SUMMARY
DNA replication in eukaryotes generates DNA supercoiling which may intertwine (braid) daughter chromatin fibers to form precatenanes, posing topological challenges during chromosome segregation. The mechanisms that limit precatenane formation remain unclear. By making direct torque measurements, we demonstrate that the intrinsic mechanical properties of chromatin play a fundamental role in dictating precatenane formation and regulating chromatin topology. Whereas a single chromatin fiber is torsionally soft, a braided fiber is torsionally stiff, indicating that supercoiling on chromatin substrates is preferentially directed in front of the fork during replication. We further show that topoisomerase II relaxation displays a strong preference for a single chromatin fiber over a braided fiber. These results suggest a synergistic coordination -the mechanical properties of chromatin inherently suppress precatenane formation during replication elongation by driving DNA supercoiling ahead of the fork, where supercoiling is more efficiently removed by topoisomerase II.
INTRODUCTION
The helical nature of double stranded DNA (dsDNA) innately promotes the generation of torsional stress during essential processes such as replication and transcription (Postow et al., 2001; Ullsperger et al., 1995; Watson and Crick, 1953). Motor proteins involved in these processes track the helical groove of dsDNA and thus twist the DNA as they forward translocate. This action creates topological impasses during chromatin replication and gene expression (Postow et al., 2001). How they are resolved remains an outstanding fundamental problem in biology.
In particular, to ensure successful cell division, the two newly replicated daughter DNA strands must fully segregate, without any intertwining. However, replication over each helical rise of dsDNA produces one (+) supercoil (Ullsperger et al., 1995), which typically cannot dissipate efficiently via DNA end rotation due to topological barriers (Dixon et al., 2012; Naughton et al., 2013) and must therefore distribute to the DNA ahead of and/or behind the replication fork, where it may then be removed by topoisomerases. Supercoiling distributed behind the fork intertwines (or ‘braids’) the two daughter strands, forming precatenanes (Champoux and Been, 1980). If left unresolved, precatenanes mature into full DNA catenanes that obstruct proper chromosome separation and cause DNA damage and genome instability (Keszthelyi et al., 2016). The detrimental nature of unresolved chromatin intertwining highlights the importance of mechanisms that limit precatenane formation and allow for the efficient resolution of DNA supercoiling. The complexity of these topological challenges in the context of chromatin, however, has posed significant barriers to experimentation. As a consequence, our mechanistic understanding of such processes has remained conspicuously limited despite their great importance to cellular viability.
The partitioning of supercoiling between the front and back of the replisome is coupled to replisome rotation (also known as fork rotation) (Champoux and Been, 1980; Postow et al., 2001). A non-rotating replisome directs supercoiling into the DNA ahead of the replication fork as it elongates, whereas a rotating replisome will distribute supercoiling behind the fork, leading to catenation of the daughter DNA strands. In vivo genetics studies examining eukaryotic fork rotation have found that rotation is largely inhibited during replication elongation by protein complexes associated with the replisome (Schalbetter et al., 2015) but occurs near termination, when two replisomes collide head-on (Seidman and Salzman, 1979; Sundin and Varshavsky, 1981; Tapper and DePamphilis, 1978), and near stable protein-DNA fragile sites (Schalbetter et al., 2015). The mechanisms by which these behaviors are accomplished and regulated remain unclear.
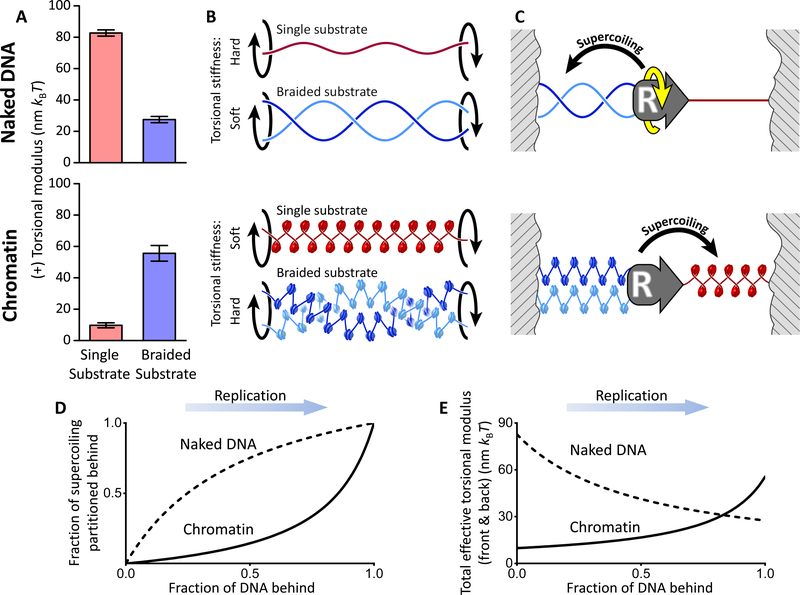
These previous studies focused on the interactions of specific proteins with the replisome in restricting fork rotation. However, the intrinsic role of the torsional mechanics of the chromatin substrates has remained largely overlooked. Fork rotation is an inherently mechanical process related to the balance of torque ahead of and behind the replisome. Despite the fact that a replisome can actively generate torsion during replication, it lacks any means of driving its substrates out of torsional equilibrium because the replisome itself experiences minimal viscous drag (Quantification and Statistical Analysis) and is not known to be tethered to any cellular scaffolding. Thus, supercoiling generated by replisome progression will partition ahead of and behind the fork so as to maintain a balance of torque (Figure 1). As a consequence, if it is easier to twist the DNA ahead of the fork, then fork rotation will be minimal and (+) supercoiling will primarily remain in front of the replisome. By contrast, if it is easier to twist the DNA behind the fork, (+) supercoiling will primarily distribute behind the replisome by fork rotation, forming precatenanes. Through this torque balance, the torsional mechanical properties of single and braided chromatin fibers play a decisive role in determining supercoiling partitioning and the occurrence of fork rotation. This, in turn, has implications for the efficient resolution of supercoiling by topoisomerases. However, these torsional mechanical properties remain largely unknown.
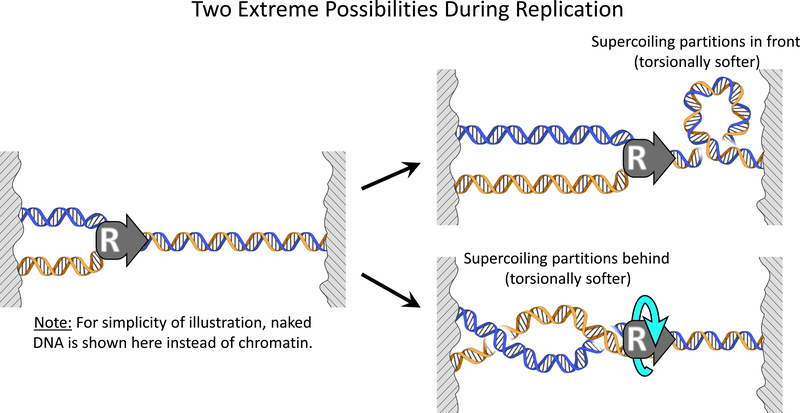
Figure 1.
Torsional stiffness of the chromatin substrates dictates supercoiling partitioning during DNA replication. For simplicity of illustration, naked DNA is shown here instead of chromatin. During replication, torque is balanced on both sides of the replisome and thus supercoiling partitions based on the ratio of the torsional stiffness of the substrate in front of the replisome to that of the substrate behind the replisome. Shown are two extreme possibilities of partitioning. If the torsional stiffness of the single substrate in front of the replisome is much smaller, replisome rotation is minimal and supercoiling partitions primarily to the front. On the other hand, if the torsional stiffness of the braided substrate behind the replisome is much smaller, the replisome rotates extensively and supercoiling partitions primarily to behind the replisome.
Despite being integral to eukaryotic replication, braided chromatin fibers have not been previously investigated mechanistically due to the complexity in creating such substrates. In this work, we have developed methods to create and benchmark both single and braided chromatin fiber substrates, determined their torsional mechanical properties, and examined how topoisomerase II differentially relaxes these substrates. Our measurements reveal that the intrinsic torsional mechanical properties of chromatin play a fundamental role in suppressing precatenation by preferentially partitioning DNA supercoiling to the single chromatin fiber in front of a replisome, which we show to be a preferred substrate for topoisomerase II. This work demonstrates the importance of chromatin torsional mechanics and its synergistic coordination with topoisomerase activity in regulating DNA topology.
RESULTS
Single and Double Chromatin Fiber Substrates
Investigation of the torsional mechanical properties of chromatin substrates required the development of methods to form both long single chromatin fiber substrates (as would be located in front of a replisome) and long double chromatin substrates (as would be located behind a replisome) and to assess the nucleosome array size and nucleosome quality for both types of substrates (Figure 2). Once established, these methods create benchmarks for torsional mechanical investigations for this study and provide broader opportunities to utilize chromatin fibers in potential future studies of processes taking place over chromatin.
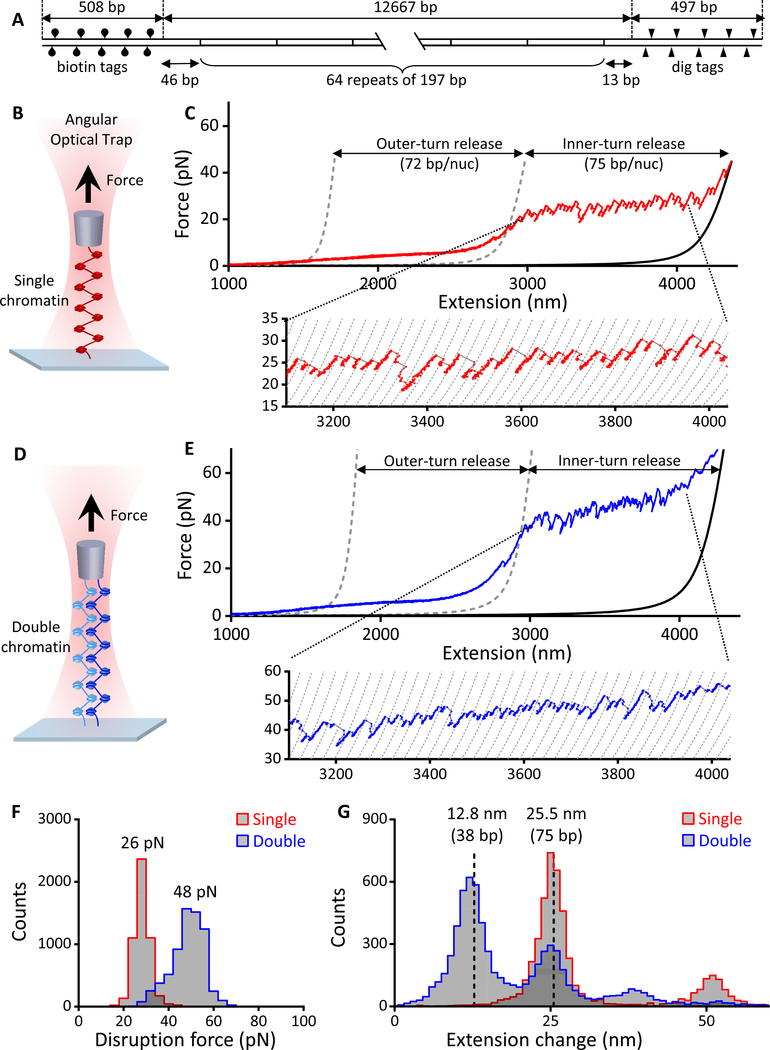
Figure 2. Stretching single and double chromatin fiber arrays.
