Abstract
Introduction
Recent studies highlight the crucial regulatory roles of transposable elements (TEs) on proximal gene expression in distinct biological contexts such as disease and development. However, computational tools extracting potential TE –proximal gene expression associations from RNA-sequencing data are still missing.
Implementation
Herein, we developed a novel R package, using a linear regression model, for studying the potential influence of TE species on proximal gene expression from a given RNA-sequencing data set. Our R package, namely TEffectR, makes use of publicly available RepeatMasker TE and Ensembl gene annotations as well as several functions of other R-packages. It calculates total read counts of TEs from sorted and indexed genome aligned BAM files provided by the user, and determines statistically significant relations between TE expression and the transcription of nearby genes under diverse biological conditions.
Availability
TEffectR is freely available at https://github.com/karakulahg/TEffectR along with a handy tutorial as exemplified by the analysis of RNA-sequencing data including normal and tumour tissue specimens obtained from breast cancer patients.
Keywords: Transposable elements, Gene regulation, Gene expression, Regression, Linear model, R package
Introduction
Transposable elements (TEs) are DNA sequences that are able to translocate themselves along a host genome (Biemont & Vieira, 2006). They were discovered by Barbara McClintock in the 1950s in maize and defined for the first time as controlling elements on the action of nearby genes (McClintock, 1956). TEs constitute a considerable portion of most eukaryotic genomes (Kazazian Jr, 2004; Kelly & Leitch, 2011; Lander et al., 2001) and are divided into two main classes according to their transposition mechanism (Wicker et al., 2007). Class I elements (also known as retrotransposons) use RNA intermediates and a reverse transcriptase whereas Class II elements (also known as DNA transposons) act through DNA intermediates for their translocation (Wicker et al., 2007). These mechanisms are also called as “copy-and-paste” and “cut-and-paste” transpositions, respectively. In addition to acting as key players in genome size expansion and evolution (Kazazian Jr, 2004), previous studies highlighted the critical roles of TEs in distinct biological contexts, such as cancer (Hancks & Kazazian Jr, 2016; Johanning et al., 2017; Lee et al., 2012), embryonic development (Yandim & Karakulah, 2019b), senescence and aging (De Cecco et al., 2019), and stress response (Rech et al., 2019).
In parallel to the advent of next generation sequencing technologies, considerable attention has been paid to elucidate the regulatory activities of TEs on gene expression on a genome-wide scale. TEs are now recognized as the natural source of diverse regulatory sequences (Trizzino et al., 2017) including the promoters (Jordan et al., 2003), transcription factor binding sites (Bourque et al., 2008; Karakulah, 2018), enhancers (Chuong et al., 2013), and silencers (Bire et al., 2016) in the host genome. For example, MER39, a human long terminal repeat (LTR), acts as an endometrium-specific promoter and plays an essential role for the expression of the prolactin gene during pregnancy (Emera et al., 2012). Similarly, an MT-C retrotransposon-derived promoter is required to produce the oocyte-specific isoform of the Dicer gene in mice and its absence leads to female infertility (Flemr et al., 2013). It has also been reported in a comprehensive computational study that the majority of primate-specific regulatory sequences are originated from TEs (Jacques, Jeyakani & Bourque, 2013). In line with this, the influence of TEs on proximal gene expression was documented both in rat (Dong et al., 2017) and maize (Makarevitch et al., 2015). Furthermore, housekeeping genes were distinguished by their distinct repetitive DNA sequence environment (Eller et al., 2007). When it comes to understanding the links between TEs and proximal genes, it is postulated that TE intermediates (DNA or RNA) may interfere with the transcription of adjacent genes either directly or through recruited factors, and that an activated or repressed TE has the potential to modulate the chromatin environment of such genes and thereby influence their expression states (Elbarbary, Lucas & Maquat, 2016; Huda et al., 2009).
Despite the above-mentioned efforts on dissecting the influence of TEs on the expressions of proximal genes, a systematic and statistically valid approach is still missing, particularly due to the fact that TEs have many copies in the genome. In other words, it is challenging to link a particular TE in a specific location to a particular gene of interest. Still, a notable effort has been devoted to developing computational methods on the matter. Among these, two online tools, PlanTEnrichment (Karakulah & Suner, 2017) and GREAM (Chandrashekar, Dey & Acharya, 2015), allow their users to determine overrepresented TEs that are located adjacently of a given list of genes in plants and mammals, respectively. RTFAdb (Karakulah, 2018), using transcription factor binding profiles of the Encyclopedia of DNA Elements (ENCODE) project (The ENCODE Project Consortium’, 2012), can be utilized for exploring the regulatory roles of TEs. TETools (Lerat et al., 2017) and RepEnrich (Criscione et al., 2014) are popular computational tools to study differential expression of TEs under different biological conditions. Additionally, RepEnrich can help to provide insights into the transcriptional regulation of TEs by linking chromatin immunoprecipitation followed by sequencing (ChIP-seq) and expression profiling data sets. However, these tools do not allow one to directly link the expression of location specific TEs to a given proximal gene. Hence, we developed a novel R (https://www.r-project.org) package, using linear regression model (LM), for dissecting significant associations between TEs and proximal genes in a given RNA-sequencing (RNA-seq) data set. Our R package, namely TEffectR, makes use of publicly available RepeatMasker TE (http://www.repeatmasker.org) and Ensembl gene annotations (https://www.ensembl.org/index.html) and calculate total read counts of TEs from sorted and indexed genome aligned BAM files. Then, it predicts the influence of TE expression on the transcription of proximal genes under diverse biological conditions. In order to demonstrate the utility of TEffectR, we examined a publicly available RNA-seq data set collected from breast cancer patients. A detailed background of LM is also given in the following section.
