Abstract
Local adaptation is assumed to occur when populations differ in a phenotypic trait or a set of traits, and such variation has a genetic basis. Here, we introduce Arabidopsis halleri and its life history as a perennial model system to study population differentiation and local adaptation. Studies on altitudinal adaptation have been conducted in two regions: Mt. Ibuki in Japan and the European Alps. Several studies have demonstrated altitudinal adaptation in ultraviolet-B (UV-B) tolerance, leaf water repellency against spring frost and anti-herbivore defences. Studies on population differentiation in A. halleri have also focused on metal hyperaccumulation and tolerance to heavy metal contamination. In these study systems, genome scans to identify candidate genes under selection have been applied. Lastly, we briefly discuss how RNA-Seq can broaden phenotypic space and serve as a link to underlying mechanisms. In conclusion, A. halleri provides us with opportunities to study population differentiation and local adaptation, and relate these to the genetic systems underlying target functional traits.
Keywords: Altitudinal adaptation, clonal propagation, common garden experiment, genome scan, life history, metalliferous habitat, perennial, RNA-Seq
One of the most fascinating observations on plants in natural habitats is the spatial co-variation between functional traits and natural environments. Here, we introduce a small perennial clonal herb, Arabidopsis halleri and its life history as a model system to study local adaptation along environmental gradients. Many studies using this species have addressed altitudinal adaptation in diverse functional traits and population differentiation in response to heavy metal contamination in soils. The species provides us with opportunities to study population differentiation and local adaptation, and relate these to the genetic systems underlying target functional traits using recent molecular tools.
Introduction
One of the most fascinating observations on plants in natural habitats is the spatial co-variation between functional traits and natural environments. In particular, when the phenotypic difference has a genetic basis, we expect that the spatial variation in selection might have shaped the co-variation between phenotype and environment. Population differentiation refers to genetic differentiation between populations in a trait or a set of traits (Linhart and Grant 1996). Most ecological traits show a certain level of phenotypic plasticity, and a growth experiment under common conditions (a common garden experiment) is required to visualize and quantify genetic-based phenotypic difference between populations (Savolainen et al. 2013). Local adaptation refers to the population differentiation that is attributable to adaptation to local biotic and abiotic environments. To confirm whether the observed population differentiation is the result of local adaptation, the presence of home-site advantage needs to be established by performing a reciprocal transplant experiment; plants from a particular local population have a higher fitness than plants from another population in the original habitat (Kawecki and Ebert 2004).
There is widespread evidence of local adaptation in plants, and there are many variations in the combination of phenotypic traits and environmental factors (Kawecki and Ebert 2004; Savolainen et al. 2013). However, in many cases, the underlying genetic mechanisms of local adaptation are unknown. Recently, using natural populations and natural accessions of model species for molecular biology and genetics, such as Arabidopsis thaliana, researchers have applied advanced techniques to dissect adaptation genetically (Savolainen et al. 2013; Weigel and Nordborg 2015). Genome-wide association studies (GWAS) have identified the genes involved in adaptation to biotic and abiotic environments in A. thaliana (reviewed in Bamba et al. 2019). For example, genome-wide single-nucleotide polymorphisms (SNPs) have been investigated for associations with the environmental conditions of the source sites (Hancock et al. 2011) and with fitness measures in a common garden experiment (Fournier-Level et al. 2011). The patterns of local adaptation in A. thaliana allowed one to interpret co-variation of multiple traits as a set of life history strategies (Takou et al. 2019), such as competitor, stress-tolerator and ruderal (CSR) strategies (Grime et al. 1988). Another advanced tool that has recently been used in ecological studies is whole-genome measures of gene expression (transcriptome) analysis with microarray and RNA-Seq (Richards et al. 2012; Alvarez et al. 2015). Variation in gene expression can be treated as phenotypes that have been analysed in conventional studies of population differentiation and local adaptation. Heritable variation in gene expression can be a target of natural selection, and differentiation in gene expression between populations can be shaped by both adaptive and non-adaptive processes (Oleksiak et al. 2002). Therefore, detection of the heritable structures of gene expression variation within and between populations will provide a clue to the underlying genetic mechanisms of local adaptation (Gould et al. 2018).
The combination of target traits and environmental factors involved in local adaptation are specific to life history and the habitat of the target species. Therefore, species in the genus Arabidopsis, and other closely related species, widen the opportunities for studying diverse traits under selection (Clauss and Koch 2006). The entire genus is now becoming a model system, which allows us to elucidate adaptation with unique combinations of traits and environments, which cannot be studied in A. thaliana (Hohmann et al. 2014; Yant and Bomblies 2017; Koch 2019). There have been several studies in which Arabidopsis relatives were used as a model system to study population differentiation and local adaptation (Turner et al. 2010; Leinonen et al. 2013). The life history of A. thaliana as an annual plant is characterized by a short lifespan, self-compatible flowers and numerous small seeds, and these characteristics are advantageous in a laboratory model plant. Perennial life history, however, will elucidate the adaptation in some important aspects of plant life history, such as outcrossing breeding systems, plant–pollinator interactions, allocation to reproductive and vegetative growth and clonal propagation. Furthermore, annual plants are rare in some habitats, often those with low productivity, e.g. alpine, high-latitude and forest floor habitats. Therefore, adaptation to these habitats requires a study system involving perennial plants.
In this review, we introduced A. halleri (L.) O'Kane & Al-Shehbaz as a perennial model system to study population differentiation and local adaptation, amongst which adaptation to altitudinal and edaphic environmental gradients has extensively been studied. This species is characterized by extensive clonal propagation through aerial rosette formation and an outcrossing breeding system by insect pollination. The plant bears leaves all year round, and vegetative parts are exposed to diverse biotic and abiotic stresses. The combination of clonal reproduction and year-round foliage allowed us to repeatedly sample leaves from the same individuals under natural conditions, and has enhanced repeated transcriptome analyses under natural conditions using RNA-Seq. We will highlight a series of RNA-Seq studies using A. halleri, and discuss the future applications of transcriptome analysis to studies of population differentiation and local adaptation.
Arabidopsis halleri
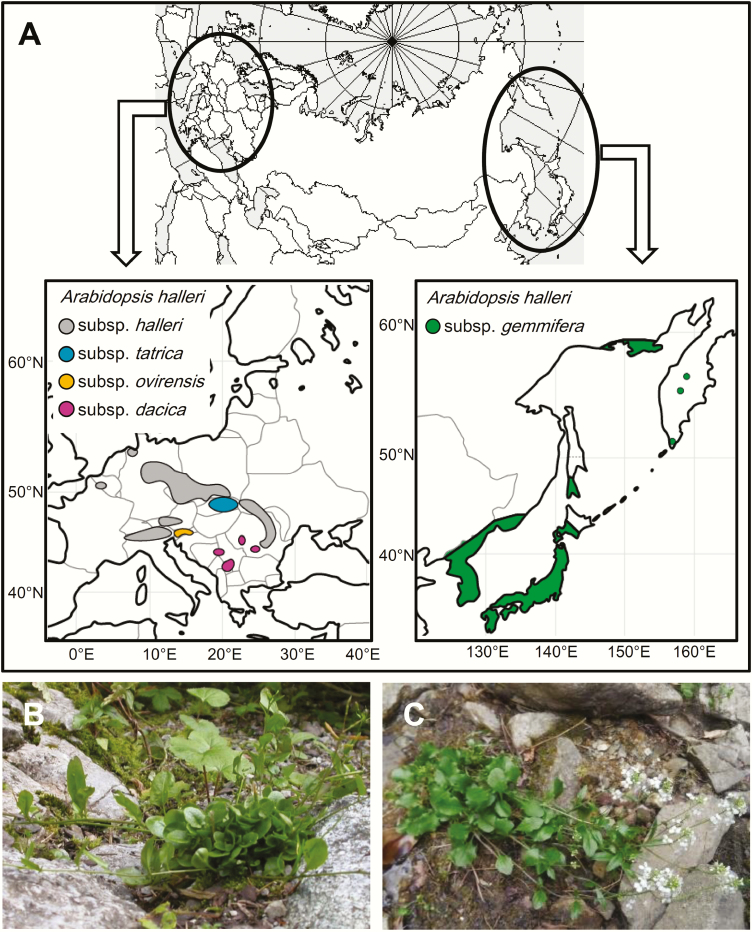
Arabidopsis halleri is found in Northern Hemisphere, showing clear disjunction into two part of Eurasia, i.e. East Asia and Europe (Fig. 1A). Four subspecies have been recognized in Europe; A. halleri subsp. halleri, A. halleri subsp. tatrica (Pawł.) Kolník (Fig. 1B), A. halleri subsp. ovirensis (Wulfen) O'Kane & Al-Shehbaz, A. halleri subsp. dacica (Heuff.) Kolník (Kolník and Marhold 2006; Hohmann et al. 2014; Šrámková-Fuxová et al. 2017). The eastern distribution is represented by a single subspecies, A. halleri subsp. gemmifera (Matsum.) O'Kane & Al-Shehbaz (Fig. 1C), which occurs in Japan, Korea, north-eastern China and the Russian Far East, including Sakhalin and Kamchatka (Kudoh et al. 2018). All subspecies of A. halleri have been reported to be diploid with a basic chromosome number = 8 (2x = 2N = 16; Al-Shehbaz and O’Kane 2002; Kolník and Marhold 2006). The genome size is estimated to be 250 Mb (Briskine et al. 2017). The genome assembly has been published and updated (Akama et al. 2014; Briskine et al. 2017), and 32 553 protein-coding genes were estimated, which is similar to the 32 670 genes of A. lyrata (L.) O'Kane & Al-Shehbaz (Hu et al. 2011) and >28 775 genes of A. thaliana (The Arabidopsis Genome Initiative 2000; Briskine et al. 2017). The divergence time from A. thaliana has been estimated as 5–18 million years (Koch et al. 2000; Ossowski et al. 2010), and 0.34–2.5 million years with the more closely related species A. lyrata (Castric et al. 2008; Roux et al. 2011).