(A) DNA template design. For details, see the “DNA Template Construction” section under Method Details.
(B) Experimental configuration to stretch a single chromatin fiber along the axial direction of the AOT to disrupt nucleosomes.
(C) An example trace of the force-extension curve of a single chromatin fiber containing ~ 53 nucleosomes, showing ~ 72 bp of smooth outer-turn DNA release and ~ 75 bp of sudden inner-turn release from each nucleosome. Outer-turn DNA release starts at ~ 2 pN and ends at ~ 15 pN, before the inner-turn DNA release starts, similar to what we have reported previously (Brower-Toland et al., 2005; Brower-Toland et al., 2002). The two gray dashed curves (Wang et al., 1997) correspond to naked DNA of lengths such that their force-extension curves cross the chromatin fiber curve at 2 pN and 15 pN and are used to characterize the amount of outer-turn DNA released. The black solid curve corresponds to a naked DNA whose number of base-pairs is the same as that of the chromatin fiber’s DNA (12,667 bp). The dashed curves of the inset are naked DNA curves with 75 bp increments in length.
(D) Experimental configuration to stretch a double chromatin fiber along the axial direction of the AOT to disrupt nucleosomes.
(E) An example trace of the resulting force-extension curve of a double chromatin fiber, each containing ~ 47 nucleosomes. The two gray dashed curves (Wang et al., 1997) are theoretical curves of two parallel naked DNA molecules of lengths such that they cross the chromatin fiber curves at 4 pN and 30 pN and are used to characterize the amount of outer-turn DNA released. The black solid curve is a theoretical force-extension curve of two parallel naked DNA molecules, each of 12,667 bp, the same as the DNA template length of the chromatin fiber. The dashed curves of the inset are naked DNA curves of two parallel molecules with 75 bp increments in length.
(F) The distributions of inner-turn disruption force for single and double chromatin fiber substrates. The peak values of these distributions are indicated.
(G) The distributions of extension change from inner-turn disruptions for single and double chromatin fiber substrates. Extension change at a disruption event was defined as the change in the DNA contour length and expressed either in units of nm or bp. The two dashed lines are located at 25.5 nm (or 75 bp) and half of this value.
See also Figures S3 and S4.
To form a chromatin fiber, we assembled nucleosomes onto a 12.7 kbp DNA molecule containing 64 repeats of a nucleosome positioning element (NPE) flanked by ~ 500 bp multi-labeled tethering adaptor at each end (Figure 2A and Figure S1; Method Details). This DNA length is comparable to the mean distance that a replisome replicates through in yeast, which is estimated to be ~ 15 kbp (Bell and Labib, 2016; Sekedat et al., 2010). Importantly, all mechanical and topoisomerase activity measurements were conducted in the same physiological buffer that contains magnesium and is compatible with topoisomerase II activity (Quantification and Statistical Analysis).
To benchmark a single chromatin substrate, we mechanically disrupted the nucleosomes in the substrate. Each chromatin fiber was torsionally constrained between two surfaces via multi-labeled tethering adaptors at the two ends. One end was anchored to the surface of a microscope coverslip while the other end was anchored to the bottom of a nanofabricated quartz cylinder held in an angular optical trap (AOT) (Figure 2B). The AOT permits direct torque measurements of a biological substrate under a controlled amount of supercoiling and also has the ability to measure forces and displacements of a trapped cylinder (Deufel et al., 2007; Forth et al., 2008; Ma et al., 2013; Ma et al., 2019; Sheinin et al., 2011). Our AOT has also been optimized for long DNA substrates (Figure S2). For this particular assay, we did not introduce any twist to the substrate; the AOT was used only to stretch the chromatin fiber along the axial direction of the AOT to disrupt nucleosomes.
The resulting force-extension curve was used to characterize the composition of the nucleosome array (e.g., nucleosomes versus subnucleosomal structures) and the number of nucleosomes on the DNA (Figure 2C). We interpret such curves based on a multi-stage disruption model of chromatin that we previously established (Brower-Toland et al., 2005; Brower-Toland and Wang, 2004; Brower-Toland et al., 2002; Hall et al., 2009; Sheinin et al., 2013). Under lower forces (2 pN < F < 15 pN, between the two dashed curves), exit and entry DNA (interacting primarily with H2A/H2B dimers) is released from histone core octamers. This “outer-turn” DNA is ~ 72 bp in length for each nucleosome. The release is gradual, occurring simultaneously over all nucleosomes with a force plateau at ~ 3 pN. By assaying the total DNA released in this low force region, we determined the number of nucleosomes initially containing a wrapped outer-turn Nout, corresponding to the number of complete octamers bound to the template. The second stage of the disruption starts as the force increases beyond 15 pN, where distinctive disruption force peaks are clearly detectable (see inset), with a mean disruption force of ~ 26 pN (Figure 2F). Each high disruption force peak corresponds to a release of ~ 75 bp of “inner-turn” DNA (interacting primarily with the H3/H4 tetramer) from a single nucleosome (Figure 2G). This stage of disruption occurs sequentially among nucleosomes. By assaying the total DNA released in this inner-turn region, we determined the number of bound tetramers Nin. Similar nucleosome disruption characteristics have been reported by several other groups for shorter single chromatin fibers (Gemmen et al., 2005; Li et al., 2016; Mack et al., 2012; Meng et al., 2015).
Notably, we also developed a method to create a double chromatin fiber. In order for this substrate to better reflect the leading and lagging strands behind a replisome, we surveyed what is known about whether each of the two strands can rotate freely around its own helical axis during replication. The DNA in the lagging strand is generally thought to be able to freely rotate due to the presence of the ssDNA regions of the Okazaki fragments (Postow et al., 2001; Ullsperger et al., 1995). For the leading strand, although some torsion accumulation may occur (Yu and Droge, 2014), free rotation may still be a reasonable approximation (Postow et al., 2001; Ullsperger et al., 1995). Torsion within the leading strand may be relaxed because two replisomes initiating from the same origin move in opposite directions, resulting in the leading strand of one replisome being the lagging strand of the other replisome (Burgers and Kunkel, 2017) and thus allowing the leading strand to rotate freely around its helical axis. In addition, DNA polymerases may exchange with others in solution (Beattie et al., 2017; Lewis et al., 2017), transiently detach from the DNA (Kurth et al., 2013), and/or rotate relative to the helicase at the replication fork (Zhou et al., 2017). Given these considerations, both chromatin fibers used to form double chromatin fiber substrates in our experiments were nicked in order to allow rotation about their respective helical axes.
To form a double chromatin fiber substrate, we tethered two nicked chromatin fibers between the coverslip and the bottom of a quartz cylinder held in the AOT (Methods Details). To benchmark the number and quality of nucleosomes of a double chromatin fiber substrate, we stretched it along the axial direction of the AOT to disrupt nucleosomes (Figure 2D). The resulting force-extension curve showed a force that was roughly double that of a single chromatin fiber because the force was applied to two nearly parallel nucleosome arrays (Figures 2E and 2F). A closer examination of the inner-turn DNA release at the high force disruption region shows that some disruptions from the two fibers occurred concurrently, giving rise to a 75 bp DNA release from each fiber, while other disruptions occurred sequentially, giving rise to half of the 75 bp DNA release (Figure 2G). Using a similar analysis as that for a single chromatin fiber, the force-extension relation allowed us to estimate Nout and Nin for each substrate.
These stretching experiments (Figure 2; Figure S3; Figure S4) were performed at the end of each torsional measurement described below. This allowed us to determine the number of nucleosomes in a single chromatin fiber substrate (Figures S3A and S3B) and the mean number of nucleosomes of each fiber in a double chromatin fiber substrate (Figures S4A and S4B). We selected chromatin fibers with Nout and Nin being comparable within measurement uncertainties, consistent with fibers containing primarily full nucleosomes (Quantification and Statistical Analysis). This minimized contributions from subnucleosomes (Nout being substantially smaller than Nin) which may be generated during nucleosome assembly or sample handling. These stretching experiments, coupled with the reversibility requirement imposed for the torsional measurement experiments described below (Figures S3E and S4D), provided a rigorous assessment to evaluate the nucleosome saturation level, integrity, and stability for both the single (Figure S3) and double (Figure S4) chromatin substrates. Only chromatin fibers that passed all of these tests were included for further analysis (Quantification and Statistical Analysis).
Single Chromatin Substrates Effectively Absorb (+) Supercoiling
Despite the importance of chromatin torsional mechanics in replication, the torsional properties of chromatin fibers remain largely unknown. To determine the torsional stiffness, both DNA supercoiling and torque must be simultaneously measured. DNA supercoiling characterizes the additional turns added to the DNA, whereas torque characterizes how difficult it is to add those turns. Although more standard methods, such as two-dimensional gel electrophoresis, are available to assay DNA supercoiling, those methods cannot be used to measure torque. In comparison, the AOT is ideally suited for direct and simultaneous measurements of these properties.
To address how a single chromatin fiber (as would be located in front of a replisome) responds to torsion, we used the AOT to lift the chromatin fiber off the coverslip and placed it under a small constant force of 0.5 pN, comparable to that estimated for what might occur in vivo (Charvin et al., 2004), and then introduced twist into the fiber in both directions. During this time, the extension and torque of the fiber were simultaneously measured, allowing for direct determination of the torsional properties of a single chromatin fiber (Figures 3A–3C). Since previous studies showed that canonical nucleosomes are stable under a moderate amount of (+) torsion (Bancaud et al., 2007) but become structurally deformed when (+) torsion becomes excessive (superhelical density, defined as the number of turns added or removed relative to the total number of turns in a relaxed DNA, σ > +0.08) (Bancaud et al., 2007), we limited the number of (+) turns added to the fiber for the single chromatin fiber (σ < +0.05) (Figure 3) and selected traces whose extension signals were reversible as turns were added and then removed (Figure S3E).
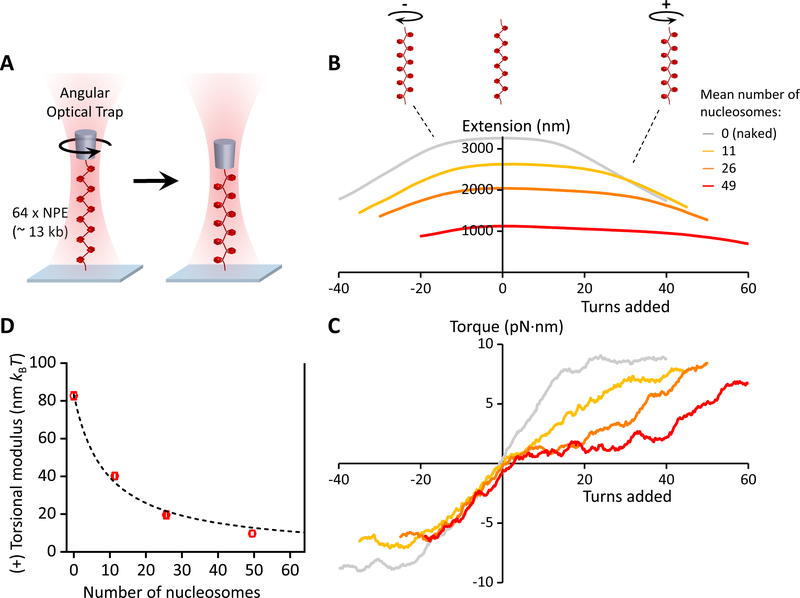
Figure 3. Torsional measurements of a single chromatin fiber.