Materials and Methods
Modeling gene expression with linear regression model
RNA-seq method yields count-type data rather than continuous measures of gene expression. Hence, generalized linear models (GLM) are used for modeling and statistical analysis of RNA-seq data sets, which are assumed to follow Poisson distribution or negative binomial distribution. In order to test differential gene expression, a number of analytical methods, including edgeR (Robinson, McCarthy & Smyth, 2010), and DESeq2 (Oshlack, Robinson & Young, 2010) use GLM where expression level of each gene is modeled as response variable while biological conditions (e.g., control vs experimental groups) are considered as explanatory variables or predictors. However, after the transformation of RNA-seq count data to log2-counts per million (logCPM) with Limma’s voom (Law et al., 2014) function, gene expression profiles can be ready for linear modelling.
LM has been used so far for modeling the regulatory effects of genetic and epigenetic factors on gene expression (Gerstung et al., 2015; Li, Liang & Zhang, 2014). For example, Li et al. developed RACER (Li, Liang & Zhang, 2014), using regression analysis approach, for exploring potential links between gene expression levels and a number of predictors, including DNA methylation level, copy number variation, transcription factor occupancy and microRNA expression level. Using a similar approach, TEffectR considers given biological conditions or covariates and TE expression levels as predictors to explain significant differences in gene abundances on a gene-by-gene basis. TEffectR assumes that each TE has a potential to influence the expression of a proximal gene. Accordingly, the expression level of a given gene can be modelled as follows:
-
•
where i denotes the genes, and Geneexpressioni represents the normalized log2(CPM) value of the ith gene.
-
•
TEni stands for the normalized log2(CPM) expression value of n th TE which is located within the upstream of the ith gene.
-
•
Covaritesmi indicates covariate effects in the model, such as tissue type, age, gender, etc.
Implementation of the TEffectR package
The TEffectR package was written in R language (v.3.5.3) and it uses the functions of diverse computational tools. To extract gene annotation data from Ensembl database and manipulation of RepeatMasker annotation files, TEffectR respectively utilizes biomaRt (Durinck et al., 2009) and biomartr (Drost & Paszkowski, 2017) packages. The GenomicRanges (Lawrence et al., 2013) tool is used to identify TE sequences that are located in the neighborhood of the gene list provided by users. For data and string manipulation steps of the TEffectR workflow, we made use of dplyr (https://dplyr.tidyverse.org/), rlist (https://renkun-ken.github.io/rlist/) and stringr (https://stringr.tidyverse.org/) packages. BEDtools (Quinlan, 2014) and Rsamtools (https://bioconductor.org/packages/release/bioc/html/Rsamtools.html) were employed for the quantification of TE-derived sequencing reads in a given list of indexed and genome aligned BAM files. Two popular differential gene expression analysis packages for RNA-seq data sets, edgeR and limma, were used for filtering, normalization and transformation of expression values of both genes and TEs. Statistical significance of each LM and covariate effect in the corresponding regression model were calculated with lm(), which is a built-in function of R.
Data collection and processing for case study
In order to demonstrate the usage of TEffectR package, we made use of a publicly available whole transcriptome sequencing dataset including normal and tumour tissue specimens obtained from 22 ER+/HER2- breast cancer patients (GEO Accession ID: GSE103001) (Wenric et al., 2017). These transcriptome libraries were particularly included as they were prepared without poly(A) selection method and thereby allowing the measurement of TE expression uniformly (Solovyov et al., 2018). We downloaded sequencing reads in FASTQ file format from Sequence Read Archive (Leinonen et al., 2011) (SRA Accession ID: SRP116023) using SRA Tool Kit v.2.9.0 with “fastq-dump –gzip –skip-technical –readids –dumpbase –clip –split-3” command. Next, sequencing reads were aligned to the human reference genome GRCh38 (Ensembl version 78) using the splice-aware aligner HISAT2 v2.1.0 (Kim, Langmead & Salzberg, 2015) with “hisat2 -p -dta -x {input.index} -1 {input_1.fq} -2 {input_2.fq} -S {out.sam}” parameters. Stringtie v1.3.5 (Pertea et al., 2015) were used with “stringtie -e -B -p -G {input.gtf} -A {output.tab} -o {output.gtf} -l {input.label}{input.bam}” parameters for expression quantification at the gene level. We considered only the uniquely mapped reads overlapping TE regions for the expression quantification of TEs. Multi-mapped reads could cause ambiguity when analysing the local effects of TEs on proximal genes as the repeats have many copies on the genome (Goerner-Potvin & Bourque, 2018; Treangen & Salzberg, 2011). To remove multi- and unmapped reads from BAM files, “samtools view -bq 60 -o {out.bam}{input.bam}” command was used. If the user would like to include the multi-mapped reads, they can skip this last command.