Figure 1.
Disjunct distributions of five subspecies of Arabidopsis halleri (A). Photographs of A. halleri subsp. tatrica in Belianske Tatry, Prešovský kraj, Slovakia (B), and A. halleri subsp. gemmifera in Monzen, Taka-cho, Hyogo Prefecture, Japan (C).
Life History
Comparisons of life history traits between A. halleri, A. lyrata, A. arenosa and A. thaliana highlight characteristics of each species (Table 1). Although A. halleri is mostly known for its heavy metal hyperaccumulation (Verbruggen et al. 2009; Krämer 2010; Stein et al. 2017), the species exhibits specific characteristics in its life history different from A. thaliana and other species in the genus (Table 1). Arabidopsis halleri, A. lyrata and A. arenosa are all perennial, but A. halleri is characterized by clonal reproduction via aerial rosette formation (Table 1). Arabidopsis lyrata has been used as a model system to study self-incompatibility (e.g. Kusaba et al. 2001; Schierup et al. 2001, 2008). Arabidopsis arenosa is often used in the study of hybrid incompatibility, because the cross between A. arenosa and A. thaliana results in viable but infertile F1 hybrids (e.g. Burkart-Waco et al. 2012), and the one tetraploid species in the genus, A. suecica, has shown to be an allotetraploid between A. arenosa and A. thaliana (O’Kane et al. 1996). The interspecies comparison of phenotypes, genomes and transcriptome data provides opportunities to elucidate underlying genetic mechanisms of adaptive traits (Clauss and Koch 2006; Baduel et al. 2016; Nikolov et al. 2019).
Table 1.
Comparisons of Arabidopsis halleri and related species, A. lyrata, A. arenosa and A. thaliana, in chromosome number, ploidy level, genome size, number of genes, distribution and reproductive and vegetative life histories. NA indicates data not available.
| Arabidopsis halleri | A. lyrata | A. arenosa | A. thaliana | |
|---|---|---|---|---|
| Chromosome number | 2n = 16 | 2n = 16 | 2n = 16 2n = 32 | 2n = 10 |
| Ploidy level | 2x | 2x | 2x, 4x | 2x |
| Genome size | 250 Mbp | 230 Mbp | NA | 125 Mbp |
| Number of genes | 32 553 | 32 670 | NA | 28 775 |
| Original distribution | Europe, East Asia, Far East Russia | Northern Europe, Arctic and Far East Russia, Alaska, Canada | Central Europe | Europe, Central Asia, North Africa |
| Life cycle | Perennial | Perennial | Perennial, occasionally annual | Annual |
| Vegetative phenology | Evergreen | Evergreen | Evergreen, winter-green | Winter-green, summer-green |
| Reproductive phenology | Spring | Spring | Spring | Spring-summer |
| Breeding system | Obligatory outbreeding | Outbreeding | Obligatory outbreeding | Predominantly inbreeding |
| Flower size | Petal length = 4–6.5 mm | Petal length = 3–8 mm | Petal length = 5–8 mm | Petal length = 2–3.5 mm |
| Self-compatibility | Incompatible | Incompatible | Compatible, incompatible | Compatible |
| Clonal propagation | Formation of aerial rosettes at shoot apical and lateral meristems | Rosettes formation by tillering | Rosettes formation by tillering | None |
Perennial life cycle
The perennial and evergreen life history of A. halleri contrasts with the annual life history of A. thaliana. Leaves can be produced all year round even in low temperatures during winter (Kudoh et al. 2018). Exposure to direct sunlight in midday during winter increases leaf temperatures to the level required for photosynthesis, which is higher than air temperature (H. Kudoh et al., unpubl. data). Recent transcriptome analyses revealed that >80 % of leaf-expressed genes are actively transcribed constantly throughout the year, and furthermore, genes associated with the avoidance of photoinhibition are upregulated during winter (Nagano et al. 2019), probably to protect photosynthetic apparatus under low temperature and high light conditions. The timing of the termination of flowering is a phenological trait specific to perennials (Aikawa et al. 2010; Satake et al. 2013; Nagahama et al. 2018), and allocation to reproduction and vegetative growth can be the adaptive traits for study using perennial systems. The perennial and evergreen habits benefit the study of diverse stress under natural conditions, because we can collect sample tissues at any time in a year, and season-specific stresses, such as heat stress during summer, frost during winter, drought during dry season, flooding during rainy season, and herbivore and pathogen attacks during warm periods (Kudoh 2016). Furthermore, it gives opportunities to make comparisons between years using the same individuals.
Outcross breeding system
This species has obligate outcrossing flowers (Kudoh et al. 2016). The flowers are large (4–6.5 mm petal length) compared with those of A. thaliana (2–3.5 mm petal length; Table 1; Al-Shehbaz et al. 2006). The flowers are self-incompatible, which is sporophytic self-incompatibility similar to other self-incompatible Brassicaceae plants (Kusaba 2001). It has been reported that polymorphisms at the S locus in populations of A. halleri are shared with those of self-incompatible populations of A. lyrata, indicating that ancestral polymorphisms have been maintained by negative frequency-dependent selection acting at the S locus (Roux et al. 2013). Because of the self-incompatibility, pollen grains need to be transferred through pollinators, and its flowers are often visited by small insects, such as solitary bees, flower flies and bee flies (Kudoh et al. 2016). A study conducted in an A. halleri subsp. halleri population using microsatellite markers estimated a 98.7 % outcrossing rate, and found high multiple paternity within maternal siblings (Llaurens et al. 2008). The results suggested that the self-incompatibility is a mechanism to maintain genetic diversity within populations.
Flowering occurs in early spring (Kudoh et al. 2018), although the actual timing of flowering in a year should vary depending on the habitat temperature regime (Satake et al. 2013). The breeding system of A. halleri provides the opportunity to study local adaptation in the traits relating to pollination success. It has been reported that there is a natural selection towards earlier flowering by a flower-feeding leaf beetle in a population of A. halleri subsp. gemmifera in central Japan (Kawagoe and Kudoh 2010). The A. halleri population is highly polymorphic because of the obligate outcrossing enforced by the self-incompatibility of the flowers (Van Rossum et al. 2004; Llaurens et al. 2008). We expect that the level of inbreeding depression should be high, but there are no measurements available due to the technical difficulties in producing offspring through inbreeding. Self-incompatibility implies some disadvantages to produce a certain set of materials, such as recombinant inbred lines (RIL) and near-isogenic lines (NIL). When the A. halleri genome was determined, selfing for five generations was conducted by bud pollinations (Briskine et al. 2017), but a more efficient procedure to produce selfing progeny is required to produce RIL and NIL. A self-compatible variant, if it is discovered, will widen the opportunities to develop these materials.
Clonal propagation via aerial rosette formation
Besides sexual reproduction, A. halleri plants propagate asexually by forming clonal rosettes (Kudoh et al. 2018). The aerial rosettes are formed from lateral meristems, along with the flowering stems at the end of flowering season, for all subspecies (Fig. 2A and B). Even apical reproductive meristems, that have produced flowers, return to vegetative meristems at the end of flowering season (Aikawa et al. 2010; Kudoh et al. 2018), and the phenomenon is known as inflorescence reversion (Tooke et al. 2005). Roots are produced at the bases of aerial rosettes when they are in the air (Fig. 2A and B). Eventually, flowering stems lay down and some of the aerial rosettes successfully establish by penetrating roots into the soil (Fig. 2C). This conspicuous mode of clonal propagation is specific to A. halleri and is in contrast with A. lyrata and A. arenosa (Table 1).
Figure 2.