(A) Experimental configuration. One end of a chromatin fiber was torsionally constrained to the surface of a microscope coverslip while the other end was torsionally constrained to the bottom of a nanofabricated quartz cylinder that was held in the angular optical trap (AOT) (Figure S2). The chromatin fiber was then placed under a constant force of 0.5 pN and twisted in both the (+) and (−) directions. At the end of this torsional measurement, the nucleosome quality and saturation level of each chromatin fiber were assayed by returning the fiber to the zero-turns state and stretching it axially to disrupt the nucleosomes (Figures 2B and 2C; Figures S3A–S3C; Quantification and Statistical Analysis). The fiber stability was assayed from the twisting data (Figures S3D and S3E).
(B & C) The measured extension and torque versus turns added, under different levels of nucleosome array saturation, indicated by the mean number of nucleosomes on the substrate (N ~ 35 traces for each curve). Extension and torque signals were smoothed by sliding windows of 1 turn and 4 turns respectively. For naked DNA, torque increases until DNA buckles to form a plectoneme, then torque plateaus. For chromatin fibers, the data suggest that an analogous structural transition may be occurring. The (+) torsional stiffness of each curve in (C) was determined from a linear fit (not shown) to the initial (pre-”buckling”) slope upon adding (+) turns.
(D) The measured torsional modulus as a function of the number of nucleosomes in the substrate. kBT is the thermal energy. Error bars were converted from the uncertainties in the slopes of the linear fits in (C). The dashed curve is a fit by a simple model, yielding a modulus of 10.2 nm kBT for a chromatin fiber with all 64 NPEs occupied (Quantification and Statistical Analysis). The solid vertical line at 64 indicates the number of NPEs on the template.
When a naked DNA molecule was twisted under identical conditions, we found that the extension decreased symmetrically for both (+) and (−) twist (Figure 3B; Figure S2C), as expected (Ma et al., 2013; Ma et al., 2019; Strick et al., 1996). Surprisingly, we found that twisting a chromatin fiber resulted in an asymmetric response, in which the fiber was able to absorb substantially more turns of (+) supercoiling before a large change in extension was induced (Figure 3B; Figures S3D–S3G). We attribute this asymmetry to the left-handedness of nucleosomes.
Importantly, the (+) torsional stiffness (i.e., the slope of the torque versus turns relationship) of chromatin was dramatically decreased relative to that of naked DNA (Figure 3C). Thus, in comparison with naked DNA, a single chromatin fiber is torsionally softer and able to more effectively resist (+) torque buildup with added twist. Earlier studies carried out under lower salt conditions suggested that chromatin serves as a topological buffer to DNA supercoiling (Bancaud et al., 2006; Celedon et al., 2009). Our studies support this notion by providing direct torque measurements under physiological conditions. This topological buffer could be a result of the ability of a nucleosome to adopt different entry and exit linker DNA configurations, allowing for efficient absorption of supercoiling (Bancaud et al., 2006).
Although not the focus of this work, these measurements also have implications for the twin-supercoiled domain model of transcription(Liu and Wang, 1987) over chromatin. The softer (+) torsional stiffness in front of the RNA polymerase may facilitate transcription elongation, whereas the relatively stiffer (−) torsional stiffness behind may facilitate nucleosome assembly.
We also converted the measured torsional stiffness into the torsional modulus (Quantification and Statistical Analysis), which is an intrinsic property of a substrate. We found that the (+) torsional modulus of a chromatin fiber decreased as nucleosome saturation was increased (Figure 3D). Thus, the ability of a single chromatin fiber to buffer (+) torsion increases with the level of nucleosome saturation.
Braided Chromatin Substrates Resist (+) Supercoiling
We next carried out torsional experiments, similar to those for a single chromatin fiber substrate described above, but with a double chromatin fiber substrate in which each fiber was nicked to prevent torsion buildup within each fiber (Method Details; Figure 4A). We mechanically braided the two fibers by twisting them to form structures akin to precatenanes while monitoring extension and torque (Figures 4B and 4C).
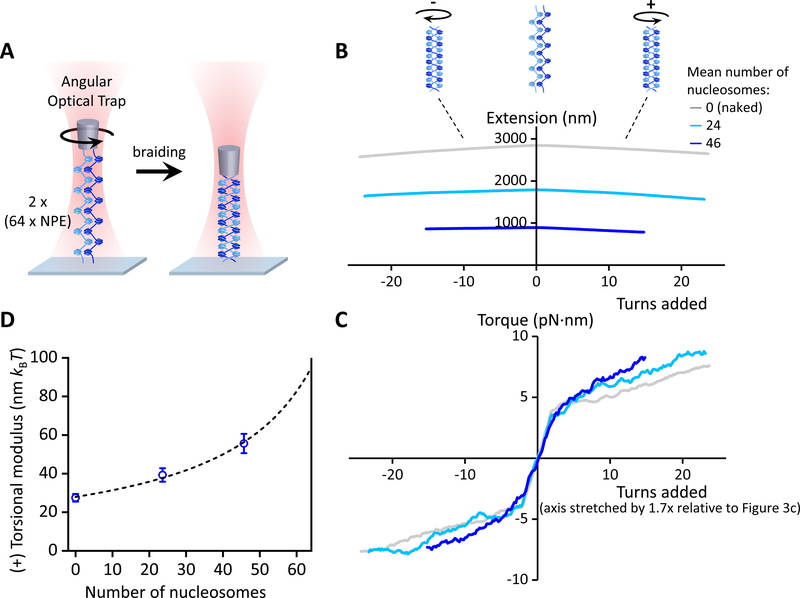
Figure 4. Torsional measurements of a braided chromatin fiber.
(A) Experimental configuration, which is similar to Figure 3A but with two nicked chromatin fibers. At the end of this torsional measurement, the nucleosome quality and saturation level of each substrate were assayed by returning the substrate to the zero-turns state and stretching it axially to disrupt the nucleosomes (Figures 2D and 2E; Figures S4A and S4B; Quantification and Statistical Analysis). The fiber stability (Figures S4C and S4D) and braid’s anchoring geometry (Figure S5) were assayed from the twisting data.
(B & C) The measured extension and torque versus turns added under different levels of nucleosome array saturation, indicated by the mean number of nucleosomes on each of the two chromatin fibers (N ~ 45 traces for each curve). Extension and torque signals were smoothed by sliding windows of 1 turn and 4 turns respectively. To retain optimal chromatin fiber integrity, only a small number of turns was added. Only traces with small anchor separations were included for analysis, and the (+) torsional stiffness was determined by the slope of a linear fit to the torque data of ≥ 2 turns (Figure S5).
(D) The measured torsional modulus as a function of the mean number of nucleosomes in each of the two chromatin fibers. Error bars were converted from the uncertainties in the slopes of the linear fits in (C). The dashed curve is a fit by a simple model, yielding a modulus of 95.0 nm kBT for a chromatin fiber braid with all 64 NPEs occupied in each fiber (Quantification and Statistical Analysis). The solid vertical line at 64 indicates the number of NPEs on the template.
In these experiments, the ends of the two chromatin fibers could assume a range of possible anchor separation distances at each surface, whereas the anchor separation of chromatin fibers at a replisome is expected to be rather small (< 100 nm) compared to the size of the replicons (Quantification and Statistical Analysis). This motivated us to develop a rigorous method to identify traces with small anchor separations (Figure S5; Quantification and Statistical Analysis). Data shown in Figure 4 correspond to those traces that passed this criterion as well as those established for fiber quality and stability (Figure S4).
Upon (+) turns being added, the extension of a braided substrate showed a mild decrease (Figure 4B; Figure S5). However, the torque signal showed an initial sharp increase, followed by a gradual increase with turns (Figure 4C; Figure S5). The initial sharp increase in torque is expected of a braiding substrate with a non-zero anchor separation and should become more discernable with an increase in end anchor separations (Charvin et al., 2005). We found that the subsequent gradual increase in torque was rather insensitive to the range of end anchor separations measured here (Figure S5). We reason that the torque slope in this region might be a better representation of the torsional stiffness experienced by a replisome (where the end anchor separation is expected to be small). Thus, we used this slope to characterize the torsional stiffness of a braid. This practice also provides a very conservative measure of this stiffness.
We found that with an increase in nucleosome saturation, the braided substrate responded to twist in a fashion that was again increasingly distinct from naked DNA, but with a trend opposite to that seen for a single chromatin fiber. In comparison to a braided naked DNA substrate, the braided chromatin substrate was torsionally stiffer; i.e., torque built up more quickly with added twist. The torsional modulus of a braided chromatin fiber notably increased with nucleosome saturation (Figure 4D). Thus, while a single chromatin fiber appears to provide a substantial buffer against the accumulation of (+) torsional stress, a braided chromatin fiber presents considerable resistance to supercoiling and can provide no such relief.
Conversion from Naked DNA to Chromatin Dramatically Alters Supercoiling Partitioning
Figures 5A and 5B directly compare data from Figures 3 and 4, highlighting the dramatic effects that chromatin has in modulating the torsional characteristics of DNA substrates. For naked DNA, the torsional modulus of a single substrate was ~ 3 times that of a braided substrate. Interestingly, this trend was reversed for chromatin; the torsional modulus of a braided chromatin substrate was ~ 5 times that of a single chromatin substrate.
Figure 5. Torsional mechanics model of replication.
(A) Comparison of the torsional moduli of single and braided substrates obtained from Figures 3D and 4D. The measured values at their highest observed numbers of nucleosomes are shown.
(B) Pictorial illustration of the torsional stiffness of different substrates.
(C) Proposed torsional mechanics model of supercoiling partitioning during replication elongation. Replication over naked DNA partitions the supercoiling primarily to behind the replisome, whereas replication over chromatin partitions the supercoiling primarily to the front of the replisome.
(D) Fraction of supercoiling partitioned to behind the replisome as a function of replication progression (see Quantification and Statistical Analysis for details).
(E) Torsional stiffness as a function of replication progression. The total effective torsional modulus Ceff of the coupled torsional spring of a single and braided substrate provides a measure of the torsional stress experienced by a replisome (see Quantification and Statistical Analysis for details). Overall, chromatin substantially reduces the torsional stress experienced by a replisome during replication in comparison to naked DNA. This advantage is only lost near termination.
This unexpected reversal in torsional modulus has significant implications for replication and leads to a torsional mechanics model for the partitioning of DNA supercoiling during replication (Figure 5C). For simplicity of illustration, consider the case where the replication fork has elongated to the middle of two torsionally constrained boundaries. When this process takes place over naked DNA, supercoiling generated by replication will partition primarily behind the replisome (where the substrate is torsionally softer), creating significant intertwining of the daughter strands. In contrast, when this process takes place over chromatin, the high torsional stiffness of a braided chromatin fiber will resist strand intertwining, suppressing fork rotation and preferentially driving supercoiling ahead of the replication fork, where the substrate is torsionally softer.
