Abstract
The need for automated and efficient systems for tracking full animal pose has increased with the complexity of behavioral data and analyses. Here we introduce LEAP (LEAP estimates animal pose), a deep-learning-based method for predicting the positions of animal body parts. This framework consists of a graphical interface for labeling of body parts and training the network. LEAP offers fast prediction on new data, and training with as few as 100 frames results in 95% of peak performance. We validated LEAP using videos of freely behaving fruit flies and tracked 32 distinct points to describe the pose of the head, body, wings and legs, with an error rate of <3% of body length. We recapitulated reported findings on insect gait dynamics and demonstrated LEAP’s applicability for unsupervised behavioral classification. Finally, we extended the method to more challenging imaging situations and videos of freely moving mice.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Connecting neural activity with behavior requires methods to parse what an animal does into its constituent components (movements of its body parts), which can then be connected with the electrical activity that generates each action. This is particularly challenging for natural behavior, which is dynamic, complex and noisy. Human classification of behavior is slow and subject to bias1,2, but speed can be increased through automation1 including methods to track and analyze animal centroids and shapes over time3–5, machine learning techniques for identifying user-defined behaviors such as fighting and courting6,7, and software to segment the acoustic signals produced by an animal8–10. However, one may not know a priori which behaviors to analyze; this is particularly true when screening mutant animals or investigating the results of neural perturbations that can alter behavior in unexpected ways.
Developments in the unsupervised clustering of postural dynamics have enabled researchers to overcome many of these challenges by analyzing the raw frames of videos in a reduced dimensional space (for example, generated via principal component analysis (PCA)). By comparing frequency spectra or fitting auto-regressive models from low-dimensional projections11,12, these methods can both define and record the occurrence of tens to hundreds of unique, stereotyped behaviors in animals such as fruit flies and mice. Such methods have been used to uncover structures in behavioral data, thereby facilitating the investigation of temporal sequences13, social interactions14, genetic mutants12,15 and the results of neural perturbation16,17.
A major drawback to the aforementioned techniques is their reliance on PCA to reduce the dimensionality of the image time series. While this produces a more manageable substrate for machine learning, it would be advantageous to directly analyze the position of each actuatable body part, as this is what is ultimately under the control of the motor nervous system. However, measuring all of the body-part positions from raw images is a challenging computer vision problem18. Previous attempts at automated body-part tracking in insects and mammals relied on physically constraining the animal and having it walk on a spherical treadmill19 or linear track20; applying physical markers to the animal19,21; or using specialized equipment such as depth cameras22–24, frustrated total internal reflection imaging19,21,25,26 or multiple cameras27. However, these techniques are all designed to work within a narrow range of experimental conditions and are not easy to adapt to disparate datasets.
To design a general algorithm capable of tracking body parts from many different kinds of experiments, we turned to deep-learning-based methods for pose estimation that have proved successful on images of humans28–34. Breakthroughs in the field have come from the adoption of fully convolutional neural network architectures for efficient training and evaluation of images35,36 and the production of a probabilistic estimate of the position of each tracked body part29,31. However, the problems of pose estimation in the typical human setting and that for laboratory animals are subtly different. Algorithms built for human images can deal with large amounts of heterogeneity in body shape, environment and image quality, but use very large labeled training sets of images37–39. In contrast, behavioral laboratory experiments are often more controlled, but the imaging conditions may be highly specific to the experimental paradigm, and labeled data, not readily available, must be generated for every experimental apparatus and animal type. One recent attempt to apply these techniques to images of behaving animals successfully used transfer learning, whereby networks initially trained for a more general object-classification task are refined by further training with relatively few samples from animal images40.
Our approach combines a GUI-driven workflow for labeling images with a simple network architecture that is easy to train and requires few computations to generate predictions. This method can automatically predict the positions of animal body parts via iterative training of deep convolutional neural networks with as few as ten frames of labeled data for initial prediction and training (training on ten frames results in 74% of estimates within a 2.5-pixel (px) error). After initial de novo training, incrementally refined predictions can be used to guide labeling in new frames, drastically reducing the time required to label sufficient examples (50 frames) to achieve a median accuracy of less than 3 px per 86 μm (distance from ground truth). Training on a workstation with a modern graphics processing unit (GPU) is efficient (<1 h) and prediction on new data is fast (up to 185 Hz after alignment). We validated the results of our method using a previously published dataset of high-quality videos of freely behaving adult fruit flies (Drosophila melanogaster11) and recapitulated a number of reported findings on insect gait dynamics as a test of its experimental validity. We then used an unsupervised behavioral classification algorithm to describe stereotyped behaviors in terms of the dynamics of individual body parts. Finally, we showed generalizability by using more challenging imaging conditions and videos from freely moving rodents.
Results
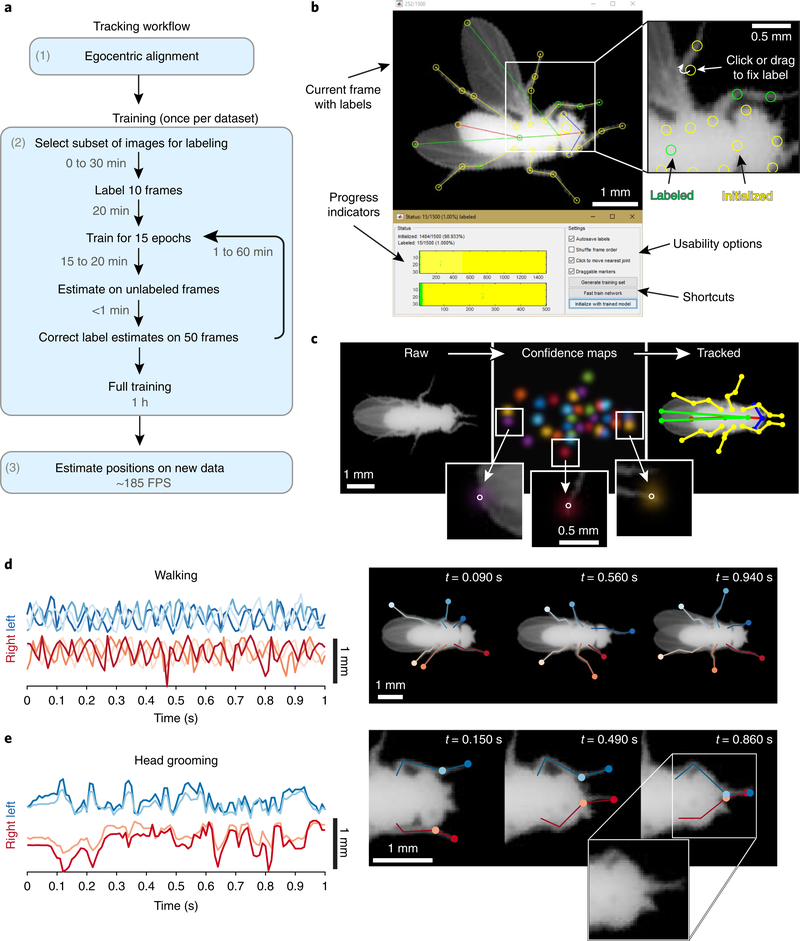
LEAP consists of three phases (see Fig. 1a and Supplementary Results for a full description). The first step is registration and alignment, in which raw video of a behaving animal is preprocessed into egocentric coordinates with an average error of 2.0°. This step increases pose estimation accuracy but can be omitted at the cost of prediction accuracy (Supplementary Fig. 1). The second step is labeling and training, in which the user provides ground truth labels to train the neural network to find body-part positions on a subset of the total images. We used cluster sampling to identify a subset of images that were representative of the complete set of poses found in a dataset (Supplementary Fig. 2). A GUI with draggable body part markers facilitated the labeling of each training image (Fig. 1b). LEAP uses a 15-layer, fully convolutional neural network that produces a set of probability distributions for the location of each body part in an image (Fig. 1c and Supplementary Fig. 3). This simple network performs equivalently to, or better than, more complicated architectures that have been used in the past (Supplementary Fig. 3b). For the fly, we tracked 32 points that define the Drosophila body joints (Supplementary Fig. 4). Labeling and training occur in an iterative procedure. Labels from the first ten images are used to train the neural network and generate body-part estimates for the rest of the training set images. Using these estimates as the initial guesses in the GUI increases the speed of labeling. This is repeated periodically, and the time to label an image drops from 2 min per frame for the first 10 frames to 6 s per frame for the last 500 frames (Supplementary Fig. 5). The third step is pose estimation, in which the network can be applied to new and unlabeled data (Fig. 1c). With minimal training, LEAP faithfully tracks all the body parts, even during challenging bouts of locomotion and in the presence of occlusion (Fig. 1d,e and Supplementary Videos 1–3). In the following sections, we demonstrate the power of this tool, using a previously published dataset of 59 male fruit flies, each recorded for 1 h at 100 Hz, for a total of > 21 million images11. All code and utilities are available at https://github.com/talmo/leap and as Supplementary Software.