Results
Overview of the TEffectR package pipeline
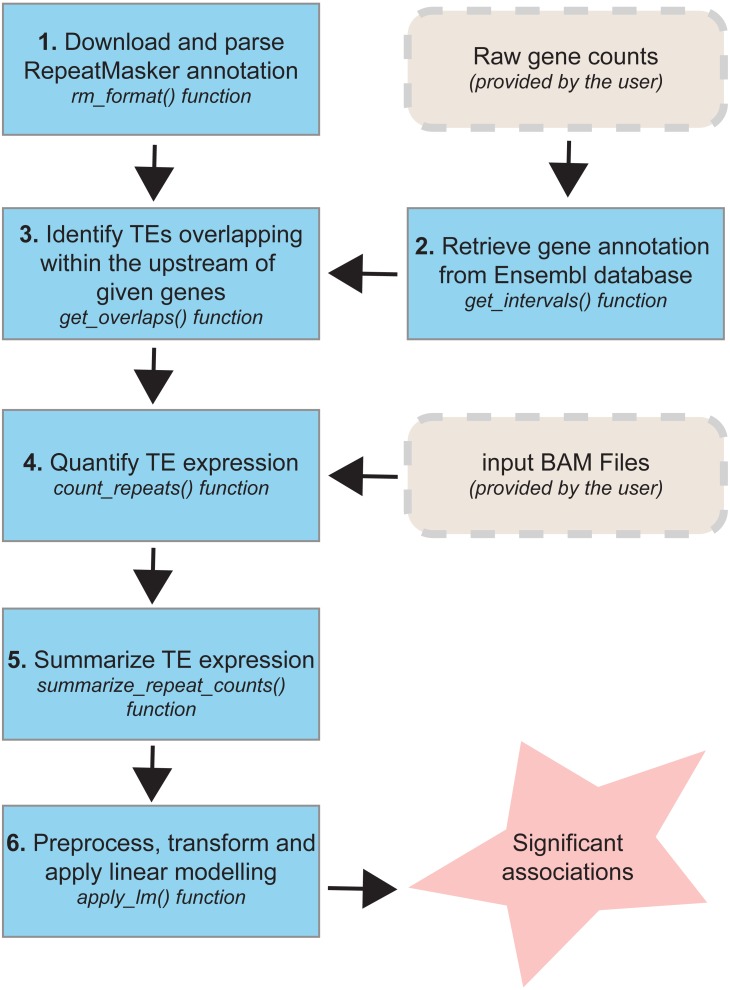
The TEffectR package includes a set of functions (Fig. 1) that allows the identification of significant associations between TEs and nearby genes for any species whose repeat annotation is publicly available at the RepeatMasker website (http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html). Currently, the complete annotation for over 60 species (from primates to nematodes) can be downloaded from this main repository and can be analyzed with our R package. The TEffectR package works initially by manipulating the repeat annotation file and make it ready for downstream analysis. In the following step, our tool takes a raw count matrix of RNA-seq dataset from the user where the first column includes the gene symbols or Ensembl IDs and the other columns contain count values of genes across samples. Then, TEffectR retrieves genomic position of each gene in the respective genome using the given count matrix. Afterwards, based on the user-defined parameters, TEffectR determines all TE species that are located within the upstream regions of each gene individually.
Figure 1. The workflow of TEffectR package.
The package contains six core functions for the identification of the potential links between TEs and nearby genes at genome-wide scale. TEffectR requires two inputs provided by the user: (i) a raw gene count matrix and (ii) genomic alignments of sequencing reads in BAM file format.
The TEffectR package contains a handy function for obtaining sequencing read counts, which are aligned to each TE region from a given list of sorted and indexed BAM files. Additionally, it can calculate the total read counts of each TE associated with a certain gene. In the following step, TEffectR merges all read count values of both genes and TEs into a single count matrix. This count matrix is then filtered, normalized with Trimmed Mean of M-values (TMM) method and transformed for linear modeling using voom() function of the limma package. In the final step, TEffectR fits a linear regression model with customized design matrix for each gene, and it calculates adjusted R-square values and significance of the model, and estimates the model parameters. The users can output the results of all calculations in tab separated values (tsv) file format to assess the contribution of each covariate (e.g., individual repeats) to the model.
Descriptions of the functions in the TEffectR package
The TEffectR package provides six unique functions for predicting the potential influence of TEs on the transcriptional activity of proximal genes in the respective genome:
TEffectR::rm_format: This function takes RepeatMasker annotation file as input and extracts the genomic location of each TE along with repeat class and family information. The output of rm_format() function is used while searching TEs that are located in the upstream region of the genes of interest.
TEffectR::get_intervals: This function is used to retrieve the genomic locations of all genes in a given read count matrix by the user. Row names of the expression matrix must be one of the following: (i) official gene symbol, (ii) Ensembl gene or (iii) transcript ID. The output of this function is utilized while determining distance between genes and TEs.
TEffectR::get_overlaps: Takes the genomic intervals of genes and TEs as input. Besides, the user also requires to provide three additional parameters: (i) the maximum distance allowed between the start sites of genes and TEs, (ii) whether genes and TEs must be located in same strand and (iii) TE family or subfamily name (e.g., SINE, LINE). This function helps to detect TEs that are localized upstream of the genes of interest. The “distance” parameter of this function could be determined by the user based on the interest of the TE localization. The user can either give a positive value, which would take the TEs localized in the upstream region of the gene; or a negative value that would correspond to the downstream of the gene. Moreover, the absolute distance is not limited; however, we used the value of “5000” to test our R package with the TEs located within the 5kb upstream regions of the genes, as previous studies confirmed that TEs located within 5kb upstream of genes provide binding sites for transcription factors and provide a possible link to nearby gene expression (Bourque et al., 2008; Nikitin et al., 2018). However, if the user aims to study long-range effects, then the distance could be set to a higher value.
TEffectR::count_repeats: This function returns a raw count matrix of the total number of reads originated from TE sequences. Only the reads exhibiting 100% overlap with given TE regions are considered and the user needs to specify individual path of each BAM file as input.
TEffectR:: summarize_repeat_counts: Takes the output of count_repeats() function as input. It is used to calculate the total number of sequencing reads derived from each TE that is located upstream of a certain gene.
TEffectR::apply_lm: This core function applies filtering (≥10 reads), TMM normalization, voom transformation and LM to the given raw count expression values, respectively. It takes four arguments: (i) raw gene counts, (ii) raw TE counts, (iii) a data frame containing user-defined covariates (e.g., tissue type, disease state), and (iv) the output of get_overlaps() function. When covariates are determined, one may include all the biological factors to see if they could explain the expression of the gene in conjunction with TE expression. However, one may as well only use the TE expression as the single predictor without the inclusion of further covariates.
The apply_lm() function returns three outputs: (i) a tsv file containing the p-value of each model, significance level of covariates and associated adjusted R squared values. The generated tab delimited file contains the list for the LM results of all genes that have at least one TE within the region of interest as given by this function. (ii) another tsv file containing log2(CPM) values of genes and TEs included in LM, and (iii) a group of diagnostic plots for each significant model (p < 0.05).