Our torsional mechanics model provides a clear mechanistic explanation for the in vivo findings which stipulated that in yeast, fork rotation is limited during replication elongation but occurs during termination (Schalbetter et al., 2015; Seidman and Salzman, 1979; Sundin and Varshavsky, 1981; Tapper and DePamphilis, 1978). This model predicts that supercoiling partitions to the front of the replisome during replication elongation, resulting in limited fork rotation. However, preferential partitioning to the front requires the presence of substantial single chromatin fiber to absorb the supercoiling. Therefore, this model also predicts that supercoiling partitions to behind the replisome as replication approaches termination (Figure 5D), leading to fork rotation, and that replisome progression encounters increasing torsional resistance (Figure 5E) as the single chromatin substrate in the front diminishes in length and thus loses its capability as a torsional buffer.
Topoisomerase II Prefers Single Chromatin Substrates
Although the torsional mechanics of chromatin assists with alleviating DNA intertwining, continued progression of a replisome will eventually lead to excessive accumulation of torsion. Thus, efficient and timely resolution of torsional stress by topoisomerases represents a critical component of the regulation of fork rotation during replication. However, it is unclear whether topoisomerase action is coordinated with the torsional mechanics of chromatin, which preferentially partitions supercoiling to the front of a replisome during elongation.
To address this question, we measured substrate-specific behaviors of budding yeast topo II on both single and braided chromatin fibers (Figure 6). It has been reported that topo II is the principal relaxase of chromatin (Salceda et al., 2006), and the enzyme is essential to the successful completion of replication in vivo (Baxter and Diffley, 2008). As a type II topoisomerase that functions by a DNA strand passage mechanism (Berger et al., 1996; Roca and Wang, 1994), topo II is known to be capable of acting ahead of the replication fork (Salceda et al., 2006) and also resolving catenation that occurs behind the fork (Baxter and Diffley, 2008; Schalbetter et al., 2015).
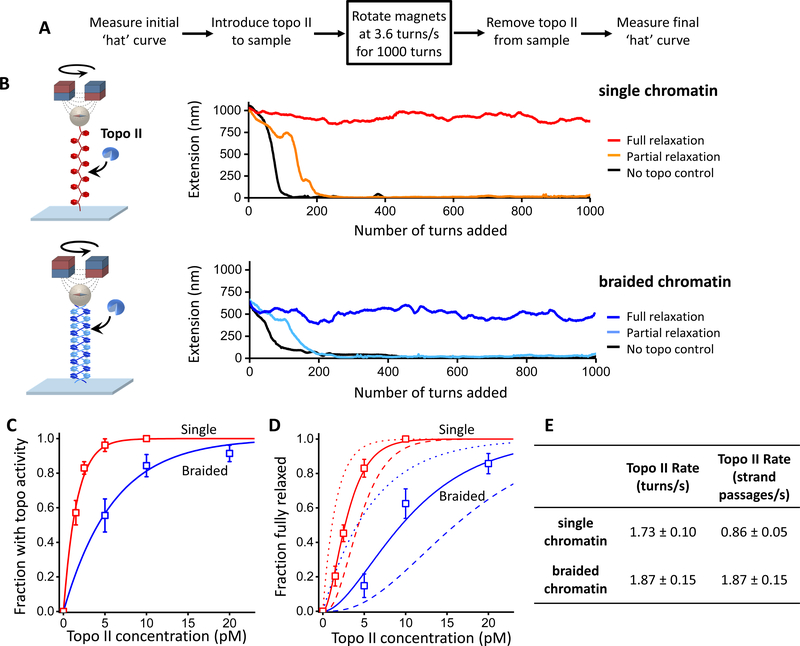
Figure 6. Topo II relaxation of single and braided chromatin fiber substrates.
Experiments were carried out using a magnetic tweezers (MT) setup with chromatin fibers each containing ~ 50 nucleosomes on average. The MT allowed supercoiling to be monitored via an extension change for multiple molecules simultaneously.
(A) Experimental scheme. Highlighted is the key step where the pair of magnets was rotated at a constant rate. The initial and final ‘hat’ (i.e., extension versus turns) curves were used to determine the saturation, stability, and geometry of arrays (Figure S6; Quantification and Statistical Analysis).
(B) Example traces of single and braided chromatin fiber substrates during the key step. A trace is defined as being fully relaxed if the mean rate of topo II relaxation is within 5% of the magnet rotation rate during the course of the key step.
(C) Fractions of traces that showed any topo II activity as a function of the concentration of topo II dimers introduced into the sample chamber. Topo II action on a substrate is assumed to follow a Poisson distribution, and thus the fraction with activity depends on topo concentration according to: with Kactive being the topo II concentration at which there is an average of one topo II molecule relaxing the substrate. Thus provides a measure of topo II’s preference for a substrate. The fits yielded Kactive = 1.6 ± 0.2 and 6.0 ± 1.0 pM (mean ± s.e.m.) for the single and braided fiber substrates, respectively.
(D) Fractions of traces that remained fully relaxed as a function of the concentration of topo II dimers introduced into the sample chamber. The Poisson distribution model predicts that the fraction of traces fully relaxed is given by: where nc is the minimum number of topo II molecules required to keep up with the rotation and Kactive was determined in (C). We found that nc = 2.1 ± 0.1 (mean ± s.e.m.) for the single substrate and 1.9 ± 0.2 for the braided substrate.
(E) Relaxation rate of a single topo II molecule on single and braided chromatin fiber substrates. From the magnet rotation rate (3.6 turns/s) and the minimum number of topoisomerase molecules required to keep up with this rotation nc obtained above, the relaxation rate for an individual topo II molecule acting on each substrate in units of turns/s was determined. Given that one strand passage event removes two self-crossings in a single substrate but one intermolecular crossing in a braided substrate (Bates and Maxwell, 2005; Vologodskii, 2007), the relaxation rates were converted to units of strand passages/s. The errors in the calculated rate were determined by considering contributions from different nc values (nc = 1, …,4) to the measured fractions.
Using a magnetic tweezers (MT) instrument, we mimicked replication fork progression by twisting chromatin substrates at a constant rate of +3.6 turns/s (Figure 6A), a value chosen based on the mean replication rate measured in vivo (Dovrat et al., 2018; Kaykov and Nurse, 2015), and then assayed the ability of topo II to remove supercoiling of either single or braided chromatin substrates under these conditions (Figure 6B). We found that, for both substrates, the fractions of traces that showed topoisomerase activity (Figure 6C) or were fully relaxed (Figure 6D) increased as topo II concentration was increased. However, the braided chromatin substrate required substantially higher concentrations of topo II to reach the same levels of torsional relaxation.
To further quantify these observations, we fit these fractions using a simple model assuming that the number of topo II molecules actively relaxing a substrate follows a Poisson distribution (see legend of Figure 6). This analysis showed that topo II’s preference for a single chromatin substrate was ~ 3.8 times higher than for a braided chromatin substrate (Figure 6C). It also showed that ~ 2 active topoisomerase enzymes were sufficient to keep up with the applied rotation rate on both single and braided chromatin substrates (Figure 6D), with each topo II molecule relaxing either a single or braided chromatin substrate at a similar rate of ~ 2 turns/s (Figure 6E). Therefore, the preferential partitioning of supercoiling into the single chromatin fiber ahead of the replication fork is congruent with topo II’s preferred substrate. These results suggest that topo II action and chromatin torsional mechanics are synergistically coordinated to prevent the buildup of replication-generated torsional stress and suppress fork rotation.
DISCUSSION
The present study highlights the important role of chromatin torsional mechanics in defining replication topology and provides quantitative explanations for observed in vivo behaviors of replication fork dynamics in terms of this topology and topo II kinetics.
Previous studies found that excessive fork rotation can result in chromosome instability, leading to an increase in the level of endogenous DNA damage and the subsequent activation of post-replication repair pathways, delaying or inhibiting mitosis, inducing chromosome mis-segregation, and resulting in aneuploidy following cell division (Baxter and Diffley, 2008; Bermejo et al., 2007; Chan et al., 2009; Debatisse et al., 2012; Keszthelyi et al., 2016; Pommier et al., 2016; Schalbetter et al., 2015). Fork rotation has previously been observed to occur not only during yeast replication termination, but also near stable protein-DNA fragile sites where the replisome encounters tightly-bound non-histone proteins, tRNA genes, and centromeres (Schalbetter et al., 2015). This has been attributed to limited accessibility for topoisomerases in front of the replication fork at these sites (Keszthelyi et al., 2016; Pommier et al., 2016). Our results suggest that the mechanics of chromatin could also play a role in fork rotation. Because tRNA genes are associated with nucleosome-free regions (Shukla and Bhargava, 2018) and centromeric nucleosomes can adapt several alternative structures (Bui et al., 2012; Furuyama and Henikoff, 2009; Henikoff et al., 2014; Shivaraju et al., 2012), it is likely that these substrates which a replisome encounters at fragile sites do not afford the same capacity to buffer DNA supercoiling as canonical nucleosomes. This limited topological buffer would force the fork to rotate more readily in order to balance the torque across the replisome. Interestingly, several regulatory proteins have been shown to be essential in limiting fork rotation in these contexts (Schalbetter et al., 2015) by interacting directly with the replisome to form a stable pausing structure (Errico et al., 2009; Gambus et al., 2006; Katou et al., 2003). Replisome pausing may be necessary to prevent excessive fork rotation by providing topoisomerases with additional time to relax torque buildup (Hodgson et al., 2007; Labib and Hodgson, 2007).
Our finding that chromatin substrates can effectively alleviate torsional stress and suppress precatenation formation underscores the importance of the torsional mechanical properties of chromatin. This alludes to the possibility that substrates may serve as a general framework for topological buffering and regulation. For instance, prokaryotes do not have true histone counterparts capable of forming left-handed particles, which may explain why fork rotation occurs more frequently with a continuous build-up of precatenation during prokaryotic replication (Cebrian et al., 2015; Peter et al., 1998). Interestingly, it has been recently demonstrated that in Caulobacter crescentus bacteria, the chromosome structuring protein GapR encircles (+) supercoiled DNA accumulated in front of the replication fork and stimulates supercoiling relaxation via type II topoisomerases, gyrase and topo IV (Guo et al., 2018). This mechanism bears resemblance to our findings that the inherent mechanical properties of chromatin drives (+) supercoiling to the front of the replication fork to be more efficiently relaxed by topo II.
Although this work focuses on the role of chromatin during replication, (+) supercoiling is also generated in the single chromatin fiber in front of transcription machineries. It has been shown that during transcription, while (+) torsion in front is able to destabilize nucleosomes (Teves and Henikoff, 2014) and thus potentially facilitate transcription through nucleosomes, excessive torsion could also create barriers to transcription (Joshi et al., 2012) by inducing RNA polymerase backtracking and pausing (Ma et al., 2013; Ma et al., 2019) and/or stimulating topo II-mediated DNA knot formation (Valdes et al., 2019). Similar (+) torsion-induced consequences could also impact replication through nucleosomes. For optimal replisome progression, the extent of torsion needs to be regulated to balance between nucleosome destabilization and minimization of daughter strand intertwining and physical barriers to the replisome.