Fig. 1 |. Body-part tracking via LEAP, a deep learning framework for animal pose estimation.
a, Overview of the tracking workflow. b, GUI for labeling images. Interactive markers denote the default or best estimate for each body part (top left). Users click or drag the markers to the correct location (top right). Colors indicate labeling progress and denote whether the marker is at the default or estimated position (yellow) or has been updated by the user (green). Progress indicators mark which frames and body parts have been labeled thus far, while shortcut buttons enable the user to export the labels to use a trained network to initialize unlabeled body parts with automated estimates. c, Data flow through the LEAP pipeline. For each raw input image (left), the network outputs a stack of confidence maps (middle). Colors in the confidence maps represent the probability distribution for each individual body part. Insets overlay individual confidence maps on the image to reveal how confidence density is centered on each body part, with the peak indicated by a circle. The peak value in each confidence map predicts the coordinate for each body part (right). d, Quantification of walking behavior using leg tip trajectories. The distance of each of the six leg tips from its own mean position during a walking bout as a function of time (left). Poses at the indicated time points (right). Blue and red traces correspond to left and right leg tips, respectively. e, Quantitative description of head grooming behavior described by leg tip trajectories. Position estimates are not confounded by occlusions when the legs pass under the head (right, inset).
Performance of LEAP: accuracy, speed, and training sample size.
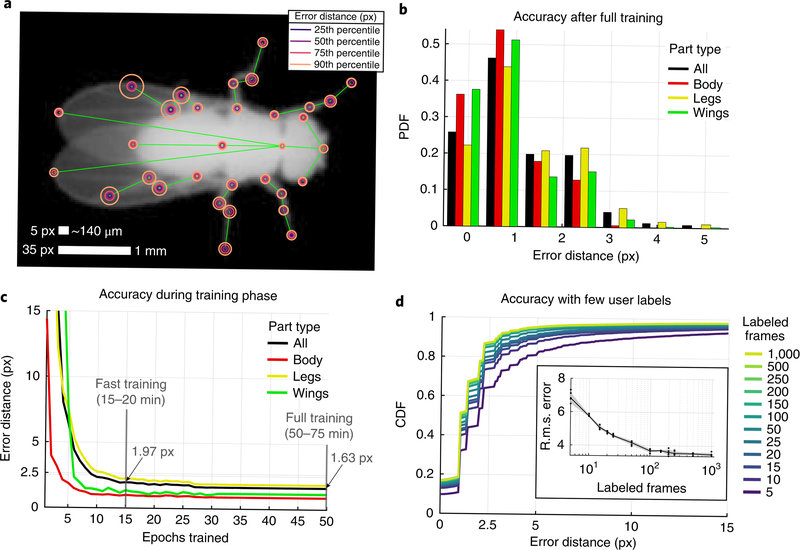
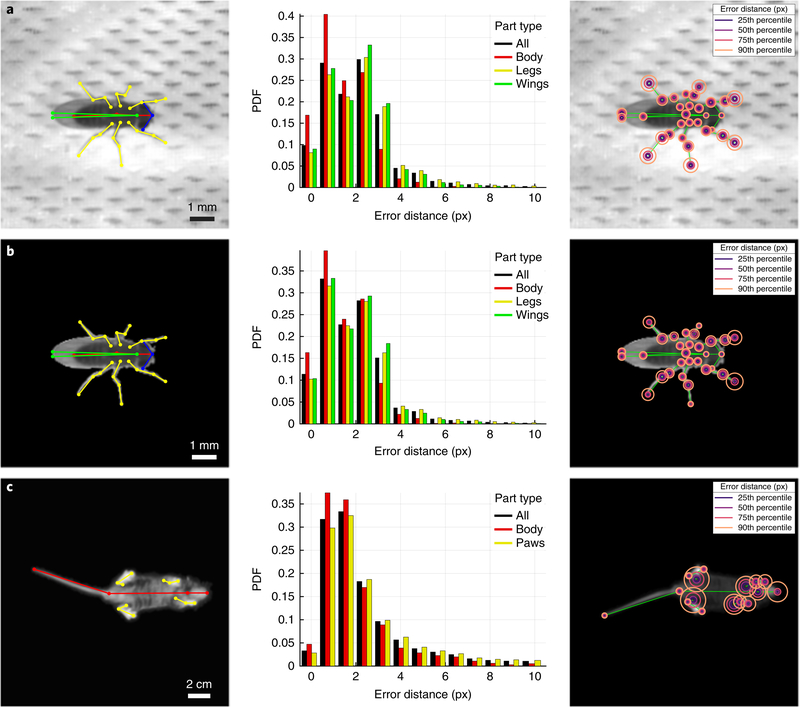
We evaluated the accuracy of LEAP after full training with 1,500 labeled images by measuring error as the Euclidean distance between estimated and ground truth coordinates of each body part on a held-out test set of 168 frames (from seven held-out flies) without augmentation. We found that the accuracy level depended on the body part being tracked, with parts that were more often occluded (for example, hind legs) resulting in slightly higher error rates (Fig. 2a). Overall, we found that error distances for all body parts were well below 3 px for the vast majority of tested images (Fig. 2b). This error was achieved rather quickly during training, with as few as 15 epochs (15–20 min of training time) required to achieve approximately 1.97 px per 56 μm overall accuracy, and less than 50 epochs (50–75 min) required for convergence to 1.63 px per 47 μm accuracy with the full training set (Fig. 2c). To measure the ground truth accuracy during the alternating labeling-training phase, we also measured the errors on the full test set as a function of the number of labeled images used for training under the fast training regime (15 epochs). We found that with as few as ten labeled images, the network was able to achieve < 2.5 px error (2–3% of body length) in 74% of the test set, while 1,000 labeled images yielded an accuracy of < 2.5 px in 87% of the test set (Fig. 2d). When examining the root-mean-square error (r.m.s. error), we found that the performance of the network plateaued at approximately 100 training frames, and labeling of only ten frames corresponded to 65% of peak performance (Fig. 2d, inset). This level of accuracy when training for few epochs with few samples contributes to the drastic reduction in time spent hand-labeling after fast training (Supplementary Fig. 5). For reference, labeling of 100 fly images with the 32-point skeleton took a total of 2 h with the LEAP GUI (with fast training performed after labeling of 10 and 50 frames), training the network took 1 h, and pose estimation on new images occurred at a rate of 185 Hz.