Aerial rosettes formed from lateral meristems along with flowering stems (A and B), and established clonal rosettes (C). Blue triangles indicate roots from aerial rosettes. An orange triangle indicates the position of the original plant. Arabidopsis halleri subsp. gemmifera in Omoide-gawa, Taka-cho, Hyogo Prefecture, Japan (A and C); A. halleri subsp. tatrica in Belianske Tatry, Prešovský kraj, Slovakia (B).
The distance between clonal rosettes is restricted, and the size of the patches, consisting of genetically identical multiple rosettes, is usually <30 cm in diameter, occasionally 30 cm to 1 m and rarely 1–2 m, as found in a population of A. halleri subsp. gemmifera (M. N. Honjo and H. Kudoh, unpubl. data). Therefore, a single population usually contains several hundred to several thousand scattered and intermingled clonal patches, each originating from a single seed. Therefore, both clonal propagation and seed production are critical for the maintenance of A. halleri populations. In a population of A. halleri subsp. halleri in France, a study using microsatellite markers reported that clonal spread occurs only at short distances <1 m, and high clonal diversity (DG > 0.9) is maintained by sexual reproduction (Van Rossum et al. 2004). Clonal population structures can be studied in further detail by analysing genome-wide SNPs and applying new techniques, such as RAD-Seq (Miller et al. 2007) and Mig-Seq (Suyama and Matsuki 2015), to estimate the kinship between clones and level of somatic mutation between clonal individuals. When conducting growth experiments, clonal propagation offers an easy way to get plants for experiments, especially taking into account the high parental diversity found in some of the A. halleri populations (Llaurens et al. 2008).
The habitat of A. halleri is generally characterized by a high frequency of natural disturbances, and therefore the turnover rate of existing rosettes is high (H. Kudoh, unpubl. data). The formation of clonal rosettes assures the continuation of clonal lineages by spreading the risk of mortality across clonally replicated rosettes (i.e. ramets of the same genet). As well as risk spreading, multiple rosettes may be physiologically integrated through connections between them, to cope with the spatial heterogeneity of environments (Gruntman et al. 2017). We expect that clonal propagation through aerial rosettes has evolved in response to habitat disturbance or habitat productivity. It has been reported that clonal spread is more extensive in less heavy metal-contaminated locations than in highly contaminated locations in a population of A. halleri subsp. halleri (Van Rossum et al. 2004).
Studies on Population Differentiation and Local Adaptation
Here, we review previous studies on population differentiation and local adaptation, frequently studied along two major environmental gradients. The first one is altitudinal adaptation, and it has been studied extensively in two regions, i.e. at Mt. Ibuki, Japan for A. halleri subsp. gemmifera and in the European Alps for A. halleri subsp. halleri. Although both studies focus on a population that occurs in an altitudinal environmental cline, the background settings largely differ. The second gradient is heavy metal concentration in the soil. Population differentiation has been studied between habitats with metalliferous and non-metalliferous soils. Because A. halleri is a representative plant that shows heavy metal hyperaccumulation, there has been a strong demand to understand its mechanisms for use in phytoremediation of heavy metal-contaminated soils, which has led to the accumulation of information on this aspect.
Altitudinal adaptation
Plant populations distributed along altitudinal gradients experience a wide range of abiotic and biotic environmental conditions, depending on altitude and aspect of slopes (Körner 2007). Plants are likely exposed to stressful environments, such as freezing temperature, drought and ultraviolet-B (UV-B), with subtle topographical variation substantially influencing plant fates. Such steep ecological and environmental gradients are likely to cause strong selection and lead to population differentiation and local adaptation. Because altitudinal gradients provide contrasting habitats within a short geographic distance, gene flow at this scale might be more effective at countering the demographic effects of populations than at larger scales. Altitudinal gradient, therefore, provides an opportunity to identify traits under selection (Gonzalo-Turpin and Hazard 2009). In the application of genome scans, outlier loci will probably represent adaptive genetic variation rather than statistical outliers, resulting from historical processes affecting the neutral genetic variation. Thus, A. halleri populations from contrasting habitats along altitudinal gradients offer excellent opportunities to study genetic differentiation and local adaptation.
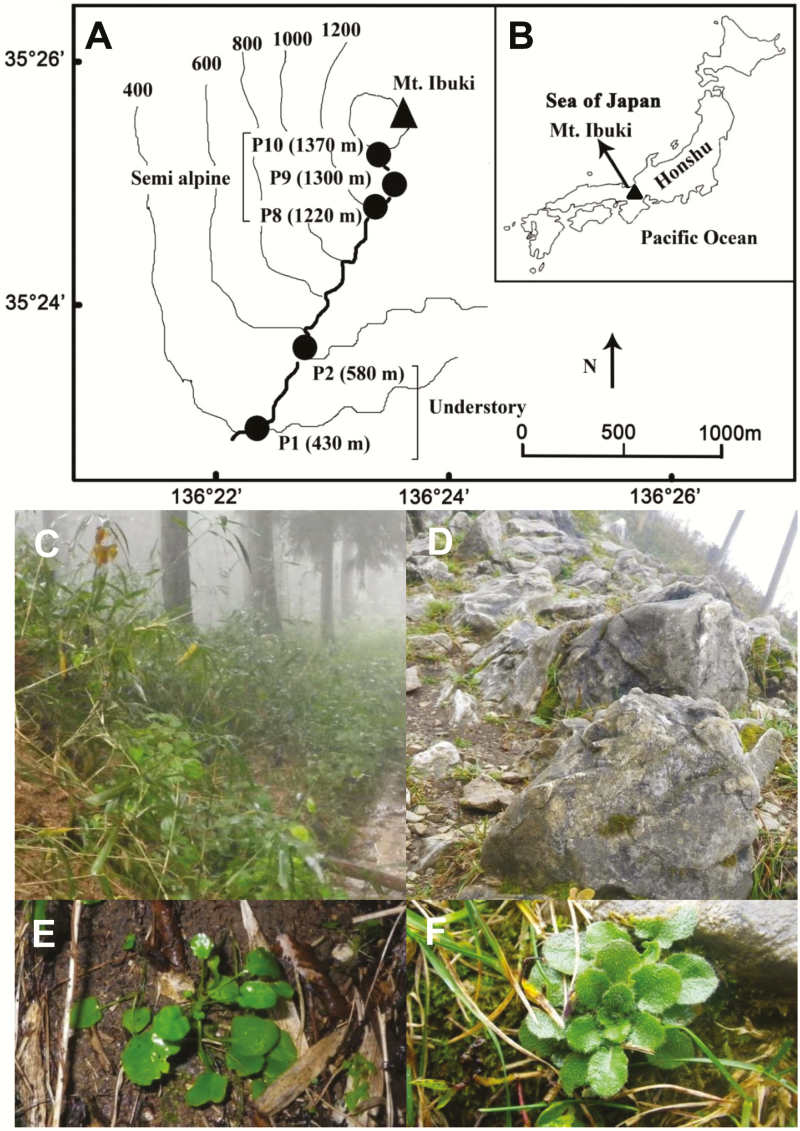
Mt. Ibuki, where A. halleri subsp. gemmifera inhabits, is located in central Japan (altitude 1377 m at the highest peak; Fig. 3A). Honshu Island, the main island of Japan, has an elongated shape but is characterized by contrasting winter weather conditions between the Sea of Japan and Pacific sides. Prevailing north-west winds from the cold Asian continental region absolve moisture when they cross the Sea of Japan and bring heavy snow to the Sea of Japan side of Honshu. Mt. Ibuki is a freestanding peak that is located on the narrowest part of Honshu Island and on the border of the Sea of Japan and Pacific weather areas (Fig. 3B). This specific location characterizes the mountain with a combination of cold wind and snow near the top and relatively warm and mild winter weather near the base.
Figure 3.
Natural populations of Arabidopsis halleri subsp. gemmifera occurring along the altitudinal gradient at Mt. Ibuki, Shiga Prefecture, Japan (A–F). A map showing locations of natural populations in low-altitude understory habitat and in high-altitude semi-alpine habitat (A). A map showing the location of Mt. Ibuki (B). Photographs representing environments of understory (C) and semi-alpine habitats (D). Rosettes of A. halleri under rainy conditions for understory (E) and semi-alpine habitats (F). In (A), a hiking route is shown by a thick line, and P1, P2, P8, P9 and P10 represent pitches 1, 2, 8, 9 and 10 along the route. Altitudes are in parentheses.
The contrasting habitats of A. halleri subsp. gemmifera on Mt. Ibuki are classified into two types: one is the edges and understory of the Cryptomeria japonica forests near the base of mountain (referred to as understory hereafter; Fig. 3C), and the other is the semi-alpine open habitat near the top (referred as to semi-alpine hereafter; Fig. 3D). In the understory of Mt. Ibuki, A. halleri plants are glabrous (Fig. 3E). Plants in the semi-alpine habitat of Mt. Ibuki have a distinctive increased density of trichomes on the leaves (Fig. 3F) compared to typical A. halleri plants (glabrous – sparsely hairy), and are sometimes treated as a variety of A. halleri subsp. gemmifera [originally described as Arabis gemmifera (Matsum.) Makino var. alpicola H. Hara (Hara 1936; Ihara 1976)]. Al-Shehbaz and O’Kane (2002) treated the name as a synonym of Arabidopsis halleri subsp. gemmifera, and Yonekura (2011) treated it as A. halleri subsp. gemmifera f. alpicola (H. Hara) Yonek.