Topoisomerases play a key role in the regulation of replication-generated torsion. We found a strong preference of topo II for a single chromatin substrate compared to a braided substrate. This preference may result from a higher degree of steric accessibility afforded by a single chromatin compared to a chromatin braid. When a single chromatin fiber is subjected to torsion, the entrance and exit DNA segments cross each other and come into close contact (Bancaud et al., 2006; Bancaud et al., 2007), and such a crossing may facilitate topo II binding and subsequent strand passage activity (Salceda et al., 2006; Sperling et al., 2011). Conversely, inter-strand DNA crossing between the two chromatin fibers in a braid may be sterically hindered due to the presence of nucleosomes. Studies of replication termination using Xenopus egg extract and a plasmid DNA (Dewar et al., 2015) found that upon completion of DNA replication, there is a significant delay before the onset of decatenation by topo II. These observations are consistent with our findings that topo II has a significantly lower preference for braided chromatin substrates compared to single substrates. In addition to topo II, topo I is thought to be able to act in the front of the replisome (Pommier et al., 2016). Topo I may associate with the replisome (Gambus et al., 2006) and help torsion relaxation without introducing DNA knots which could hinder replisome progression.
Our results show that chromatin is not just a substrate to be acted upon, nor is it only important for DNA packaging and epigenetic information storage. By providing a buffer against torsional stress, passively regulating fork rotation, and facilitating the efficient relaxation of supercoiling by topoisomerases, the unique mechanical properties of chromatin are vital participants in replication. By converting naked DNA into chromatin, eukaryotes have accomplished something that is rather remarkable: although replication through such a substrate would, at first glance, seem to hinder fork progression, chromatin in fact simplifies the torsional dynamics, limits precatenation formation, and ultimately facilitates chromatin segregation.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
The Lead Contact is Michelle D. Wang (mwang@physics.cornell.edu). Please direct any questions to the Lead Contact. All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacteria strains: The plasmid p197NRL-64EX (a.k.a. pMDW108) was generated in this study by cloning 64 tandem repeats of a 197bp sequence containing a 601 nucleosome positioning element(NPE) and 50 bp random sequence into a commercial pUC19 vector (New England Biolabs). This plasmid was transformed into New England Biolabs Stable Competent E. coli (High Efficiency), genotype: F’ proA+B+ lacIq A(lacZ)M15 zzf::Tn10 (TetR) Δ(ara-leu) 7697 araD139 fhuA ΔlacX74 galK16 galE15 e14-Φ8OdlacZΔM15 recAl relAl endAl nupG rpsL (StrR) rph spoTl Δ(mrr-hsdRMS-mcrBC). Cells were grown in Super Broth with 100 μg/mL ampicillin at 30°C for 17–24 hours. The plasmid pNFRTC (a.k.a. pMDW111) was generated in this study by cloning a 450-bp low nucleosome affinity sequence into a commercial pUC57 vector (Genscript). This plasmid was transformed into New England Biolabs 5-alpha Competent E. coli (High Efficiency), genotype: fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17. Cells were grown in Luria Broth with 100 μg/mL ampicillin at 37°C for 17 hours.
Yeast strain: Topoisomerase II was expressed and isolated from in a BCY123-derived strain MATa pep4::HIS3 prb1::LEU2 bar1::HISG lys2::GAL1/10-GAL4 can1 ade2 trp1 ura3 his3 leu2–3112. This strain is a protease-deficient strain with a GALl promoter-linked GAIA gene and originated from the James C. Wang lab (Harvard University). A starter culture was grown to saturation overnight in Complete Supplement Mixture-Ura media supplemented with 2% dextrose, 2% lactic acid, and 1.5% glycerol at 30 °C. The starter culture was then diluted 10-fold in Yeast Peptone media with 2% lactic acid and 1.5% glycerol, and grown to an O.D. of 1.0–1.3 at 30 °C (12–15 hours), at which point protein expression was induced by the addition of 2% galactose for 6 hours at 30 °C.
Cell line: Histones were isolated from human HeLa-S3 cells. Pelleted cells were obtained from the National Cell Culture Center, and histones were purified directly from these pellets. HeLa-S3 cells are derived from human female cervical tissue.
METHOD DETAILS
DNA Template Construction
The single molecule DNA construct is composed of a 64mer DNA of 12,667 bp flanked by ~ 500 bp multi-labeled tethering adaptor at each end (Figure 2A). The 64mer DNA is 64 tandem repeats of a 197 bp sequence, created using a method similar to that previously described (Wu et al., 2016). The 197 bp repeat sequence contains a 601 nucleosome positioning element (NPE) (Lowary and Widom, 1998) flanked by a total of 50 bp of random sequences at the two ends (Key Resources Table). To construct a plasmid with 64 repeats, a “parental plasmid” was created by inserting into pUC19 (New England Biolabs) the 197 bp sequence flanked by BglI and AdeI recognition sites with identical overhangs and a BstXI recognition site 13-bp downstream of the AdeI site. This parental plasmid was either digested by BstXI and BglI and treated with alkaline phosphatase, or digested with AdeI and BstXI, in two separate reactions. Both reaction products were co-purified using the same DNA purification spin column (Invitrogen) and treated with T4 ligase (New England Biolabs). Transformation of the ligation product resulted in a plasmid with doubled insert flanked by the same combination of restriction enzyme recognition sites as the parental plasmid. The final 64mer plasmid, p197NRL-64EX (a.k.a. pMDW108), was created by running this doubling protocol a total of 6 times. The 64mer insert was then restriction cut with BstXI and BglI and was purified from the plasmid.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-digoxigenin (from sheep) | Roche | Cat# 11333089001, RRID:AB_514496 |
| Bacterial and Virus Strains | ||
| NEB 5-alpha Competent E. coli (High Efficiency) | NEB | Cat# C2987H |
| NEB Stable Competent E. coli (High Efficiency) | NEB | Cat# C3040H |
| Biological Samples | ||
| Topoisomerase yeast strain MATa pep4::HIS3 prb1::LEU2 bar1::HISG lys2::GAL1/10-GAL4 can1 ade2 trp1 ura3 his3 leu2-3112 | J. C. Wang Lab (Harvard University) | N/A |
| HeLa-S3 cells | National Cell Culture Center | HA.48 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collodion solution | Sigma-Aldrich | Cat# 09986 |
| Silicone high-vacuum grease | Dow Corning | Part # 146355 D |
| Biotin-14-dATP | Invitrogen | Cat# 19524016 |
| Digoxigenin-11-dUTP | Roche | Cat# 11093088910 |
| dNTP Set 100 mM Solutions | Thermo Scientific | Cat# R0181 |
| ATP | Roche | Cat# 11140965001 |
| T4 DNA ligase | NEB | Cat# M0202S |
| Taq DNA Polymerase | NEB | Cat# M0273S |
| FastDigest BglI | Thermo Scientific | Cat# FD0074 |
| FastDigest BstXI | Thermo Scientific | Cat# FD1024 |
| Nt.BsmAI | NEB | Cat# R0121S |
| Beta-casein from bovine milk | Sigma-Aldrich | Cat# C6905 |
| Hela histone core | Brennan et al., 2016 | N/A |
| S. Cerevisiae topoisomerase II | This study | N/A |
| Oligonucleotides | ||
| Forward primer for making the biotin-labeled and digoxigenin-labeled adapters pUC57-F: GTAAAACGACGGCCAGTG |
IDT | N/A |
| Reverse primer for making the biotin-labeled and digoxigenin-labeled adapters pUC57-R: GGAAACAGCTATGACCATG |
IDT | N/A |
| The 197-bp repeat sequence of p197NRL-64ex with the 601 sequence underlined: GGGCGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTATTGAACACACCCCCTAACACACTACGACACCCC |
This study | N/A |
| The 482-bp low nucleosome affinity sequence of pNFRTC: GCCTGGGTGGCTTCATTCGTTCTTTTGTTCCTTATTTTGTTC CTTACTTAGTTGGTTATTTGCTTGTTTGGTTATTTATTTCGT TGGTTATTTGGTTAATTCCTT CTTTGCTTTCTTCATTCCTTT CTTGCTTTATTCCTTGTTTTTTTGGTTTCTTAGTTTCCTTTTC CCTAGAGGTAGCCAAAGTCTTTGCAACTAT ACTTTCAGCT CTGACAAATTTGTTCTTATTACIICICIIIIIIIIGATTTGT TCTTCCCTCTTTTTCTTAGCTAATTCTTGTCTTTCGATTCTA GTTCTATCAGCATTTCTTTATAAATCTATTTTTTTTTTTTTTT CGACACAAAATGTCTATTTCTTGGAGTGCTTACTCTTCTTT TTGTTTTTACCTTGTTTCAACTCGTTTAATCTATCAACTTTT TCCTTGATCCTTTCCAAAGATAATTTGACATCACCTTTTTG GCACTAGGTGCCACCGATGTGG |
This study | N/A |
| Recombinant DNA | ||
| p197NRL-64ex containing the 64 repeats of the Widom 601 sequence | This study | N/A |
| pNFRTC for making the multiple-label DNA end adapters | This study | N/A |
| pUC19 | New England Biolabs | Cat# N3041S |
| Software and Algorithms | ||
| LabVIEW VIs for instrument control, data acquisition, and data analysis | This study | N/A |
| MATLAB scripts for data analysis | This study | N/A |
| SeqBuilder (for DNA primer design) | DNASTAR | Version 11.2.1 (29) |
| Other | ||
| Microscope cover glass | Fisher Scientific | Cat# 12–544-B |
| Microscope slides | Fisher Scientific | Cat# 12-550-10 |
| Angular optical trap setup | Deufel et al., 2007 | N/A |
| Magnetic tweezers setup | This study | N/A |
| Dynabeads MyOne Streptavidin T1 | Invitrogen | Cat# 65601 |
| Streptavidin-coated quartz cylinder | Deufel et al., 2007 and this study | N/A |
| 4” Quartz Wafer | Precision MicroOptics | Cat# PWQB-131332 |
| PureLink Quick PCR Purification Kit | Invitrogen | Cat# K310001 |
The torsionally constrained 64mer DNA construct was formed by ligating ~ 500 bp multi-labeled adaptors to each end of the 64mer DNA. To create the adapters, a 482-bp DNA sequence with low nucleosome affinity was cloned into pUC57 (Genscript), resulting in the plasmid pNFRTC (a.k.a. pMDW111). The 482-bp DNA sequence insert was created by concatenating three 150-bp DNA sequences with low nucleosome affinity identified by a previous large-scale nucleosome occupancy study (Kaplan et al., 2009) and flanking it with BglI and BstXI cutting sites (Key Resources Table). This sequence insert was then amplified and labelled by performing a polymerase chain reaction (PCR) with 25% of dATP replaced by biotin-14-dATP (Invitrogen) or 25% of dTTP replaced by digoxigenin-11-dUTP (Sigma-Aldrich). The PCR products were digested by BglI and BstXI to create overhangs for the biotin-and dig-adaptors, respectively. The final DNA construct was formed via ligation of the 64mer DNA with these two multi-labeled adaptors.
Protein purification
Histone octamers were purified from nuclei extracted from HeLa-S3 cell pellets purchased from the National Cell Culture Center using hydroxyapatite precipitation (Brennan et al., 2016; Brower-Toland et al., 2005; Brower-Toland and Wang, 2004; Li et al., 2015). In brief, nuclei were extracted in Nuclear Pellet Prep Lysis Buffer (20mM HEPES pH 7.5), 3 mM MgCl2, 250 mM sucrose, 0.5% (v/v), IGEPAL CA-630 (NP-40) nonionic detergent, 1 tablet per 50 ml Complete Protease Inhibitor Cocktail (Roche) and 3 mM 2-mercaptoethanol (Schnitzler, 2001). Core histones were purified using a hydroxyapatite Bio-gel HTP gel (Bio-Rad Laboratories) slurry, according to methods by Wolffe and Ura (Wolffe and Ura, 1997) but with the omission of MNase digestion before fractionation. Aliquots of purified histones were stored at −80 °C.