A case example of TEffectR analysis using the RNA-seq data obtained from healthy and tumor tissues of ER+/HER2- breast cancer patients
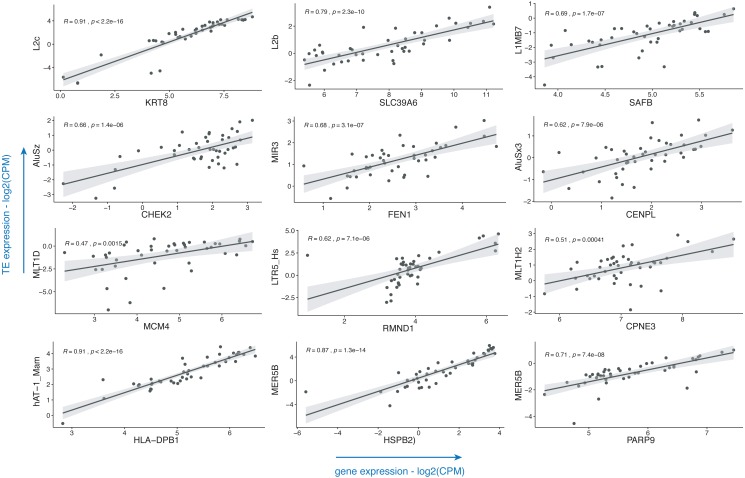
Breast cancer pathogenesis was associated with genomic instability (Kwei et al., 2010), which often presents itself with the aberrant expression of TEs (Aguilera & Garcia-Muse, 2013; Burns, 2017). TEs including LINE, SINE and LTR elements were already shown to be dysregulated in this disease (Yandım & Karakülah, 2019a; Bakshi et al., 2016; Bratthauer, Cardiff & Fanning, 1994; Johanning et al., 2017); with little or no information on the subtypes of these repeats. Also, there is a paucity of information on the impact of such dysregulatory events on the expressions of genes. To demonstrate the usage of the TEffectR package on a real case example, we ran the TEffectR package on the transcriptome RNA-seq data set of ER+/HER2- breast cancer patients. Table 1 summarizes a group of significant genes involved in breast cancer pathogenesis, diagnosis and prognosis (Abdel-Fatah et al., 2014; Chung et al., 2015; Dunning et al., 2016; Forero et al., 2016; Hammerich-Hille et al., 2010; Heinrich et al., 2010; Heo et al., 2013; Kasper et al., 2005; Kwok et al., 2015; Li et al., 2014; Storm et al., 1995; Tang et al., 2018; Tishchenko et al., 2016; Wei et al., 2011), where TEffectR presented a linear regression model that shows the associations between the expressions of these genes and that of the uniquely mapped TE sequences located within their upstream 5 kb flanking regions (Fig. 2). These genes were only given to present a contextual example and were selected based on breast cancer literature from the tab-delimited file that contains all genes with at least one TE in their upstream regions. The LM could explain the effect of these TEs on the variation in gene expression along with other covariates such as the type of tissue (i.e., healthy or tumor) or the individual patients. For example, two of the dependent variables; the LINE element “L2b” and “Tissue type” (p = 0.0013) could statistically significantly predict 70.30% of the expression of the “SLC39A6 (LIV-1)” gene whereas the covariate “patient” (p = 0.1024) could not explain the variation in this particular gene’s expression in this statistically significant model (p < 0.001). On the other hand, even though the expression of LTR5_Hs could predict 38.92% of “RMND1” expression statistically significantly (p < 0.001), neither the type of tissue (healthy or tumor; p = 0.4280) nor the individual patient (p = 0.1193) could explain the expression of this gene. From the perspective of a molecular biologist, these results may imply that SLC39A6 gene could potentially be involved in the tumorigenesis of the breast whereas this was not the case for RMND1, and the relevant repeat motif upstream of both genes could be suitable for further experimental investigation in terms of its potential to influence the expression of the proximal gene. These results may have implications on the biological roles of TEs (e.g., L2b) on breast cancer-related gene expressions (e.g., SLC39A6) and could indicate their potential roles in the carcinogenesis of the breast where the tissue type (healthy vs. tumor) p-value of the LM result is less than a significant threshold (i.e., p < 0.05).
Table 1. Examples of significant associations of LINE, SINE, LTR and DNA transposons with genes that were previously linked to breast cancer as TEffectR outputs along with multiple covariates.
Expression levels of TEs within the upstream 5 kb regions of the given genes and other covariates such as the tissue type (healthy vs. tumor) or patient number were included in the linear regression model. The p-value of the model indicates the significance of the linear model. P-values for each covariate indicate whether these factors have significant associations with the expression of the given gene. Adjusted r-square score indicates the precision of the model with significant covariate associations in terms of predicting the expression of the gene. For example, an adjusted R square of 0.8422 indicates that the linear model could explain 84.22% of the gene’s expression.