It has been reported that the level of genome-wide genetic differentiation between the low- and high-altitude populations on Mt. Ibuki was low (Fst = 0.017) in the amplified fragment length polymorphism (AFLP) analysis (Ikeda et al. 2010). The analysis using 19 microsatellite loci for 41 populations from a wide-ranging area also resulted in a close genetic similarity for the populations on Mt. Ibuki and the surrounding area (Sato and Kudoh 2014). The altitudinal gradient on Mt. Ibuki, therefore, has often been selected as a model system to study population differentiation and local adaptation.
Ultraviolet-B (280–315 nm in wave length) radiation is a critical abiotic factor increasing ecological stress in plant populations at high elevations. Although the peak of Mt. Ibuki is not high, the habitat of A. halleri in the semi-alpine area is exposed to much more direct sunlight, compared with the understory habitat in lower altitudes. Wang et al. (2016) conducted a growth chamber experiment and found an altitudinal differentiation in the response to UV-B damage. Plants from seeds collected at 380, 760 and 1300 m in elevation on Mt. Ibuki were grown with and without supplemental UV-B. They used the level of cyclobutane pyrimidine dimer (CPD) as an indicator of the level of DNA damage. They applied a UV-B treatment to plants grown for 22 days after seedling transplantation. Thereafter, CDP levels were measured in leaves after 10 and 40 days of the treatment (early and late stages, respectively). At the early stage, plants from the lowest altitude exhibited a higher CPD level and greater inhibition in biomass production, indicating that low-altitude plants were more sensitive to increased UV-B than high-altitude plants. In contrast, at a later stage, CPD for plants from the lowest altitude decreased, and CPD level and growth inhibition became similar across plants that originated from different altitudes. The response to UV stress was concluded to be constitutive in highland ecotypes, but more inducible in lowland ecotypes (Wang et al. 2016). The results suggested the population differentiation occurred under the constraint of a trade-off between growth and UV tolerance (Wang et al. 2016).
Leaf wettability is an indicator of the water affinity/repellency of the leaf surface, and measured by the contact angle of a droplet when a designated amount of water is applied to the leaf surface. When leaf wettability is high water spreads on the leaf surface, while water forms a droplet on the non-wettable leaves, and the leaf surface is kept dry in the rain and fog. Aryal et al. (2018) found cauline-leaf-specific genetic differentiation in leaf wettability between high- and low-altitudinal plants on Mt. Ibuki. Arabidopsis halleri produces two types of leaves, i.e. rosette and cauline leaves. Rosette leaves are produced all year round and the adaxial (upper) surface is exposed. Cauline leaves are produced prior to flower-bud formation and later become leaves on the flowering stems. During early spring, cauline leaves wrap young flower buds, and the abaxial surfaces are exposed. In the results of a field survey and a common garden experiment using a growth chamber, cauline leaves of semi-alpine plants, especially on abaxial surfaces, turned out to be non-wettable, whereas cauline leaves of low-altitudinal understory plants were wettable (Aryal et al. 2018). There were no altitudinal differences in the leaf wettability of rosette leaves. As a candidate gene determining the leaf wettability pattern, the gene expression of a key gene for alkane biosynthesis, AhgCER1, was examined, and the gene was highly upregulated in cauline leaves of high-altitudinal plants (Aryal et al. 2018). Alkane is known to be a highly hydrophobic molecule that is accumulated in the epidermal wax of Arabidopsis leaves (Jetter and Kunst 2008). Dry surfaces of cauline leaves are hypothesized to protect flower buds from morning frost at the higher altitudes of Mt. Ibuki in early spring (Aryal et al. 2018).
Although local adaptation has not yet been rigorously tested, an edaphic factor may be an important cause of population differentiation. High-altitudinal habitats are characterized by calcareous soil, whereas habitats with non-calcareous soil exist at lower altitudes. Wang et al. (2019) reported altitudinal population differentiation of A. halleri at Mt. Ibuki in response to soil nitrogen availability. It has been previously reported that naturally growing plants at high altitude on Mt. Ibuki accumulated less zinc than plants growing in non-calcareous habitats at lower altitudes (Kosugi et al. 2015).
Taking advantage of the small genome size, and being closely related to A. thaliana, genome scans have been applied to identify SNPs that show differentiation associated with habitat environments (Kubota et al. 2015). Genome scans have been used to identify the footprints of selection across the genome to pinpoint the loci under selection. Kubota et al. (2015) applied the method to plants from different altitudes of Mt. Ibuki, as well as another mountain nearby with a similar altitude at the highest peak, Mt. Fujiwara. They conducted individual-base re-sequencing of 56 plants and analysed 527 225 SNPs. For genome-wide SNPs, Gst = 0.043–0.048, indicating low population differentiation at the whole genome level. They successfully identified candidate genes for altitudinal differentiation, and their functional annotations involved those related to altitudinal adaptation, such as cold and freezing tolerance. Moreover, two genes involved in the convergent evolution on the two mountains were identified: homologs of GLUCAN SYNTHASE-LIKE 8 (GSL8) and PBA1. The former is related to growth form and the latter encodes proteasome subunit beta, which is related to diverse types of stresses (Kubota et al. 2015). Quantifying the allelic effects of these genes on fitness by common garden and reciprocal transplant experiments will be required to understand how selection has acted on this particular genome region.
Altitudinal variation in local herbivore pressure is predicted to be a selection agent that causes population differentiation in traits related to plant defence (Løe et al. 2007; Halbritter et al. 2018). This aspect has been studied using populations of A. halleri subsp. halleri from wide-ranging altitudes of the European Alps (Buckley et al. 2019). They found a lower degree of herbivore damage at higher altitudes in the field populations, occurring at different altitudes ranging from 300 to 2300 m. In a common garden experiment, plants showed a cline of population differentiation in association with indole glucosinolates, which imparted constitutive chemical defence. They revealed that relative quantities of indole glucosinolates significantly increased with elevation, and were negatively correlated with herbivore damage in the field. In oviposition preference assays, females of a white butterfly, Pieris brassicae, laid fewer eggs on plants obtained from high-elevation populations (Buckley et al. 2019). These results suggest that A. halleri plants at high altitude are genetically well defended against herbivory.
A genome scan study has been conducted using five populations of A. halleri in the Alps, ranging from 790 to 2308 m in altitude, to characterize genetic variation and identify genes associated with climatic variation (Fischer et al. 2013). They pooled population samples (20 individual plants per population) for whole-genome sequencing (Pool-Seq) and estimated allele frequency of populations at SNP loci. Highly differentiated genomic regions and SNPs were identified from two million SNPs using Fst-based analyses (Fischer et al. 2013). They found that the most strongly differentiated genomic regions in A. halleri were typically small and widely dispersed across the genome. By applying partial Mantel tests, 175 genes were identified to be highly associated with one or more of the five topo-climatic factors, i.e. temperature, precipitation, solar radiation, angle of slope and water balance (Fischer et al. 2013). Allele frequencies of two genes [homologs of P-GLYCOPROTEIN 1 (PGP1) and GLUTAMATE RECEPTOR 3.6 (GLR3.6)] were strongly associated with solar radiation, and those of two other genes [homologs of LOW OSMOTIC STRESS 5 (LOS5) and GLUTATHIONE PEROXIDASE 3 (GPX3)] were strongly associated with water balance. Furthermore, their gene annotations were related to responses to the associated environmental factors (Fischer et al. 2013). Later, larger scale analyses with 444 plants from 18 populations of A. halleri (ranging from 309 to 2305 m) were reported (Rellstab et al. 2017). Using two data sets, they narrowed down environmentally associated genes to 11 loci, and concluded that these genes play a significant role in large-scale adaptation to the abiotic environment. One of the examples was a homolog of SYNC1, which was related to seed dormancy, and two SNPs in the locus showed an association with precipitation (Rellstab et al. 2017).
Adaptation to metalliferous soils
Metalliferous habitats exert strong selection pressure on plants by the accumulation of heavy metals at toxic concentrations in plant tissues, and interferences with the plant metabolism (Antonovics et al. 1971; Macnair 1987). Under such circumstances, one expects to observe population differentiation and local adaptation in heavy metal tolerance between metalliferous and non-metalliferous (referred to as M and NM hereafter) sites. The rapid evolution of heavy metal tolerance in response to recent exposure to anthropogenic metal polluted areas is a classic example of local adaptation (Antonovics et al. 1971; Baker and Brooks 1989; Pollard et al. 2002). In these cases, species originally sensitive to heavy metals evolve, via local adaptation in populations exposed to high concentrations of heavy metals, to be resistant or tolerant to this toxicity (Antonovics et al. 1971; Macnair 1987).