Full length, wild type S. cerevisiae topoisomerase II (topo II) was purified from S. cerevisiae strain BCY123 via His-tags which were removed by TEV protease as previously described (Lee et al., 2017). In brief, the yeast topo II gene was cloned into the 12UraB vector (Addgene), which contains a galactose-inducible promoter and a TEV-cleavable N-terminal His6-tag. His6-tagged yeast topo II was expressed in the S. cerevisiae strain BCY123. Cells were harvested by centrifugation, flash frozen dropwise in liquid nitrogen, and lysed by cryogenic grinding. His6-topo II was purified from lysate by Ni-affinity purification followed by cation exchange. The His6-tag was then removed by incubation with His6-tagged TEV protease. The cleavage reaction was repassed over a Ni-affinity column to removed His6-TEV and uncleaved protein. Lastly, the protein was purified on a size exclusion column.
Nucleosome Assembly
Nucleosome arrays were assembled onto the 64mer DNA construct by gradient NaCI dialysis from 2.0 M to 0.6 M over 18 hours at 4°C at different molar ratios (0.25:1 to 2.5:1) of histone octamers to 601 DNA repeats (Brower-Toland et al., 2005; Huynh et al., 2005). An equal mass of 147-bp random-sequence competitor DNA to the 64mer DNA construct was added to the reconstitution reactions to avoid nucleosome over-assembly. The quality and saturation level of the nucleosome arrays were assayed by gel electrophoresis in 0.7% native agarose (Figure S1) and by stretching the nucleosome arrays using the AOT (Figure 2; Figures S3A–S3C; Figures S4A and S4B).
Single Molecule Sample Chamber Preparation
Microscope coverslips used to make single molecule sample chambers were cleaned with 95% ethanol, coated with a thin nitrocellulose film using a solution of one part 4–8% collodion solution (Sigma, 09986) and three parts amyl acetate, and dried at 80 °C for 10 minutes. This is similar to a previous method involving nitrocellulose (Meng et al., 2015). A coated coverslip and a glass slide were assembled into a microfluidic sample chamber formed using inert silicone high vacuum grease and stored in a clean plastic container for typically 24 hours or more before use.
Prior to an experiment, a sample chamber was first incubated with 20 ng/μL anti-digoxygenin for 30 minutes. For MT experiments, fiducial marker beads (Dynabeads MyOne Streptavidin T1, 65601) coated with biotin and digoxygenin labeled DNA were bound to the surface. The surface was then passivated by flushing the chamber with 25 mg/mL β-casein (Sigma, C6905) and incubated for 3 hours. β-casein has previously been shown to be an effective reagent for surface passivation (Nicholas et al., 2014).
To form single chromatin tethers, nucleosome arrays were anchored to the surface of the coverslip of a sample chamber and then attached to streptavidin-coated quartz cylinders for the AOT experiments or to 1-μm superparamagnetic beads (Dynabeads MyOne Streptavidin T1, 65601) for the MT experiments.
Quartz cylinders (diameter: 549 ± 56 nm (SD), height: 916 ± 48 nm (SD)) were optically birefringent and nanofabricated based on protocols previously described (Deufel et al., 2007) with some modifications (Ha et al., 2016). In brief, a 4” quartz wafer (Precision Micro-Optics) was coated with 100 nm of sputtered chromium for an etch mask. ~ 500 nm wide pillars were patterned in a periodic hexagonal array, with 1 μm spacing between nearest neighbors, using the negative photoresist UNV2300–0.5 exposed with deep ultraviolet lithography. The quartz pillars were etched using CHF3 and Argon plasma at 50 °C with the chromium etch mask. The ~ μm tall etched pillars were then coated with ~ 3 μm of SPR-3000 photoresist, and oxygen plasma was used to remove the top ~ 2 μm of the photoresist until only the tops of the cylinders were exposed. The tops of the pillars only were functionalized with amine groups using APTMS gas in a molecular vapor deposition tool, and then the remaining photoresist was chemically stripped off the wafer in Microposit Remover 1165. Finally, the quartz pillars were cleaved with a razor blade, and the resulting quartz cylinders were functionalized in solution with streptavidin.
To form double chromatin tethers, nucleosome arrays and streptavidin-coated particles were incubated together for ~ 30 min in a buffer containing 10 mM Tris-HCl pH 8.0, 50 mM NaCI, 1 mM EDTA, and 1.5 mg/mL β-casein and then introduced to a sample chamber. The incubation condition was tuned such that the majority of tethers were single and/or double tethers, with minimal tethers forming higher multi-tethers which can be differentiated based on stretching curves or the shape of hat curves. To prevent the torsion buildup within each DNA molecule of a braided substrate, we nicked the tethers in situ using a nicking enzyme (Nt.BsmAI), achieving > 95% nicking efficiency.
All experiments were carried out in the topo reaction buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 50 mM KCl, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5 mM TCEP, 1 mM ATP, and 1.5 mg/mL β casein) in a soundproof room at a temperature of 23 °C.
QUANTIFICATION AND STATISTICAL ANALYSIS
Rotational viscous drag on a replisome. Related to Figures 1 and 5
The viscous drag torque experienced by a eukaryotic replisome during replication may be estimated based on the replisome’s dimensions. We estimate the maximum possible size of the replisome to be 80 nm in diameter, based on summing the diameters of the leading strand DNA polymerase (~ 10 nm) (Langston et al., 2014), helicase (or CMG) (~ 10 nm) (Langston et al., 2014), the lagging strand DNA polymerase (~ 10 nm) (Johansson et al., 2001), and the Okazaki fragment) (~ 50 nm) (Smith and Whitehouse, 2012). This value may somewhat overestimate the replisome size.
For simplicity, we treat the replisome as a sphere of radius R < 40 nm in which case the viscous torque is given by We estimate that the angular velocity ω ≤ 4 turn/s given the speed of replication ~ 40 bp/s (Dovrat et al., 2018; Kaykov and Nurse, 2015). The viscosity of rotational mobility measured in vivo to 4 × 10−3 Pa·s (Williams et al., 1997; Ye et al., 2018). Therefore, the replisome should experience a maximum viscous torque τ∼ 0.08 to 0.15 pN·nm.
To put this value in perspective, we must compare this value with the torque that DNA polymerase can generate. Although DNA polymerase’s torque generation capacity has not been measured, the torque that an RNA polymerase motor can generate has been measured to be 11–19 pN nm (Ma et al., 2013; Ma et al., 2019). Both DNA polymerase and RNA polymerase convert chemical energy derived from polymerization reactions to generate forces and torques, and both work against DNA torsional stress from DNA supercoiling as well as roadblocks from other bound proteins. Therefore, it is likely that the torque generation capacity of the two types of motors are of the same order of magnitude. This suggests that the rotational viscous drag experienced by a replisome plays a minimal role in restricting its rotation since the viscous drag is expected to be negligible compared to the torque generation capacity of the polymerase.
Torsional measurements with the AOT. Related to Figures 3 and 4
Single molecule torsional measurements were carried out using an AOT (La Porta and Wang, 2004). In contrast to a conventional optical trap, an AOT can also rotate a trapped nanofabricated quartz cylinder about its cylindrical axis by the rotation of the trapping beam’s linear polarization, and the torque exerted on the cylinder can be measured by the change in the angular momentum of the transmitted beam. Thus an AOT allows simultaneous control and measurement of rotation, torque, displacement, and force of the trapped cylinder (Deufel et al., 2007; Forth et al., 2008; Ma et al., 2013; Ma et al., 2019; Sheinin et al., 2011).
On the AOT, while a single or double chromatin substrate was held under 0.5 pN using a force clamp with the laser power input to the objective held at 30 mW, the substrate was twisted at 4 turns/s and the torque and extension were simultaneously measured (Figures 3B and 3C; Figures S3D–S3G; Figures 4B and 4C; Figures S4C–S4D). At the end of the experiment, the substrate was returned to the initial zero-turns state. Subsequently, it was stretched axially (along the direction of laser propagation) to disrupt the nucleosomes on the DNA in order to quantify nucleosome composition and the number of nucleosomes on the DNA (Figure 2; Figures S3A–S3C; Figures S4A and S4B). During the stretching, the coverslip was moved away from the trap center at a constant speed of 400 nm/s with the laser power input to the objective at 30 mW, until the cylinder was 350 nm from trap center. Subsequent stretching was carried out by increasing the DNA extension at a constant rate of 400 nm/s by clamping the cylinder position relative to the trap center through modulating laser intensity. Data were analog filtered to 5 kHz, digitized at 10 kHz, and further filtered to 20 Hz and 500 Hz for twisting and stretching data respectively.
Topoisomerase Assays. Related to Figure 6
Topoisomerase experiments were carried out on a custom built MT setup based on previous designs (Lipfert et al., 2009; Seol and Neuman, 2011; Strick et al., 1996). In brief, the magnetic field was generated with a pair of 0.25” cube neodymium magnets (K&J Magnetics B444) which were arranged with their dipoles oriented in opposing directions and parallel to the optical axis of the microscope and with a separation gap of 0.5 mm. Magnetic bead images were collected by a Nikon 40x objective lens (Plan Apo 40× 0.95 NA) onto a 2.3 MP camera (Basler acA1920–155um) at a frame rate of 10 fps and an exposure time of 0.5 ms. The bead positions were tracked in three dimensions using an algorithm implemented in LabVIEW based on the source code available on Omar Saleh’s website (Lansdorp et al., 2013).
We carried out a control experiment to determine the relaxation rate of yeast topo II on a single naked DNA substrate. We adapted a previous method (Strick et al., 2000) to assay budding yeast topo II. In this experiment, each naked DNA molecule (same as that used in Figures 2–6) was torsionally anchored between a coverslip and a magnetic bead and held at 0.5 ± 0.08 pN (mean ± SD) using the MT instrument. Topo II at very low concentration (1 pM) was introduced into the sample chamber prior to the start of the measurement. Subsequently, the DNA was mechanically twisted to form a (+) plectoneme and the magnet position was then held fixed. Subsequent topo II relaxation was reflected as an increase in the DNA extension. We determined that the rate of topo II to be 1.8 ± 0.6 turns/s (mean ± SD), comparable to those previously obtained for eukaryotic topoisomerase II of different species on naked DNA substrates (Seol et al., 2013; Strick et al., 2000).
For each experiment shown in Figure 6, once tethers were formed in a sample chamber, tethers were held under 0.50 ± 0.08 pN (mean ± SD) of force on the MT. While tethers of a single (Figure S6A) or a braided substrate (Figure S6D) were twisted, the extensions were monitored. At the end of the experiment, the magnet was returned to the initial zero-turns state. This process established the “initial hat curve” for each tether. Subsequently, S. cerevisiae topo II, diluted in the topo reaction buffer, was introduced into the sample chamber and incubated for 2 minutes. The magnet was then rotated at 3.6 turns/s for 1000 turns while the extension of each tether was tracked. Immediately afterwards, topo flushing buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 50 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5 mM TCEP, and 1.5 mg/mL β-casein) was flushed through the sample chamber to remove unbound topo II and inactivate bound topo II. A “final hat curve” of each tether was acquired in the same fashion as that of the “initial hat curve” for comparison. At this step, the tether was wound to the surface to obtain a height offset for an absolute length measurement.
