications for drug development, J Neurosci Res 80(2) (2005) 151–9. [DOI] [PubMed] [Google Scholar]
- [58].Schettini G, Govoni S, Racchi M, Rodriguez G, Phosphorylation of APP-CTF-AICD domains and interaction with adaptor proteins: signal transduction and/or transcriptional role--relevance for Alzheimer pathology, J Neurochem 115(6) (2010) 1299–308. [DOI] [PubMed] [Google Scholar]
- [59].Lai A, Sisodia SS, Trowbridge IS, Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain, J Biol Chem 270(8) (1995) 3565–73. [PubMed] [Google Scholar]
- [60].Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH, Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42, J Biol Chem 274(27) (1999) 18851–6. [DOI] [PubMed] [Google Scholar]
- [61].Shariati SA, De Strooper B, Redundancy and divergence in the amyloid precursor protein family, FEBS Lett 587(13) (2013) 2036–45. [DOI] [PubMed] [Google Scholar]
- [62].Cao X, Sudhof TC, A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60, Science 293(5527) (2001) 115–20. [DOI] [PubMed] [Google Scholar]
- [63].Cao X, Sudhof TC, Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation, J Biol Chem 279(23) (2004) 24601–11. [DOI] [PubMed] [Google Scholar]
- [64].Artavanis-Tsakonas S, Rand MD, Lake RJ, Notch signaling: cell fate control and signal integration in development, Science 284(5415) (1999) 770–6. [DOI] [PubMed] [Google Scholar]
- [65].Bray SJ, Notch signalling: a simple pathway becomes complex, Nat Rev Mol Cell Biol 7(9) (2006) 678–89. [DOI] [PubMed] [Google Scholar]
- [66].Kovall RA, Gebelein B, Sprinzak D, Kopan R, The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force, Dev Cell 41(3) (2017) 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A, A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE, Mol Cell 5(2) (2000) 207–16. [DOI] [PubMed] [Google Scholar]
- [68].Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R, A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1, Mol Cell 5(2) (2000) 197–206. [DOI] [PubMed] [Google Scholar]
- [69].Kopan R, Ilagan MX, The canonical Notch signaling pathway: unfolding the activation mechanism, Cell 137(2) (2009) 216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lieber T, Kidd S, Young MW, kuzbanian-mediated cleavage of Drosophila Notch, Genes Dev 16(2) (2002) 209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R, Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization, J Biol Chem 279(49) (2004) 50864–73. [DOI] [PubMed] [Google Scholar]
- [72].Schroeter EH, Kisslinger JA, Kopan R, Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain, Nature 393(6683) (1998) 382–6. [DOI] [PubMed] [Google Scholar]
- [73].Weng AP, Ferrando AA, Lee W, Morris J.P.t, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC, Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia, Science 306(5694) (2004) 269–71. [DOI] [PubMed] [Google Scholar]
- [74].Aster JC, Pear WS, Blacklow SC, The Varied Roles of Notch in Cancer, Annu Rev Pathol 12 (2017) 245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, Maric D, Eberhart CG, Fine HA, Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation, Cancer Res 65(6) (2005) 2353–63. [DOI] [PubMed] [Google Scholar]
- [76].Kovall RA, Blacklow SC, Mechanistic insights into Notch receptor signaling from structural and biochemical studies, Curr Top Dev Biol 92 (2010) 31–71. [DOI] [PubMed] [Google Scholar]
- [77].Nam Y, Weng AP, Aster JC, Blacklow SC, Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex, J Biol Chem 278(23) (2003) 21232–9. [DOI] [PubMed] [Google Scholar]
- [78].Wilson JJ, Kovall RA, Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA, Cell 124(5) (2006) 985–96. [DOI] [PubMed] [Google Scholar]
- [79].Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, Handford PA, Downing AK, Structural and functional properties of the human notch-1 ligand binding region, Structure 12(12) (2004) 2173–83. [DOI] [PubMed] [Google Scholar]
- [80].Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E, Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells, J Immunol 158(11) (1997) 5083–6. [PubMed] [Google Scholar]
- [81].Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH, Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells, Nature 391(6668) (1998) 703–7. [DOI] [PubMed] [Google Scholar]