Metal hyperaccumulation is the ability to allocate extraordinarily large amounts of metals to shoots, without showing any toxicity symptoms (Antonovics et al. 1971). Hyperaccumulation is widely observed in plants; however, the levels of quantitative variation among populations and between individuals within populations can vary considerably (Gonneau et al. 2014; Stein et al. 2017). This is true for A. halleri, for which heavy metal hyperaccumulation and heavy metal tolerance seem to be the rule, irrespective of the provenance of populations (i.e. M or NM habitats) (Meyer et al. 2015; Stein et al. 2017). Therefore, evolution of heavy metal tolerance in A. halleri is likely to have a long history that involves multiple complex mechanisms (Pauwels et al. 2006). The population differentiation of metal hyperaccumulation and local adaptation of heavy metal tolerance in A. halleri have been reported as a form of enhanced tolerance; plants from M populations are more tolerant than those from NM populations to high Zn concentrations (Pauwels et al. 2006; Meyer et al. 2010) and Cd (Meyer et al. 2015) in population averages. Generally, a large standing variation seems to be maintained both for M and NM populations (Bert et al. 2000, 2002; Stein et al. 2017).
To evaluate Zn tolerance of scattered European populations of A. halleri in M and NM sites, Pauwels et al. (2006) measured survival rates experimentally under conditions of Zn concentration ranging from 1 to 2000 µM. They found Zn tolerance in all examined A. halleri populations. Plants from the M populations were the most tolerant; however, the ranges of variation in tolerance overlapped between NM and M populations. Moreover, relatively high levels of tolerance were detected in some NM populations. They considered that highly tolerant plants have been maintained in some populations without encountering heavy metal exposure, and they termed the phenomenon constitutive tolerance. Later phylogenetic analyses revealed the existence of two genetic groups in Europe that originated from geographic isolation during the glacial period, and the enhancement of heavy metal tolerance in anthrophonic contaminated sites occurred independently in each genetic group (Pauwels et al. 2012). We also should consider current gene flow between M and NM populations, which may make a contribution to the existence of enhanced tolerance in NM population.
Meyer et al. (2010) examined genetic variation in Zn tolerance between seven M and five NM populations in Poland and Slovakia. To test the hypothesis that divergent selection has shaped this polymorphism, the morphological and physiological traits of shoots and roots were measured to quantify the response of A. halleri to Zn exposure. On average, tolerance levels measured were higher in plants from the M populations than those of the NM populations. Phenotypic variability was high and mostly accounted for the differences between individuals within populations. Genetic differentiation (QST) for the photosystem II yield of leaves (a measure of photosynthetic efficiency) was large in contrast to the small differentiation (FST) estimated from 10 microsatellite loci and thus was probably shaped by divergent selection. They discussed how Zn tolerance is increased in M populations by selection, which acts on the standing genetic variation within an ancestral NM population.
Babst-Kostecka et al. (2018) investigated the population genetic structure of eight M and six NM populations of A. halleri in Poland using 10 microsatellite loci. Different geographical groups, rather than M and NM separation, were identified. Genetic variation in Zn hyperaccumulation was detected in all the investigated populations, suggesting that Zn hyperaccumulation can respond to selection. Moreover, they suggested that the current distribution of A. halleri in southern Poland could be a result of habitat fragmentation caused by climatic shifts after the last glacial period, rather than due to the recent colonization of industrially polluted sites. In addition, some lowland NM populations may have been derived from M populations. They concluded that Zn hyperaccumulation evolved both ways, towards higher levels at NM sites and lower levels at M sites.
In most of the reported studies, Zn accumulation level in shoots was higher in plants from NM populations than that in plants from M populations under the common experimental conditions (Bert et al. 2000; Stein et al. 2017; Babst-Kostecka et al. 2018). A similar pattern has been reported for Cd accumulation (Stein et al. 2017). Therefore, the function of Zn and Cd hyperaccumulation of A. halleri in NM habitats remains an intriguing question. Multiple hypotheses regarding the function of the metal hyperaccumulation have been discussed (Boyd and Martens 1992). Initially, because absolute metallophytes often show hyperaccumulation, it was considered to be a part of physiological tolerance against the high level of metals in soil (Baker 1981). However, it remained unsolved how metal hyperaccumulation enhances plant fitness. More recently, the ‘elemental defence hypothesis’, in which high metal concentration in plant tissues acts as a defence against certain herbivores or pathogens, has been tested in many studies (Boyd 2004, 2007; Fones et al. 2010; Rascio and Navari-Izzo 2011; Kazemi-Dinan et al. 2014; Stolpe et al. 2017). Assuming that similar levels of metal concentration are required for defence in M and NM populations, one may expect that increased hyperaccumulation ability for plants in the latter populations.
As we have seen in the studies of altitudinal adaptation on Mt. Ibuki and in the Alps, a genome scan by whole-genome re-sequencing of population pools (Pool-Seq) was conducted to identify SNPs that associate with heavy metal concentration in natural habitats (Sailer et al. 2018). They used two M and two NM populations (N = 119 plants) and identified 57 SNPs in 19 genes significantly associated with soil adaptation. These genes included those related to transmembrane transport and responses to stress. One example is METAL TOLERANCE PROTEIN A2 (MTPA2). Therefore, both translocation of heavy metals and detoxification processes are likely to be important in the adaptation to M and NM soils.
Quantitative trait locus (QTL) mapping, transcriptome studies and functional analyses have resulted in critical progression of our understanding of the molecular mechanisms underlying metal tolerance and accumulation in A. halleri (reviewed by Verbruggen et al. 2009; Krämer 2010). For Zn tolerance and accumulation in A. halleri, constitutive high expression levels of genes involved in root uptake (members of the ZIP gene family, a metal transporter family first identified in plants), root-to-shoot translocation [HEAVY METAL ATPAse 4 (HMA4) and NICOTIANAMINE SYNTHASE 2 (NAS2)] and vacuolar sequestration [METAL TOLERANCE PROTEIN 1 (MTP1)] play a central role. For Cd tolerance, HMA4 also has an important function in root-to-shoot metal translocation and metal distribution in shoots (Hanikenne et al. 2008, 2013). Furthermore, HMA4 has been reported to be triplicated in A. halleri (Hanikenne et al. 2008, 2013). The knowledge on the actual genes involved in hyperaccumulation and tolerance could provide us with a great advantage in studying population differentiation and local adaptation in A. halleri.
RNA-Seq – Widening Phenotype Space
Next-generation sequencing (NGS) is being used increasingly for the study of population differentiation and local adaptation. In the above examples, as in the method to analyse population differentiation using whole genome SNPs, genome scans using individual-based re-sequencing and the re-sequencing of population pools (Pool-Seq) have been applied in the study of A. halleri. Here, we introduce RNA-Seq methods that can be used in future studies of population differentiation and local adaptation of A. halleri.
High-throughput RNA-Seq
Transcriptome analyses using RNA-Seq have become a standard method in molecular biology to compare multiple genotypes (e.g. wild type vs. mutant) and to evaluate environmental responses. Thus, application of RNA-Seq in the study of population differentiation and local adaptation is expected. The quantitative measures of gene expression can be treated as conventional measures of phenotypic traits for samples from field populations, common garden experiments and reciprocal transplant experiments (Gould et al. 2018). Therefore, the key innovation is whether it is applicable for multiple samples. Development of high-throughput and cost-effective methods for RNA-Seq library preparation, and the application of these methods to target species is required. Several methods have been already developed (Wang et al. 2011; Townsley et al. 2015).
Application of high-throughput RNA-Seq has been done previously in a natural population of A. halleri subsp. gemmifera in central Japan (Kudoh et al. 2018). As a molecular phenology study in the population (Kudoh 2016), leaf transcriptome data from six A. halleri plants were obtained weekly for 2 years, and bi-hourly for 2 days on spring and autumn equinoxes, and summer and winter solstices (Nagano et al. 2019). It has been reported that 16.7 and 41.8 % of the leaf-expressed genes (2879 and 7185 out of 17 205, respectively) show seasonal and diurnal oscillations (Nagano et al. 2019). Although the study did not aim to analyse population differentiation and local adaptation, it reveals an important property of gene expression when we use it as a phenotypic trait; i.e. the sampling time in a day needs to be strictly controlled, because many genes exhibit diurnal changes in their expression levels. The tissue, time and environment specificity in gene expression will be important to interpret the observed variation in gene expressions between and within populations.