| Link to breast cancer | Gene name | TE name | r squared | Adjusted r-squared | Model p-value | Individual p-values |
|---|---|---|---|---|---|---|
| Biomarkera | KRT8 (CK8) | L2c (LINE) | 0.8532 | 0.8422 | 1.026E–16 | L2c: 1.356E–13 Tissue type: 0.0332 Patient: 0.7974 |
| Prognosisb | SLC39A6 (LIV-1) | L2b (LINE) |
0.7231 | 0.7023 | 3.100E–11 | L2b: 7.536E–08 Tissue type: 0.0013 Patient: 0.1024 |
| Molecular pathogenesisc | SAFB | L1MB7 (LINE) | 0.5131 | 0.4766 | 2.107E–06 | L1MB7: 1.114E–07 Tissue type: 0.6112 Patient: 0.1394 |
| Susceptibilitydand prognosise | CHEK2 | AluJb, AluSx AluS (SINE) |
0.6362 | 0.5883 | 1.645E–07 | AluJb: 0.0433 AluSx: 0.0426 AluSz: 0.0005 Tissue type: 0.0023 Patient: 0.0033 |
| Susceptibilityfand prognosisg | FEN1 | MIR3 (SINE) |
0.5545 | 0.5211 | 3.703E–07 | MIR3: 2.572E–06 Tissue type: 0.0122 Patient: 0.3886 |
| Molecular genetics and pathogenesish | CENPL | AluSx3, AluY (SINE) |
0.5489 | 0.5027 | 2.118E–06 | AluSx3: 0.0007 AluY: 0.2000 Tissue type: 0.0066 Patient: 0.2467 |
| Prognosisi | MCM4 | MLT1D (LTR) |
0.5733 | 0.5413 | 1.587E–07 | MLT1D: 0.0012 Tissue type: 1.544E–06 Patient: 0.1674 |
| Susceptibilityj | RMND1 | LTR5_Hs (LTR) |
0.4318 | 0.3892 | 4.279E–05 | LTR5_Hs: 1.782E–05 Tissue type: 0.4280 Patient: 0.1193 |
| Molecular pathogenesis and prognosisk | CPNE3 | MLT1H2 (LTR) |
0.3910 | 0.3453 | 0.0002 | MLT1H2: 0.0002 Tissue type: 0.0055 Patient: 0.9407 |
| Biomarker and prognosisl | HLA-DPB1 | hAT-1_Mam (DNA) |
0.8318 | 0.8192 | 1.548E–15 | hAT-1_Mam: 1.092E–14 Tissue type: 0.5467 Patient: 0.2850 |
| Molecular pathogenesismand biomarkern | HSPB2 (HSP27) | MER5B (DNA) |
0.7756 | 0.7587 | 4.791E–13 | MER5B: 8.050E–07 Tissue type: 0.5756 Patient: 0.1733 |
| Molecular pathogenesiso | PARP9 | MER5B (DNA) |
0.5929 | 0.5624 | 6.287E–08 | MER5B: 1.141E–06 Tissue type: 0.0054 Patient: 0.5464 |
Notes.
Figure 2. Scatter plots that demonstrate the correlations of normalized read counts of genes given in Table 1 with the normalized read counts of TEs present in their upstream 5-kb regions.
(CPM: counts per million).
Discussion
Repetitive DNA and its regulatory effects on chromatin environment and gene expression have been recognized well since the early years that follow the discovery of chromatin modifications (He et al., 2019; Huda et al., 2009; Martens et al., 2005). Dynamic expression patterns of TEs during distinct stages of human embryonic development (Garcia-Perez, Widmann & Adams, 2016; Grow et al., 2015; Yandim & Karakulah, 2019b), where the whole genome is tightly regulated in a highly orchestrated manner, and the power of TEs to modify gene expression patterns by various routes (Garcia-Perez, Widmann & Adams, 2016), highlight the importance of studying the links of TE expression with proximal gene transcription. TEffectR does not only provide a linear regression model between the expression of TEs and a gene in a given genomic location, but it also presents a platform to make this information traverse through biological contexts such as cancer, treatment, age, etc. The option of adding a desired number of covariates along with TEs that are present in a desired distance interval from a given gene allows one to study the associations between TEs and genes along with multiple factors. Substantial studies suggest that some human gene promoters are derived from TEs (Cohen, Lock & Mager, 2009) and that some TEs could act as distal enhancers (Kunarso et al., 2010). Still, it should be noted that significant associations documented via TEffectR do not necessarily mean that a given TE indeed has an influential effect on the transcription of the proximal gene. Conversely, transcriptional activation of a given gene could also influence the expression of the nearby TE, and the TE might not have an effect on gene expression at all. This is why functional experiments should always be performed to clearly answer crucial biological questions regarding this matter, where TEffectR acts as a useful guideline to point out significant associations.
Conclusion
The highly complex interactions among the regulatory networks of the genome are at the center of attention of many areas of molecular biology, developmental biology and epigenetics. Here, we present TEffectR, an R package, which elaborately dissects the associations between the expressions of genes and the transposable elements nearby them in a unified linear regression model. The inclusion of a desired number of factors as covariates allows a biologist to study such associations in a broader context.
Funding Statement
The authors received no funding for this work.
Contributor Information
Gökhan Karakülah, Email: gokhan.karakulah@deu.edu.tr.
Aslı Suner, Email: asli.suner@ege.edu.tr.
Additional Information and Declarations
Competing Interests
Gökhan Karakülah and Aslı Suner are Academic Editors for PeerJ.
Author Contributions
Gökhan Karakülah conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Nazmiye Arslan conceived and designed the experiments, performed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Cihangir Yandım conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Aslı Suner conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The analysis pipeline is available at https://github.com/karakulahg/TEffectR.