Fig. 2 |. LEAP is accurate and requires little training or labeled data.
a, Part-wise accuracy distribution after full training. Circles are plotted on a reference image to indicate the fraction of held-out testing data (168 images from seven held-out flies) for which estimated positions of the particular body part are closer to the ground truth than the radii. Scale bars indicate image and physical size; 35 px is equivalent to 1 mm at this resolution. b, Accuracy summary on held-out test set after full training. PDF, probability density function. c, Accuracy as a function of training time. In the ‘fast training’ regime, n = 1,215 labeled frames were used for training. Lines and shaded area (smaller than line width) indicate the mean and s.e.m. for all held-out test images pooled over five runs. Run time estimates based on high-end consumer or enterprise GPUs. d, Accuracy as a function of the number of training examples. Distributions indicate estimation errors in a held-out test set (n = 168 frames) with varying numbers of labeled images used for training, pooled over five ‘fast training’ runs. CDF, cumulative distribution function. Inset: median overall r.m.s. error over these five replicates at each sample size.
Leg tracking with LEAP recapitulates previously described gait structure.
To evaluate the usefulness of our pose estimator for producing experimentally valid measurements, we used it to analyze the gait dynamics of freely moving flies. Previous work on Drosophila gait relied on imaging systems that use a combination of optical touch sensors and high-speed video recording to follow fly legs as they walk25. Such systems cannot track the limbs when they are not in contact with the surface (during swing). Other methods to investigate gait dynamics use a semi-automated approach to label fly limbs18,41 and require manual correction of automatically generated predictions; these semi-automated approaches therefore typically utilize smaller datasets.
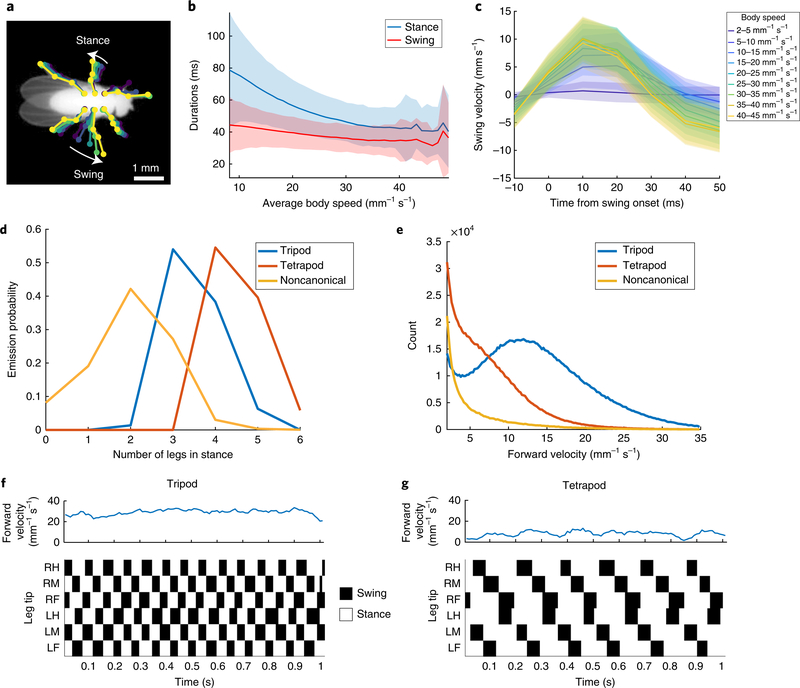
We evaluated our network on a dataset of 59 adult male fruit flies11 and extracted the predicted positions of each leg tip in each of 21 million frames. For every frame in which the fly was moving forward (7.2 h per 2.6 million frames total), we encoded each leg as either in swing or in stance, depending on whether the leg was moving forward or backward relative to the fly’s direction of motion (Fig. 3a). Using this encoding, we measured the relationship between the fly’s speed and the duration of stance and swing (Fig. 3b). Similar to previous work, we found that swing duration was relatively constant across walking speeds, whereas stance duration decreased with walking speed25. Because our methods allowed us to estimate animal pose during both stance and swing (versus only during stance25), we had the opportunity to investigate the dynamics of leg motion during the swing phase. We found that swing velocity increased with body speed, in agreement with previous results25 (Fig. 3c). We also found that fly leg velocities followed a parabolic trajectory parameterized by body speed (Fig. 3c).
Fig. 3 |. LEAP recapitulates known gait patterning in flies.
a, Schematic of swing and stance encoding. Stance is defined by a negative horizontal velocity in egocentric coordinates. b, Duration of swing and stance as a function of average body speed. These data comprise approximately 7.2 h in which the fly was moving forward (2.6 million frames). Shaded regions indicate 1 s.d. c, Swing velocity as a function of time from swing onset, and binned by body speed (n = 1,868,732 swing bouts across all legs). Shaded regions indicate 1 s.d. d, Emission probabilities of numbers of legs in stance for each hidden state in the HMM (Methods). Hidden state emissions resemble tripod, tetrapod and noncanonical gaits. e, Distributions of velocities for each hidden state. f,g, Examples of tripod (f) and tetrapod (g) gaits identified by the HMM. RH, right hind leg tip; RM, right mid; RF, right fore; LH, left hind; LM, left mid; LF, left fore.
We then trained a three-state hidden Markov model (HMM) to capture the different gait modes exhibited by Drosophila41. The emission probabilities from the model of the resulting hidden states were indicative of tripod, tetrapod and noncanonical/wave gaits (Fig. 3d). As expected, we observed tripod gait at high body velocities and tetrapod or noncanonical gaits at intermediate and low velocities, in accordance with previous work25,41,42 (Fig. 3e–g). These results demonstrate that our pose estimator is able to effectively capture the dynamics of known complex behaviors, such as locomotion.
Body dynamics reveal structure in the fly behavioral repertoire.
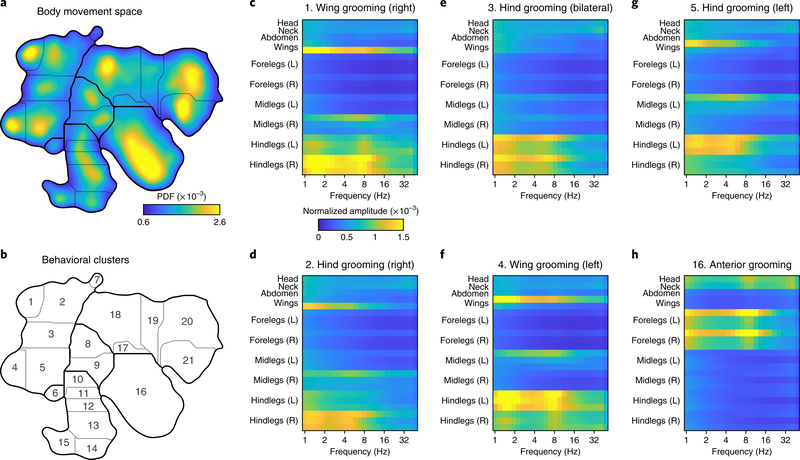
We next used the output of LEAP as the first step in an unsupervised analysis of the fly behavioral repertoire11. We calculated the position of each body part relative to the center of the fly thorax for each point in time and then computed a spectrogram for each of these time series via the continuous wavelet transform (CWT). We then concatenated these spectrograms and embedded the resulting feature vectors for each time point into a two-dimensional (2D) manifold we term a behavior space (Fig. 4a). The feature vectors represent the dynamics of each body part across different time scales, and as has been shown previously, the distribution of embedded time points in this space is concentrated into a number of strong peaks that represent stereotyped behaviors seen across time and in multiple individuals11.