Transcriptome data have three innovative properties that give us advantages in the study of population differentiation and local adaptation; first, a comprehensive phenotype set, second, a linker between the phenotype of ecological traits and underlying genetic basis, and third, a trait set that gives a clue to identifying the causal nucleotide substitution(s). As per the first property, RNA-Seq data include genes that are involved in wide range of morphological and physiological traits. Because of this comprehensiveness, we can explore in which traits populations are differentiated without targeting particular traits in advance. As per the second property, gene expression data are more closely linked to underlying genetic mechanisms than phenotypes of ecological traits. Therefore, the detection of differentially expressed genes (DEGs) between populations allows us to identify which part of the gene regulation network is under selection and/or is responsive to selection. As per the last point, the causal nucleotide substitution(s) may be detected in RNA-Seq sequences when they exist in transcribed regions. Causal substitution(s) might be located in the regulatory region of most upstream DEGs in the regulatory network of targeted phenotypic traits. With the combination of SNP analyses on the genome, it could help elucidate the causal nucleotide substitution(s) responsible for population differentiation and local adaptation.
Dual RNA-Seq
In RNA-Seq, NGS determines all RNA reads in a prepared library, and therefore not only the RNAs of the target tissues, but also RNAs derived from endogenous organisms, can be subjected to further analyses. Simultaneous analysis of hosts and parasites has been termed as dual RNA-Seq (Westermann et al. 2012), and the idea can be applied to any endogenous organisms including pathogens, parasites and symbionts. In the previous examples, transcriptomes of pathogenic endogenous fungi and host plants were analysed simultaneously (Tierney et al. 2012; Westermann et al. 2012, 2016). The method can be applied to the study of population differentiation and local adaptation for both host and endogenous organisms.
A method to identify all infected plant viruses using RNA-Seq has been developed (Nagano et al. 2015). Because this method targets total RNA, except plant rRNA, in contrast to commonly used methods, which target mRNA, a broad range of viruses can be detected. It has been reported that five viruses, Turnip mosaic virus (TuMV), Brassica yellows virus (BrYV), Cucumber mosaic virus (CMV), Pelagonium zonate spot virus (PZSV) and Arabidopsis halleri partitivirus 1 (AhPV1), can infect naturally growing A. halleri subsp. gemmifera (Kamitani et al. 2016, 2017). Endogenous organisms might act as environmental factors that can result in divergent selection between populations. The TuMV infection rates were compared between multiple natural populations, and ranged from 0 to 57 % (Kamitani et al. 2019). We still have limited knowledge on the regional variation in virus infection rates and whether this geographic variation is translated into differentiation among A. halleri populations.
An original dual RNA-Seq for the simultaneous analysis of host and pathogen transcriptomes (Westermann et al. 2012), virus accumulation and host transcriptome can be examined simultaneously in the plant virus–host system (Kamitani et al. 2016). Dual RNA-Seq employed for TuMV–A. halleri systems has revealed that the level of virus accumulation changes dynamically within the host and virus infection affects host transcriptomes by altering the expression of genes related to defence responses and flavonoid biosynthesis during long-term persistent infections (Honjo et al. 2019). When the relationship between the host and endogenous organisms is persistent, we may expect co-evolution between host plants and endogenous organisms. It has been considered that there exists a trade-off between virulence and transmission rates. How such relationships between host and endogenous organisms affect population differentiation and local adaptation is an open question that can be addressed in future using A. halleri as a target species.
Epigenetic modifications
Epigenetic modifications, such as DNA methylation and post-transcribed histone modification, determine chromatin states that alter gene expression, and can play a critical role in the regulation of phenotypes in response to local environments. Some classes of epigenetic modification have been reported to show environmental responses for long periods, sometimes even across generations (Verhoeven et al. 2016). As DNA methylation and histone modification can be measured at a single nucleotide and a couple of 100 bp, respectively, measurements for each gene can be utilized to interpret the results of RNA-Seq. It is noteworthy that methods to analyse the epigenome of A. halleri under field conditions have become available; analyses on histone modifications using chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) or next-generation sequencing (ChIP-Seq), and DNA methylation using bisulphite sequencing (Nishio et al. 2016; Ito et al. 2019).
Conclusions
In conclusion, A. halleri provided us with opportunities to study population differentiation and local adaptation, especially along altitudinal gradients and non-metalliferous–metalliferous soil gradients. In the former examples, diverse factors such as temperature, UV-B, frost, soil nutrients and herbivores have been identified as selection agents. In the latter examples, A. halleri plants show heavy metal hyperaccumulation irrespective of the origins, and plants from metalliferous habitats exhibited enhanced heavy metal tolerance. Therefore, in either adaptation to high-altitudinal environments or to heavy metal-contaminated soils, alternation in multiple genetic pathways is likely to be involved in the adaptation processes. The results of genome scans also supported this hypothesis, and owing to the close relatedness to A. thaliana, most of the candidate genes under selection have been annotated, making it easier to infer their function. By combination with rigorous common garden experiments, growth chamber experiments and reciprocal transplant experiments, gene expression data could provide critical information to elucidate genetic systems underlying observed population differentiation and local adaptation. Because we now have lists of the candidate genes responsible for local adaptation, the effort of connecting expression differences in these genes with fitness outcomes in natural habitats has become critically important.
Sources of Funding
This study was supported by Japan Science and Technology Agency (JST), CREST no. JPMJCR15O1 and Japan Society for the Promotion of Science (JSPS) Grant-in Aid for Scientific Research (A) no. 19H01001.
Contributions by the Authors
M.N.H. and H.K. conceived the idea, collected literature and wrote the manuscript.
Conflict of Interest
None declared.
Acknowledgements
We thank Dr K. Marhold for the cooperative field trips in Europe during which photographs of A. halleri were taken and T. Kato for the assistance with literature survey.
Literature Cited
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. 2010. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proceedings of the National Academy of Sciences of the United States of America 107:11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama S, Shimizu-Inatsugi R, Shimizu KK, Sese J. 2014. Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Research 42:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA, Arai K, Ohba H. 2006. Cruciferae. In: Iwatsuki K, Boufford DE, Ohba H, eds. Flora of Japan, volume IIa. Tokyo: Kodansha Ltd, 454–511. [Google Scholar]
- Al-Shehbaz IA, O’Kane SL. 2002. Taxonomy and phylogeny of Arabidopsis (Brassicaceae). In: Somerville C, Meyerowitz E, eds. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Schrey AW, Richards CL. 2015. Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Molecular Ecology 24:710–725. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Bradshaw AD, Turner RG. 1971. Heavy metal tolerance in plants. Advances in Ecological Research 7:1–85. [Google Scholar]
- Aryal B, Shinohara W, Honjo MN, Kudoh H. 2018. Genetic differentiation in cauline-leaf-specific wettability of a rosette-forming perennial Arabidopsis from two contrasting montane habitats. Annals of Botany 121:1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst-Kostecka A, Schat H, Saumitou-Laprade P, Grodzinska K, Bourceaux A, Pauwels M, Frerot H. 2018. Evolutionary dynamics of quantitative variation in an adaptive trait at the regional scale: the case of zinc hyperaccumulation in Arabidopsis halleri. Molecular Ecology 27:3257–3273. [DOI] [PubMed] [Google Scholar]
- Baduel P, Arnold B, Weisman CM, Hunter B, Bomblies K. 2016. Habitat-associated life history and stress-tolerance variation in Arabidopsis arenosa. Plant Physiology 171:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AJM. 1981. Accumulators and excluders—strategies in the response of plants to heavy metals. Journal of Plant Nutrition 3:643–54. [Google Scholar]
- Baker AJM, Brooks RR. 1989. Terrestrial higher plants which hyperaccumulate metallic elements, a review of their distribution, ecology and phytochemistry. Biorecovery 1:81–126. [Google Scholar]
- Bamba M, Kawaguchi YW, Tsuchimatsu T. 2019. Plant adaptation and speciation studied by population genomic approaches. Development, Growth & Differentiation 61:12–24. [DOI] [PubMed] [Google Scholar]