Evaluation of chromatin fiber integrity and saturation. Related to Figures 2, 3, 4, and 6
Here we provide a detailed description of our criteria for determining the number of nucleosomes in a substrate and for evaluating nucleosome composition, stability, and substrate geometry.
During each experiment using the AOT, we first twisted a chromatin substrate under a constant force (0.5 pN) by both introducing turns and removing turns. This process not only yielded data on extension and torque versus turns from the substrate (Figures 3B and 3C; Figures 4B and 4C), but also allowed for evaluation of reversibility and stability of the substrate (Figures S3Eand S4D), and in the case of a braided substrate, the anchor separation of the substrate (Figure S5). To further examine the composition and level of saturation of a chromatin substrate, we stretched the substrate to a high force of ~ 50 pN for a single substrate (Figure 2C) and ~ 80 pN for a braided substrate (Figure 2E). During data analysis, we determine if each trace meets the selection criteria detailed below for inclusion for further analysis in main text Figures 3 and 4.
Prior to each topoisomerase experiment of a chromatin substrate using the MT, we also performed a twisting experiment under a constant force (0.5 pN), similar to that of the AOT, by both introducing turns and removing turns from the substrate. The resulting hat curve allowed for evaluation of reversibility and stability of the substrate (Figures S6B and S6E), an estimate of the number of nucleosomes on the substrate, and in the case of a braided substrate, an estimate of the anchor separation of the substrate (Figure S6F). During data analysis, we determine if each trace meets the selection criteria detailed below for inclusion for further analysis in main text Figure 6.
Single chromatin fiber substrate on AOT
-
-
In the stretching experiment performed subsequent to the twisting experiment, the measured contour length of DNA after nucleosome disruption at 40–45 pN must agree to within 10% of the theoretical value for that of a 12,667 bp DNA construct (Figure 2C). This step removes shorter DNA templates that occasionally occur due to sequence repeat deletion during bacterial transformation (Bzymek and Lovett, 2001).
-
-
In the stretching experiment performed subsequent to the twisting experiment, analysis must show that where Nout is the number of outer turns released and Nin is the number of inner turns released (Figures 2C and S3B). This optimizes the chance that each array contains primarily complete and canonical nucleosomes with minimal contributions from other structures.
-
-
In the twisting experiments, the mean difference in extension between the hat curves of adding turns and removing turns must be < 50 nm (Figure S3E). This minimal hysteresis requirement ensures that the substrate is stable during the course of torsional measurements.
Braided chromatin fiber substrate on AOT
-
-
In the stretching experiment performed subsequent to the twisting experiment, the measured contour length of DNA after nucleosome disruption at 70–75 pN must agree to within 10% of the theoretical value for that of a double 12,667 bp substrate (Figure 2E). As with the single substrates, this step removes shorter DNA templates that occasionally occur due to sequence repeat deletion during bacterial transformation (Bzymek and Lovett, 2001).
-
-
In the stretching experiment performed subsequent to the twisting experiments, analysis must show that (Figures 2E and S4B). When the two arrays in a double substrate contain substantially different numbers of nucleosomes, our method for detecting the boundaries of inner-turn and outer-turn releases will result in a detected Nin closer to that of the array with a larger number of nucleosomes and the detected Nout closer to that of the array with a smaller number of nucleosomes. Therefore, the detected becomes large and will naturally be excluded from further analysis. Thus this selection criterion simultaneously selects substrates with two arrays both being similar in the number of nucleosomes and containing primarily canonical nucleosomes.
-
-
In the twisting experiments, we require that the mean difference in extension between the hat curves of adding turns and removing turns must be < 50 nm (Figure S4D). This step selects traces that are reversible and stable during the course of torsional measurements.
-
-
An important consideration for a braiding substrate is the geometry of the two molecules in the substrate, more specifically anchor separations at both ends. As discussed in the figure legend of Figure S5, to select traces with small anchor separations, we normalized each hat curve by the maximum extension of the trace and fit the normalized extension between −10 and +10 turns to a parabola. We require that the rise of the normalized extension above this fit be smaller than 0.015 and the quadratic term magnitude of the fit be smaller than 5 × 10−4/turns2.
Single chromatin fiber substrate on MT
-
-
In the twisting assay performed before experiments, we require that the mean difference in extension between the hat curves of adding turns and removing turns must be < 50 nm (Figure S6B). This step selects traces that are stable over time.
-
-
From the hat curve’s maximum extension (extension at zero turns), we calculated the number of nucleosomes on the substrate using the linear relationship between the extension at zero turns versus number of nucleosomes established on the AOT (Figure S3C). We selected traces with an extension consistent with 50 ± 6 nucleosomes.
-
-
We require the hat curve’s (+) transition width (Figure S6C) to be within 20% of the expected value established on the AOT (Figure S3G). This selection criterion selects traces with good nucleosome composition. This procedure additionally removes tethers that were partially stuck to the surface because they will exhibit short and narrow hat curves.
Braided chromatin fiber substrate on MT
-
-
In the twisting assay performed before experiments, we require that the mean difference in extension between the hat curves of adding turns and removing turns must be < 50 nm (Figure S6E). This step selects traces that are stable over time.
-
-
From the extension at zero turns, we calculated the number of nucleosomes on the substrate using the linear relationship between extension at zero turns and array saturation established on the AOT (Figure S4E). We selected traces with an extension consistent with 50 ± 6 nucleosomes.
-
-
To select traces with small anchor separations at both ends, we used similar selection criteria as those for the AOT data, where the rise of the normalized extension above the parabola fit must be smaller than 0.07 and the quadratic term of the fit must be smaller than 1.4 × 10−3/turns2 (Figure S6F). These criteria were somewhat relaxed from those for the corresponding AOT data in order to accommodate the larger noise in extension measurements with the MT.
Torsional modulus determination. Related to Figures 3 and 4.
The torsional modulus of a substrate C may be obtained from the torsional stiffness of the substrate k for a given contour length of DNA L: C = kL. In all experiments described in this work, the DNA template has a contour length L of ~ 4300 nm.
At each level of nucleosome saturation (number of nucleosomes on the substrate), the torsional stiffness was obtained from the initial (+) slope of the measured torque (τ) versus turns (n) relation (Figure 3C; Figure 4C; Figure S5): which was then used to obtain the torsional modulus. We then plotted torsional modulus C as a function of number of nucleosomes on the substrate (Figure 3D; Figure 4D).
We model a partially occupied nucleosome array as two linear torsional springs in series, with one taking on the torsional properties of a fully occupied array (torsional modulus Cchrom) while the other taking on the torsional properties of naked DNA (torsional modulus CDNA). In this case, the inverse torsional modulus of a partially occupied array should be the sum of the inverse moduli of both parts, weighted by their respective length fraction ( for fully occupied array and for naked DNA):
Thus by fitting this expression to the relationships in Figure 3D and Figure 4D, we obtained the torsional modulus for a fully occupied array Cchrom for fully occupied single and braided chromatin substrates, respectively.
It is important to note that this is an overly simplified model that will likely not fully capture the behaviors of partially occupied chromatin substrates. Therefore, we only used this model to provide some intuition on how torsional modulus might depend on the nucleosome occupancy and to guide our eyes in main text Figures 3C and 4C. For the direct comparison of single and braided chromatin substrates presented in main text Figure 5, we instead used values measured at the highest attainable numbers of nucleosomes occupied, in order not to overstate our conclusions. We anticipate that the contrast of torsional moduli between single and braided chromatin substrates would be more dramatic with fully occupied chromatin substrates (Figure 5, see legend).
Note that direct measurements of moduli of fully occupied chromatin substrates are exceedingly difficult. In vitro assembly of saturated nucleosome arrays often leads to formation of subnucleosomal structures such as tetrasomes in the linker DNA regions. Those arrays would not pass our selection criteria.
Supercoiling partitioning and torque build up during replication. Related to Figure 5.
Supercoiling may partition ahead of or behind a replisome. Supercoiling partitioning is dictated by the torsional stiffness of the single substrate in front of the replisome kfront versus that of the braided substrate behind the replisome kbehind. The torsional stiffness of a substrate k relates torque τ to rotation through a rotation angle θ according to The torsional stiffness k is related to the torsional modulus C of the substrate by the contour length of DNA
The fraction of supercoiling partitioned behind of the replisome p (Figure 5D) as a function of replication progression can be calculated as the followed. Let the DNA supercoiling angle generated by replication be which must partition to the behind Because the torque in the substrate behind the replisome is balanced by the torque in the front, we have Consequently, This can be re-written in terms of the fraction of the total contour length in the double substrate behind: with f = 0 at initiation and f = 1 near termination. Thus the fraction of supercoiling behind is: During elongation (e.g., f = 0.5), for naked DNA, thus supercoiling partitions primarily behind during replication. In contrast, for chromatin substrates, therefore supercoiling partitions primarily to the front during replication elongation.
Torque build up during replication progression can be characterized by a total effective torsional modulus Ceff of the two coupled substrates in front of and behind a replication fork (Figure 5E).: Thus torque in the substrate is: where σ is the supercoiling density. Ceff provides a measure of the torsional stress experience by a replisome. Overall, chromatin substantially reduces the torsional stress experienced by a replisome during replication in comparison to naked DNA. This advantage is only lost near termination
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Wang Laboratory and J.P. Sethna, D.B. Liarte, V. Gahdi, E.E. Alani, and J.E. Peters for helpful discussion and comments, J.E. Baker for initial data analysis software of the MT, and L.D. Brennan for purification of histones. This work was supported by HHMI (to M.D.W.) and NIH (T32GM008267 to M.D.W, F31CA224896 to J.H.L., and R01CA077373 to J.M.B.). The quartz cylinder fabrication was performed at the Cornell NanoScale Science & Technology Facility (CNF), a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by NSF (Grant NNCI-1542081).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures and can be found with this article online.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
For custom programs and scripts used in this study, please contact Dr. Michelle Wang (mwang@physics.cornell.edu).