Fig. 4 |. Unsupervised embedding of body position dynamics.
a, Density of freely moving fly body-part trajectories, after projection of their spectrograms into two dimensions via unsupervised nonlinear manifold embedding11. The distribution shown was generated from 21.1 million frames. Regions in the space with higher density correspond to stereotyped movement patterns, whereas low-density regions form natural divisions between distinct dynamics. A watershed algorithm was used to separate the peaks in the probability distribution (Methods). b, Cluster boundaries from a with cluster numbers indicated. c–h, Average spectrograms for the indicated body parts from time points that fall within the dominant grooming clusters; cluster numbers are indicated in b. Qualitative labels for each cluster based on visual inspection are provided for convenience. Color map corresponds to normalized power for each body part.
We identified clusters in the behavior space distribution by grouping together regions of high occupancy and stereotypy (Fig. 4b). This distribution was qualitatively similar to what we found previously by using a PCA-based compression of the images (Supplementary Fig. 6). A major advantage to using pose estimation over PCA-based image compression is the ability to describe stereotyped behaviors by the dynamics of each body part. We calculated the average concatenated spectrogram for each cluster and found that specific behaviors were recapitulated in the motion power spectrum for each body part (Fig. 4c–h).
This method can be used to accurately describe grooming, a class of behaviors that is highly represented in our dataset. Posterior grooming behaviors exhibited a distinctly symmetric topology (Fig. 4b–g), revealing both bilateral (Fig. 4e) and unilateral grooming of the wings (Fig. 4c,f) and the rear of the abdomen (Fig. 4d,g). These behaviors involve unilateral, broadband (1–8 Hz) motion of the hind legs on one side of the body and a slower (~1.5 Hz) folding of the wing on the same side of the body. In contrast, anterior grooming is characterized by broadband motions of both front legs with a peak at ~9 Hz, representing the legs rubbing against each other (Fig. 4h).
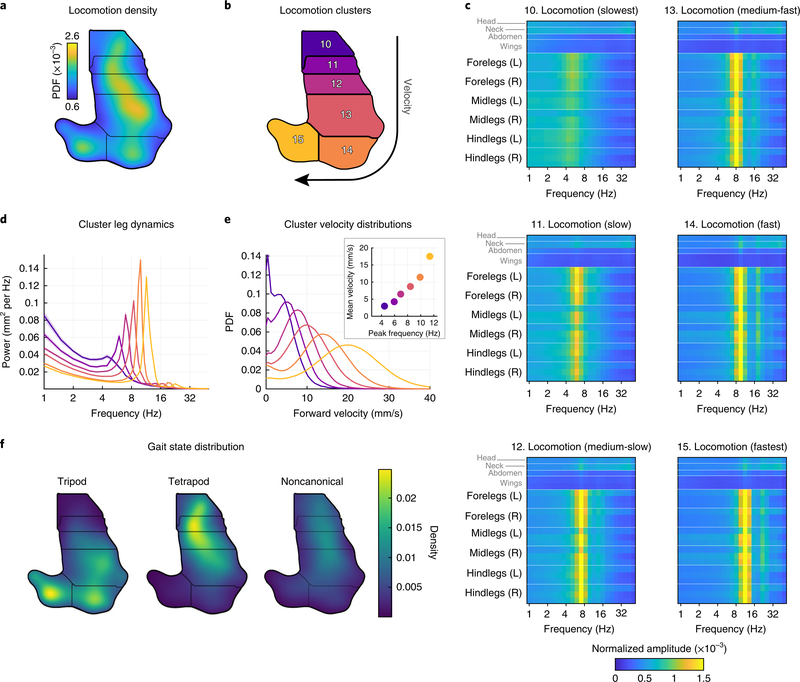
We also discovered a number of unique clusters related to locomotion (Fig. 5a,b). The slowest state (cluster 10) involved several frequencies with a broad peak centered at 5.1 Hz (Fig. 5c–e). This can be seen in both the concatenated spectrograms (Fig. 5c) and the power spectrum averaged over all leg positions (Fig. 5d). The fly center-of-mass velocity distribution for this behavior is shown in Fig. 5e. As the fly speeds up (clusters 10–15, Fig. 5e), the peak frequency for the legs increases monotonically to 11.5 Hz (cluster 15). We next asked whether the tripod and tetrapod gaits we found in our previous analysis (Fig. 3) were represented by distinct regions in the behavior space. We found that tripod gait was used predominantly in the three fastest locomotion behaviors, whereas the tetrapod (and to a lesser extent the noncanonical) gait was used for the three slower locomotion behaviors (Fig. 5f).
Fig. 5 |. Locomotor clusters in behavior space separate distinct gait modes.
a,b, Density (a) and cluster (b) labels of locomotion clusters (from the same behavioral space shown in Fig. 4a). c, Average spectrograms (similar to Fig. 4c–h) quantifying the dynamics in each cluster. d, Average power spectra calculated from the leg joint positions for each cluster in c. Colors correspond to the cluster numbers in b. e, The distribution of forward locomotion velocity as a function of cluster number. Colors correspond to cluster numbers in b. Inset, forward locomotion velocity as a function of peak leg frequency. f, Gait modes identified by HMM from swing/stance state correspond to distinct clusters.
LEAP generalizes to images with complex backgrounds or of other animals.
To test the robustness and generalizability of our approach under more varied imaging conditions, we evaluated the performance of LEAP on a dataset in which pairs of flies were imaged against a nonuniform and low-contrast background of porous mesh (~4.2 million frames, ~11.7 h of video) (Fig. 6a). We first labeled only the male flies from these images, and, using the same workflow as in the first dataset, we found that the pose estimator was able to reliably recover body-part positions with high accuracy despite poorer illumination and a complex background that was at times indistinguishable from the fly (Fig. 6a and Supplementary Video 4). We then evaluated the performance of the network when the background was masked out14 (Fig. 6b). Even with substantial errors in the masking (for example, leg or wing segmentation artifacts), we found that the accuracy improved slightly when the background pixels were excluded from the images compared with that achieved with the raw images (Fig. 6b and Supplementary Video 4). We also tested whether a single network trained on both male and female images performed better or worse than the network trained on only male images. We found that the overall performance was similar (Supplementary Fig. 7) but that the network trained on only male images performed slightly better. This discrepancy is due largely to body parts that are used in very different ways by males and females (for example, the wings, which generate song in males but never in females), and can be overcome with additional training. Finally, we tested the applicability of our framework to animals with different morphology by tracking videos of freely behaving mice (Mus musculus) imaged from below in an open arena (Fig. 6c). We observed comparable accuracy in these mice despite considerable occlusion during behaviors such as rearing (Fig. 6c and Supplementary Video 5).
Fig. 6 |. LEAP generalizes to images with complex backgrounds or of other animals.
a, LEAP estimates on a separate dataset of 42 freely moving male flies, each imaged against a heterogeneous background of mesh and microphones, with side illumination (~4.2 million frames, ~11.7 h). 32 body parts (Supplementary Fig. 4) were tracked, and 1,530 labeled frames were used for training. Error rates for position estimates were calculated on a held-out test set of 400 frames (center) and were comparable to those achieved for images with higher signal to noise (compare with Fig. 2b). Part-wise error distances (right). b, LEAP estimates on masked images from the dataset described in a. Background was subtracted using standard image processing algorithms (Methods) to reduce the effect of background artifacts. c, LEAP estimates on a dataset of freely moving mice imaged from below (~3 million frames, ~4.8 h). Three points are tracked per leg, in addition to the tip of the snout, neck, and base and tip of the tail (left)—1,000 labeled frames were used for training. Accuracy rates on a held-out test set (of 242 frames) (center).