- Bert V, Bonnin I, Saumitou-Laprade P, De Laguérie P, Petit D. 2002. Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytologist 155:47–57. [DOI] [PubMed] [Google Scholar]
- Bert V, MacNair MR, De Laguerie P, Saumitou-Laprade P, Petit D. 2000. Zinc tolerance and accumulation in metallicolous and non metallicolous populations of Arabidopsis halleri (Brassicaceae). New Phytologist 146:225–233. [DOI] [PubMed] [Google Scholar]
- Boyd RS. 2004. Ecology of metal hyperaccumulation. New Phytologist 162:563–567. [DOI] [PubMed] [Google Scholar]
- Boyd RS. 2007. The defense hypothesis of elemental hyperaccumulation: status, challenges and new directions. Plant and Soil 293:153–176. [Google Scholar]
- Boyd RS, Martens SN. 1992. The raison d’être for metal hyperaccumulation by plants. In: Baker AJM, Proctor J, Reeves RD, eds. The ecology of ultramafic serpentine soils. Andover, UK: Intercept, 279–289. [Google Scholar]
- Briskine RV, Paape T, Shimizu-Inatsugi R, Nishiyama T, Akama S, Sese J, Shimizu KK. 2017. Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Molecular Ecology Resources 17:1025–1036. [DOI] [PubMed] [Google Scholar]
- Buckley J, Pashalidou FG, Fischer MC, Widmer A, Mescher MC, De Moraes CM. 2019. Divergence in glucosinolate profiles between high- and low-elevation populations of Arabidopsis halleri correspond to variation in field herbivory and herbivore behavioral preferences. International Journal of Molecular Sciences 20:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart-Waco D, Josefsson C, Dilkes B, Kozloff N, Torjek O, Meyer R, Altmann T, Comai L. 2012. Hybrid incompatibility in Arabidopsis is determined by a multiple-locus genetic network. Plant Physiology 158:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X. 2008. Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genetics 4:e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA. 2006. Poorly known relatives of Arabidopsis thaliana. Trends in Plant Science 11:449–459. [DOI] [PubMed] [Google Scholar]
- Fischer MC, Rellstab C, Tedder A, Zoller S, Gugerli F, Shimizu KK, Holderegger R, Widmer A. 2013. Population genomic footprints of selection and associations with climate in natural populations of Arabidopsis halleri from the Alps. Molecular Ecology 22:5594–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fones H, Davis CA, Rico A, Fang F, Smith JA, Preston GM. 2010. Metal hyperaccumulation armors plants against disease. PLoS Pathogens 6:e1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. 2011. A map of local adaptation in Arabidopsis thaliana. Science 334:86–89. [DOI] [PubMed] [Google Scholar]
- Gonneau C, Genevois N, Frérot H, Sirguey C, Sterckeman T. 2014. Variation of trace metal accumulation, major nutrient uptake and growth parameters and their correlations in 22 populations of Noccaea caerulescens. Plant and Soil 384:271–287. [Google Scholar]
- Gonzalo-Turpin H, Hazard L. 2009. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. Journal of Ecology 97:742–751. [Google Scholar]
- Gould BA, Chen Y, Lowry DB. 2018. Gene regulatory divergence between locally adapted ecotypes in their native habitats. Molecular Ecology 27:4174–4188. [DOI] [PubMed] [Google Scholar]
- Grime JP, Hodgson JG, Hunt R. 1988. Comparative plant ecology: a functional approach to common British species. London: Unwin Hyman. [Google Scholar]
- Gruntman M, Anders C, Mohiley A, Laaser T, Clemens S, Hӧreth S, Tielbӧrger K. 2017. Clonal integration and heavy-metal stress: responses of plants with contrasting evolutionary backgrounds. Evolutionary Ecology 31:305–316. [Google Scholar]
- Halbritter AH, Fior S, Keller I, Billeter R, Edwards PJ, Holderegger R, Karrenberg S, Pluess AR, Widmer A, Alexander JM. 2018. Trait differentiation and adaptation of plants along elevation gradients. Journal of Evolutionary Biology 31:784–800. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334:83–86. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Kroymann J, Trampczynska A, Bernal M, Motte P, Clemens S, Krämer U. 2013. Hard selective sweep and ectopic gene conversion in a gene cluster affording environmental adaptation. PLoS Genetics 9:e1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. 2008. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453:391–395. [DOI] [PubMed] [Google Scholar]
- Hara H. 1936. Arabis gemmifera var. alpicola (Koidz.) Hara. Journal of Japanese Botany 12:900–901. [Google Scholar]
- Hohmann N, Schmickl R, Chiang TY, Lučanová M, Kolář F, Marhold K, Koch MA. 2014. Taming the wild: resolving the gene pools of non-model Arabidopsis lineages. BMC Evolutionary Biology 14:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo MN, Emura N, Kawagoe T, Sugisaka J, Kamitani M, Nagano AJ, Kudoh H. 2019. Seasonality of interactions between a plant virus and its host during persistent infection in a natural environment. The ISME Journal. doi: 10.1038/s41396-019-0519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KF, Van de Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K. 1976. Mode of local differentiation in Arabis lyrata and A. gemmifera Cruciferae in Japan. Journal of the Faculty of Science, The University of Tokyo, Section III, Botany 12:1–36. [Google Scholar]
- Ikeda H, Setoguchi H, Morinaga S-I. 2010. Genomic structure of lowland and highland ecotypes of Arabidopsis halleri subsp. gemmifera (Brassicaceae) on Mt. Ibuki. Acta Phytotaxonomica et Geobotanica 61:21–26. [Google Scholar]
- Ito T, Nishio H, Tarutani Y, Emura N, Honjo MN, Toyoda A, Fujiyama A, Kakutani T, Kudoh H. 2019. Seasonal stability and dynamics of DNA methylation in plants in a natural environment. Genes 10:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R, Kunst L. 2008. Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. The Plant Journal 54:670–683. [DOI] [PubMed] [Google Scholar]
- Kamitani M, Nagano AJ, Honjo MN, Kudoh H. 2016. RNA-Seq reveals virus-virus and virus-pant interactions in nature. FEMS Microbiology Ecology 92:fiw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani M, Nagano AJ, Honjo MN, Kudoh H. 2017. First report of Pelargonium zonate spot virus from wild Brassicaceae plants in Japan. Journal of General Plant Pathology 83:329–332. [Google Scholar]
- Kamitani M, Nagano AJ, Honjo MN, Kudoh H. 2019. A survey on plant viruses in natural Brassicaceae communities using RNA-seq. Microbial Ecology 78:113–121. [DOI] [PubMed] [Google Scholar]
- Kawagoe T, Kudoh H. 2010. Escape from floral herbivory by early flowering in Arabidopsis halleri subsp. gemmifera. Oecologia 164:713–720. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecology Letters 7:1225–1241. [Google Scholar]
- Kazemi-Dinan A, Thomaschky S, Stein RJ, Krämer U, Müller C. 2014. Zinc and cadmium hyperaccumulation act as deterrents towards specialist herbivores and impede the performance of a generalist herbivore. New Phytologist 202:628–639. [DOI] [PubMed] [Google Scholar]
- Koch MA. 2019. The plant model system Arabidopsis set in an evolutionary, systematic, and spatio-temporal context. Journal of Experimental Botany 70:55–67. [DOI] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Molecular Biology and Evolution 17:1483–1498. [DOI] [PubMed] [Google Scholar]
- Kolník M, Marhold K. 2006. Distribution, chromosome numbers and nomenclature conspect of Arabidopsis halleri (Brassicaceae) in the Carpathians. Biologia 61:41–50. [Google Scholar]
- Körner C. 2007. The use of ‘altitude’ in ecological research. Trends in Ecology & Evolution 22:569–574. [DOI] [PubMed] [Google Scholar]
- Kosugi A, Tamaru J, Gotou K, Furihata HY, Shimizu A, Kawabe A, Harada E. 2015. Metal accumulation by Arabidopsis halleri subsp. gemmifera at a limestone mining site. Australian Journal of Botany 63:134–140. [Google Scholar]
- Krämer U. 2010. Metal hyperaccumulation in plants. Annual Review of Plant Biology 61:517–534. [DOI] [PubMed] [Google Scholar]
- Kubota S, Iwasaki T, Hanada K, Nagano AJ, Fujiyama A, Toyoda A, Sugano S, Suzuki Y, Hikosaka K, Ito M, Morinaga S. 2015. A genome scan for genes underlying microgeographic-scale local adaptation in a wild Arabidopsis species. PLoS Genetics 11:e1005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh H. 2016. Molecular phenology in plants: in natura systems biology for the comprehensive understanding of seasonal responses under natural environments. New Phytologist 210:399–412. [DOI] [PubMed] [Google Scholar]
- Kudoh H, Honjo MN, Nishio H, Sugisaka J. 2018. The long-term “In Natura” study sites of Arabidopsis halleri for plant transcription and epigenetic modification analyses in natural environments. Methods in Molecular Biology 1830:41–57. [DOI] [PubMed] [Google Scholar]
- Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME. 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. The Plant Cell 13:627–643. [PMC free article] [PubMed] [Google Scholar]
- Leinonen PH, Remington DL, Leppälä J, Savolainen O. 2013. Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata. Molecular Ecology 22:709–723. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Grant MC. 1996. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics 27:237–277. [Google Scholar]
- Llaurens V, Castric V, Austerlitz F, Vekemans X. 2008. High paternal diversity in the self-incompatible herb Arabidopsis halleri despite clonal reproduction and spatially restricted pollen dispersal. Molecular Ecology 17:1577–1588. [DOI] [PubMed] [Google Scholar]