- [82].Smith KM, Wu J, Bakker AB, Phillips JH, Lanier LL, Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors, J Immunol 161(1) (1998) 7–10. [PubMed] [Google Scholar]
- [83].Feng J, Call ME, Wucherpfennig KW, The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site, PLoS Biol 4(5) (2006) e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Shi Y, Holtzman DM, Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight, Nat Rev Immunol 18(12) (2018) 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Call ME, Wucherpfennig KW, Chou JJ, The structural basis for intramembrane assembly of an activating immunoreceptor complex, Nat Immunol 11(11) (2010) 1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K, Variant of TREM2 associated with the risk of Alzheimer’s disease, N Engl J Med 368(2) (2013) 107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Atagi Y, Liu CC, Painter MM, Chen XF, Verbeeck C, Zheng H, Li X, Rademakers R, Kang SS, Xu H, Younkin S, Das P, Fryer JD, Bu G, Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2), J Biol Chem 290(43) (2015) 26043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bailey CC, DeVaux LB, Farzan M, The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E, J Biol Chem 290(43) (2015) 26033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M, TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia, Neuron 91(2) (2016) 328–40. [DOI] [PubMed] [Google Scholar]
- [90].Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, Holtzman DM, Cirrito JR, Colonna M, TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model, Cell 160(6) (2015) 1061–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kober DL, Brett TJ, TREM2-Ligand Interactions in Health and Disease, J Mol Biol 429(11) (2017) 1607–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bouchon A, Hernandez-Munain C, Cella M, Colonna M, A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells, J Exp Med 194(8) (2001) 1111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Daws MR, Lanier LL, Seaman WE, Ryan JC, Cloning and characterization of a novel mouse myeloid DAP12-associated receptor family, Eur J Immunol 31(3) (2001) 783–91. [DOI] [PubMed] [Google Scholar]
- [94].Hedley PL, Jorgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M, The genetic basis of long QT and short QT syndromes: a mutation update, Hum Mutat 30(11) (2009) 1486–511. [DOI] [PubMed] [Google Scholar]
- [95].Jentsch TJ, Neuronal KCNQ potassium channels: physiology and role in disease, Nat Rev Neurosci 1(1) (2000) 21–30. [DOI] [PubMed] [Google Scholar]
- [96].Peroz D, Rodriguez N, Choveau F, Baro I, Merot J, Loussouarn G, Kv7.1 (KCNQ1) properties and channelopathies, J Physiol 586(7) (2008) 1785–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Robbins J, KCNQ potassium channels: physiology, pathophysiology, and pharmacology, Pharmacol Ther 90(1) (2001) 1–19. [DOI] [PubMed] [Google Scholar]
- [98].Schmitt N, Schwarz M, Peretz A, Abitbol I, Attali B, Pongs O, A recessive C-terminal Jervell and Lange-Nielsen mutation of the KCNQ1 channel impairs subunit assembly, EMBO J 19(3) (2000) 332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT, Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2, Circulation 102(10) (2000) 1178–85. [DOI] [PubMed] [Google Scholar]
- [100].Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT, Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias, Nat Genet 12(1) (1996) 17–23. [DOI] [PubMed] [Google Scholar]
- [101].Long SB, Campbell EB, Mackinnon R, Crystal structure of a mammalian voltage-dependent Shaker family K+ channel, Science 309(5736) (2005) 897–903. [DOI] [PubMed] [Google Scholar]