Discussion
Here we present a pipeline (LEAP) that uses a deep neural network to track the body parts of a behaving animal in all frames of a movie via labeling of a small number of images from across the dataset. LEAP does not use a single trained ‘generalist’ network to analyze pose across datasets, as is done in the case of human pose estimation. Rather, we present a framework that uses an active GUI and simple network architecture that can be quickly trained on any new image dataset for which pre-existing labels are not available.
Tracking only the centroid of an animal and its change in position or heading over time is probably an insufficient level of description for determining how the nervous system controls most behaviors. Previous studies have addressed the issue of pose estimation through centroid tracking3, pixel-wise correlations11,12 or specialized apparatus for tracking body parts19,22,25,41,43. For the last, applying markers to an animal can limit natural behavior, and systems that track particular body parts are not in general scalable to all body parts or animals with a very different body plan.
We demonstrate the value of LEAP by showing how it can be applied to the study of locomotor gait dynamics and unsupervised behavioral mapping in Drosophila. Previous studies of gait dynamics have been limited to short stretches of locomotor bouts that were captured with a specialized imaging system25 or to the number of behavioral frames that could be hand-labeled41. We show that LEAP not only recapitulates previous findings on locomotor gait, but also discovers new aspects of the behavior. Body-part tracking provides a solution to a major shortcoming in existing approaches, namely, that researchers have to interpret identified behaviors simply by watching videos11,12. When LEAP is used as the first step in such unsupervised algorithms, each discovered behavior can be interpreted through analysis of the dynamics of each body part.
There are a number of applications for this pipeline beyond those demonstrated here. Because the network learns body positions from a small number of labeled frames, the network can probably be trained to track a wide variety of animal species and classes of behavior. Further, LEAP could be extended to tracking of body parts in three dimensions with the use of either multiple cameras or depth-sensitive devices. This will probably be useful for tracking body parts of head-fixed animals moving on an air-supported treadmill with simultaneous neural recording44,45. Such experiments would be particularly suited to our approach, as the videos from head-fixed animals are inherently recorded in egocentric coordinates. Body-part positions could then be used to decode neural activity, with mapping onto a substrate that approximates muscle coordinates. Additionally, we note that the fast prediction performance of our method might make it compatible with closed-loop experimentation, where joint positions may be computed in real time to control experimental parameters such as stimuli presented to the animal or optogenetic modulation. Lastly, through the addition of a segmentation step for analyzing videos of multiple animals3,14,46, LEAP can potentially estimate poses for multiple interacting individuals.
An important aspect of LEAP is the active training framework that identifies useful images for labeling and provides a GUI for iterative labeling, training and evaluation of network performance. We highlight that this framework can be used with any network architecture. Although we use a relatively simple network that trains quickly, other networks, such as those that utilize transfer learning40 or stacked hourglasses with skip connections and intermediate supervision47, can also be implemented within the LEAP framework and may increase performance for other kinds of data.
In summary, we present a method for tracking body-part positions of freely moving animals with little manual effort and without the use of physical markers. We anticipate that this tool will reduce the technical barriers to addressing a broad range of previously intractable questions in ethology and neuroscience through quantitative analysis of the dynamic changes in the full pose of an animal over time.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41592-018-0234-5.
Methods
Datasets.
Details on the dataset of 59 adult male Drosophila can be found in refs. 11,13. Animals were allowed to move freely in a backlit 100-mm-diameter circular arena covered by a 2-mm-tall clear polyethylene terephthalate glycol dome. Videos were captured from the top with a Point Grey Gazelle camera at a resolution of ~35 px per mm at 100 frames per second (FPS) for 1 h for each fly, totaling ~21 million frames for the dataset. To calculate the spatial resolution for these videos, we assumed a mean male fly length of 2.82 mm (ref. 48).
The second fly dataset reported here (Fig. 5) consists of 42 videos of freely moving pairs of virgin male and female fruit flies (NM91 strain) 3–5 d post-eclosion. Only males from these videos were analyzed in this study. Flies moved freely within a 30-mm-diameter circular arena with a 2-mm-tall clear polyethylene terephthalate glycol dome against a white mesh floor covering an array of microphones, resulting in an inhomogeneous image background. Videos were captured from above with a Point Grey Flea3 camera at a resolution of ~25 px per mm at 100 FPS, totaling ~4.2 million frames.
The mouse dataset for Fig. 5 consisted of 29 videos of C57BL/6 strain mice (Mus musculus), 15 weeks (108 d) old. Animals moved freely in a 45.7 × 45.7 cm open field arena with a clear acrylic floor for 10 min each. Videos were captured from below with infrared illumination using a Point Grey Blackfly S camera at a resolution of 1.95 px per mm at 170 FPS, totaling ~3 million frames. Experimental procedures were approved by the Princeton University Institutional Animal Care and Use Committee and conducted in accordance with the National Institutes of Health guidelines for the humane care and use of laboratory animals. Mice used in this study were ordered through The Jackson Laboratory and had at least 1 week of acclimation to the Princeton Neuroscience Institute vivarium before experimental procedures were performed. Mice were kept in group cages with food and water ad libitum under a reversed 12/12-h dark-light cycle (light, 19:30–07:30).
Preprocessing and alignment to generate egocentric images for labeling and training in LEAP.
For the main fly dataset (59 males), we used the alignment algorithm from ref. 11. The raw videos consisted of unoriented bounding boxes around the flies from a closed-loop camera tracking system. This technique relies on videos in which the animal remains visible and in focus. We then aligned individual frames to a template image of an oriented fly by matching the peak of the radon transformed fly image to recover the orientation and then computing the cross-correlation to center the fly. The centroid and orientation parameters were used to crop a 192 × 192-px oriented bounding box in each frame. Code for alignment is available in the repository accompanying the original paper: https://github.com/gordonberman/MotionMapper.
For the second fly dataset (42 males), we adapted a previously published method for tracking and segmentation of videos of courting fruit flies14. We first modeled the mesh background of the images by fitting a normal distribution to each pixel in the frame across time with a constant variance to account for camera shot noise. The posterior was evaluated at each pixel of each frame and then thresholded to segment the foreground pixels. Because of the inhomogeneity of the arena floor mesh, substanial segmentation artifacts were introduced, particularly when translucent or very thin body parts (that is, wings and legs) could not be disambiguated from the dark background mesh holes. The subsequent steps of histogram thresholding, morphological filtering and ellipse fitting were performed as described previously in ref. 14. We developed a simple GUI for proofreading the automated ellipse tracking before extracting 192 × 192-px oriented bounding boxes. We extracted bounding boxes for both animals in each frame and saved both the raw pixels containing the background mesh and the foreground-only images that contain segmentation artifacts. This pipeline was implemented in MATLAB, and the code is available in the code repository accompanying this paper.