References
- Abdel-Fatah et al. (2014).Abdel-Fatah TM, Russell R, Albarakati N, Maloney DJ, Dorjsuren D, Rueda OM, Moseley P, Mohan V, Sun H, Abbotts R, Mukherjee A, Agarwal D, Illuzzi JL, Jadhav A, Simeonov A, Ball G, Chan S, Caldas C, Ellis IO, Wilson 3rd DM, Madhusudan S. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Molecular Oncology. 2014;8:1326–1338. doi: 10.1016/j.molonc.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera & Garcia-Muse (2013).Aguilera A, Garcia-Muse T. Causes of genome instability. Annual Review of Genetics. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Bakshi et al. (2016).Bakshi A, Herke SW, Batzer MA, Kim J. DNA methylation variation of human-specific Alu repeats. Epigenetics. 2016;11:163–173. doi: 10.1080/15592294.2015.1130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont & Vieira (2006).Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bire et al. (2016).Bire S, Casteret S, Piegu B, Beauclair L, Moire N, Arensbuger P, Bigot Y. Mariner transposons contain a silencer: possible role of the polycomb repressive complex 2. PLOS Genetics. 2016;12:e1005902. doi: 10.1371/journal.pgen.1005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque et al. (2008).Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Research. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratthauer, Cardiff & Fanning (1994).Bratthauer GL, Cardiff RD, Fanning TG. Expression of LINE-1 retrotransposons in human breast cancer. Cancer. 1994;73:2333–2336. doi: 10.1002/1097-0142(19940501)73:9<2333::aid-cncr2820730915>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Burns (2017).Burns KH. Transposable elements in cancer. Nature Reviews Cancer. 2017;17:415–424. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- Chandrashekar, Dey & Acharya (2015).Chandrashekar DS, Dey P, Acharya KK. GREAM: a web server to short-list potentially important genomic repeat elements based on over-/under-representation in specific chromosomal locations, such as the gene neighborhoods, within or across 17 mammalian species. PLOS ONE. 2015;10:e0133647. doi: 10.1371/journal.pone.0133647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung et al. (2015).Chung L, Onyango D, Guo Z, Jia P, Dai H, Liu S, Zhou M, Lin W, Pang I, Li H, Yuan YC, Huang Q, Zheng L, Lopes J, Nicolas A, Chai W, Raz D, Reckamp KL, Shen B. The FEN1 E359K germline mutation disrupts the FEN1-WRN interaction and FEN1 GEN activity, causing aneuploidy-associated cancers. Oncogene. 2015;34:902–911. doi: 10.1038/onc.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong et al. (2013).Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nature Genetics. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, Lock & Mager (2009).Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Criscione et al. (2014).Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco et al. (2019).De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, Le O, Beausejour C, Ambati J, Ambati K, Simon M, Seluanov A, Gorbunova V, Slagboom PE, Helfand SL, Neretti N, Sedivy JM. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al. (2017).Dong Y, Huang Z, Kuang Q, Wen Z, Liu Z, Li Y, Yang Y, Li M. Expression dynamics and relations with nearby genes of rat transposable elements across 11 organs, 4 developmental stages and both sexes. BMC Genomics. 2017;18:666. doi: 10.1186/s12864-017-4078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost & Paszkowski (2017).Drost HG, Paszkowski J. Biomartr: genomic data retrieval with R. Bioinformatics. 2017;33:1216–1217. doi: 10.1093/bioinformatics/btw821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning et al. (2016).Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, Healey CS, Kar S, Pooley KA, Lopez-Knowles E, Dicks E, Barrowdale D, Sinnott-Armstrong NA, Sallari RC, Hillman KM, Kaufmann S, Sivakumaran H, Moradi Marjaneh M, Lee JS, Hills M, Jarosz M, Drury S, Canisius S, Bolla MK, Dennis J, Wang Q, Hopper JL, Southey MC, Broeks A, Schmidt MK, Lophatananon A, Muir K, Beckmann MW, Fasching PA, Dos-Santos-Silva I, Peto J, Sawyer EJ, Tomlinson I, Burwinkel B, Marme F, Guenel P, Truong T, Bojesen SE, Flyger H, Gonzalez-Neira A, Perez JI, Anton-Culver H, Eunjung L, Arndt V, Brenner H, Meindl A, Schmutzler RK, Brauch H, Hamann U, Aittomaki K, Blomqvist C, Ito H, Matsuo K, Bogdanova N, Dork T, Lindblom A, Margolin S, Kosma VM, Mannermaa A, Tseng CC, Wu AH, Lambrechts D, Wildiers H, Chang-Claude J, Rudolph A, Peterlongo P, Radice P, Olson JE, Giles GG, Milne RL, Haiman CA, Henderson BE, Goldberg MS, Teo SH, Yip CH, Nord S, Borresen-Dale AL, Kristensen V, Long J, Zheng W, Pylkas K, Winqvist R, Andrulis IL, Knight JA, Devilee P, Seynaeve C, Figueroa J, Sherman ME, Czene K, Darabi H, Hollestelle A, Van den Ouweland AM, Humphreys K, Gao YT, Shu XO, Cox A, Cross SS, Blot W, Cai Q, Ghoussaini M, Perkins BJ, Shah M, Choi JY, Kang D, Lee SC, Hartman M, Kabisch M, Torres D, Jakubowska A, Lubinski J, Brennan P, Sangrajrang S, Ambrosone CB, Toland AE, Shen CY, Wu PE, Orr N, Swerdlow A, McGuffog L, Healey S, Lee A, Kapuscinski M, John EM, Terry MB, Daly MB, Goldgar DE, Buys SS, Janavicius R, Tihomirova L, Tung N, Dorfling CM, Van Rensburg EJ, Neuhausen SL, Ejlertsen B, Hansen TV, Osorio A, Benitez J, Rando R, Weitzel JN, Bonanni B, Peissel B, Manoukian S, Papi L, Ottini L, Konstantopoulou I, Apostolou P, Garber J, Rashid MU, Frost D, Embrace, Izatt L, Ellis S, Godwin AK, Arnold N, Niederacher D, Rhiem K, Bogdanova-Markov N, Sagne C, Stoppa-Lyonnet D, Damiola F, Collaborators GS. Sinilnikova OM, Mazoyer S, Isaacs C, Claes KB, De Leeneer K, De la Hoya M, Caldes T, Nevanlinna H, Khan S, Mensenkamp AR, Hebon, Hooning MJ, Rookus MA, Kwong A, Olah E, Diez O, Brunet J, Pujana MA, Gronwald J, Huzarski T, Barkardottir RB, Laframboise R, Soucy P, Montagna M, Agata S, Teixeira MR, kConFab I, Park SK, Lindor N, Couch FJ, Tischkowitz M, Foretova L, Vijai J, Offit K, Singer CF, Rappaport C, Phelan CM, Greene MH, Mai PL, Rennert G, Imyanitov EN, Hulick PJ, Phillips KA, Piedmonte M, Mulligan AM, Glendon G, Bojesen A, Thomassen M, Caligo MA, Yoon SY, Friedman E, Laitman Y, Borg A, Von Wachenfeldt A, Ehrencrona H, Rantala J, Olopade OI, Ganz PA, Nussbaum RL, Gayther SA, Nathanson KL, Domchek SM, Arun BK, Mitchell G, Karlan BY, Lester J, Maskarinec G, Woolcott C, Scott C, Stone J, Apicella C, Tamimi R, Luben R, Khaw KT, Helland A, Haakensen V, Dowsett M, Pharoah PD, Simard J, Hall P, Garcia-Closas M, Vachon C, Chenevix-Trench G, Antoniou AC, Easton DF, Edwards SL. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1 RMND1 and CCDC170. Nature Genetics. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck et al. (2009).Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary, Lucas & Maquat (2016).Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351(6274):aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller et al. (2007).Eller CD, Regelson M, Merriman B, Nelson S, Horvath S, Marahrens Y. Repetitive sequence environment distinguishes housekeeping genes. Gene. 2007;390:153–165. doi: 10.1016/j.gene.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera et al. (2012).Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Molecular Biology and Evolution. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- Flemr et al. (2013).Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Forero et al. (2016).Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, Downs-Kelly E, Burwell TC, Vaklavas C, Buchsbaum DJ, Myers RM, LoBuglio AF, Varley KE. Expression of the MHC Class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunology Research. 2016;4:390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez, Widmann & Adams (2016).Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development. 2016;143:4101–4114. doi: 10.1242/dev.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung et al. (2015).Gerstung M, Pellagatti A, Malcovati L, Giagounidis A, Porta MG, Jadersten M, Dolatshad H, Verma A, Cross NC, Vyas P, Killick S, Hellstrom-Lindberg E, Cazzola M, Papaemmanuil E, Campbell PJ, Boultwood J. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nature Communications. 2015;6:5901. doi: 10.1038/ncomms6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerner-Potvin & Bourque (2018).Goerner-Potvin P, Bourque G. Computational tools to unmask transposable elements. Nature Reviews Genetics. 2018;19:688–704. doi: 10.1038/s41576-018-0050-x. [DOI] [PubMed] [Google Scholar]
- Grow et al. (2015).Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RA, Wysocka J. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerich-Hille et al. (2010).Hammerich-Hille S, Kaipparettu BA, Tsimelzon A, Creighton CJ, Jiang S, Polo JM, Melnick A, Meyer R, Oesterreich S. SAFB1 mediates repression of immune regulators and apoptotic genes in breast cancer cells. Journal of Biological Chemistry. 2010;285:3608–3616. doi: 10.1074/jbc.M109.066431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks & Kazazian Jr (2016).Hancks DC, Kazazian Jr HH. Roles for retrotransposon insertions in human disease. Mobile DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2019).He J, Fu X, Zhang M, He F, Li W, Abdul MM, Zhou J, Sun L, Chang C, Li Y, Liu H, Wu K, Babarinde IA, Zhuang Q, Loh YH, Chen J, Esteban MA, Hutchins AP. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nature Communications. 2019;10:34. doi: 10.1038/s41467-018-08006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich et al. (2010).Heinrich C, Keller C, Boulay A, Vecchi M, Bianchi M, Sack R, Lienhard S, Duss S, Hofsteenge J, Hynes NE. Copine-III interacts with ErbB2 and promotes tumor cell migration. Oncogene. 2010;29:1598–1610. doi: 10.1038/onc.2009.456. [DOI] [PubMed] [Google Scholar]
- Heo et al. (2013).Heo CK, Hwang HM, Ruem A, Yu DY, Lee JY, Yoo JS, Kim IG, Yoo HS, Oh S, Ko JH, Cho EW. Identification of a mimotope for circulating anti-cytokeratin 8/18 antibody and its usage for the diagnosis of breast cancer. International Journal of Oncology. 2013;42:65–74. doi: 10.3892/ijo.2012.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda et al. (2009).Huda A, Marino-Ramirez L, Landsman D, Jordan IK. Repetitive DNA elements, nucleosome binding and human gene expression. Gene. 2009;436:12–22. doi: 10.1016/j.gene.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, Jeyakani & Bourque (2013).Jacques PE, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLOS Genetics. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanning et al. (2017).Johanning GL, Malouf GG, Zheng X, Esteva FJ, Weinstein JN, Wang-Johanning F, Su X. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Scientific Reports. 2017;7:41960. doi: 10.1038/srep41960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan et al. (2003).Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends in Genetics. 2003;19:68–72. doi: 10.1016/S0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Karakulah (2018).Karakulah G. RTFAdb: a database of computationally predicted associations between retrotransposons and transcription factors in the human and mouse genomes. Genomics. 2018;110:257–262. doi: 10.1016/j.ygeno.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Karakulah & Suner (2017).Karakulah G, Suner A. PlanTEnrichment: a tool for enrichment analysis of transposable elements in plants. Genomics. 2017;109:336–340. doi: 10.1016/j.ygeno.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Kasper et al. (2005).Kasper G, Weiser AA, Rump A, Sparbier K, Dahl E, Hartmann A, Wild P, Schwidetzky U, Castanos-Velez E, Lehmann K. Expression levels of the putative zinc transporter LIV-1 are associated with a better outcome of breast cancer patients. International Journal of Cancer. 2005;117:961–973. doi: 10.1002/ijc.21235. [DOI] [PubMed] [Google Scholar]
- Kazazian Jr (2004).Kazazian Jr HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kelly & Leitch (2011).Kelly LJ, Leitch IJ. Exploring giant plant genomes with next-generation sequencing technology. Chromosome Research. 2011;19:939–953. doi: 10.1007/s10577-011-9246-z. [DOI] [PubMed] [Google Scholar]
- Kim, Langmead & Salzberg (2015).Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso et al. (2010).Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nature Genetics. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Kwei et al. (2010).Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Molecular Oncology. 2010;4:255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok et al. (2015).Kwok HF, Zhang SD, McCrudden CM, Yuen HF, Ting KP, Wen Q, Khoo US, Chan KY. Prognostic significance of minichromosome maintenance proteins in breast cancer. American Journal of Cancer Research. 2015;5:52–71. [PMC free article] [PubMed] [Google Scholar]