References
- Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, Mozziconacci J, Lavelle C, Croquette V, Victor JM, Prunell A, et al. (2006). Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol 13, 444–450. [DOI] [PubMed] [Google Scholar]
- Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. (2007). Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell 27, 135–147. [DOI] [PubMed] [Google Scholar]
- Bates AD, and Maxwell A (2005). DNA topology, 2nd edn (Oxford ; New York, Oxford University Press; ). [Google Scholar]
- Baxter J, and Diffley JF (2008). Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell 30, 790–802. [DOI] [PubMed] [Google Scholar]
- Beattie TR, Kapadia N, Nicolas E, Uphoff S, Wollman AJ, Leake MC, and Reyes-Lamothe R (2017). Frequent exchange of the DNA polymerase during bacterial chromosome replication. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, and Labib K (2016). Chromosome Duplication in Saccharomyces cerevisiae. Genetics 203, 1027–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, and Wang JC (1996). Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, and Foiani M (2007). Top1-and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev 21, 1921–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan LD, Forties RA, Patel SS, and Wang MD (2016). DNA looping mediates nucleosome transfer. Nat Commun 7, 13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, and Wang MD (2005). Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol 346, 135–146. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, and Wang MD (2004). Use of optical trapping techniques to study single-nucleosome dynamics. Methods Enzymol 376, 62–72. [DOI] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, and Wang MD (2002). Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A 99, 1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, and Dalal Y (2012). Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PMJ, and Kunkel TA (2017). Eukaryotic DNA Replication Fork. Annu Rev Biochem 86, 417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, and Lovett ST (2001). Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A 98, 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian J, Castan A, Martinez V, Kadomatsu-Hermosa MJ, Parra C, Fernandez-Nestosa MJ, Schaerer C, Hernandez P, Krimer DB, and Schvartzman JB (2015). Direct Evidence for the Formation of Precatenanes during DNA Replication. J Biol Chem 290, 13725–13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon A, Nodelman IM, Wildt B, Dewan R, Searson P, Wirtz D, Bowman GD, and Sun SX (2009). Magnetic tweezers measurement of single molecule torque. Nano Lett 9, 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ, and Been MD (1980). Topoisomerases and the swivel problem (Academic Press; ). [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, and Hickson ID (2009). Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11, 753–760. [DOI] [PubMed] [Google Scholar]
- Charvin G, Allemand JF, Strick TR, Bensimon D, and Croquette V (2004). Twisting DNA: single molecule studies. Contemporary Physics 45, 383–403. [Google Scholar]
- Charvin G, Vologodskii A, Bensimon D, and Croquette V (2005). Braiding DNA: experiments, simulations, and models. Biophys J 88, 4124–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, and Brison O (2012). Common fragile sites: mechanisms of instability revisited. Trends Genet 28, 22–32. [DOI] [PubMed] [Google Scholar]
- Deufel C, Forth S, Simmons CR, Dejgosha S, and Wang MD (2007). Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods 4, 223–225. [DOI] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, and Walter JC (2015). The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovrat D, Dahan D, Sherman S, Tsirkas I, Elia N, and Aharoni A (2018). A Live-Cell Imaging Approach for Measuring DNA Replication Rates. Cell Rep 24, 252–258. [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, and Luger K (2004). Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375, 23–44. [DOI] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, and Costanzo V (2009). Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J 28, 3681–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, and Wang MD (2008). Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys Rev Lett 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, and Henikoff S (2009). Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, and Labib K (2006). GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8, 358–366. [DOI] [PubMed] [Google Scholar]
- Gemmen GJ, Sim R, Haushalter KA, Ke PC, Kadonaga JT, and Smith DE (2005). Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J Mol Biol 351, 89–99. [DOI] [PubMed] [Google Scholar]
- Guo MS, Haakonsen DL, Zeng WJ, Schumacher MA, and Laub MT (2018). A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication. Cell 175, 583-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Janissen R, Ussembayev YY, van Oene MM, Solano B, and Dekker NH (2016). Tunable top-down fabrication and functional surface coating of single-crystal titanium dioxide nanostructures and nanoparticles. Nanoscale 8, 10739–10748. [DOI] [PubMed] [Google Scholar]
- Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, and Wang MD (2009). High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol 16, 124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ramachandran S, Krassovsky K, Bryson TD, Codomo CA, Brogaard K, Widom J, Wang JP, and Henikoff JG (2014). The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. Elife 3, e01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, and Labib K (2007). Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell 18, 3894–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, and Rhodes D (2005). A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol 345, 957–968. [DOI] [PubMed] [Google Scholar]
- Johansson E, Majka J, and Burgers PM (2001). Structure of DNA polymerase delta from Saccharomyces cerevisiae. J Biol Chem 276, 43824–43828. [DOI] [PubMed] [Google Scholar]
- Joshi RS, Pina B, and Roca J (2012). Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res 40, 7907–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, and Shirahige K (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kaykov A, and Nurse P (2015). The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res 25, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi A, Minchell NE, and Baxter J (2016). The Causes and Consequences of Topological Stress during DNA Replication. Genes (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Georgescu RE, and O’Donnell ME (2013). A solution to release twisted DNA during chromosome replication by coupled DNA polymerases. Nature 496, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Porta A, and Wang MD (2004). Optical torque wrench: Angular trapping, rotation, and torque detection of quartz microparticles. Phys Rev Lett 92. [DOI] [PubMed] [Google Scholar]
- Labib K, and Hodgson B (2007). Replication fork barriers: pausing for a break or stalling for time? EMBO Rep 8, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, and O’Donnell ME (2014). CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A 111, 15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp BM, Tabrizi SJ, Dittmore A, and Saleh OA (2013). A high-speed magnetic tweezer beyond 10,000 frames per second. Rev Sci Instrum 84, 044301. [DOI] [PubMed] [Google Scholar]
- Lee JH, Wendorff TJ, and Berger JM (2017). Resveratrol: A novel type of topoisomerase II inhibitor. J Biol Chem 292, 21011–21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, Horan NP, Duderstadt KE, Cox MM, Robinson A, Dixon NE, et al. (2017). Single-molecule visualization of fast polymerase turnover in the bacterial replisome. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, et al. (2015). Dynamic regulation of transcription factors by nucleosome remodeling. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen P, Yu J, Dong L, Liang D, Feng J, Yan J, Wang PY, Li Q, Zhang Z, et al. (2016). FACT Remodels the Tetranucleosomal Unit of Chromatin Fibers for Gene Transcription. Mol Cell 64, 120–133. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Hao X, and Dekker NH (2009). Quantitative modeling and optimization of magnetic tweezers. Biophys J 96, 5040–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, and Wang JC (1987). Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276, 19–42. [DOI] [PubMed] [Google Scholar]
- Ma J, Bai L, and Wang MD (2013). Transcription Under Torsion. Science 340, 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Tan C, Gao X, Fulbright RM Jr., Roberts JW, and Wang MD (2019). Transcription factor regulation of RNA polymerase’s torque generation capacity. Proc Natl Acad Sci U S A 116, 2583–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack AH, Schlingman DJ, Ilagan RP, Regan L, and Mochrie SG (2012). Kinetics and thermodynamics of phenotype: unwinding and rewinding the nucleosome. J Mol Biol 423, 687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF (2007). Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E Stat Nonlin Soft Matter Phys 76, 021926. [DOI] [PubMed] [Google Scholar]
- Meng H, Andresen K, and van Noort J (2015). Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res 43, 3578–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, and Gilbert N (2013). Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 20, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MP, Rao L, and Gennerich A (2014). Covalent immobilization of microtubules on glass surfaces for molecular motor force measurements and other single-molecule assays. Methods Mol Biol 1136, 137–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Ullsperger C, Hiasa H, Marians KJ, and Cozzarelli NR (1998). The structure of supercoiled intermediates in DNA replication. Cell 94, 819–827. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, and Cozzarelli NR (2001). Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci U S A 98, 8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, and Wang JC (1994). DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77, 609–616. [DOI] [PubMed] [Google Scholar]
- Salceda J, Fernandez X, and Roca J (2006). Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J 25, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalbetter SA, Mansoubi S, Chambers AL, Downs JA, and Baxter J (2015). Fork rotation and DNA precatenation are restricted during DNA replication to prevent chromosomal instability. Proc Natl Acad Sci U S A 112, E4565–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler GR (2001). Isolation of histones and nucleosome cores from mammalian cells. Curr Protoc Mol Biol Chapter 21, Unit 21 25. [DOI] [PubMed] [Google Scholar]
- Seidman MM, and Salzman NP (1979). Late replicative intermediates are accumulated during simian virus 40 DNA replication in vivo and in vitro. J Virol 30, 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat MD, Fenyo D, Rogers RS, Tackett AJ, Aitchison JD, and Chait BT (2010). GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol 6, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol Y, Gentry AC, Osheroff N, and Neuman KC (2013). Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIalpha. J Biol Chem 288, 13695–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol Y, and Neuman KC (2011). Magnetic tweezers for single-molecule manipulation. Methods Mol Biol 783, 265–293. [DOI] [PubMed] [Google Scholar]
- Sheinin MY, Forth S, Marko JF, and Wang MD (2011). Underwound DNA under Tension: Structure, Elasticity, and Sequence-Dependent Behaviors. Phys Rev Lett 107, 108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin MY, Li M, Soltani M, Luger K, and Wang MD (2013). Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun 4, 2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, and Gerton JL (2012). Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150, 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, and Bhargava P (2018). Regulation of tRNA gene transcription by the chromatin structure and nucleosome dynamics. Biochim Biophys Acta Gene Regul Mech 1861, 295–309. [DOI] [PubMed] [Google Scholar]
- Smith DJ, and Whitehouse I (2012). Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Jeong KS, Kitada T, and Grunstein M (2011). Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc Natl Acad Sci U S A 108, 12693–12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick TR, Allemand JF, Bensimon D, Bensimon A, and Croquette V (1996). The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- Strick TR, Croquette V, and Bensimon D (2000). Single-molecule analysis of DNA uncoiling by a type topoisomerase. Nature 404, 901–904. [DOI] [PubMed] [Google Scholar]
- Sundin O, and Varshavsky A (1981). Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell 25, 659–669. [DOI] [PubMed] [Google Scholar]
- Tapper DP, and DePamphilis ML (1978). Discontinuous DNA replication: accumulation of Simian virus 40 DNA at specific stages in its replication. J Mol Biol 120, 401–422. [DOI] [PubMed] [Google Scholar]
- Teves SS, and Henikoff S (2014). Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullsperger CJ, Vologodskii AV, and Cozzarelli NR (1995). Unlinking of DNA by Topoisomerases During DNA Replication In Nucleic Acids and Molecular Biology, Eckstein F, and Lilley MJ, eds. (Springer-Verlag Berlin Heidelberg; ), pp. 115–142. [Google Scholar]
- Valdes A, Coronel L, Martinez-Garcia B, Segura J, Dyson S, Diaz-Ingelmo O, Micheletti C, and Roca J (2019). Transcriptional supercoiling boosts topoisomerase II-mediated knotting of intracellular DNA. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A (2007). Monte Carlo simulation of DNA topological properties. Topology in Molecular Biology, 23–41. [Google Scholar]
- Wang MD, Yin H, Landick R, Gelles J, and Block SM (1997). Stretching DNA with optical tweezers. Biophys J 72, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, and Crick FH (1953). Genetical implications of the structure of deoxyribonucleic acid. Nature 171, 964–967. [DOI] [PubMed] [Google Scholar]
- Williams SP, Haggie PM, and Brindle KM (1997). 19F NMR measurements of the rotational mobility of proteins in vivo. Biophys J 72, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, and Ura K (1997). Transcription of dinucleosomal templates. Methods 12, 10–19. [DOI] [PubMed] [Google Scholar]
- Wu C, Read C, McGeehan J, and Crane-Robinson C (2016). The construction of customized nucleosomal arrays. Anal Biochem 496, 71–75. [DOI] [PubMed] [Google Scholar]
- Ye Y, Wu Q, Zheng W, Jiang B, Pielak GJ, Liu M, and Li C (2018). Quantification of size effect on protein rotational mobility in cells by (19)F NMR spectroscopy. Anal Bioanal Chem 410, 869–874. [DOI] [PubMed] [Google Scholar]
- Yu H, and Droge P (2014). Replication-induced supercoiling: a neglected DNA transaction regulator? Trends Biochem Sci 39, 219–220. [DOI] [PubMed] [Google Scholar]
- Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, Diffley JFX, and Costa A (2017). CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci U S A 114, 4141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.