For the mouse videos, a separate preprocessing pipeline was developed. Raw videos were processed in three stages: (1) animal tracking, (2) segmentation from background and (3) alignment to the body centroid and tail–body interface. In stage (1), we tracked the mouse’s torso centroid by subtracting a background image (median calculated at each pixel value across that video), retrieving pixels with a brightness above a chosen threshold from background (mice were brighter than background) and using morphological opening to eliminate noise and the mouse’s appendages. The largest contiguous region reliably captured the mouse’s torso (referred to below as the torso mask) and was used to fit an ellipse whose center was used to approximate the center of the animal. In stage (2), a similar procedure as in stage (1) was employed to retrieve a full body mask. In this stage, a more permissive threshold and smaller morphological opening radius were used than in stage (1) to capture the mouse’s body edges, limbs and tail while still eliminating noise. The pixels outside of this body mask were set to zero. In stage (3) each segmented video frame was translated and rotated such that frame’s center coincided with the center of the animal and the x-axis lay on the line connecting the center and tail-body attachment point. The tail-body attachment point was defined as the center of a region overlapping between the torso mask and a dilated tail mask. The tail mask was defined as the largest region remaining after subtraction of the torso mask from the full body mask and application of a morphological opening. After applying these masks to segment the raw images, we extracted bounding boxes by using the ellipse center and orientation.
Oriented bounding boxes were cropped to 192 × 192 px for all datasets to ensure consistency in output image size after repeated pooling and upsampling steps in the neural network. These data were stored in self-describing HDF5 files.
Sampling diverse images for labeling and training in LEAP.
To ensure diversity in image and pose space when operating at low sample sizes, we used a multistage cluster sampling technique. First, we sampled n0 images uniformly from each dataset by using a fixed stride over time to minimize correlations being temporally adjacent samples. We then used PCA to reduce their dimensionality and projected the images down to the first D principal components. After dimensionality reduction, the images were grouped via k-means clustering into k subgroups from which n images were randomly sampled from each group. To minimize the time necessary for the network to generalize to images from all groups, we sorted the dataset such that consecutive samples cycled through the groups. This way, uniform sampling was maintained even at the early phases of user labeling, ensuring that even a network trained on only the first few images would be optimized to estimate body-part positions for a diversity of poses. We used n0 = 500, yielding 29,500 initial samples; D = 50, which is sufficient to explain 80% of the variance in the data (Supplementary Fig. 2); and k = 10 and n = 150 to produce a final dataset of 1,500 frames for labeling and training.
LEAP neural network design and implementation.
We based our network architecture on previous designs of neural networks for human pose estimation29,31,47. We adopted a fully convolutional architecture that learns a mapping from raw images to a set of confidence maps. These maps are images that can be interpreted as the 2D probability distribution (that is, a heat map) centered at the spatial coordinates of each body part within the image. We trained the network to output one confidence map per body part stacked along the channel axis.
Our network consists of 15 layers of repeated convolutions and pooling (Supplementary Fig. 3a). The convolution block consists of ×3 convolution layers (64 filters, 3 × 3 kernel size, 1 × 1 stride, ReLU activation). The full network consists of ×1 convolution block, ×1 max pooling across channels (2 × 2 pooling size, 2 × 2 stride), × 1 convolution block (128 filters), × 1 max pooling (2 × 2 pooling size, 2 × 2 stride), × 1 convolution block (256 filters), ×1 transposed convolution (128 filters, 3 × 3 kernel size, 2 × 2 stride, ReLU activation, Glorot normal initialization), × 2 convolution (128 filters, 3 × 3 kernel size, 1 × 1 stride, ReLU activation), and × 1 transposed convolution (128 filters, 3 × 3 kernel size, 2 × 2 stride, linear activation, Glorot normal initialization).
We base our decisions of these hyperparameters on the idea that repeated convolutions and strided max pooling enable the network to learn feature detectors across spatial scales. This allows the network to learn how to estimate confidence maps using a global image structure that provides contextual information that can be used to improve estimates, even for occluded parts29,31. Despite the loss of resolution from pooling, the upsampling learned through transposed convolutions is sufficient to recover the spatial precision in the confidence maps. We do not use skip connections, residual modules, stacked networks, regression networks or affinity fields in our architecture as used in other approaches of human pose estimation29,31–33,47.
For comparison, we also implemented the stacked hourglass network47. We tested both the single hourglass version and × 2 stacked hourglass with intermediate supervision. The hourglass network consisted of ×4 residual bottleneck modules (64 output filters) with max pooling (2 × 2 pool, 2 × 2 stride), followed by their symmetric upsampling blocks and respective skip connections. The stacked version adds intermediate supervision in the form of a loss term on the output of the first network in addition to the final output.
We implemented all versions of neural networks in Python via Keras and TensorFlow, popular deep learning packages that allow transparent GPU acceleration and easy portability across operating systems and platforms. All Python code was written for Python v.3.6.4. Required libraries were installed via the pip package manager: numpy (v.1.14.1), h5py (v.2.7.1), TensorFlow-gpu (v.1.6.0), keras (v.2.1.4). We tested our code on machines running either Windows 10 (v.1709) and a RedHat-based Linux distribution (Springdale 7.4) with no additional steps required to port the software other than installing the required libraries. All networks were compared using the same aligned dataset so as to remove complications due to differences in preprocessing.
Code for all network implementations is available in the main repository accompanying this paper (https://github.com/talmo/leap) and Supplementary Software.
LEAP training procedure.
Prior to training, we generated an augmented dataset from the user-provided labels and corresponding images. We first doubled the number of images by mirroring the images along the body symmetric axis (defined from the preprocessing) and adjusting the body-part coordinates accordingly, including swapping left/right body part labels (for example, legs). Then, we generated confidence maps for each body part in each image by rendering the 2D Gaussian probability distribution centered at the ground truth body-part coordinates, μ = (x, y), and fixed covariance, Σ = diag(σ) with a constant σ = 5 px. These were pre-generated and cached to disk to minimize the necessary processing time during training.
Once confidence maps were computed for each image, we split the dataset into training, validation and test sets. The training set was used for backpropagation of the loss for updating network weights, the validation set was used to estimate performance and adjust the learning rate over epochs, and the test set was held out for analysis. For the fast training, the dataset was split into only training (90%) and validation (10%) sets to make the best use of data when training with very few labels. For full training, the dataset was split into training (76.5%), validation (13.5%) and testing (10%) sets. All analyses reported here share the same held-out test set to ensure it is never trained against for any replicate.
All training was done using the Adam optimizer with default parameters as described in the original paper49. We started with a learning rate of 1e-3 but used a scheduler to reduce it by a factor of 0.1 when the validation loss failed to improve by a minimum threshold of 1e-5 for three epochs. The loss function optimized against is simply the mean squared error between estimated and ground truth confidence maps.
During training, we considered an epoch to be a set of 50 batches of 32 images, which were drawn sequentially from the training set, cycling back to the first image if there were less than 50 × 32 = 1,600 images. Images were then augmented by application of a small random rotation (−15–15°) to the input image and the corresponding ground truth confidence maps (Supplementary Fig. 1a). At the end of 50 batches of training, 10 batches were sampled from the separate validation set, augmented and evaluated, and the loss was used for learning rate scheduling described above. Training and validation sets are shuffled at the end of each epoch. An epoch was evaluated in 60–90 s, including all augmentation, forward and reverse passes, and the validation forward pass when running on a modern GPU (NVIDIA GeForce GTX 1080 Ti or P100). We ran this entire procedure for 15 epochs during the fast training stage and for 50 epochs during the full training stage. For analyses, a minimum of five replicates were fully trained on each dataset to estimate the stability of optimization convergence. We evaluated the performance of the network on a held-out test set of images without augmentation.
Pose estimation from confidence maps.