- [102].Sun J, MacKinnon R, Cryo-EM Structure of a KCNQ1/CaM Complex Reveals Insights into Congenital Long QT Syndrome, Cell 169(6) (2017) 1042–1050 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL Jr., Vanoye CG, Kim HJ, Sanders CR, Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel, Biochemistry 47(31) (2008) 7999–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kroncke BM, Van Horn WD, Smith J, Kang C, Welch RC, Song Y, Nannemann DP, Taylor KC, Sisco NJ, George AL Jr., Meiler J, Vanoye CG, Sanders CR, Structural basis for KCNE3 modulation of potassium recycling in epithelia, Sci Adv 2(9) (2016) e1501228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Panaghie G, Tai KK, Abbott GW, Interaction of KCNE subunits with the KCNQ1 K+ channel pore, J Physiol 570(Pt 3) (2006) 455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lundby A, Tseng GN, Schmitt N, Structural basis for K(V)7.1-KCNE(x) interactions in the I(Ks) channel complex, Heart Rhythm 7(5) (2010) 708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wrobel E, Tapken D, Seebohm G, The KCNE Tango - How KCNE1 Interacts with Kv7.1, Front Pharmacol 3 (2012) 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Barrese V, Stott JB, Greenwood IA, KCNQ-Encoded Potassium Channels as Therapeutic Targets, Annu Rev Pharmacol Toxicol 58 (2018) 625–648. [DOI] [PubMed] [Google Scholar]
- [109].Jepps TA, Carr G, Lundegaard PR, Olesen SP, Greenwood IA, Fundamental role for the KCNE4 ancillary subunit in Kv7.4 regulation of arterial tone, J Physiol 593(24) (2015) 5325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Nakajo K, Kubo Y, KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel, J Gen Physiol 130(3) (2007) 269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Huang H, Kuenze G, Smith JA, Taylor KC, Duran AM, Hadziselimovic A, Meiler J, Vanoye CG, George AL Jr., Sanders CR, Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations, Sci Adv 4(3) (2018) eaar2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bezanilla F, How membrane proteins sense voltage, Nat Rev Mol Cell Biol 9(4) (2008) 323–32. [DOI] [PubMed] [Google Scholar]
- [113].Lu Z, Klem AM, Ramu Y, Ion conduction pore is conserved among potassium channels, Nature 413(6858) (2001) 809–13. [DOI] [PubMed] [Google Scholar]
- [114].Long SB, Campbell EB, Mackinnon R, Voltage sensor of Kv1.2: structural basis of electromechanical coupling, Science 309(5736) (2005) 903–8. [DOI] [PubMed] [Google Scholar]
- [115].Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE, Mechanism of voltage gating in potassium channels, Science 336(6078) (2012) 229–33. [DOI] [PubMed] [Google Scholar]
- [116].Papazian DM, Shao XM, Seoh SA, Mock AF, Huang Y, Wainstock DH, Electrostatic interactions of S4 voltage sensor in Shaker K+ channel, Neuron 14(6) (1995) 1293–301. [DOI] [PubMed] [Google Scholar]
- [117].Peng D, Kim JH, Kroncke BM, Law CL, Xia Y, Droege KD, Van Horn WD, Vanoye CG, Sanders CR, Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel, Biochemistry 53(12) (2014) 2032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM, Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits, Biophys J 72(4) (1997) 1489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sachyani D, Dvir M, Strulovich R, Tria G, Tobelaim W, Peretz A, Pongs O, Svergun D, Attali B, Hirsch JA, Structural basis of a Kv7.1 potassium channel gating module: studies of the intracellular c-terminal domain in complex with calmodulin, Structure 22(11) (2014) 1582–94. [DOI] [PubMed] [Google Scholar]
- [120].Liu W, Devaux JJ, Calmodulin orchestrates the heteromeric assembly and the trafficking of KCNQ2/3 (Kv7.2/3) channels in neurons, Mol Cell Neurosci 58 (2014) 40–52. [DOI] [PubMed] [Google Scholar]
- [121].Wiener R, Haitin Y, Shamgar L, Fernandez-Alonso MC, Martos A, Chomsky-Hecht O, Rivas G, Attali B, Hirsch JA, The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction, J Biol Chem 283(9) (2008) 5815–30. [DOI] [PubMed] [Google Scholar]
- [122].Alaimo A, Alberdi A, Gomis-Perez C, Fernandez-Orth J, Bernardo-Seisdedos G, Malo C, Millet O, Areso P, Villarroel A, Pivoting between calmodulin lobes triggered by calcium in the Kv7.2/calmodulin complex, PLoS One 9(1) (2014) e86711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Gamper N, Li Y, Shapiro MS, Structural requirements for differential sensitivity of KCNQ K+ channels to modulation by Ca2+/calmodulin, Mol Biol Cell 16(8) (2005) 3538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Haitin Y, Attali B, The C-terminus of Kv7 channels: a multifunctional module, J Physiol 586(7) (2008) 1803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]