- Løe G, Toräng P, Gaudeul M, Ågren J. 2007. Trichome production and spatiotemporal variation in herbivory in the perennial herb Arabidopsis lyrata. Oikos 116:134–142. [Google Scholar]
- Macnair MR. 1987. Heavy metal tolerance in plants: a model evolutionary system. Trends in Ecology & Evolution 2:354–359. [DOI] [PubMed] [Google Scholar]
- Meyer CL, Juraniec M, Huguet S, Chaves-Rodriguez E, Salis P, Isaure MP, Goormaghtigh E, Verbruggen N. 2015. Intraspecific variability of cadmium tolerance and accumulation, and cadmium-induced cell wall modifications in the metal hyperaccumulator Arabidopsis halleri. Journal of Experimental Botany 66:3215–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CL, Kostecka AA, Saumitou-Laprade P, Créach A, Castric V, Pauwels M, Frérot H. 2010. Variability of zinc tolerance among and within populations of the pseudometallophyte species Arabidopsis halleri and possible role of directional selection. New Phytologist 185:130–142. [DOI] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research 17:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama A, Kubota Y, Satake A. 2018. Climate warming shortens flowering duration: a comprehensive assessment of plant phenological responses based on gene expression analyses and mathematical modeling. Ecological Research 33:1059–1068. [Google Scholar]
- Nagano AJ, Honjo MN, Mihara M, Sato M, Kudoh H. 2015. Detection of plant viruses in natural environments by using RNA-Seq. Methods in Molecular Biology 1236:89–98. [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Kawagoe T, Sugisaka J, Honjo MN, Iwayama K, Kudoh H. 2019. Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nature Plants 5:74–83. [DOI] [PubMed] [Google Scholar]
- Nikolov LA, Shushkov P, Nevado B, Gan X, Al-Shehbaz IA, Filatov D, Bailey CD, Tsiantis M. 2019. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytologist 222:1638–1651. [DOI] [PubMed] [Google Scholar]
- Nishio H, Buzas DM, Nagano AJ, Suzuki Y, Sugano S, Ito M, Morinaga S, Kudoh H. 2016. From the laboratory to the field: assaying histone methylation at FLOWERING LOCUS C in naturally growing Arabidopsis halleri. Genes & Genetic Systems 91:15–26. [DOI] [PubMed] [Google Scholar]
- O’Kane SL, Schaal BA, Al-Shehbaz IA. 1996. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Systematic Botany 21:559–566. [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. 2002. Variation in gene expression within and among natural populations. Nature Genetics 32:261–266. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M, Frérot H, Bonnin I, Saumitou-Laprade P. 2006. A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study: Arabidopsis halleri (Brassicaceae). Journal of Evolutionary Biology 19:1838–1850. [DOI] [PubMed] [Google Scholar]
- Pauwels M, Vekemans X, Godé C, Frérot H, Castric V, Saumitou-Laprade P. 2012. Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytologist 193:916–928. [DOI] [PubMed] [Google Scholar]
- Pollard AJ, Powell KD, Harper FA, Smith JAC. 2002. The genetic basis of metal hyperaccumulation in plants. Critical Reviews in Plant Sciences 21:539–566. [Google Scholar]
- Rascio N, Navari-Izzo F. 2011. Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting? Plant Science 180:169–181. [DOI] [PubMed] [Google Scholar]
- Rellstab C, Fischer MC, Zoller S, Graf R, Tedder A, Shimizu KK, Widmer A, Holderegger R, Gugerli F. 2017. Local adaptation (mostly) remains local: reassessing environmental associations of climate-related candidate SNPs in Arabidopsis halleri. Heredity 118:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Rosas U, Banta J, Bhambhra N, Purugganan MD. 2012. Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genetics 8:e1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Castric V, Pauwels M, Wright SI, Saumitou-Laprade P, Vekemans X. 2011. Does speciation between Arabidopsis halleri and Arabidopsis lyrata coincide with major changes in a molecular target of adaptation? PLoS One 6:e26872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Pauwels M, Ruggiero MV, Charlesworth D, Castric V, Vekemans X. 2013. Recent and ancient signature of balancing selection around the S-locus in Arabidopsis halleri and A. lyrata. Molecular Biology and Evolution 30:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer C, Babst-Kostecka A, Fischer MC, Zoller S, Widmer A, Vollenweider P, Gugerli F, Rellstab C. 2018. Transmembrane transport and stress response genes play an important role in adaptation of Arabidopsis halleri to metalliferous soils. Scientific Reports 8:16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake A, Kawagoe T, Saburi Y, Chiba Y, Sakurai G, Kudoh H. 2013. Forecasting flowering phenology under climate warming by modelling the regulatory dynamics of flowering-time genes. Nature Communications 4:2303. [DOI] [PubMed] [Google Scholar]
- Sato Y, Kudoh H. 2014. Fine-scale genetic differentiation of a temperate herb: relevance of local environments and demographic change. AoB Plants 6:plu070; doi: 10.1093/aobpla/plu070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nature Reviews Genetics 14:807–820. [DOI] [PubMed] [Google Scholar]
- Schierup MH, Bechsgaard JS, Christiansen FB. 2008. Selection at work in self-incompatible Arabidopsis lyrata. II. Spatial distribution of S haplotypes in Iceland. Genetics 180:1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup MH, Mable BK, Awadalla P, Charlesworth D. 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158:387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šrámková-Fuxová G, Záveská E, Kolář F, Lučnová M, Španiel S, Marhold K. 2017. Range-wide genetic structure of Arabidopsis halleri (Brassicaceae): glacial persistence in multiple refugia and origin of the Northern Hemisphere disjunction. Botanical Journal of the Linnean Society 185:321–342. [Google Scholar]
- Stein RJ, Höreth S, de Melo JR, Syllwasschy L, Lee G, Garbin ML, Clemens S, Krämer U. 2017. Relationships between soil and leaf mineral composition are element-specific, environment-dependent and geographically structured in the emerging model Arabidopsis halleri. New Phytologist 213:1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe C, Krämer U, Müller C. 2017. Heavy metal hyperaccumulation in leaves of Arabidopsis halleri is accompanied by a reduced performance of herbivores and shifts in leaf glucosinolate and element concentrations. Environmental and Experimental Botany 133:78–86. [Google Scholar]
- Suyama Y, Matsuki Y. 2015. MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Scientific Reports 5:16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takou M, Wieters B, Kopriva S, Coupland G, Linstädter A, De Meaux J. 2019. Linking genes with ecological strategies in Arabidopsis thaliana. Journal of Experimental Botany 70:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Tierney L, Linde J, Müller S, Brunke S, Molina JC, Hube B, Schöck U, Guthke R, Kuchler K. 2012. An interspecies regulatory network inferred from simultaneous RNA-seq of Candida albicans invading innate immune cells. Frontiers in Microbiology 3:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke F, Ordidge M, Chiurugwi T, Battey N. 2005. Mechanisms and function of flower and inflorescence reversion. Journal of Experimental Botany 56:2587–2599. [DOI] [PubMed] [Google Scholar]
- Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR. 2015. BrAD-seq: breath adapter directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Frontiers in Plant Science 6:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics 42:260–263. [DOI] [PubMed] [Google Scholar]
- Van Rossum F, Bonnin I, Fenart S, Pauwels M, Petit D, Saumitou-Laprade P. 2004. Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-incompatible and heavy-metal-tolerant species. Molecular Ecology 13:2959–2967. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. Molecular mechanisms of metal hyperaccumulation in plants. New Phytologist 181:759–776. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJ, vonHoldt BM, Sork VL. 2016. Epigenetics in ecology and evolution: what we know and what we need to know. Molecular Ecology 25:1631–1638. [DOI] [PubMed] [Google Scholar]
- Wang QW, Daumal M, Nagano S, Yoshida N, Morinaga SI, Hikosaka K. 2019. Plasticity of functional traits and optimality of biomass allocation in elevational ecotypes of Arabidopsis halleri grown at different soil nutrient availabilities. Journal of Plant Research 132:237–249. [DOI] [PubMed] [Google Scholar]
- Wang Q-W, Nagano S, Ozaki H, Morinaga S-I, Hidema J, Hikosaka K. 2016. Functional differentiation in UV-B-induced DNA damage and growth inhibition between highland and lowland ecotypes of two Arabidopsis species. Environmental and Experimental Botany 131:110–119. [Google Scholar]
- Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP. 2011. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One 6:e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Nordborg M. 2015. Population genomics for understanding adaptation in wild plant species. Annual Review of Genetics 49:315–338. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Gorski SA, Vogel J. 2012. Dual RNA-seq of pathogen and host. Nature Reviews Microbiology 10:618–630. [DOI] [PubMed] [Google Scholar]
- Yant L, Bomblies K. 2017. Genomic studies of adaptive evolution in outcrossing Arabidopsis species. Current Opinion in Plant Biology 36:9–14. [DOI] [PubMed] [Google Scholar]
- Yonekura K. 2011. Taxonomic notes on vascular plants in Japan and its adjacent regions II. Journal of Japanese Botany 86:230–241. [Google Scholar]