Predictions of body-part positions were computed directly on the GPU. We implement a channel-wise global maximum operation to convert the confidence maps into image coordinates as a TensorFlow function, further improving runtime prediction performance by avoiding the costly transfer of large confidence map arrays. All prediction functions including normalization and saving were implemented as a self-contained Python script with a command-line interface for ease of batch processing.
Computing hardware.
All performance tests were conducted on a high-end consumer-grade workstation equipped with an Intel Core i7–5960X CPU, 128 GB DDR4 RAM, NVMe solid state drives and a single NVIDIA GeForce 1080 GTX Ti (12 GB) GPU. We also used Princeton University’s High Performance Computing cluster with nodes equipped with NVIDIA P100 GPUs for batch processing. These higher-end cards afford a speed-up of ~1.5× in processing runtime during the training phase.
Accuracy analysis.
For all analyses of accuracy (Figs. 2 and 6 and Supplementary Figs. 3 and 5), we trained at least five replicates of the network with the same training/validation/testing datasets. All analyses were performed in MATLAB R2018a (MathWorks). We used the gramm toolbox for figure plotting50.
Gait analysis.
We translated the body position coordinates to egocentric coordinates by subtracting the predicted location of the intersection between the thorax and abdomen from all other body-position predictions for each frame. We then calculated the instantaneous velocity along the rostrocaudal axis of each leg tip within these truly egocentric reference coordinates. The speed of each body part was smoothed using a Gaussian filter with a five-frame moving window. For each leg tip, instances in which the smoothed velocity was greater than zero were defined as swing, while those with velocity less than zero were defined as stance. Information from this egocentric axis was combined with allocentric tracking data to incorporate speed and orientation information. The centroids and orientations of the flies were smoothed using a moving mean filter with a five-frame window to find the instantaneous speed and forward velocity. To remove idle bouts and instances of backward walking, all gait analyses were limited to times when the fly was moving in the forward direction at a velocity greater than 2 mm s−1 (approximately one body length per second) unless otherwise noted. The analyses relating stance and swing duration to body velocity were limited to forward velocities greater than 7.2 mm s−1, to remain in line with previous work25.
To measure gait modes, we trained an HMM to model gait as described previously41. The training data consisted of a vector denoting the number of legs in stance for bouts in which the fly was moving forward at a velocity greater than 2 mm s−1 lasting longer than 0.5 s. Training data were sampled such that up to 3,000 frames were taken from each video, resulting in a total of 159,270 frames. We trained a three-state HMM using the Baum–Welch algorithm and randomly initialized transition and emission probabilities51. We designated each hidden state as tripod, tetrapod or noncanonical in accordance with the estimated emission probabilities. We then used the Viterbi algorithm along with our estimated transition and emission matrices to predict the most probable sequence of hidden states from which the observed stance vectors for the entire dataset would emerge52.
Unsupervised embedding of body-part dynamics.
In order to create a map of motor behaviors described by body-part movements, we used a previously described method for discovering stereotypy in postural dynamics11. First, body-part positions were predicted for each frame in our dataset to yield a set of 32 time series of egocentric trajectories in image coordinates for each video. We recentered these time series by subtracting the thorax coordinate at each time point and rescaled them to comparable ranges by z-scoring each time series. The time series were then expanded into spectrograms by application of the CWT parametrized by the Morlet wavelet as the mother wavelet and 25 scales chosen to match dyadically spaced center frequencies spanning 1–50 Hz. This time-frequency representation augments the instantaneous representation of pose at each time point to one that captures oscillations across many time scales. The instantaneous spectral amplitudes of each body part were then concatenated into a single vector of length 2(J − 1)F, where J is the number of body parts before subtraction of the body part used as a reference (that is, the thorax) and doubled to account for both x and y coordinates, and F is the number of frequencies being measured via CWT. In our data, this resulted in a 1,550-dimensional representation at each time point (frame).
Finally, we performed nonlinear dimensionality reduction on these high-dimensional vectors by using a nonlinear manifold embedding algorithm53. We first selected representative time points via importance sampling, wherein a random sampling of time points in each video is embedded into a 2D manifold via t-distributed stochastic neighbor embedding (t-SNE) and clustered via the watershed transform. This allowed us to choose a set of time points from each video that were representative of their local clusters—that is, spanning the space of postural dynamics. We then computed a final behavior space distribution by embedding the selected representative time points using t-SNE to produce the full manifold of postural dynamics in two dimensions.
After projecting all remaining time points in the dataset into this manifold, we computed their 2D distribution and smoothed with a Gaussian kernel with σ = 0.65 to approximate the probability density function of this space. We clipped the range of this density map to the range 0.5 × 10−3 to 2.75 × 10−3 to exclude low-density regions and merge very high-density regions. We then clustered similar points by segmenting the space into regions of similar body-part dynamics by applying the watershed transform to the density. Although both the manifold coordinates representation of each time point are not immediately meaningful, we were able to derive an intuitive interpretation of each cluster by referring to the high-dimensional representation of their constituent time points. To do this, we sampled time points from each cluster and averaged their corresponding high-dimensional feature vector, which we could then visualize by reshaping it into a body-part-frequency matrix (Fig. 4).
Supplementary Material
Acknowledgements
The authors acknowledge J. Pillow for discussions; B.C. Cho for contributions to the acquisition and preprocessing pipeline for mouse experiments; P. Chen for a previous version of a neural network for pose estimation that was useful in designing our method; H. Jang, M. Murugan, and I. Witten for feedback on the GUI and other discussions; G. Guan for assistance maintaining flies; and the Murthy, Shaevitz and Wang labs for general feedback. This work was supported by the NIH R01 NS104899–01 BRAIN Initiative Award and an NSF BRAIN Initiative EAGER Award (to M.M. and J.W.S.), NIH R01 MH115750 BRAIN Initiative Award (to S.S.-H.W. and J.W.S.), the Nancy Lurie Marks Family Foundation and NIH R01 NS045193 (to S.S.-H.W.), an HHMI Faculty Scholar Award (to M.M.), NSF GRFP DGE-1148900 (to T.D.P.), and the Center for the Physics of Biological Function sponsored by the National Science Foundation (NSF PHY-1734030).
Footnotes
Competing interests
T.D.P., D.E.A., J.W.S. and M.M. are named as inventors on US provisional patent no. 62/741,643 filed by Princeton University.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-018-0234-5.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Code availability. The code for running LEAP, as well as all accompanying GUIs, trained networks, labeled data and analysis code for figure reproduction, can be found in the Supplementary Software and in the following repository: https://github.com/talmo/leap.
Data availability
The entire primary dataset of 59 aligned, high-resolution behavioral videos is made available online for reproducibility or further studies based off of this method, as well as labeled data to train and ground-truth the networks, pre-trained networks used for all analyses, and estimated body-part positions for all 21 million frames. This dataset (~170 GiB) is freely available at http://arks.princeton.edu/ark:/88435/dsp01pz50gz79z. Data from additional fly and mouse datasets used in Fig. 6 can be made available upon reasonable request.