- Lander et al. (2001).Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, De la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, De Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Law et al. (2014).Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence et al. (2013).Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLOS Computational Biology. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2012).Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette 3rd LJ, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ, Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen et al. (2011).Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Research. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat et al. (2017).Lerat E, Fablet M, Modolo L, Lopez-Maestre H, Vieira C. TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Research. 2017;45:e17. doi: 10.1093/nar/gkw953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2014).Li C, Bai J, Hao X, Zhang S, Hu Y, Zhang X, Yuan W, Hu L, Cheng T, Zetterberg A, Lee MH, Zhang J. Multi-gene fluorescence in situ hybridization to detect cell cycle gene copy number aberrations in young breast cancer patients. Cell Cycle. 2014;13:1299–1305. doi: 10.4161/cc.28201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Liang & Zhang (2014).Li Y, Liang M, Zhang Z. Regression analysis of combined gene expression regulation in acute myeloid leukemia. PLOS Computational Biology. 2014;10:e1003908. doi: 10.1371/journal.pcbi.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch et al. (2015).Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLOS Genetics. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens et al. (2005).Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO Journal. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock (1956).McClintock B. Controlling elements and the gene. Cold Spring Harbor Symposia on Quantitative Biology. 1956;21:197–216. doi: 10.1101/SQB.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Nagel et al. (2012).Nagel JH, Peeters JK, Smid M, Sieuwerts AM, Wasielewski M, De Weerd V, Trapman-Jansen AM, Van den Ouweland A, Brüggenwirth H, Van I Jcken WF, Klijn JG, Van der Spek PJ, Foekens JA, Martens JW, Schutte M, Meijers-Heijboer H. Gene expression profiling assigns CHEK2 1100delC breast cancers to the luminal intrinsic subtypes. Breast Cancer Research and Treatment. 2012;132(2):439–448. doi: 10.1007/s10549-011-1588-x. [DOI] [PubMed] [Google Scholar]
- Nikitin et al. (2018).Nikitin D, Penzar D, Garazha A, Sorokin M, Tkachev V, Borisov N, Poltorak A, Prassolov V, Buzdin AA. Profiling of human molecular pathways affected by retrotransposons at the level of regulation by transcription factor proteins. Frontiers in Immunology. 2018;9:30. doi: 10.3389/fimmu.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack, Robinson & Young (2010).Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biology. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea et al. (2015).Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan (2014).Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Current Protocols in Bioinformatics. 2014;47:11–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech et al. (2019).Rech GE, Bogaerts-Marquez M, Barron MG, Merenciano M, Villanueva-Canas JL, Horvath V, Fiston-Lavier AS, Luyten I, Venkataram S, Quesneville H, Petrov DA, Gonzalez J. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLOS Genetics. 2019;15:e1007900. doi: 10.1371/journal.pgen.1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, McCarthy & Smyth (2010).Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyov et al. (2018).Solovyov A, Vabret N, Arora KS, Snyder A, Funt SA, Bajorin DF, Rosenberg JE, Bhardwaj N, Ting DT, Greenbaum BD. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Reports. 2018;23:512–521. doi: 10.1016/j.celrep.2018.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm et al. (1995).Storm FK, Gilchrist KW, Warner TF, Mahvi DM. Distribution of Hsp-27 and HER-2/neu in in situ and invasive ductal breast carcinomas. Annals of Surgical Oncology. 1995;2:43–48. doi: 10.1007/BF02303701. [DOI] [PubMed] [Google Scholar]
- Tang et al. (2018).Tang X, Zhang H, Long Y, Hua H, Jiang Y, Jing J. PARP9 is overexpressed in human breast cancer and promotes cancer cell migration. Oncology Letters. 2018;16:4073–4077. doi: 10.3892/ol.2018.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium’ (2012).The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishchenko et al. (2016).Tishchenko I, Milioli HH, Riveros C, Moscato P. Extensive transcriptomic and genomic analysis provides new insights about luminal breast cancers. PLOS ONE. 2016;11:e0158259. doi: 10.1371/journal.pone.0158259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen & Salzberg (2011).Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature Reviews Genetics. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino et al. (2017).Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. Transposable elements are the primary source of novelty in primate gene regulation. Genome Research. 2017;27:1623–1633. doi: 10.1101/gr.218149.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2011).Wei L, Liu TT, Wang HH, Hong HM, Yu AL, Feng HP, Chang WW. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-kappaB. Breast Cancer Research. 2011;13:R101. doi: 10.1186/bcr3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenric et al. (2017).Wenric S, ElGuendi S, Caberg JH, Bezzaou W, Fasquelle C, Charloteaux B, Karim L, Hennuy B, Freres P, Collignon J, Boukerroucha M, Schroeder H, Olivier F, Jossa V, Jerusalem G, Josse C, Bours V. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Scientific Reports. 2017;7:17452. doi: 10.1038/s41598-017-17811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker et al. (2007).Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yandım & Karakülah (2019a).Yandım C, Karakülah G. Dysregulated expression of repetitive DNA in ER+/HER2- breast cancer. Cancer Genetics. 2019a;239:36–45. doi: 10.1016/j.cancergen.2019.09.002. [DOI] [PubMed] [Google Scholar]
- Yandim & Karakulah (2019b).Yandim C, Karakulah G. Expression dynamics of repetitive DNA in early human embryonic development. BMC Genomics. 2019b;20:439. doi: 10.1186/s12864-019-5803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
The analysis pipeline is available at https://github.com/karakulahg/TEffectR.