References
- 1.Anderson DJ & Perona P Toward a science of computational ethology. Neuron 84, 18–31 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Szigeti B, Stone T & Webb B Inconsistencies in C. elegans behavioural annotation. Preprint at bioRxiv https://www.biorxiv.org/content/early/2016/07/29/066787 (2016). [Google Scholar]
- 3.Branson K, Robie AA, Bender J, Perona P & Dickinson MH High-throughput ethomics in large groups of Drosophila. Nat. Methods 6, 451–457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swierczek NA, Giles AC, Rankin CH & Kerr RA High-throughput behavioral analysis in C. elegans. Nat. Methods 8, 592–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y, Coen P, Sun M & Shaevitz JW Efficient multiple object tracking using mutually repulsive active membranes. PLoS ONE 8, e65769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dankert H, Wang L, Hoopfer ED, Anderson DJ & Perona P Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 6, 297–303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabra M, Robie AA, Rivera-Alba M, Branson S & Branson K JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Arthur BJ, Sunayama-Morita T, Coen P, Murthy M & Stern DL Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 11, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson SE, Dave AS & Margoliash D Template-based automatic recognition of birdsong syllables from continuous recordings. J. Acoust. Soc. Am. 100, 1209–1219 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Tachibana RO, Oosugi N & Okanoya K Semi-automatic classification of birdsong elements using a linear support vector machine. PLoS ONE 9, e92584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman GJ, Choi DM, Bialek W & Shaevitz JW Mapping the stereotyped behaviour of freely moving fruit flies. J. R. Soc. Interface 11, 20140672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiltschko AB et al. Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman GJ, Bialek W & Shaevitz JW Predictability and hierarchy in Drosophila behavior. Proc. Natl Acad. Sci. USA 113, 11943–11948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klibaite U, Berman GJ, Cande J, Stern DL & Shaevitz JW An unsupervised method for quantifying the behavior of paired animals. Phys. Biol. 14, 015006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q et al. The PSI-U1 snRNP interaction regulates male mating behavior in Drosophila. Proc. Natl Acad. Sci. USA 113, 5269–5274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein JT et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344, 386–392 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Cande J et al. Optogenetic dissection of descending behavioral control in Drosophila. eLife 7, e34275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlmann V, Ramdya P, Delgado-Gonzalo R, Benton R & Unser M FlyLimbTracker: an active contour based approach for leg segment tracking in unmarked, freely behaving Drosophila. PLoS ONE 12, e0173433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kain J et al. Leg-tracking and automated behavioural classification in Drosophila. Nat. Commun. 4, 1910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machado AS, Darmohray DM, Fayad J, Marques HG & Carey MR A quantitative framework for whole-body coordination reveals specific deficits in freely walking ataxic mice. eLife 4, e07892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nashaat MA et al. Pixying behavior: a versatile real-time and post hoc automated optical tracking method for freely moving and head fixed animals. eNeuro 4, e34275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanjappa A et al. Mouse pose estimation from depth images. arXiv Preprint at https://arxiv.org/abs/1511.07611 (2015). [Google Scholar]
- 23.Nakamura A et al. Low-cost three-dimensional gait analysis system for mice with an infrared depth sensor. Neurosci. Res. 100, 55–62 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Mirbozorgi SA & Ghovanloo M An automated behavior analysis system for freely moving rodents using depth image. Med. Biol. Eng. Comput. 56, 1807–1821 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Mendes CS, Bartos I, Akay T, Márka S & Mann RS Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster. eLife 2, e00231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendes CS et al. Quantification of gait parameters in freely walking rodents. BMC Biol. 13, 50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrou G & Webb B Detailed tracking of body and leg movements of a freely walking female cricket during phonotaxis. J. Neurosci. Methods 203, 56–68 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Toshev A & Szegedy C DeepPose: human pose estimation via deep neural networks. arXiv Preprint at https://arxiv.org/abs/1312.4659 (2013). [Google Scholar]
- 29.Tompson JJ, Jain A, LeCun Y & Bregler C Joint training of a convolutional network and a graphical model for human pose estimation In Advances in Neural Information Processing Systems Vol. 27 (eds Ghahramani Z, Welling M, Cortes C, Lawrence ND. & Weinberger KQ) 1799–1807 (Curran Associates, Inc., Red Hook, 2014). [Google Scholar]
- 30.Carreira J, Agrawal P, Fragkiadaki K & Malik J Human pose estimation with iterative error feedback. arXiv Preprint at https://arxiv.org/abs/1507.06550 (2015). [Google Scholar]
- 31.Wei S-E, Ramakrishna V, Kanade T & Sheikh Y Convolutional pose machines. arXiv Preprint at https://arxiv.org/abs/1602.00134 (2016). [Google Scholar]
- 32.Bulat A & Tzimiropoulos G Human pose estimation via convolutional part heatmap regression. arXiv Preprint at https://arxiv.org/abs/1609.01743 (2016). [Google Scholar]
- 33.Cao Z, Simon T, Wei S-E & Sheikh Y Realtime multi-person 2D pose estimation using part affinity fields. arXiv Preprint at https://arxiv.org/abs/1611.08050 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Tome D, Russell C & Agapito L Lifting from the deep: convolutional 3D pose estimation from a single image. arXiv Preprint at https://arxiv.org/abs/1701.00295 (2017). [Google Scholar]
- 35.Shelhamer E, Long J & Darrell T Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 640–651 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Ronneberger O, Fischer P & Brox T U-Net: convolutional networks for biomedical image segmentation In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 234–241 (Springer International Publishing, Cham, Switzerland, 2015). [Google Scholar]
- 37.Lin T-Y et al. Microsoft COCO: common objects in context In Computer Vision – ECCV 2014 740–755 (Springer International Publishing, Cham, Switzerland, 2014). [Google Scholar]
- 38.Andriluka M, Pishchulin L, Gehler P & Schiele B 2D human pose estimation: new benchmark and state of the art analysis. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 3686–3693 (IEEE Computer Society, 2014). [Google Scholar]
- 39.Güler RA, Neverova N & Kokkinos I DensePose: dense human pose estimation in the wild. arXiv Preprint at https://arxiv.org/abs/1802.00434 (2018). [Google Scholar]
- 40.Mathis A et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Isakov A et al. Recovery of locomotion after injury in Drosophila melanogaster depends on proprioception. J. Exp. Biol. 219, 1760–1771 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Wosnitza A, Bockemühl T, Dübbert M, Scholz H & Büschges A Inter-leg coordination in the control of walking speed in Drosophila. J. Exp. Biol. 216, 480–491 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Qiao B, Li C, Allen VW, Shirasu-Hiza M & Syed S Automated analysis of long-term grooming behavior in Drosophila using a k-nearest neighbors classifier. eLife 7, e34497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dombeck DA, Khabbaz AN, Collman F, Adelman TL & Tank DW Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron 56, 43–57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seelig JD & Jayaraman V Neural dynamics for landmark orientation and angular path integration. Nature 521, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Escudero A, Vicente-Page J, Hinz RC, Arganda S & de Polavieja GG idTracker: tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 11, 743–748 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Newell A, Yang K & Deng J Stacked hourglass networks for human pose estimation. arXiv Preprint at https://arxiv.org/abs/1603.06937 (2016). [Google Scholar]
- 48.Chyb S & Gompel N Atlas of Drosophila Morphology: Wild-type and Classical Mutants (Academic Press, London, Waltham and San Diego, 2013). [Google Scholar]
- 49.Kingma DP & Ba J Adam: a method for stochastic optimization. arXiv Preprint at https://arxiv.org/abs/1412.6980 (2014). [Google Scholar]
- 50.Morel P Gramm: grammar of graphics plotting in MATLAB. J. Open Source Softw. 3, 568 (2018). [Google Scholar]
- 51.Baum LE, Petrie T, Soules G & Weiss N A maximization technique occurring in the statistical analysis of probabilistic functions of markov chains. Ann. Math. Stat. 41, 164–171 (1970). [Google Scholar]
- 52.Viterbi A Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory 13, 260–269 (1967). [Google Scholar]
- 53.van der Maaten L & Hinton G Visualizing data using t-SNE. J. Mach.Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.